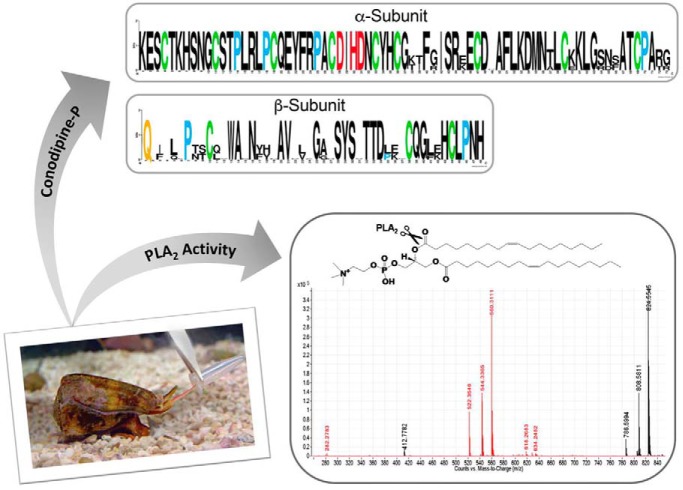
Cone snail venom is a wide source of active molecules that have potential pharmacological and biotechnological applications. Several proteins have been reported in the venom of cone snails. Here we describe the isolation and characterization of the sPLA2 Conodipines P1-3 from the injected venom of Conus purpurascens. We employed a combined proteotranscriptome approach to obtain the full sequences these Conodipines. The activity of Conodipine-P1 was assessed by a mass spectrometry-based method, which provides the first detailed Conodipine activity analysis.
Keywords: Enzyme Kinetics*, Enzyme catalysis*, Venoms, Proteogenomics, Tandem Mass Spectrometry, Mass Spectrometry, cone snails, conodipines, enzymatic activity, phospholipase A2, venomics
Graphical Abstract
Highlights
Three novel Conodipines P1-3 in the injected venom of Conus purpurascens.
Conodipines P1-3 have consensus catalytic characteristics of sPLA2.
We determined multiple modification sites in Conodipines P1-3.
Evaluated the activity of Conohyal-P1 by a MS-based method.
Abstract
The phospholipase A2 (PLA2s) superfamily are ubiquitous small enzymes that catalyze the hydrolysis of phospholipids at the sn-2 ester bond. PLA2s in the venom of cone snails (conodipines, Cdpi) are composed of two chains termed as alpha and beta subunits. Conodipines are categorized within the group IX of PLA2s. Here we describe the purification and biochemical characterization of three conodipines (Cdpi-P1, -P2 and -P3) isolated from the injected venom of Conus purpurascens. Using proteomics methods, we determined the full sequences of all three conodipines. Conodipine-P1-3 have conserved consensus catalytic domain residues, including the Asp/His dyad. Additionally, these enzymes are expressed as a mixture of proline hydroxylated isoforms. The activities of the native Conodipine-Ps were evaluated by conventional colorimetric and by MS-based methods, which provide the first detailed cone snail venom conodipine activity monitored by mass spectrometry. Conodipines can have medicinal applications such inhibition of cancer proliferation, bacterial and viral infections among others.
The phospholipase A2 (PLA2)1 superfamily (EC 3.1.1.4) comprises a broad range of small enzymes that catalyze the hydrolysis of phospholipids at the sn-2 ester bond to produce lysophospholipids and fatty acids (1, 2). The fatty acid produced by the PLA2s catalysis can be potent mediators of inflammation and signal transduction and could act either as a second messenger or as a precursor of eicosanoids, (3). Additionally, the lysophospholipids exert a wide variety of biological effects on cells and tissues (4–6). Fifteen groups (I to XV) and various subgroups of PLA2 have been identified including five distinct types of enzymes: secreted PLA2s (sPLA2s), cytosolic PLA2s (cPLA2s), Ca2+-independent PLA2s (iPLA2s), platelet-activating factor acetyl hydrolases/oxidized lipid lipoprotein associated PLA2s (LpPLA2s), adipose PLA2s (AdPLA2s), and lysosomal PLA2s (LPPLA2s). Most of these enzymes have been extensively studied as they play a major role in the regulation of phospholipid turnover, membrane trafficking and fluidity, cell maturation, apoptosis, and the production of hormones such as prostaglandins (7–10).
