In-depth proteome profiling of primary human CD138-positive plasma cells, derived from bone marrow biopsies from patients with different stages of multiple myeloma, have been performed on a Q Exactive orbitrap. Analysis of the 6218 identified proteins using label-free quantification with the MaxQuant software revealed strategies adopted by myeloma cells to overcome limitations imposed by hypoxic conditions in the bone marrow microenvironment, including specific immune evasion mechanisms and metabolic adaptations.
Keywords: Mass Spectrometry, Tandem Mass Spectrometry, Metabolomics, Immunohistochemistry, Cancer Biology*, Hypoxia, Immune evasion, Metabolic adaptations, Multiple Myeloma, Primary human myeloma cells
Graphical Abstract
Highlights
In-depth proteome profiling of primary human myeloma cells
Characteristics of myeloma cells are related to hypoxic bone marrow conditions
Myeloma cells show specific immune evasion strategies
Metabolic adaptations involve tumor and stroma cells
Abstract
Multiple Myeloma (MM) is an incurable plasma cell malignancy primarily localized within the bone marrow (BM). It develops from a premalignant stage, monoclonal gammopathy of undetermined significance (MGUS), often via an intermediate stage, smoldering MM (SMM). The mechanisms of MM progression have not yet been fully understood, all the more because patients with MGUS and SMM already carry similar initial mutations as found in MM cells. Over the last years, increased importance has been attributed to the tumor microenvironment and its role in the pathophysiology of the disease. Adaptations of MM cells to hypoxic conditions in the BM have been shown to contribute significantly to MM progression, independently from the genetic predispositions of the tumor cells. Searching for consequences of hypoxia-induced adaptations in primary human MM cells, CD138-positive plasma cells freshly isolated from BM of patients with different disease stages, comprising MGUS, SMM, and MM, were analyzed by proteome profiling, which resulted in the identification of 6218 proteins. Results have been made fully accessible via ProteomeXchange with identifier PXD010600. Data previously obtained from normal primary B cells were included for comparative purposes. A principle component analysis revealed three clusters, differentiating B cells as well as MM cells corresponding to less and more advanced disease stages. Comparing these three clusters pointed to the alteration of pathways indicating adaptations to hypoxic stress in MM cells on disease progression. Protein regulations indicating immune evasion strategies of MM cells were determined, supported by immunohistochemical staining, as well as transcription factors involved in MM development and progression. Protein regulatory networks related to metabolic adaptations of the cells became apparent. Results were strengthened by targeted analyses of a selected panel of metabolites in MM cells and MM-associated fibroblasts. Based on our data, new opportunities may arise for developing therapeutic strategies targeting myeloma disease progression.
Multiple myeloma (MM)1 is a hematological tumor localized primarily in the bone marrow (BM), characterized by the proliferation of malignant antibody-secreting plasma cells. The disease is accompanied by severe health problems such as bone lesions, hypercalcemia, renal failure, anemia and immunodeficiency (1). Even though several new therapeutic strategies extending the survival time of patients have been developed over the last years, MM remains an incurable disease (2). Further, genetic alterations in MM cells are heterogeneous and it is difficult to predict disease course or therapeutic response for individual patients (3). Even though first-line treatments may be successful, relapses are to be expected and tumors then typically become more aggressive and more difficult to treat. MM develops from a premalignant stage, monoclonal gammopathy of undetermined significance (MGUS), and typically progresses through an intermediary stage, smoldering MM (SMM). It is still not completely understood what drives this progression, as patients with MGUS and SMM already carry similar mutations as found in MM cells, suggesting that these mutations are necessary but not enough for tumor development (4–6).
Over the last years, the tumor microenvironment has been recognized to play an important role in MM progression, as well as in immune evasion and drug resistance of MM cells (7, 8). New therapeutic approaches have thus been developed, targeting not only the tumor itself but also tumor-supporting stromal cells such as fibroblasts and endothelial cells, as well as cells of the immune system (9, 10). Further, significance has been attributed to hypoxic conditions in the BM and the role of hypoxia in MM biology, offering novel possibilities for treatment strategies (11–13). MM cells encountering hypoxic environments in the BM apparently enhance pathways that allow them to survive and proliferate under these conditions. Consequently, processes such as angiogenesis and anti-apoptotic survival and proliferation strategies may be induced, and alternative metabolic pathways may be activated in the tumor cells (14–16). As a result, hypoxia-surviving MM cells may become more aggressive and more resistant to therapies (17).
Recent advances in proteomics technologies have given us the opportunity to better understand cellular processes involved in development and progression of diseases (18–22). Proteomics has already been used by others to assess specific aspects of MM such as development of drug resistance (23–26). In line with a recent work on chronic lymphocytic leukemia (CLL) B cells (22), in the present study we have investigated primary human multiple myeloma (MM) cells. The aim was to apply proteome profiling to get deeper insights into pathways MM cells follow in response to hypoxic conditions in the BM microenvironment. Hypoxia-related proteome signatures of MM cells, associated with survival, proliferation and mechanisms to escape apoptosis and immune response were investigated, as well as new targets for therapeutic interventions. To this end, we analyzed primary human CD138-positive plasma cells freshly isolated from the BM of patients with different disease stages, comprising MGUS, SMM, and MM. Another aim was to find out if, like chronic lymphocytic leukemia cells (22), MM cells of individual donors, despite MM being associated with a rather heterogeneous genotype, would show consistent protein patterns indicating pathways commonly adapted by MM cells. The study design should also allow investigating whether there were differences in the proteome of cells from premalignant and earlier stages compared with more advanced stages of the disease, which could provide insights into the pathogenesis and progression of multiple myeloma.
EXPERIMENTAL PROCEDURES
Study Cohort and Bone Marrow Sampling
For proteome profiling experiments, bone marrow (BM) samples from thirteen patients with different stages of multiple myeloma (MM), including monoclonal gammopathy of undetermined significance (MGUS), smoldering MM (SMM) and diagnosed MM, were obtained from routinely taken BM aspirates at the Vienna General Hospital. Written informed consent was obtained from all patients, as well as approval of the Ethics Committee of the Medical University of Vienna (application nr. 1181/2013). In the same way another five BM aspirates were taken from patients with diagnosed MM for targeted analyses of a selected panel of metabolites (described below). From three of these samples, plasma cells were isolated. From the two other samples, as well as from two biopsies also used for proteomics experiments (indicated in Table I), BM fibroblasts were prepared as described below. Both, BM plasma cells and BM fibroblasts were subjected to the analysis of selected metabolites.
Table I. Clinical parameters for patients included in the present study and type of experiment performed with the respective biopsy sample.
| Patient # | Diagnosis | MM-specific treatment | Experiments |
|||
|---|---|---|---|---|---|---|
| Proteomics | PC-Metabolites | Fib-Metabolites | IHC | |||
| MM1 | cyclin D1-neg. MGUS; IR 5% | no | x | |||
| MM2 | κ-pos., cyclin D1-neg. IgA-SMM; IR 50% | no | x | |||
| MM3 | λ-pos., cyclin D1-neg., IgH-MAF/t(14,16)-pos., monosomy 14 & 16-pos. IgA-SMM; IR 40% | no | x | |||
| MM4 | λ-pos., cyclin D1-neg. plasma cell myeloma recidive; IR 20% | yes | x | |||
| MM5 | λ-pos., cyclin D1-neg. plasma cell myeloma recidive; IR 10–15% | yes | x | |||
| MM6 | λ-pos., cyclin D1-pos. secondary plasma cell myeloma after ASCT; IR 70% | yes | x | |||
| MM7 | κ-pos., cyclin D1-neg., low-secretory plasma cell myeloma recidive after ASCT; IR 60% | yes | x | |||
| MM8 | λ-pos., cyclin D1-neg. newly diagnosed plasma cell myeloma; IR 70% | no | x | |||
| MM9 | λ-pos., IgA-positive, newly diagnosed plasma cell myeloma; IR 70% | no | x | |||
| MM10 | κ-pos., cyclin D1-neg., IgH-MAF/t(14,16)-pos., TP53 Deletion-pos., polysomy 4, 14 & 16-pos., trisomy 17-pos. newly diagnosed light-chain plasma cell myeloma; IR 85% | no | x | x | ||
| MM11 | λ-pos., cyclin D1-neg., polysomy 14-pos., trisomy 17-pos., newly diagnosed light-chain plasma cell myeloma; IR >90% | no | x | x | ||
| MM12 | κ-pos., bcl1-pos., low-secretory plasma cell myeloma; IR 80% | yes | x | |||
| MM13 | λ-pos., heterozygous IgH and p53 deletion-pos., ATM amplification-pos., newly diagnosed plasma cell myeloma; IR 50% | no | x | |||
| MM14 | λ-pos., cyclin D1-neg. newly diagnosed plasma cell myeloma; IR 70% | no | x | |||
| MM15 | IgG-κ-pos. plasma cell myeloma; IR 40% | yes | x | |||
| MM16 | IgG-λ-pos., bcl1-pos. plasma cell myeloma; advanced stage; IR N/A | yes | x | |||
| MM17 | λ-pos., cyclin D1-neg., plasma cell myeloma; IR 50% | yes | x | |||
| MM18 | IgD-λ-pos., cyclin D1-neg., newly diagnosed plasma cell myeloma; IR 50% | no | x | |||
| MM19 | κ-pos, cyclin-D1-neg., newly diagnosed IgG-myeloma; IR:10% | no | x | |||
| MM20 | κ-pos, cyclin-D1-neg.,13q14 deletion, newly diagnosed IgA-myeloma; IR:15% | no | x | |||
| MM21 | λ-pos, cyclin-D1-neg., newly diagnosed IgA-myeloma; IR:20% | no | x | |||
| MM22 | λ-pos, cyclin-D1-neg., newly diagnosed IgA-myeloma; IR:40% | no | x | |||
| MM23 | κ-pos, cyclin-D1-neg., 13q14 deletion, trisomy 3,9,11, newly diagnosed IgA-myeloma; IR:60% | no | x | |||
| MM24 | λ-pos, cyclin-D1-neg., newly diagnosed light chain-myeloma; IR:70% | no | x | |||
| MM25 | κ-pos, cyclin-D1-neg., 13q14-delation, TP53-deletion, newly diagnosed IgA-myeloma; IR:70% | no | x | |||
| MM26 | λ-pos, cyclin-D1-neg., newly diagnosed IgG-myeloma; IR:80% | no | x | |||
| MM27 | κ-pos, cyclin-D1-neg.,13q14-deletion, 17p-deletion, newly diagnosed IgG-myeloma; IR:80% | no | x | |||
PC-Metabolites, targeted analyses of amino acids performed with BM plasma cells (PCs); Fib- Metabolites, targeted analyses of amino acids performed with MM-associated BM fibroblasts; IHC, immunohistochemistry; IR, BM infiltration rate with PCs; ASCT, autologous stem cell transplantation.
For immunohistochemical staining, BM biopsies from eight patients with different stages of MM were used. These biopsies were obtained for routine diagnostics at the University Hospital of Regensburg. No additional biopsies for research purposes were taken. After routine diagnostics, remaining material was used for immunohistochemistry (IHC) with approval of the Ethics Committee of the University of Regensburg (application nr. 051097). Clinical parameters, as well as if patients have obtained MM-specific treatments or not, are indicated in Table I for all patients included in the present study.
Plasma Cell Isolation
Bone marrow aspirates were filtered through 40 μm filter (40 μm Nylon Cell Stainer, BD Falcon), diluted 1:2 with PBS, overlaid on Ficoll Paque (GE Healthcare, Bio-Sciences AB, Uppsala, Sweden), and centrifuged at 720 × g for 20 min at room temperature with minimal acceleration and deceleration settings. Mononuclear cells were collected from the resulting interface, were washed with PBS, then resuspended in PBS. Plasma cells were isolated using magnetic activated cell sorting (MACS). To this end, mononuclear cells were incubated at 4 °C for 15 min, after adding magnetic bead-coupled anti-CD138 antibody (Miltenyi Biotec, Bergisch Gladbach, Germany). After another PBS washing step, cells were resuspended in MACS buffer (1× PBS, 0.5% FBS, 2 mm EDTA) and pipetted onto a preconditioned MACS LS column mounted on a magnetic holder (both Miltenyi Biotec, Bergisch Gladbach, Germany). Cells were washed with MACS buffer and CD138-positive plasma cells eluted by removing the MACS LS column from the magnet and pressing 4 ml of MACS buffer through the column. The plasma cell-containing eluate was diluted to 10 ml and cell number as well as viability was determined using the MOXI Z Mini Automated Cell Counter (ORFLO Technologies, Ketchum, ID). Cells were then pelleted by centrifugation at 590 × g for 5 min at 4 °C.
Cell Lysis and Subcellular Fractionation of Primary Human Bone Marrow Plasma Cells
Cell lysis and subcellular fractionation were performed applying a previously established protocol (27). In short, CD138-positive cells were resuspended in lysis buffer supplemented with protease inhibitors at 4 °C to achieve cell lysis. After centrifugation, the cytoplasmic fraction was collected in the supernatant. The pellet was dissolved in 500 mm NaCl solution and subsequently diluted in NP40-buffer; after centrifugation, nuclear protein extracts were collected in the supernatant. Cytoplasmic and nuclear proteins were precipitated in ice-cold ethanol overnight and solubilized in sample buffer (7.5 m urea, 1.5 m thiourea, 4% CHAPS. 0.05% SDS, 100 mm DTT). Protein concentrations were assessed by applying a Bradford assay (Bio-Rad-Laboratories, Vienna, Austria).
Proteolytic Digestion and Sample Clean-up for LC-MS/MS Analysis
Protein fractions were subjected to a filter-assisted proteolytic digestion with a modified version of the FASP protocol (28, 29). In short, 20 μg of proteins were loaded onto a prewetted MWCO filter (Pall Austria Filter GmbH, Vienna, Austria) with a pore size of 3 kDa, followed by reduction of disulfide bonds with dithiothreitol (DTT), alkylation with iodoacetamide (IAA) and washing steps with 50 mm ammonium bicarbonate buffer. Digestion of proteins was achieved by applying two times Trypsin/Lys-C with Mass Spec Grade quality (Promega, Mannheim, Germany), at first overnight, and in a second step for 4 h. Resulting peptides were eluted through the filter by centrifugation, and clean-up was performed using C-18 spin columns (Pierce, Thermo Fisher Scientific, Austria).
LC-MS/MS Analysis
For LC-MS/MS analyses, samples were reconstituted in 5 μl 30% formic acid (FA), supplemented with four synthetic peptide standards for internal quality control, and diluted with 40 μl mobile phase A (97.9% H2O, 2% ACN, 0.1% FA). Of this solution 10 μl were injected into a Dionex Ultimate 3000 nano LC-system coupled to a Q Exactive orbitrap mass spectrometer equipped with a nanospray ion source (Thermo Fisher Scientific, Austria). All samples were analyzed as technical replicates. As a preconcentration step, peptides were loaded on a 2 cm × 75 μm C18 Pepmap100 pre-column (Thermo Fisher Scientific, Austria) at a flow rate of 10 μl/min using mobile phase A. Elution from the precolumn to a 50 cm × 75 μm Pepmap100 analytical column (Thermo Fisher Scientific, Austria) and subsequent separation was achieved at a flow rate of 300 nl/min using a gradient of 8% to 40% mobile phase B (79.9% ACN, 2% H2O, 0.1% FA) over 235 min with a total chromatographic run time of 280 min. For mass spectrometric detection, MS scans were performed in the range from m/z 400–1400 at a resolution of 70000 (at m/z = 200). MS/MS scans of the eight most abundant ions were achieved through HCD fragmentation at 30% normalized collision energy and analyzed in the orbitrap at a resolution of 17,500 (at m/z = 200).
Data Analysis
The MaxQuant software (version 1.6.0.1), including the Andromeda search engine, was used for data analysis (30). For positive protein identification, as a minimum two peptides, at least one of them being unique, had to be detected. Trypsin/P was specified in the digestion mode. Peptide mass tolerance was set to 50 and 25 ppm for the first and the main search, respectively. The false discovery rate (FDR) was set to 0.01 both on peptide and protein level. The database applied for the search was the human Uniprot database (version 06/2017, with 20100 reviewed entries and 22088 isoforms). Carbamidomethylation was set as fixed modification, methionine oxidation and N-terminal acetylation as variable modifications. Each peptide could have a maximum of two missed cleavages and two modifications. “Match between runs” was enabled and the alignment and match time window set to 25 and 1 min, respectively.
In parallel, to allow data submission to the ProteomeXchange Consortium via the PRIDE partner repository (31), raw files were analyzed using Proteome Discoverer 1.4 (Thermo Fisher Scientific, Austria) using Mascot 2.5 (Matrix Science, UK). Protein identification was performed by searching raw files against the SwissProt Database (version 11/2015 with 20 193 entries), applying a mass tolerance of 50 ppm at MS1 level and 100 mmu at MS2 level, and allowing for up to two missed cleavages per peptide. Fixed and variable modifications were set in the same way as for the MaxQuant search described above. Resulting data were submitted to the ProteomeXchange Consortium via the PRIDE partner repository and can be accessed via www.proteomeexchange.org with the identifier PXD010600.
Experimental Design and Statistical Rationale
Plasma cells isolated from bone marrow of ten patients with multiple myeloma (MM) and three patients with premalignant stages of MM (MGUS or SMM), as indicated in Table I, were used for proteome profiling experiments. Only patients diagnosed according to the guidelines provided by the International Myeloma Working Group (32) were included. Patients suffering from severe comorbidities were excluded. In addition to the MM cell data set, data previously obtained from six biological replicates of peripheral B cells (22), isolated from six healthy donors by applying magnetic activated cell sorting with anti-CD19 antibodies, were included for comparative purposes. B cells were used as reference cell system, representing precursors of plasma cells which have hardly experienced hypoxia. All biological samples were measured as technical replicates with LC-MS/MS, which resulted in 19 independent data sets and a total of 38 measurements. The MaxQuant software (version 1.6.0.1), including the Andromeda search engine, was used for data analysis (30). For statistical data evaluation, the Perseus software (version 1.6.0.2) was used (30, 33). Reverse sequences, potential contaminants as well as proteins identified only by site were removed. Additionally, the normal distribution of data points was manually checked via the histogram function implemented in the Perseus software. Label-free quantification (LFQ) values were logarithmized to base 2, and technical replicates were averaged. Protein groups were filtered for valid values, keeping only those identified in at least 70% of B cell or MM cell samples. Missing values were then replaced from a normal distribution with a down shift of 1.8 and a width of 0.3 in order to enable t-testing and volcano plots. A principle component analysis (PCA), based on label-free quantification of cytoplasmic proteins, was performed. To determine protein groups significantly regulated between resulting clusters, two-sided t-tests employing multiparameter correction with a q-value ≤0.01 (permutation-based FDR correction with s0 = 0.5) were applied. For selected proteins, heat maps representing LFQ values determined in each sample were generated by a custom R (https://www.r-project.org) script. Protein groups found to be significantly regulated were further submitted to the oPOSSUM software (version 3.0) (34), which allowed the detection of over-represented conserved transcription factor binding sites in the corresponding sets of genes.
Targeted Analyses of a Selected Panel of Metabolites
For targeted analyses of a selected panel of metabolites, one million of CD138-positive plasma cells isolated from BM biopsy material of three patients with diagnosed MM (Table I) was suspended in lysis buffer (10 mm phosphate buffer in 85% ethanol) and lysed by three freeze-thaw cycles. The obtained cell lysates were analyzed with the AbsoluteIDQ p180 kit (Biocrates Life Sciences AG, Innsbruck, Austria), as described previously (20). In short, LC-MS and flow injection (FIA)-MS analyses were carried out on a 4000 QTRAP MS system (AB Sciex, Framingham, MA) coupled to a 1200 RR HPLC system (Agilent, Palo Alto, CA), using the Analyst 1.6.2 software (also AB SCIEX). Data evaluation was performed with the software supplied with the kit (MetIDQ, version 5–4-8-DB100-Boron-2607, Biocrates Life Sciences). Results were again compared with data obtained previously from the analysis of B cells (22).
In parallel, from BM samples of four MM patients (Table I), fibroblasts were prepared as previously described (8). In short, BM samples were filtered through a 40 μm mesh (40 μm Nylon Cell Stainer, BD Falcon), the residue in the filter was transferred into a culture flask with fibroblast basal medium (FBM, Lonza Clonetics, # CC-3131) supplemented with one FGM BulletKit (Lonza Clonetics, # CC-3130) and 10% FCS. The culture flask was placed in an incubator at 37 °C in a humidified atmosphere containing 5% CO2, refreshing the medium after 24 h. For comparative purposes, three biological replicates of human mesenchymal stem cells (hMSC, passage 4 to 5; Lonza), cultured in mesenchymal stem cell growth medium (Lonza) supplemented with the associated Bulletkit and 100 U/ml penicillin/streptomycin (ATCC/LGC Standards, London, UK), were used. After reaching 75% confluence, fibroblasts and hMSC were detached by trypsin-EDTA (Sigma-Aldrich) treatment. Cells were washed twice with PBS and one million of each of these cells was processed and used for targeted analyses of a selected panel of metabolites as described above for CD138-positive plasma cells.
Immunohistochemistry
Immunohistochemical stainings were performed on 3 μm thick formalin-fixed, paraffin-embedded tissue sections. Antigen retrieval was performed with Ventana cell conditioning solution 1 (Tris-EDTA buffer pH 8 at 100 °C 32 min) (Ventana Medical Systems, Tucson, AZ). Subsequently, the slides were incubated with primary antibodies against SDC1 (CD138; mouse monoclonal, dilution 1:100, Dako, Denmark), SIGIRR (rabbit polyclonal, dilution 1:500, abcam, United Kingdom), CD46 (rabbit monoclonal, dilution 1:750, abcam, United Kingdom), SLAM7 (rabbit polyclonal, dilution 1:150, Sigma Aldrich, Germany) for 32 min at 36 °C. The immunoreactivity was visualized with the Optiview DAB IHC Detection Kit (Ventana Medical Systems). Slides were counterstained with hematoxylin.
RESULTS
In-depth Proteome Profiling of Primary Human Myeloma Cells
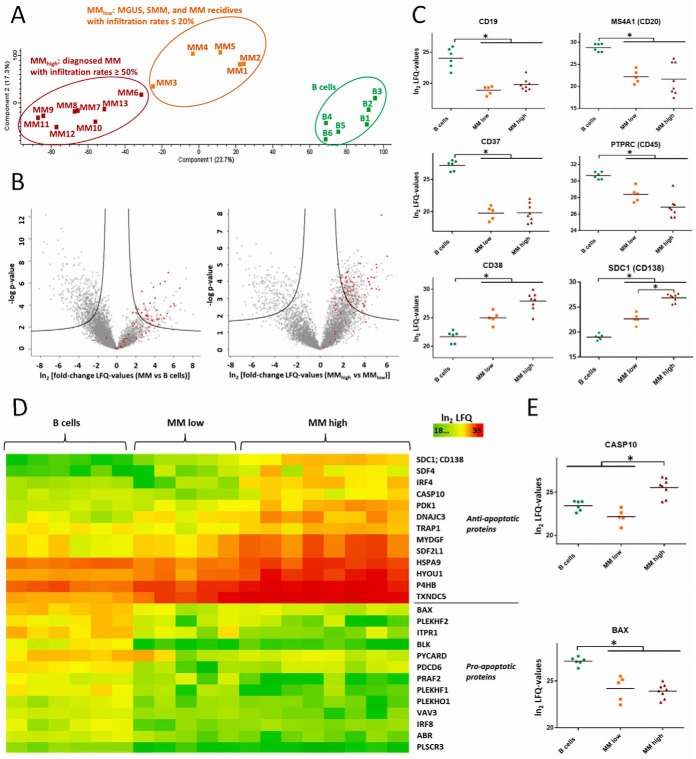
Primary human CD138-positive plasma cells were isolated from freshly isolated bone marrow (BM) biopsy material of ten patients with diagnosed MM, one patient with monoclonal gammopathy of undetermined significance (MGUS), the premalignant stage of MM, and two patients with smoldering multiple myeloma (SMM), an intermediary stage of MM (Table I). Cells were fractionated into cytoplasm and nuclear extracts and analyzed by in-depth proteome profiling using a Q Exactive orbitrap. In total, 6038 and 3415 proteins were identified in cytoplasmic and nuclear fractions of the cells, respectively, (supplemental Tables S1 and S2) by means of the MaxQuant Andromeda search engine (30). A principle component analysis (PCA), based on label-free quantification of cytoplasmic proteins, distinguished three groups: (1) B cells; (2) premalignant stages of MM, and diagnosed MM with BM plasma cell infiltration rates up to 20%, termed MMlow; (3) diagnosed and more advanced stages of MM which match to BM plasma cell infiltration rates of at least 40%, termed MMhigh (Fig. 1A). Remarkably, in both groups, MMhigh and MMlow, patients who had obtained MM-specific therapies and patients who had not been specifically treated before were included (indicated in Table I). Therefore, it was particularly relevant to observe that proteome profiles were that homogeneous in each of these groups, and displayed more commonalities than differences, indicating that protein expression profiles of MM cells were more dependent on the disease stage and hardly on patient treatments. In total, 637 and 605 cytoplasmic proteins were found significantly regulated between MM cells and B cells, and between MMhigh and MMlow cells, respectively, illustrated by volcano plots in Fig. 1B and listed in supplemental Table S1. In the nuclear extracts, 414 proteins were found significantly regulated between MM cells and B cells, and 158 proteins between MMhigh and MMlow cells (supplemental Table S2). MGUS, SMM, and MM cells, when compared all together to B cells, were referred to as “MM cells.” Several well-known differentiation markers of plasma cells, such as CD19, MS4A1 (CD20) and CD38, as well as marker proteins characteristic for myeloma cells, such as CD37, PTPRC (CD45), and SDC1 (CD138) (35, 36), were found regulated as expected in MM versus B cells (Fig. 1C).