Animal venom contains a wide range of sPLA2s, which are of interest because they are potent toxins with effects on prey such as hemorrhage, myotoxicity, neurotoxicity, cardiotoxicity, and inflammation (5, 11–15). sPLA2s from venomous animals such as snakes (2), scorpions (16), bees (17), corals (18), and lizards (19) have been widely described. They have also been found in the venom of marine invertebrates such as starfish (20–22), sponges (23), sea anemones (24, 25), jellyfish (26), and lionfish (27). Venom sPLA2s are enzymes with molecular mass between 13 and 18 kDa and their hydrolytic activity is Ca2+ dependent.
Venom-derived sPLA2s can be either monomeric or (noncovalent or covalent) homo- or hetero- dimers or larger oligomers. Despite this structural diversity, the mechanism of hydrolysis appears to be similar. Venom-derived sPLA2s are nonselective toward the fatty acid type at the sn-2 position of phospholipids. The catalytic cleft of the venom sPLA2s is surrounded by hydrophobic residues that in the presence of Ca2+ bind the substrate. Four key residues are essential for catalysis sPLA2s: the His/Asp dyad, Tyr and Asp (5, 28–30).
The sPLA2s in the venom of cone snails (conodipines) expand the reach of these enzymes. Conodipines (Cdpi) are composed of two chains (alpha and beta), and are classified within PLA2 group IX. The first conodipine described was a partial sequence for conodipine-M (Cdpi-M), isolated from the dissected venom of C. magus (31). Recently, conodipines were reported from the transcriptome of C. consors (32), C. victorae (33), C. tribblei (34), C. lenavati (UniProt A0A0K8TTR8),C. monile (UniProt A0A161J284), C. ermineus (35) and C. geographus (36). Using sequences from tryptic peptides and the transcriptome, ten conodipines were found in the dissected venom of C. geographus, which differed in sequence and inter-cysteine loop spacing. Conodipines have a molecular mass near 14 kDa with their two subunits joined by one or more disulfide bonds. However, there is poor sequence homology between conodipines and other sPLA2s, despite having the conserved Asp/His catalytic dyad.
Here, we report the isolation and characterization of three conodipines, Cdpi-P1, -P2 and -P3 found the injected venom of C. purpurascens, the only fish-hunting cone snail species that inhabits the tropical Eastern Pacific region (Graphical Abstract Fig.). These are the first PLA2s reported from the injected venom of a cone snail. We have used combined proteomic-transcriptomic approaches, and have determined the full sequences of these Cdpi-Ps. Additionally, we assessed the activities Cdpi-P1 and injected venom using mass spectrometry-based methods and conventional PLA2 assays.
EXPERIMENTAL PROCEDURES
Extraction of Injected Venom
Specimens of C. purpurascens (20–50 mm) were collected from intertidal areas at the shores of the Republic of Ecuador and kept alive in aquaria for venom extraction. Extraction of injected venom of C. purpurascens was carried out according to the procedure of Hopkins et al. (37) with modifications (38).
Experimental Design and Statistical Rationale
The purpose of this study was to characterize PLA2s (conodipines) in the venom proteome of C. purpurascens. Therefore, we employed methods to maximize the number of conodipines identified, including strategies to identify parts of their sequences (Edman degradation, transcriptomic data mining; MALDI-MS of HPLC fractions, LC-MS/MS of HPLC fractions). The design and statistical rationale for each of the experiments conducted have been described in each subsection. The injected predatory venom was collected and screen from 27 individuals. The specimens with most conodipine content in their venom were selected for further analysis. Conodipine extraction was made based on produced biological quintuplicates. The eventual dataset includes 5 technical replicates of selected HPLC fractions, two technical replicates of reduced and alkylated but undigested venom, and two replicates of reduced, alkylated, and digested venom. The remaining samples are neither biological nor technical replicates but are subsets of venom components fractionated to maximize the potential of LC-MS/MS for conodipine identification.