Fig. 1.
Proteome profiling of primary human MM cells and comparison to B cells. The Perseus software (30, 33) was used to perform statistical data analyses. A, PCA based on label-free quantification (LFQ) of 6038 cytoplasmic proteins detected in B cells and MM cells, showing three groups: B cells; MMlow cells from patients with MGUS or SMM, as well as from patients with MM with BM infiltration rates with MM cells ≤ 20%; and MMhigh cells from patients with diagnosed MM and BM infiltration rates ≥ 40%. B, Volcano plots comparing the abundance levels of the same proteins in MM versus B cells (left panel) and in MMhigh versus MMlow cells (right panel). Proteins, which were found significantly up- or downregulated, are delineated in each plot in the right and in the left area above the black curves, respectively. Proteins involved in translational processes, according to GO terms, are highlighted in red. C, Levels of typical markers for B cell differentiation and/or for MM cells. D, Heat maps representing LFQ values of proteins involved in pro- or anti-apoptotic processes. E, Levels of two proteins involved in apoptosis. Significant regulations are indicated by asterisks.
Proteins Involved in Apoptosis Regulated in MM Cells
To determine biological processes regulated in MM cells and possibly related to the hallmarks of cancer (37), we used different resources such as DAVID functional annotation tool (38, 39), UniProt information about proteins (40), and performed thorough research of scientific literature. In this way, first, we determined pathways that point to suppression of apoptosis in MM cells, including upregulation of anti-apoptotic proteins and downregulation of pro-apoptotic proteins (Fig. 1D). Some of these proteins were found regulated in all MM cells relative to B cells, exemplified by apoptosis regulator BAX, whereas others were only regulated at advanced stages of the disease, such as, for example, CASP10 (Fig. 1E).
Proteins Involved in ER Activities Regulated in MM Cells
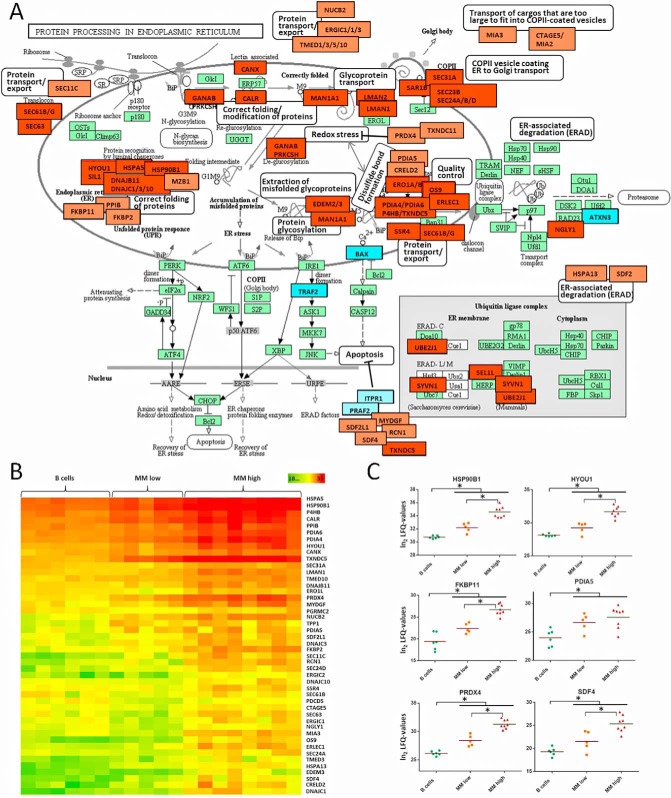
Further, several proteins related to biological activities in the endoplasmic reticulum (ER) such as folding and modification of newly synthesized proteins, as well as degradation of incorrectly folded proteins, and transport processes from the ER to the Golgi apparatus, were found at significantly elevated levels in MM cells (Fig. 2A). Most of these proteins were upregulated in a progressive way between B and MMlow cells, and further between MMlow and MMhigh cells (Fig. 2B and 2C), consistent with the increasing amounts of immunoglobulins produced by malignant plasma cells on disease progression. This was further accompanied by a significant upregulation of proteins involved in translational processes, according to GO terms (highlighted in Fig. 1B). Similarly, upregulation of proteins involved in control of redox stress caused by disulfide bond formation, such as PRDX4 and TXNDC11 (41, 42), was observed (Figs. 2A–2C). In parallel, pathways that limit ER stress-induced apoptosis seemed to be enhanced especially in MMhigh cells, involving downregulation of BAX, TRAF2, ITPR1, and PRAF2 (43–45) and upregulation of SDF2L1, MYDGF, SDF4, RCN1, and TXNDC5 (46–49) (Figs. 1D, 1E, 2A and 2C).
Fig. 2.
Protein processing in the in endoplasmic reticulum (ER) of MM cells. A, Based on the KEGG database for pathway analysis, proteins significantly upregulated in MM cells and related to ER activities were highlighted in red, downregulated proteins in blue. Lighter colors were used when proteins were associated to ER activities based on other references such as the UniProt database (40). Reproduction of the KEGG pathway map image “Protein processing in endoplasmic reticulum” (hsa04141) (127) with permission of Kanehisa Laboratories. B, Heat maps representing LFQ values of ER related proteins, demonstrating a progressive upregulation from B cells to MMlow and moreover to MMhigh cells. C, For selected proteins, corresponding LFQ values are represented as dot plots. Significant regulations are indicated by asterisks.
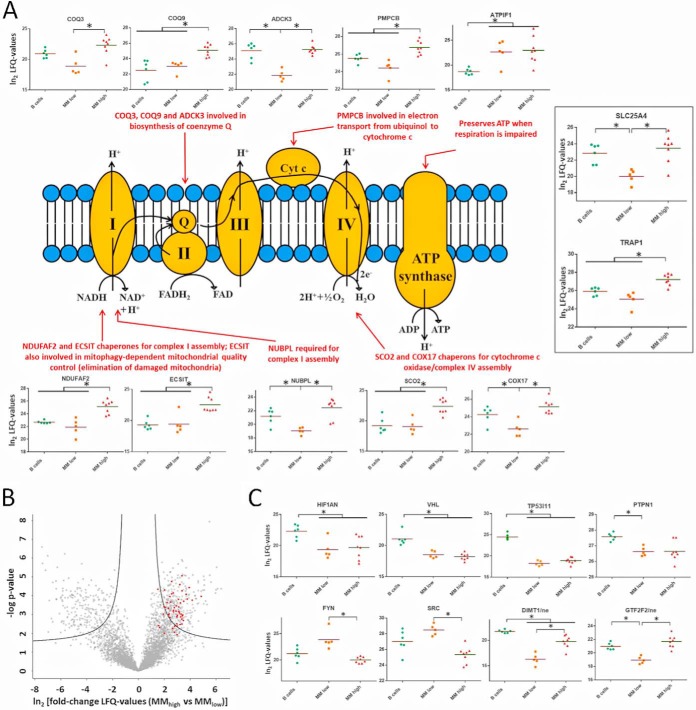
Regulation of Proteins Related to Respiratory Activities and to Hypoxia
Numerous mitochondrial proteins were found deregulated in MM cells. Although reduced levels of proteins of the respiratory chain, such as COX6C, MT-CO2, or ATP5L, were determined in all MM cells compared with B cells (supplemental Table S1), several other proteins of the respiratory chain, especially chaperones necessary for the assembly of respiratory complexes, were found at significant higher levels in MMhigh than in B cells and MMlow cells (Fig. 3A). These findings may point to a downshift of respiratory activities in MMlow cells and a reactivation in MMhigh cells. In line with this, proteins involved in mitochondrial translation, according to GO terms, appeared to be significantly upregulated in MMhigh versus MMlow cells (highlighted in Fig. 3B). Additionally, several proteins related to hypoxia were found regulated. HIF1AN and VHL (Fig. 3C), known to prevent activation of hypoxia-inducible factor (14, 50–53), were determined at reduced levels in MM cells, similarly to TP53I11 and PTPN1, described to be targets of hypoxia-induced micro-RNA-210 (54). Remarkably, DIMT1, described to be downregulated by micro-RNA-210 likewise (16), was found at lower levels only in MMlow cells, but was upregulated again in MMhigh cells. Similarly, higher levels of FYN and SRC, two tyrosine-protein kinases of the same family, were determined at higher levels only in MMlow cells; hypoxia has been shown to activate FYN either by phosphorylation or by upregulation of protein expression (55, 56). Also consistent with this, GTF2F2, reported to be downregulated under hypoxic conditions (57), was found at lower levels in MMlow than in MMhigh cells. These findings indicate that MM cells activate mechanisms in response to hypoxic conditions in the BM and that adaptation processes take place in MMhigh cells.
Fig. 3.
Regulation of proteins related to respiration and protein regulation by hypoxia. A, Levels of selected proteins related to the respiratory chain are represented, and biological functions are indicated for some proteins. B, Volcano plot comparing the protein levels in MMhigh versus MMlow cells. Proteins involved in mitochondrial translation processes, according to GO terms, are highlighted in red. C, Levels of selected proteins regulated by hypoxia, detected in the nuclear (ne) or in the cytoplasmic fraction (not specified). Significant regulations are indicated by asterisks.
Regulation of Proteins Involved in Metabolic Processes
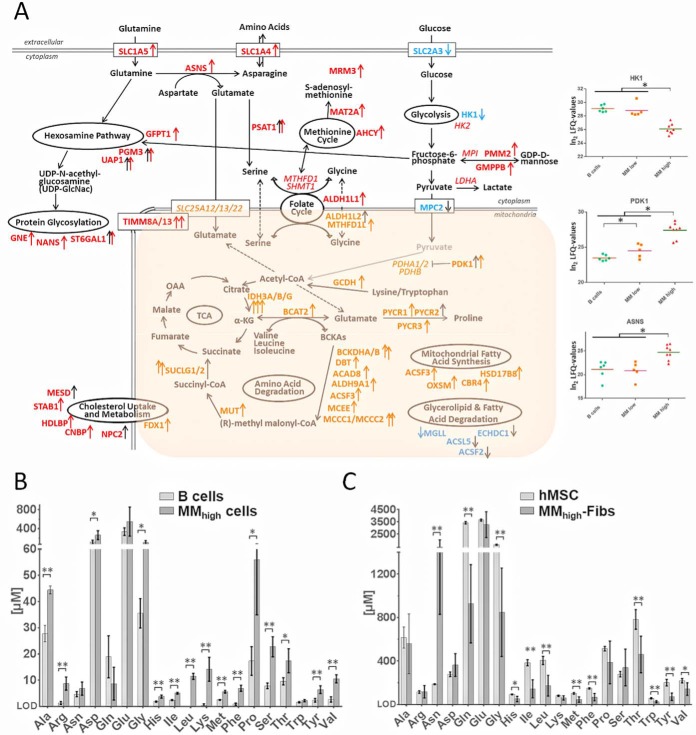
In addition, mitochondrial and cytoplasmic proteins involved in several metabolic processes appeared to be deregulated in myeloma cells. Fig. 4A shows metabolic pathways in which proteins were found significantly up- or downregulated between B cells, MMlow cells and MMhigh cells, the most important regulatory events occurring between MMlow and MMhigh cells. Most strikingly, the hexosamine pathway, amino acid interconversion reactions and degradation processes, the folate and the methionine cycle, cholesterol uptake and metabolism, and mitochondrial fatty acid synthesis were found upregulated in myeloma cells of advanced disease stages. These pathways seem thus to play an important role in the progression of multiple myeloma.
Fig. 4.
Regulation of proteins related to metabolic processes taking place in MM cells and results of metabolomics experiments. A, Metabolic pathways for which enzymes were found significantly up- or downregulated in MM versus B cells (black arrows) and in MMhigh versus MMlow cells (red and orange arrows for proteins upregulated in the cytoplasm and in mitochondria, respectively, blue arrows for downregulated proteins). Some proteins found at high levels but not regulated are indicated in italics. For three selected proteins, determined LFQ values are represented as dot plots. B, Levels of amino acids in B and MMhigh cells. C, Levels of amino acids in MM-associated BM fibroblasts (MM-Fibs) and in mesenchymal cells of the BM (hMSC) that were used as reference system. Significant regulations are indicated by asterisks. For metabolomics experiments: ** p-values < 0.01; * p-values < 0.05.
To support these results, we also performed targeted analyses of amino acids with both, plasma cells and MM-associated fibroblasts isolated from the BM of patients with advanced disease stages, using B cells and human mesenchymal stem cells of the BM (hMSCs) as reference cell system, respectively. In this way, higher amounts of several amino acids were determined in MMhigh cells compared with B cells (Fig. 4B), strengthening our proteomics data, which suggest that MM cells may use amino acids not primarily for the synthesis of immunoglobulins, but possibly also for other purposes such as catabolism. In parallel, MM-associated fibroblasts, essential supporters of MM cells in the BM microenvironment (8), proved to have rather low levels of amino acids, except for asparagine, which was present at extremely high levels in these cells when compared with hMSCs (Fig. 4C). Fibroblasts seem thus to be deregulated in a concerted fashion to MM cells.
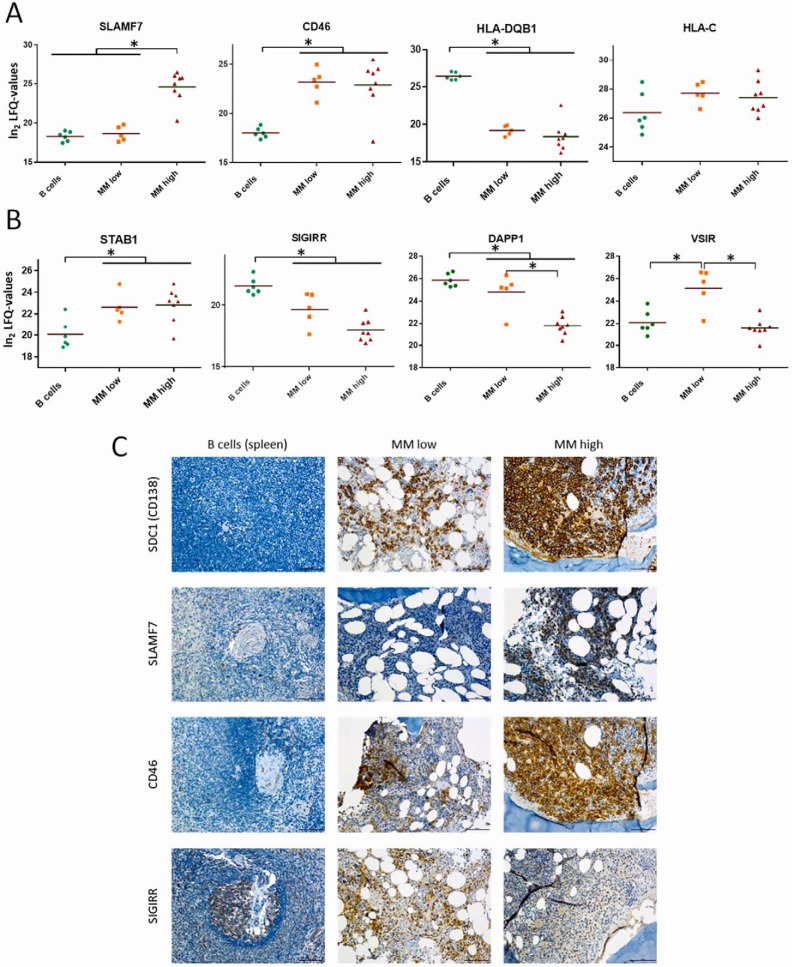
Protein Regulation Associated with Immune Evasion Strategies
Several pathways leading to immune evasion are known for MM cells. Our data independently confirmed upregulation of SLAMF7 and CD46 (10, 58), and downregulation of MHC-IIs such as HLA-DQB1 (35), together with positive expression of MHC-I molecules such as HLA-C in MM cells (Fig. 5A). Moreover, we were able to determine new candidates possibly involved in immune escape mechanisms of MM cells likewise. Regulation of such candidates, including STAB1, SIGIRR, DAPP1 as well as VSIR (Fig. 5B), have been described to suppress immune responses and to be involved in the patho-mechanisms of different diseases (59–63). Immunohistochemistry independently verified the relative expression levels of the proteins SLAM7, CD46, and SIGIRR together with SDC1 (CD138), which depend on the disease stage (Fig. 5C).
Fig. 5.
Regulation of proteins related to immune evasion strategies of MM cells. A, Levels of selected proteins known to be involved in immune evasion strategies of MM cells. B, Levels of selected proteins potentially contributing to immune evasion strategies of MM cells likewise. C, Selected proteins were verified by immunohistochemistry (IHC). Stainings were performed on BM biopsies of patient MM19–27, MM19–21 representing MMlow and MM22–27 MMhigh cases. Infiltration rates (IR) were assessed by evaluation of the entire tissue section. Because of focal infiltrates, the IR might vary within one section. Depicted image sections resemble representative areas. B cells in germinal centers of the spleen were used as reference system.
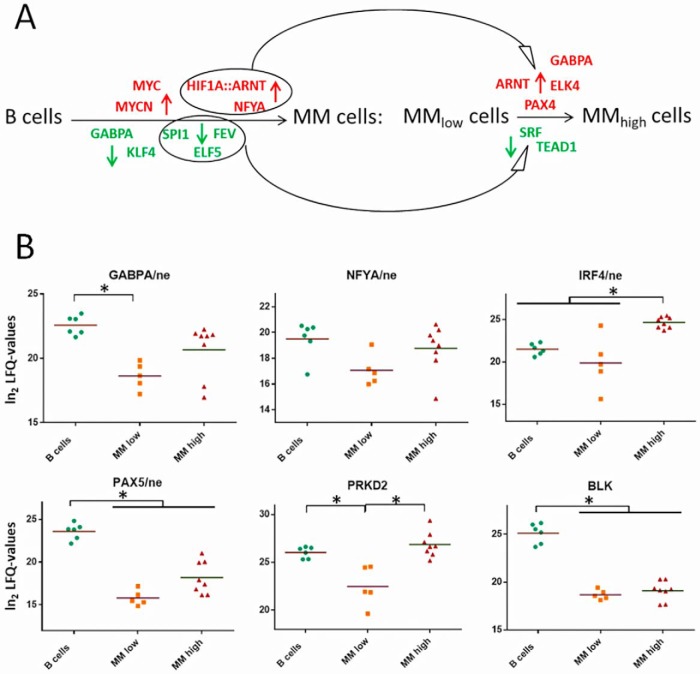
Transcription Factors Involved in Regulating Proteins in Primary Human Myeloma Cells
Finally, in order to determine over-represented conserved transcription factor binding sites and, thus, the main transcription factors responsible for the observed proteome alterations in MM cells, the corresponding sets of genes were submitted to the oPOSSUM software version 3.0 (34). This allowed us to find out transcription factors apparently playing an important role in the development and/or progression of multiple myeloma. Besides transcription factors known to be activated in MM, such as MYC, HIF1A, and ARNT (13, 64, 65), or to be suppressed in MM, such as SPI1 (PU.1) and KLF4 (66, 67), we were able to determine also NFYA, GABPA, ELK4, and PAX4 as transcription factors that may play an important role in the pathogenesis of MM (Fig. 6A). Levels of GABPA and NFYA determined in the nuclear fractions of MM cells are represented in Fig. 6B, together with the nuclear levels of two other transcriptions factors, PAX5 and IRF4, known to be involved in MM likewise. Downregulation of PAX5 is commonly associated with plasma cell differentiation and was related to multiple myeloma cell survival (68, 69). IRF4, originally identified as the product of a proto-oncogene involved in chromosomal translocations in multiple myeloma (70), has been described to be transcribed by MYC, whereas SPI1 seems to suppress the expression of IRF4 (64, 66). Finally, levels of two target proteins, PRKD2 and BLK are also shown in Fig. 6B. PRKD2 expression depends on the activity of transcription factor GABPA (71). BLK has been shown to be suppressed by MYC via downregulation of PAX5 (72).
Fig. 6.
Tanscription factors (TFs) mainly responsible for the observed up- and downregulations of proteins in MM cells. A, Well-known and newly determined TFs that may play an important role in MM development and/or progression. Red, TFs involved in upregulation of proteins; green, TFs involved in downregulation of proteins. HIF1A, ARNT, FYA, SPI1, FEV and ELF5 may be responsible for both, regulation of proteins in MM relative to B cells and in MMhigh relative to MMlow cells. B, Levels of TFs detected in nuclear fractions (ne), and of two known TF targets, PRKD2 and BLK, detected in cytoplasmic fractions. Significant regulations are indicated by asterisks.
DISCUSSION
Myeloma Cells are Adapted to Hypoxic Conditions in the Bone Marrow Microenvironment
It is generally recognized that the bone marrow (BM) is rather hypoxic compared with peripheral blood (13, 73). Silva and Gatenby have demonstrated using computer simulation that adaptation to hypoxic conditions in the BM microenvironment is essential for myeloma cells to progress to an aggressive stage (74). HIF1A and ARNT have been described to be upregulated by tumor cells including myeloma cells under low oxygen conditions (13, 65). Both transcription factors seem to be responsible for the induction of relevant proteins in MM cells (Fig. 6A). The observed downregulation of HIF1AN and VHL (Fig. 3C) also points to hypoxic conditions in the BM microenvironment of the cells (11, 14). Remarkably, even though expression of HIF1AN and VHL may also be affected by typical anti-myeloma treatments such as Bortezomid or Dexamethasone (52, 75), levels of these proteins were rather similar in all MM cells and apparently independent on patient treatments. Further, reduced levels of proteins of the respiratory chain, such as COX6C, MT-CO2 or ATP5L (supplemental Table S1) indicate a limited oxygen consumption rate in MM cells. In line with this, ATPase inhibitory factor ATPIF1 was found highly induced in MM cells (Fig. 3A); this protein is necessary to preserve ATP at the expense of the mitochondrial membrane potential when respiration is impaired, and protects cells from ATP depletion in response to hypoxia and glucose deprivation (76, 77).
However, a trend of increased synthesis of proteins of the respiratory complexes in MMhigh relative to MMlow cells was also observed, together with upregulation of the ADP/ATP translocase SLC25A4 (Fig. 3A). Further, proteins involved in mitochondrial translation, as well as TRAP1, an important mitochondrial chaperon that directly interacts with respiratory complexes and contributes to their stability and activity (78) were found at significantly elevated levels in MMhigh cells (Figs. 3A and 3B). Although its role in carcinogenesis is controversial, TRAP1 has been found highly expressed in several cancers, as reviewed by Matassa et al. (79), providing tumor cells the ability to use spare respiratory capacity when oxygen and glucose are limited (80). Additionally, mitochondrial enzymes of the folate cycle, highly relevant for mitochondrial translation and described to be strongly upregulated in proliferating lymphocytes (81), were found upregulated in MMhigh cells as well (Fig. 4A). Thus, it seems that, at advanced stages of the disease, myeloma cells get access to more oxygen, consistent with the “angiogenic switch” necessary for MM progression, as claimed by Silva and Gatenby (74), and with the pro-angiogenic phenotype associated with MM-related endothelial cells (15).
Metabolic Adaptations in MM Cells as Response to Hypoxic Conditions in the Bone Marrow
Myeloma cells have to adapt their metabolic pathways to be able to survive hypoxia. PDK1 is one of the key enzymes regulating metabolic adaptations in response to hypoxia, allowing cell proliferation under these conditions (82). We have found this enzyme significantly upregulated in MM versus B cells, and further in MMhigh versus MMlow cells (Fig. 4A). This kinase prevents the formation of acetyl-coenzyme A from pyruvate by inhibiting the activity of pyruvate dehydrogenase, thus disconnecting glycolysis and citric acid cycle. This strategy is well-known as Warburg-effect, a strategy commonly used by tumor cells, as reviewed by Nissim Hay (83). Inhibition of PDK1 has been proposed as target for MM therapy (84). Further, we found mitochondrial pyruvate carrier MPC2 significantly downregulated in MM cells (Fig. 4A); loss of MPC has been associated with carcinogenicity as well as with a stem cells phenotype (85, 86). Consistent with this, hypoxia has been shown to induce a stem cell-like phenotype in MM cells (87), and GABPA and its target PRKD2 (Figs. 6A and 6B) have been described to control proliferation of hematopoietic stem cells as well as development of leukemia (71).
Moreover, glucose import and the glycolytic enzyme HK1 appeared to be significantly downregulated in MMhigh cells (Fig. 4A), pointing to a reduced glycolysis rate in MM cells of advanced stages of the disease. This is in accordance with the already discussed upregulation of ATPIF1 and TRAP1 in MMhigh cells. On the other hand, uptake of glutamine seems to be highly increased in MMhigh cells (Fig. 4A), consistent with the observed glutamine dependence of MM cells (88). Glutamine is required for the hexosamine pathway, which is highly upregulated in MMhigh cells (Fig. 4A). This pathway leads to protein glycosylation as part of the completion process of newly synthesized proteins. Multiple myeloma is characterized by aberrantly glycosylated immunoglobulins (89), which are synthesized in high amounts by MM cells. Most importantly, glutamine can also be interconverted into asparagine, which might be used for the import of other amino acids via reciprocal exchange (90); levels of glutamine-hydrolyzing asparagine synthetase ASNS and transporter proteins involved in these processes, SLC1A4 and SLC1A5, were found significantly increased in MMhigh cells (Fig. 4A). Results of the targeted analyses of amino acids further point to the likelihood that BM fibroblasts may supply MM cells with several of the necessary amino acids, probably in exchange for asparagine (Figs. 4B and 4C). This view is supported by the fact that BM stromal cells such as fibroblasts are widely used as feeder layers for MM cells in cell culture experiments (91). Remarkably, asparagine synthesis has been associated with survival of glutamine-dependent tumor cells, and expression of ASNS was correlated with poor prognosis in several tumors (92). Studies have indeed suggested good anti-myeloma activity of asparaginase (93).
Glutamate arising from the interconversion of glutamine to asparagine may be used for the synthesis of serine, which feeds into the folate cycle (Fig. 4A). Interfering with the serine metabolism has been proposed to improve anti-myeloma therapies (94). Alternatively, glutamate may be transported into mitochondria to be channeled into the citric acid cycle, directly or via interconversion into other amino acids (Fig. 4A). As depicted above, it appears that myeloma cells of advanced stages of the disease are capable of increasing respiration again. Our data indicate that fuels for oxidative phosphorylation in MMhigh cells may arise from degradation of amino acids (Fig. 4A). Remarkably, we also observed increased mitochondrial fatty acid synthesis and upregulation of proteins involved in the methionine cycle (Fig. 4A). These two pathways give rise to lipoic acid, a cofactor of the enzyme BCKDH, which is necessary for branched chain amino acid degradation (95) and was found upregulated in MMhigh cells (Fig. 4A). There is, furthermore, evidence that mitochondrial fatty acid synthesis is essential for respiration and mitochondrial biogenesis (96).
Two proteins involved in mitochondrial fusion, CLPP and CLPX (97), were also found significantly upregulated in MMhigh cells, whereas OMA1, which prevents fusion (98) and SLC25A46, which is involved in mitochondrial fission (99) were found significantly downregulated in MM cells (supplemental Table S1). Mitochondria, which are metabolically challenged when cells switch to use amino acids or fatty acids for ATP production, have been described to undergo active fusion to avoid oxidative stress and mitochondrial damage (100). Enlarged mitochondria in multiple myeloma cells have already been described in the 1980s by Ghadially et al. (101). Further, we also found upregulation of ECSIT in MMhigh cells relative to MMlow and B cells (Fig. 3A); this protein, besides being involved in the assembly of mitochondrial complex I, also plays an essential role in mitophagy-dependent mitochondrial quality control, preventing accumulation of damaged mitochondria (102).
In addition, cholesterol uptake as well as cholesterol metabolic processes were found upregulated in MM cells and especially in MMhigh cells (Fig. 4A). This is in accordance with observations about hypocholesterinemia in the plasma of MM patients, associated with increased LDL clearance and use of cholesterol by MM cells, which seem to be necessary for MM cell survival (103, 104).
Responsible for deregulation of metabolic processes in MMhigh versus MMlow cells may be the activation of transcription factor NFYA, which appeared to be involved in protein regulation in MM cells and showed a trend of upregulation in MMhigh cells (Figs. 6A and 6B). This transcription factor promotes the expression of proteins fueling metabolic pathways commonly altered in cancer cells, such as the serine, the one carbon, the glycine, the glutamine pathways and the cholesterol pathway (105). Further, MYC is involved in regulating glutamine metabolism as well. Very recently, Gonsalves et al. have demonstrated that higher levels of MYC in smoldering myeloma cells correlate with an increased use of glutamine for the citric acid cycle and a shorter time to progression to MM (106). This is consistent with our data indicating an increased uptake of glutamine in MMhigh cells, which apparently channel this amino acid into the citric acid cycle (Fig. 4A).
Myeloma Cells Adapt ER Activities to Hypoxic Conditions in the Bone Marrow
Stress proteins of the endoplasmic reticulum (ER) such as glucose-regulated and heat-shock proteins have been shown to be involved in resistance against environmental stress such as that caused by hypoxia, in avoidance of stress-induced apoptosis, and in cancer development (107, 108). It has also been demonstrated that protein-disulfide isomerases are upregulated in response to hypoxia and are critical for cell viability under these conditions (109). In line with this, we found massive upregulation of HSP90B1 (GRP94), HSPA5 (GRP78), HYOU1 (GRP170), and HSPA13 in MM versus B cells, and moreover in MMhigh versus MMlow cells; the same trend was observed for protein-disulfide isomerases such as P4HB, PDIA4, PDIA5, and PDIA6 (Figs. 2A–2C). Azoitei et al. have shown that signals from hypoxia and HSP90 pathways are interconnected via PRKD2 (Figs. 6A and 6B), which is apparently necessary to prevent apoptosis and promote tumor angiogenesis and tumor growth (110). Activation of transcription factor PAX4 (Fig. 6A), proposed as candidate oncogene in hematologic malignancies (111), has also shown to contribute to protect against ER stress-induced apoptosis (112).
Myeloma Cells Have Strategies to Prevent Apoptosis Under Hypoxic and Normoxic Conditions
Ikeda et al. have demonstrated that myeloma cells are compatible with both hypoxic and normoxic conditions (16). Under hypoxic conditions, HIF apparently activates micro-RNA-210 that blocks the DIMT1-IRF4 axis and leads to glycolysis and to quiescence of the cells. Under normoxic conditions, active DIMT1 and IRF4 induce myeloma cell maturation and proliferation. Remarkably, we found IRF4 and DIMT1 to be present at significantly higher levels in MMhigh than in MMlow cells (Figs. 3C and 6B). This fits to our model in which MMlow cells are confronted with more hypoxic conditions than MMhigh cells, and that glycolysis is rather downregulated in MMhigh versus MMlow cells. Most importantly, myeloma cells are obviously capable of preventing apoptosis under both conditions (16). In line with this, we were able to determine several proteins that may support cell death escaping mechanisms in MMlow and MMhigh cells (Fig. 1D). In MMlow cells hypoxia-induced anti-apoptotic factors such as SRC and FYN (55, 113) were found upregulated (Fig. 3C). This was accompanied by downregulation of pro-apoptotic proteins such as hypoxia-regulated proteins BAX (114), TP53I11 (115), and BLK (116) (Figs. 1E, 3C, and 6B). In MMhigh cells, elevated levels of anti-apoptotic proteins such as HYOU1 (117), SDF4 (47, 118), TRAP1 (119), and the ADP/ATP translocase SLC25A4 (120) (Figs. 1D, 2C, and 3A) were detected, together with IRF4 (121), which may also act indirectly in an anti-apoptotic way via induction of CASP10 (122) (Figs. 1E and 6B).
Myeloma Cells Develop Hypoxia-driven Strategies to Escape Immune Response
Besides evading intrinsic apoptosis, MM cells have pathways that limit extrinsic apoptosis induced by the immune system, involving for example upregulation of CD38, CD46, SLAMF7 (CS1), and MHC-I molecules (Figs. 1C, 5A, and 5C) (10, 58). In case of CD38 and SLAM7 therapeutic antibodies are already tested in clinical trials for combination therapies (123). Remarkably, SLAM7, showed different expression levels depending on the disease stage (Figs. 5A and 5C), and may thus be applied as stratification marker of use in antibody therapy. Reduced levels of MHC-II molecules, as well as MHC-II-associated proteins such as CD37 (Figs. 1C and 5A), may also contribute to immune evasion by impeding the antigen presentation capacity of MM cells and their interactions with T cells (124, 125). Upregulation of MYC (Fig. 6A) has been shown to disrupt MHC-II-mediated immune recognition of human B cell tumors (126).
The presently applied untargeted method also revealed novel candidates potentially involved in immune escape mechanisms of MM cells, such as DAPP1 and STAB1 (Fig. 5B) (59, 62). Further, VSIR (Fig. 5B) may negatively regulate T cell functions, at least at earlier stages of the disease. This protein has been found highly expressed in tumor microenvironments where it seems to suppress the proliferation of T cells without affecting B cells, as described by Lines et al. (63). Reduced levels of SIGIRR (Figs. 5B and 5C) may also be involved in immune escape mechanisms of MM cells. This protein has also been found downregulated in chronic lymphocytic leukemia (61). It is however worth mentioning that, when comparing immune evasion strategies of MM cells with those of CLL cells described in a previous study of our group (22), more differences than similarities became evident.
Remarkably, hypoxic conditions in the BM microenvironment may induce mechanisms leading to immune evasion of MM cells. As mentioned before, Ikeda et al. have shown that under hypoxic conditions micro-RNA-210 is activated in MM cells (16). Noman et al. have demonstrated that, in lung cancer and melanoma, high levels of micro-RNA-210 dramatically decrease tumor cell susceptibility to cytotoxic T cell-mediated lysis via coordinate silencing of PTPN1, HOXA1 and TP53I11 (54). Our data indicate that PTPN1 is significantly reduced in MMlow relative to B cells and a similar trend was observed for MMhigh versus B cells (Fig. 3C). HOXA1 was not detected in any sample investigated in the present study. TP53I11 was found significantly downregulated in both MMlow and MMhigh cells (Fig. 3C), and the higher oxygen availability of MMhigh cells seems not to be sufficient to restore high levels of this protein.
Conclusion and Outlook
The present proteome profiling data strongly support that adaptation of MM cells to hypoxia accompanies myeloma disease progression. Our results demonstrate that in-depth proteome profiling is very well suited to give deep insights into processes that are contributing to tumor development and progression. The data clearly reproduced established knowledge on myeloma cells, but also present novel findings and causal relations of relevant pathways taking place in these tumor cells. Importantly, it seems that strategies exploited by myeloma cells to allow survival and proliferation, including immune evasion mechanisms and metabolic adaptations, are more dependent on the disease state than on the genetic background. This study may thus support the development of improved stratification and anti-myeloma treatment strategies.
Data availability
Proteomics data were submitted to the ProteomeXchange Consortium via the PRIDE partner repository. Results are fully accessible via www.proteomeexchange.org with the identifier PXD010600.
Supplementary Material
Acknowledgments
We thank Guenter Walder for assisting cell culture experiments and Raphael Ambros for managing patient data.
Footnotes
* This work was supported by the “Jubilaeumsfond der Oesterreichischen Nationalbank” (project number 16703) and further support by the Faculty of Chemistry of the University of Vienna.
 This article contains supplemental Tables.
This article contains supplemental Tables.
The authors declare no competing financial interests.
1 The abbreviations used are:
- MM
- multiple myeloma
- BM
- bone marrow
- ER
- endoplasmic reticulum
- FASP
- filter aided sample preparation
- GO
- gene ontology
- HCD
- high-energy collisional dissociation
- IR
- infiltration rate
- LDL
- low density lipoprotein
- LFQ
- label-free quantification
- MACS
- magnetic activated cell sorting
- MGUS
- monoclonal gammopathy of undetermined significance
- MHC
- major histocompatibility complex
- MWCO
- molecular weight cut off
- PCA
- principle component analysis
- SMM
- smoldering multiple myeloma
- TF
- transcription factor.
REFERENCES
- 1. Kyle R. A., and Rajkumar S. V. (2008) Multiple myeloma. Blood 111, 2962–2972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Terpos E., and Soc I. M. (2018) Multiple myeloma: clinical updates from the American Society of Hematology Annual Meeting, 2017. Cl. Lymph. Myelom. Leuk. 18, 321–334 [DOI] [PubMed] [Google Scholar]
- 3. du Pont S. R., Cleynen A., Fontan C., Attal M., Munshi N., Corre J., and Avet-Loiseau H. (2017) Genomics of multiple myeloma. J. Clin. Oncol. 35, 963–967 [DOI] [PubMed] [Google Scholar]
- 4. van de Donk N. W. C. J., Mutis T., Poddighe P. J., Lokhorst H. M., and Zweegman S. (2016) Diagnosis, risk stratification and management of monoclonal gammopathy of undetermined significance and smoldering multiple myeloma. Int. J. Lab. Hematol. 38, 110–122 [DOI] [PubMed] [Google Scholar]
- 5. Bianchi G., and Munshi N. C. (2015) Pathogenesis beyond the cancer clone (s) in multiple myeloma. Blood 125, 3049–3058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dutta A. K., Fink J. L., Grady J. P., Morgan G. J., Mullighan C. G., To L. B., Hewett D. R., and Zannettino A. C. W. (2018) Subclonal evolution in disease progression from MGUS/SMM to multiple myeloma is characterised by clonal stability. Leukemia 33, 457–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mitsiades C. S., Mitsiades N. S., Munshi N. C., Richardson P. G., and Anderson K. C. (2006) The role of the bone microenvironment in the pathophysiology and therapeutic management of multiple myeloma: Interplay of growth factors, their receptors and stromal interactions. Eur. J. Cancer 42, 1564–1573 [DOI] [PubMed] [Google Scholar]
- 8. Slany A., Haudek-Prinz V., Meshcheryakova A., Bileck A., Lamm W., Zielinski C., Gerner C., and Drach J. (2014) Extracellular matrix remodeling by bone marrow fibroblast-like cells correlates with disease progression in multiple myeloma. J. Proteome Res. 13, 844–854 [DOI] [PubMed] [Google Scholar]
- 9. Gooding S., and Edwards C. M. (2016) New approaches to targeting the bone marrow microenvironment in multiple myeloma. Curr. Opin. Pharmacol. 28, 43–49 [DOI] [PubMed] [Google Scholar]
- 10. Pittari G., Vago L., Festuccia M., Bonini C., Mudawi D., Giaccone L., and Bruno B. (2017) Restoring natural killer cell immunity against multiple myeloma in the era of new drugs. Front. Immunol. 8, 1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hu J., Van Valckenborgh E., Menu E., De Bruyne E., and Vanderkerken K. (2012) Understanding the hypoxic niche of multiple myeloma: therapeutic implications and contributions of mouse models. Dis. Model Mech. 5, 763–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Filippi I., Saltarella I., Aldinucci C., Carraro F., Ria R., Vacca A., and Naldini A. (2018) Different adaptive responses to hypoxia in normal and multiple myeloma endothelial cells. Cell Physiol. Biochem. 46, 203–212 [DOI] [PubMed] [Google Scholar]
- 13. Colla S., Storti P., Donofrio G., Todoerti K., Bolzoni M., Lazzaretti M., Abeltino M., Ippolito L., Neri A., Ribatti D., Rizzoli V., Martella E., and Giuliani N. (2010) Low bone marrow oxygen tension and hypoxia-inducible factor-1 alpha overexpression characterize patients with multiple myeloma: role on the transcriptional and proangiogenic profiles of CD138(+) cells. Leukemia 24, 1967–1970 [DOI] [PubMed] [Google Scholar]
- 14. Martin S. K., Diamond P., Gronthos S., Peet D. J., and Zannettino A. C. (2011) The emerging role of hypoxia, HIF-1 and HIF-2 in multiple myeloma. Leukemia 25, 1533–1542 [DOI] [PubMed] [Google Scholar]
- 15. Vacca A., Ria R., Semeraro F., Merchionne F., Coluccia M., Boccarelli A., Scavelli C., Nico B., Gernone A., Battelli F., Tabilio A., Guidolin D., Petrucci M. T., Ribatti D., and Dammacco F. (2003) Endothelial cells in the bone marrow of patients with multiple myeloma. Blood 102, 3340–3348 [DOI] [PubMed] [Google Scholar]
- 16. Ikeda S., Kitadate A., Abe F., Saitoh H., Michishita Y., Hatano Y., Kawabata Y., Kitabayashi A., Teshima K., Kume M., Takahashi N., and Tagawa H. (2017) Hypoxia-inducible microRNA-210 regulates the DIMT1-IRF4 oncogenic axis in multiple myeloma. Cancer Sci. 108, 641–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bhaskar A., and Tiwary B. N. (2016) Hypoxia inducible factor-1 alpha and multiple myeloma. Int. J. Adv. Res. 4, 706–715 [PMC free article] [PubMed] [Google Scholar]
- 18. Groessl M., Slany A., Bileck A., Gloessmann K., Kreutz D., Jaeger W., Pfeiler G., and Gerner C. (2014) Proteome profiling of breast cancer biopsies reveals a wound healing signature of cancer-associated fibroblasts. J. Proteome Res. 13, 4773–4782 [DOI] [PubMed] [Google Scholar]
- 19. Slany A., Bileck A., Kreutz D., Mayer R. L., Muqaku B., and Gerner C. (2016) Contribution of human fibroblasts and endothelial cells to the hallmarks of inflammation as determined by proteome profiling. Mol. Cell. Proteomics 15, 1982–1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tahir A., Bileck A., Muqaku B., Niederstaetter L., Kreutz D., Mayer R. L., Wolrab D., Meier S. M., Slany A., and Gerner C. (2017) Combined proteome and eicosanoid profiling approach for revealing implications of human fibroblasts in chronic inflammation. Anal. Chem. 89, 1945–1954 [DOI] [PubMed] [Google Scholar]
- 21. Muqaku B., Eisinger M., Meier S. M., Tahir A., Pukrop T., Haferkamp S., Slany A., Reichle A., and Gerner C. (2017) Multi-omics analysis of serum samples demonstrates reprogramming of organ functions via systemic calcium mobilization and platelet activation in metastatic melanoma. Mol. Cell. Proteomics 16, 86–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mayer R. L., Schwarzmeier J. D., Gerner M. C., Bileck A., Mader J. C., Meier-Menches S. M., Gerner S. M., Schmetterer K. G., Pukrop T., Reichle A., Slany A., and Gerner C. (2018) Proteomics and metabolomics identify molecular mechanisms of aging potentially predisposing for chronic lymphocytic leukemia. Mol. Cell. Proteomics 17, 290–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dytfeld D., Luczak M., Wrobel T., Usnarska-Zubkiewicz L., Brzezniakiewicz K., Jamroziak K., Giannopoulos K., Przybylowicz-Chalecka A., Ratajczak B., Czerwinska-Rybak J., Nowicki A., Joks M., Czechowska E., Zawartko M., Szczepaniak T., Grzasko N., Morawska M., Bochenek M., Kubicki T., Morawska M., Tusznio K., Jakubowiak A., and Komarnicki M. (2016) Comparative proteomic profiling of refractory/relapsed multiple myeloma reveals biomarkers involved in resistance to bortezomib-based therapy. Oncotarget 7, 56726–56736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fernando R. C., de Carvalho F., Mazzotti D. R., Evangelista A. F., Braga W. M. T., de Lourdes Chauffaille M., Leme A. F. P., and Colleoni G. W. B. (2015) Multiple myeloma cell lines and primary tumors proteoma: protein biosynthesis and immune system as potential therapeutic targets. Genes Cancer 6, 462–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Xie Z., Gunaratne J., Cheong L. L., Liu S. C., Koh T. L., Huang G., Blackstock W. P., and Chng W. J. (2013) Plasma membrane proteomics identifies biomarkers associated with MMSET overexpression in T(4;14) multiple myeloma. Oncotarget 4, 1008–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ge F., Xiao C. L., Yin X. F., Lu C. H., Zeng H. L., and He Q. Y. (2010) Phosphoproteomic analysis of primary human multiple myeloma cells. J. Proteomics 73, 1381–1390 [DOI] [PubMed] [Google Scholar]
- 27. Haudek-Prinz V. J., Klepeisz P., Slany A., Griss J., Meshcheryakova A., Paulitschke V., Mitulovic G., Stockl J., and Gerner C. (2012) Proteome signatures of inflammatory activated primary human peripheral blood mononuclear cells. J Proteomics 76, Spec No., 150–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bileck A., Kreutz D., Muqaku B., Slany A., and Gerner C. (2014) Comprehensive Assessment of Proteins Regulated by Dexamethasone Reveals Novel Effects in Primary Human Peripheral Blood Mononuclear Cells. J. Proteome Res. 13, 5989–6000 [DOI] [PubMed] [Google Scholar]
- 29. Wisniewski J. R., Zougman A., Nagaraj N., and Mann M. (2009) Universal sample preparation method for proteome analysis. Nat. Methods 6, 359–362 [DOI] [PubMed] [Google Scholar]
- 30. Cox J., and Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 31. Vizcaino J. A., Csordas A., del-Toro N., Dianes J. A., Griss J., Lavidas I., Mayer G., Perez-Riverol Y., Reisinger F., Ternent T., Xu Q. W., Wang R., and Hermjakob H. (2016) 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 44, D447–D456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Palumbo A., Avet-Loiseau H., Oliva S., Lokhorst H. M., Goldschmidt H., Rosinol L., Richardson P., Caltagirone S., Lahuerta J. J., Facon T., Bringhen S., Gay F., Attal M., Passera R., Spencer A., Offidani M., Kumar S., Musto P., Lonial S., Petrucci M. T., Orlowski R. Z., Zamagni E., Morgan G., Dimopoulos M. A., Durie B. G., Anderson K. C., Sonneveld P., San Miguel J., Cavo M., Rajkumar S. V., and Moreau P. (2015) Revised international staging system for multiple myeloma: A report from international myeloma working group. J. Clin. Oncol. 33, 2863–2869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cox J., and Mann M. (2012) 1D and 2D annotation enrichment: a statistical method integrating quantitative proteomics with complementary high-throughput data. BMC Bioinformatics 13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ho Sui S. J., Mortimer J. R., Arenillas D. J., Brumm J., Walsh C. J., Kennedy B. P., and Wasserman W. W. (2005) oPOSSUM: identification of over-represented transcription factor binding sites in co-expressed genes. Nucleic Acids Res. 33, 3154–3164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lohmeyer J., Hadam M., Santoso S., Forster W., Schulz A., and Pralle H. (1988) Establishment and characterization of a permanent human Iga2 kappa-myeloma cell-line. Brit. J. Haematol. 69, 335–343 [DOI] [PubMed] [Google Scholar]
- 36. Rawstron A. C., Owen R. G., Davies F. E., Johnson R. J., Jones R. A., Richards S. J., Evans P. A., Child J. A., Smith G. M., Jack A. S., and Morgan G. J. (1997) Circulating plasma cells in multiple myeloma: Characterization and correlation with disease stage. Brit. J. Haematol. 97, 46–55 [DOI] [PubMed] [Google Scholar]
- 37. Hanahan D., and Weinberg R. A. (2011) Hallmarks of cancer: The next generation. Cell 144, 646–674 [DOI] [PubMed] [Google Scholar]
- 38. Huang D. W., Sherman B. T., and Lempicki R. A. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 [DOI] [PubMed] [Google Scholar]
- 39. Huang D. W., Sherman B. T., and Lempicki R. A. (2009) Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bateman A., Martin M. J., O'Donovan C., Magrane M., Alpi E., Antunes R., Bely B., Bingley M., Bonilla C., Britto R., Bursteinas B., Bye Jee -A. -H., Cowley A., Da Silva A., De Giorgi M., Dogan T., Fazzini F., Castro L. G., Figueira L., Garmiri P., Georghiou G., Gonzalez D., Hatton-Ellis E., Li W. Z., Liu W. D., Lopez R., Luo J., Lussi Y., MacDougall A., Nightingale A., Palka B., Pichler K., Poggioli D., Pundir S., Pureza L., Qi G. Y., Rosanoff S., Saidi R., Sawford T., Shypitsyna A., Speretta E., Turner E., Tyagi N., Volynkin V., Wardell T., Warner K., Watkins X., Zaru R., Zellner H., Xenarios I., Bougueleret L., Bridge A., Poux S., Redaschi N., Aimo L., Argoud-Puy G., Auchincloss A., Axelsen K., Bansal P., Baratin D., Blatter M. C., Boeckmann B., Bolleman J., Boutet E., Breuza L., Casal-Casas C., de Castro E., Coudert E., Cuche B., Doche M., Dornevil D., Duvaud S., Estreicher A., Famiglietti L., Feuermann M., Gasteiger E., Gehant S., Gerritsen V., Gos A., Gruaz-Gumowski N., Hinz U., Hulo C., Jungo F., Keller G., Lara V., Lemercier P., Lieberherr D., Lombardot T., Martin X., Masson P., Morgat A., Neto T., Nouspikel N., Paesano S., Pedruzzi I., Pilbout S., Pozzato M., Pruess M., Rivoire C., Roechert B., Schneider M., Sigrist C., Sonesson K., Staehli S., Stutz A., Sundaram S., Tognolli M., Verbregue L., Veuthey A. L., Wu C. H., Arighi C. N., Arminski L., Chen C. M., Chen Y. X., Garavelli J. S., Huang H. Z., Laiho K., McGarvey P., Natale D. A., Ross K., Vinayaka C. R., Wang Q. H., Wang Y. Q., Yeh L. S., Zhang J., and Consortium U. (2017) UniProt: the universal protein knowledgebase. Nucleic Acids Res. 45, D158–D169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Shimizu Y., and Hendershot L. M. (2009) Oxidative folding: cellular strategies for dealing with the resultant equimolar production of reactive oxygen species. Antioxid. Redox Sign. 11, 2317–2331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Demasi A. P., Martinez E. F., Napimoga M. H., Freitas L. L., Vassallo J., Duarte A. S., Soares A. B., Araujo N. S., and Araujo V. C. (2013) Expression of peroxiredoxins I and IV in multiple myeloma: association with immunoglobulin accumulation. Virchows Arch. 463, 47–55 [DOI] [PubMed] [Google Scholar]
- 43. Scorrano L., Oakes S. A., Opferman J. T., Cheng E. H., Sorcinelli M. D., Pozzan T., and Korsmeyer S. J. (2003) BAX and BAK regulation of endoplasmic reticulum Ca2+: A control point for apoptosis. Science 300, 135–139 [DOI] [PubMed] [Google Scholar]
- 44. Zeng T., Peng L. F., Chao H. C., Xi H. B., Fu B., Wang Y. B., Zhu Z. W., and Wang G. X. (2015) IRE1 alpha-TRAF2-ASK1 complex-mediated endoplasmic reticulum stress and mitochondrial dysfunction contribute to CXC195-induced apoptosis in human bladder carcinoma T24 cells. Biochem. Bioph. Res. Co. 460, 530–536 [DOI] [PubMed] [Google Scholar]
- 45. Vento M. T., Zazzu V., Loffreda A., Cross J. R., Downward J., Stoppelli M. P., and Iaccarino I. (2010) Praf2 is a novel Bcl-xL/Bcl-2 interacting protein with the ability to modulate survival of cancer cells. Plos One 5, e15636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tiwari A., Schuiki I., Zhang L. L., Allister E. M., Wheeler M. B., and Volchuk A. (2013) SDF2L1 interacts with the ER-associated degradation machinery and retards the degradation of mutant proinsulin in pancreatic beta-cells. J. Cell Sci. 126, 1962–1968 [DOI] [PubMed] [Google Scholar]
- 47. Chen L., Xu S., Liu L., Wen X., Xu Y., Chen J., and Teng J. (2014) Cab45S inhibits the ER stress-induced IRE1-JNK pathway and apoptosis via GRP78/BiP. Cell Death Dis. 5, e1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Xu S., Xu Y., Chen L., Fang Q., Song S., Chen J., and Teng J. (2017) RCN1 suppresses ER stress-induced apoptosis via calcium homeostasis and PERK-CHOP signaling. Oncogenesis 6, e304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Peng F. S., Zhang H. L., Du Y. H., and Tan P. Q. (2018) Cetuximab enhances cisplatin-induced endoplasmic reticulum stress-associated apoptosis in laryngeal squamous cell carcinoma cells by inhibiting expression of TXNDC5. Mol. Med. Reports 17, 4767–4776 [DOI] [PubMed] [Google Scholar]
- 50. Lando D., Peet D. J., Gorman J. J., Whelan D. A., Whitelaw M. L., and Bruick R. K. (2002) FIH-1 is an asparaginyl hydroxylase enzyme that regulates the transcriptional activity of hypoxia-inducible factor. Gene Dev. 16, 1466–1471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Maxwell P. H., Wiesener M. S., Chang G. W., Clifford S. C., Vaux E. C., Cockman M. E., Wykoff C. C., Pugh C. W., Maher E. R., and Ratcliffe P. J. (1999) The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature 399, 271–275 [DOI] [PubMed] [Google Scholar]
- 52. Shin D. H., Chun Y. S., Lee D. S., Huang L. E., and Park J. W. (2008) Bortezomib inhibits tumor adaptation to hypoxia by stimulating the FIH-mediated repression of hypoxia-inducible factor-1. Blood 111, 3131–3136 [DOI] [PubMed] [Google Scholar]
- 53. Mahon P. C., Hirota K., and Semenza G. L. (2001) FIH-1: a novel protein that interacts with HIF-1alpha and VHL to mediate repression of HIF-1 transcriptional activity. Genes Dev. 15, 2675–2686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Noman M. Z., Buart S., Romero P., Ketari S., Janji B., Mari B., Mami-Chouaib F., and Chouaib S. (2012) Hypoxia-inducible miR-210 regulates the susceptibility of tumor cells to lysis by cytotoxic T cells. Cancer Res. 72, 4629–4641 [DOI] [PubMed] [Google Scholar]
- 55. Hu H., Takano N., Xiang L., Gilkes D. M., Luo W., and Semenza G. L. (2014) Hypoxia-inducible factors enhance glutamate signaling in cancer cells. Oncotarget 5, 8853–8868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Razorenova O. V., Finger E. C., Colavitti R., Chernikova S. B., Boiko A. D., Chan C. K., Krieg A., Bedogni B., LaGory E., Weissman I. L., Broome-Powell M., and Giaccia A. J. (2011) VHL loss in renal cell carcinoma leads to up-regulation of CUB domain-containing protein 1 to stimulate PKC{delta}-driven migration. Proc. Natl. Acad. Sci. U.S.A. 108, 1931–1936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Minchenko O. H., Tsymbal D. O., Minchenko D. O., and Kubaychuk O. O. (2016) Hypoxic regulation of MYBL1, MEST, TCF3, TCF8, GTF2B, GTF2F2 and SNAI2 genes expression in U87 glioma cells upon IRE1 inhibition. Ukr. Biochem. J. 88, 52–62 [DOI] [PubMed] [Google Scholar]
- 58. Sherbenou D. W., Aftab B. T., Su Y., Behrens C. R., Wiita A., Logan A. C., Acosta-Alvear D., Hann B. C., Walter P., Shuman M. A., Wu X. B., Atkinson J. P., Wolf J. L., Martin T. G., and Liu B. (2016) Antibody-drug conjugate targeting CD46 eliminates multiple myeloma cells. J. Clin. Invest. 126, 4640–4653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Palani S., Elima K., Ekholm E., Jalkanen S., and Salmi M. (2016) Monocyte stabilin-1 suppresses the activation of Th1 lymphocytes. J. Immunol. 196, 115–123 [DOI] [PubMed] [Google Scholar]
- 60. Karikoski M., Marttila-Ichihara F., Elima K., Rantakari P., Hollmen M., Kelkka T., Gerke H., Huovinen V., Irjala H., Holmdahl R., Salmi M., and Jalkanen S. (2014) Clever-1/stabilin-1 controls cancer growth and metastasis. Clin Cancer Res. 20, 6452–6464 [DOI] [PubMed] [Google Scholar]
- 61. Vilia M. G., Fonte E., Rodriguez T. V., Tocchetti M., Ranghetti P., Scarfo L., Papakonstantinou N., Ntoufa S., Stamatopoulos K., Ghia P., and Muzio M. (2017) The inhibitory receptor toll interleukin-1R 8 (TIR8/IL-1R8/SIGIRR) is downregulated in chronic lymphocytic leukemia. Leukemia Lymphoma 58, 2419–2425 [DOI] [PubMed] [Google Scholar]
- 62. Al-Alwan M., Hou S., Zhang T. T., Makondo K., and Marshall A. J. (2010) Bam32/DAPP1 promotes B cell adhesion and formation of polarized conjugates with T cells. J. Immunol. 184, 6961–6969 [DOI] [PubMed] [Google Scholar]
- 63. Lines J. L., Pantazi E., Mak J., Sempere L. F., Wang L., O'Connell S., Ceeraz S., Suriawinata A. A., Yan S., Ernstoff M. S., and Noelle R. (2014) VISTA is an immune checkpoint molecule for human T cells. Cancer Res. 74, 1924–1932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Shaffer A. L., Emre N. C. T., Lamy L., Ngo V. N., Wright G., Xiao W. M., Powell J., Dave S., Yu X., Zhao H., Zeng Y. X., Chen B. Z., Epstein J., and Staudt L. M. (2008) IRF4 addiction in multiple myeloma. Nature 454, 226–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Hassen W., Kassambara A., Reme T., Sahota S., Seckinger A., Vincent L., Cartron G., Moreaux J., Hose D., and Klein B. (2015) Drug metabolism and clearance system in tumor cells of patients with multiple myeloma. Oncotarget 6, 6431–6447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Ueno N., Nishimura N., Ueno S., Endo S., Tatetsu H., Hirata S., Hata H., Matsuoka M., Mitsuya H., and Okuno Y. (2017) PU.1 acts as tumor suppressor for myeloma cells through direct transcriptional repression of IRF4. Oncogene 36, 4481–4497 [DOI] [PubMed] [Google Scholar]
- 67. Schoenhals M., Kassambara A., Veyrune J. L., Moreaux J., Goldschmidt H., Hose D., and Klein B. (2013) Kruppel-like factor 4 blocks tumor cell proliferation and promotes drug resistance in multiple myeloma. Haematologica 98, 1442–1449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Nera K. P., Kohonen P., Narvi E., Peippo A., Mustonen L., Terho P., Koskela K., Buerstedde J. M., and Lassila O. (2006) Loss of Pax5 promotes plasma cell differentiation. Immunity 24, 283–293 [DOI] [PubMed] [Google Scholar]
- 69. Proulx M., Cayer M. P., Drouin M., Laroche A., and Jung D. (2010) Overexpression of PAX5 induces apoptosis in multiple myeloma cells. Int. J. Hematol. 92, 451–462 [DOI] [PubMed] [Google Scholar]
- 70. Iida S., Rao P. H., Butler M., Corradini P., Boccadoro M., Klein B., Chaganti R. S., and Dalla-Favera R. (1997) Deregulation of MUM1/IRF4 by chromosomal translocation in multiple myeloma. Nat. Genet. 17, 226–230 [DOI] [PubMed] [Google Scholar]
- 71. Yang Z. F., Zhang H. J., Ma L. Y., Peng C., Chen Y. Y., Wang J. L., Green M. R., Li S. G., and Rosmarin A. G. (2013) GABP transcription factor is required for development of chronic myelogenous leukemia via its control of PRKD2. Proc. Natl. Acad. Sci. U.S.A. 110, 2312–2317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Zhang H. J., Peng C., Hu Y. G., Li H. W., Sheng Z., Chen Y. Y., Sullivan C., Cerny J., Hutchinson L., Higgins A., Miron P., Zhang X. Q., Brehm M. A., Li D. G., Green M. R., and Li S. G. (2012) The Blk pathway functions as a tumor suppressor in chronic myeloid leukemia stem cells. Nat. Genet. 44, 861–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Harrison J. S., Rameshwar P., Chang V., and Bandari P. (2002) Oxygen saturation in the bone marrow of healthy volunteers. Blood 99, 394–394 [DOI] [PubMed] [Google Scholar]
- 74. Silva A. S., and Gatenby R. A. (2011) Adaptation to survival in germinal center is the initial step in onset of indolent stage of multiple myeloma. Mol. Pharm. 8, 2012–2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Vettori A., Greenald D., Wilson G. K., Peron M., Facchinello N., Markham E., Sinnakaruppan M., Matthews L. C., McKeating J. A., Argenton F., and van Eeden F. J. M. (2017) Glucocorticoids promote Von Hippel Lindau degradation and Hif-1alpha stabilization. Proc. Natl. Acad. Sci. U.S.A. 114, 9948–9953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Campanella M., Parker N., Tan C. H., Hall A. M., and Duchen M. R. (2009) IF(1): setting the pace of the F(1)F(o)-ATP synthase. Trends Biochem. Sci. 34, 343–350 [DOI] [PubMed] [Google Scholar]
- 77. Campanella M., Casswell E., Chong S., Farah Z., Wieckowski M. R., Abramov A. Y., Tinker A., and Duchen M. R. (2008) Regulation of mitochondrial structure and function by the F1Fo-ATPase inhibitor protein, IF1. Cell Metab. 8, 13–25 [DOI] [PubMed] [Google Scholar]
- 78. Altieri D. C., Stein G. S., Lian J. B., and Languino L. R. (2012) TRAP-1, the mitochondrial Hsp90. Biochim. Biophys. Acta 1823, 767–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Matassa D. S., Agliarulo I., Avolio R., Landriscina M., and Esposito F. (2018) TRAP1 regulation of cancer metabolism: dual role as oncogene or tumor suppressor. Genes 9, 195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Chae Y. C., Angelin A., Lisanti S., Kossenkov A. V., Speicher K. D., Wang H., Powers J. F., Tischler A. S., Pacak K., Fliedner S., Michalek R. D., Karoly E. D., Wallace D. C., Languino L. R., Speicher D. W., and Altieri D. C. (2013) Landscape of the mitochondrial Hsp90 metabolome in tumours. Nat. Commun. 4, 2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Morscher R. J., Ducker G. S., Li S. H. J., Mayer J. A., Gitai Z., Sperl W., and Rabinowitz J. D. (2018) Mitochondrial translation requires folate-dependent tRNA methylation. Nature 554, 128–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Goto M., Miwa H., Suganuma K., Tsunekawa-Imai N., Shikami M., Mizutani M., Mizuno S., Hanamura I., and Nitta M. (2014) Adaptation of leukemia cells to hypoxic condition through switching the energy metabolism or avoiding the oxidative stress. BMC Cancer 14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Hay N. (2016) Reprogramming glucose metabolism in cancer: can it be exploited for cancer therapy? Nat. Rev. Cancer 16, 635–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Fujiwara S., Kawano Y., Yuki H., Okuno Y., Nosaka K., Mitsuya H., and Hata H. (2013) PDK1 inhibition is a novel therapeutic target in multiple myeloma. Brit. J. Cancer 108, 170–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Li X. L., Ji Y. S., Han G. Y., Li X. R., Fan Z. R., Li Y. Q., Zhong Y. L., Cao J., Zhao J., Zhang M. Z., Wen J. G., Goscinski M. A., Nesland J. M., and Suo Z. H. (2016) MPC1 and MPC2 expressions are associated with favorable clinical outcomes in prostate cancer. BMC Cancer 16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Schell J. C., Olson K. A., Jiang L., Hawkins A. J., Van Vranken J. G., Xie J. X., Egnatchik R. A., Earl E. G., DeBerardinis R. J., and Rutter J. (2014) A role for the mitochondrial pyruvate carrier as a repressor of the Warburg effect and colon cancer cell growth. Molecular Cell 56, 400–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Muz B., de la Puente P., Azab F., Luderer M., and Azab A. K. (2014) Hypoxia promotes stem cell-like phenotype in multiple myeloma cells. Blood Cancer J 4, e262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Bolzoni M., Chiu M., Accardi F., Vescovini R., Airoldi I., Storti P., Todoerti K., Agnelli L., Missale G., Andreoli R., Bianchi M. G., Allegri M., Barilli A., Nicolini F., Cavalli A., Costa F., Marchica V., Toscani D., Mancini C., Martella E., Dall'Asta V., Donofrio G., Aversa F., Bussolati O., and Giuliani N. (2016) Dependence on glutamine uptake and glutamine addiction characterize myeloma cells: a new attractive target. Blood 128, 667–679 [DOI] [PubMed] [Google Scholar]
- 89. Aurer I., Lauc G., Dumic J., Rendic D., Matisic D., Milos M., Heffer-Lauc M., Flogel M., and Labar B. (2007) Aberrant glycosylation of Igg heavy chain in multiple myeloma. Coll. Antropol. 31, 247–251 [PubMed] [Google Scholar]
- 90. Krall A. S., Xu S., Graeber T. G., Braas D., and Christofk H. R. (2016) Asparagine promotes cancer cell proliferation through use as an amino acid exchange factor. Nat. Commun. 7, 11457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Degrassi A., Hilbert D. M., Rudikoff S., Anderson A. O., Potter M., and Coon H. G. (1993) In vitro culture of primary plasmacytomas requires stromal cell feeder layers. Proc. Natl. Acad. Sci. U.S.A. 90, 2060–2064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Zhang J., Fan J., Venneti S., Cross J. R., Takagi T., Bhinder B., Djaballah H., Kanai M., Cheng E. H., Judkins A. R., Pawel B., Baggs J., Cherry S., Rabinowitz J. D., and Thompson C. B. (2014) Asparagine plays a critical role in regulating cellular adaptation to glutamine depletion. Mol. Cell 56, 205–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Agrawal N. R., Bukowski R. M., Rybicki L. A., Kurtzberg J., Cohen L. J., and Hussein M. A. (2003) A Phase I-II trial of polyethylene glycol-conjugated L-asparaginase in patients with multiple myeloma. Cancer 98, 94–99 [DOI] [PubMed] [Google Scholar]
- 94. Zaal E. A., Wu W., Jansen G., Zweegman S., Cloos J., and Berkers C. R. (2017) Bortezomib resistance in multiple myeloma is associated with increased serine synthesis. Cancer Metab 5, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Hiltunen J. K., Schonauer M. S., Autio K. J., Mittelmeier T. M., Kastaniotis A. J., and Dieckmann C. L. (2009) Mitochondrial fatty acid synthesis type II: more than just fatty acids. J. Biol. Chem. 284, 9011–9015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Kastaniotis A. J., Autio K. J., Keratar J. M., Monteuuis G., Makela A. M., Nair R. R., Pietikainen L. P., Shvetsova A., Chen Z. J., and Hiltunen J. K. (2017) Mitochondrial fatty acid synthesis, fatty acids and mitochondrial physiology. Bba-Mol. Cell. Biol. L 1862, 39–48 [DOI] [PubMed] [Google Scholar]
- 97. Deepa S. S., Bhaskaran S., Ranjit R., Qaisar R., Nair B. C., Liu Y., Walsh M. E., Fok W. C., and Van Remmen H. (2016) Down-regulation of the mitochondrial matrix peptidase ClpP in muscle cells causes mitochondrial dysfunction and decreases cell proliferation. Free Radic. Biol. Med. 91, 281–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Anand R., Wai T., Baker M. J., Kladt N., Schauss A. C., Rugarli E., and Langer T. (2014) The i-AAA protease YME1L and OMA1 cleave OPA1 to balance mitochondrial fusion and fission. J. Cell Biol. 204, 919–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Abrams A. J., Hufnagel R. B., Rebelo A., Zanna C., Patel N., Gonzalez M. A., Campeanu I. J., Griffin L. B., Groenewald S., Strickland A. V., Tao F., Speziani F., Abreu L., Schule R., Caporali L., La Morgia C., Maresca A., Liguori R., Lodi R., Ahmed Z. M., Sund K. L., Wang X., Krueger L. A., Peng Y., Prada C. E., Prows C. A., Schorry E. K., Antonellis A., Zimmerman H. H., Abdul-Rahman O. A., Yang Y., Downes S. M., Prince J., Fontanesi F., Barrientos A., Nemeth A. H., Carelli V., Huang T., Zuchner S., and Dallman J. E. (2015) Mutations in SLC25A46, encoding a UGO1-like protein, cause an optic atrophy spectrum disorder. Nat. Genet. 47, 926–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Lee J. Y., Kapur M., Li M., Choi M. C., Choi S., Kim H. J., Kim I., Lee E., Taylor J. P., and Yao T. P. (2014) MFN1 deacetylation activates adaptive mitochondrial fusion and protects metabolically challenged mitochondria. J. Cell Sci. 127, 4954–4963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Ghadially F. N. (1985) Diagnostic electron microscopy of tumours, 2nd ed. Ed., Butterworths, London [Google Scholar]
- 102. Carneiro F. R. G., Lepelley A., Seeley J. J., Hayden M. S., and Ghosh S. (2018) An essential role for ECSIT in mitochondrial complex I assembly and mitophagy in macrophages. Cell Rep. 22, 2654–2666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Tirado-Velez J. M., Benitez-Rondan A., Cozar-Castellano I., Medina F., and Perdomo G. (2012) Low-density lipoprotein cholesterol suppresses apoptosis in human multiple myeloma cells. Ann. Hematol. 91, 83–88 [DOI] [PubMed] [Google Scholar]
- 104. Yavasoglu I., Tombuloglu M., Kadikoylu G., Donmez A., Cagirgan S., and Bolaman Z. (2008) Cholesterol levels in patients with multiple myeloma. Ann. Hematol. 87, 223–228 [DOI] [PubMed] [Google Scholar]
- 105. Benatti P., Chiaramonte M. L., Lorenzo M., Hartley J. A., Hochhauser D., Gnesutta N., Mantovani R., Imbriano C., and Dolfini D. (2016) NF-Y activates genes of metabolic pathways altered in cancer cells. Oncotarget 7, 1633–1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Gonsalves W. I., Ramakrishnan V., Hitosugi T., Ghosh T., Jevremovic D., Dutta T., Sakrikar D., Petterson X. M., Wellik L., Kumar S. K., and Nair K. S. (2018) Glutamine-derived 2-hydroxyglutarate is associated with disease progression in plasma cell malignancies. JCI Insight 3, e94543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Garrido C., Gurbuxani S., Ravagnan L., and Kroemer G. (2001) Heat shock proteins: endogenous modulators of apoptotic cell death. Biochem. Biophys. Res. Commun. 286, 433–442 [DOI] [PubMed] [Google Scholar]
- 108. Lee A. S. (2001) The glucose-regulated proteins: stress induction and clinical applications. Trends Biochem. Sci. 26, 504–510 [DOI] [PubMed] [Google Scholar]
- 109. Tanaka S., Uehara T., and Nomura Y. (2000) Up-regulation of protein-disulfide isomerase in response to hypoxia/brain ischemia and its protective effect against apoptotic cell death. J. Biol. Chem. 275, 10388–10393 [DOI] [PubMed] [Google Scholar]
- 110. Azoitei N., Diepold K., Brunner C., Rouhi A., Genze F., Becher A., Kestler H., van Lint J., Chiosis G., Koren J., Frohling S., Scholl C., and Seufferlein T. (2014) HSP90 supports tumor growth and angiogenesis through PRKD2 protein stabilization. Cancer Res. 74, 7125–7136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Li Y. H., Nagai H., Ohno T., Ohashi H., Murohara T., Saito H., and Kinoshita T. (2006) Aberrant DNA demethylation in promoter region and aberrant expression of mRNA of PAX4 gene in hematologic malignancies. Leukemia Res. 30, 1547–1553 [DOI] [PubMed] [Google Scholar]
- 112. Mellado-Gil J. M., Jimenez-Moreno C. M., Martin-Montalvo A., Alvarez-Mercado A. I., Fuente-Martin E., Cobo-Vuilleumier N., Lorenzo P. I., Bru-Tari E., Herrera-Gomez I. D., Lopez-Noriega L., Perez-Florido J., Santoyo-Lopez J., Spyrantis A., Meda P., Boehm B. O., Quesada I., and Gauthier B. R. (2016) PAX4 preserves endoplasmic reticulum integrity preventing beta cell degeneration in a mouse model of type 1 diabetes mellitus. Diabetologia 59, 755–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Mahbub Hasan A. K., Ijiri T., and Sato K. (2012) Involvement of Src in the adaptation of cancer cells under microenvironmental stresses. J. Signal Transduct. 2012, 483796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Eischen C. M., Roussel M. F., Korsmeyer S. J., and Cleveland J. L. (2001) Bax loss impairs Myc-induced apoptosis and circumvents the selection of p53 mutations during Myc-mediated lymphomagenesis. Mol. Cell. Biol. 21, 7653–7662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Wu Y., Liu X. M., Wang X. J., Zhang Y., Liang X. Q., and Cao E. H. (2009) PIG11 is involved in hepatocellular carcinogenesis and its over-expression promotes Hepg2 cell apoptosis. Pathol. Oncol. Res. 15, 411–416 [DOI] [PubMed] [Google Scholar]
- 116. Crivellaro S., Carra G., Panuzzo C., Taulli R., Guerrasio A., Saglio G., and Morotti A. (2016) The non-genomic loss of function of tumor suppressors: an essential role in the pathogenesis of chronic myeloid leukemia chronic phase. BMC Cancer 16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Montesi M., Jahn K., Bonewald L., Stea S., Bordini B., and Beraudi A. (2016) Hypoxia mediates osteocyte ORP150 expression and cell death in vitro. Mol. Med. Rep. 14, 4248–4254 [DOI] [PubMed] [Google Scholar]
- 118. Delcourt N., Quevedo C., Nonne C., Fons P., O'Brien D., Loyaux D., Diez M., Autelitano F., Guillemot J. C., Ferrara P., Muriana A., Callol C., Herault J. P., Herbert J. M., Favre G., and Bono F. (2015) Targeted identification of sialoglycoproteins in hypoxic endothelial cells and validation in zebrafish reveal roles for proteins in angiogenesis. J. Biol. Chem. 290, 3405–3417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Kang B. H., Plescia J., Dohi T., Rosa J., Doxsey S. J., and Altieri D. C. (2007) Regulation of tumor cell mitochondrial homeostasis by an organelle-specific Hsp90 chaperone network. Cell 131, 257–270 [DOI] [PubMed] [Google Scholar]
- 120. Winter J., Klumpe I., Heger J., Rauch U., Schultheiss H. P., Landmesser U., and Dorner A. (2016) Adenine nucleotide translocase 1 overexpression protects cardiomyocytes against hypoxia via increased ERK1/2 and AKT activation. Cell Signal 28, 152–159 [DOI] [PubMed] [Google Scholar]
- 121. Lin F. R., Kuo H. K., Ying H. Y., Yang F. H., and Lin K. I. (2007) Induction of apoptosis in plasma cells by B lymphocyte-induced maturation protein-1 knockdown. Cancer Res. 67, 11914–11923 [DOI] [PubMed] [Google Scholar]
- 122. Lamy L., Ngo V. N., Emre N. C., Shaffer A. L. 3rd, Yang Y., Tian E., Nair V., Kruhlak M. J., Zingone A., Landgren O., and Staudt L. M. (2013) Control of autophagic cell death by caspase-10 in multiple myeloma. Cancer Cell 23, 435–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Ishida T. (2018) Therapeutic antibodies for multiple myeloma. Jpn. J. Clin. Oncol. 48, 957–963 [DOI] [PubMed] [Google Scholar]
- 124. Knobeloch K. P., Wright M. D., Ochsenbein A. F., Liesenfeld O., Lohler J., Zinkernagel R. M., Horak I., and Orinska Z. (2000) Targeted inactivation of the tetraspanin CD37 impairs T-cell-dependent B-cell response under suboptimal costimulatory conditions. Mol. Cell. Biol. 20, 5363–5369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Rocha-Perugini V., Sanchez-Madrid F., and del Hoyo G. M. (2016) Function and dynamics of tetraspanins during antigen recognition and immunological synapse formation. Front. Immunol. 6, 653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. God J. M., Cameron C., Figueroa J., Amria S., Hossain A., Kempkes B., Bornkamm G. W., Stuart R. K., Blum J. S., and Haque A. (2015) Elevation of c-MYC disrupts HLA class II-mediated immune recognition of human B cell tumors. J. Immunol. 194, 1434–1445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Kanehisa M., Sato Y., Kawashima M., Furumichi M., and Tanabe M. (2016) KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 44, D457–D462 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Proteomics data were submitted to the ProteomeXchange Consortium via the PRIDE partner repository. Results are fully accessible via www.proteomeexchange.org with the identifier PXD010600.