Fractionation and Identification of Cdpi-Ps
Five to ten injected venom samples (∼80 μl) were pooled and dissolved in 0.1% (v/v) TFA and separated by reversed phase HPLC (RP-HPLC) using a 4.6 mm × 50 mm, 2.6-μm-particle diameter, 100-Å-pore size Kinetex C8 column (Phenomenex, Torrance, CA) with a flow of 1 ml/min. The mobile phases were 0.1% (v/v) TFA (Solution A) and 0.1% (v/v) TFA in 60% (v/v) acetonitrile (Solution B). The venom was separated with an incremental linear gradient of 1% (v/v) B/min. All fractions were manually collected, lyophilized, and kept at −80 °C before further use. The fractions corresponding to the conodipines were resuspended in 50 μl of buffer; the conodipine concentrations were determined using an EpochTM 2 microplate spectrophotometer (BioTeck®, Winooski, VT) operating at λ = 280 nm using the calculated extinction coefficient from ProtParam (39).
MALDI-TOF Mass Spectrometry of the Cdpi-Ps Fractions
Positive ion MALDI-TOF mass spectrometry was carried out on an Applied Biosystems Voyager-DE Pro spectrometer. Samples were dissolved in 0.1% TFA (v/v)/60% (v/v) acetonitrile, and applied on 4-hydroxy-3,5-dimethoxycinnamic acid (sinapinic acid) matrix. Mass spectra were obtained in the linear and reflector mode (M/ΔM resolution ∼10000) using Calmix 1 and Calmix 2 (Applied Biosystems, Foster City, CA) as external calibration standards. An aliquot (1 μl) of each RP-HPLC fraction was subjected for mass spectrometry.
Cdpi-P1 Partial Sequencing by Edman Degradation
An aliquot corresponding to fraction II was dried, redissolved in 0.1 m Tris-HCl (pH 6.2), 5 mm EDTA, 0.1% (v/v) sodium azide, and reduced with 10 mm DTT. Following incubation at 60 °C for 30 min, the sample was alkylated at a final volume of 15 μl with 20 mm iodoacetamide and 2 μl of NH4OH (pH 10.5) at room temperature for 1 h in the dark. The reduced/alkylated subunits were separated by RP-HPLC and were dried for furthered N terminus sequence by Edman degradation on an Applied Biosystems Procise model 491A sequencer.
RNA Extraction, RNA-Sequencing, Transcriptome Assembly and Conodipines-P1–6 Sequencing
The sequences of the precursors of Cdpi-P1–6 were determined from the transcriptome of the dissected venom of C. purpurascens as described by Möller et al., 2017 (40). Briefly, a single adult specimen of C. purpurascens was dissected, the venom duct removed and placed in TRIZOL (Invitrogen, Carlsbad, CA). The RNA was extracted from venom duct and prepared utilizing Illumina Poly(A)-Truseq preparation protocol. Samples were sequenced on Illumina Next-Seq 500 sequencing platform to produce 21 million forward and reverse reads. The paired-end data was then assembled the de novo assembler Trinity (41) to produce a final FASTA dataset containing 96,000 independent assembled contig sequences. The N terminus sequence of the Cdpi-P1 obtained by Edman degradation was used to identify the protein from the BLAST (Basic Local Alignment Search Tool) database generated from the transcriptome (42). The signal region and predictions of the cleavage sites were determined using SignalP 4.0 (43), NetChop 3.1 (44) and ProP 1.0 server (45). Sequence alignments were performed by Clustal Omega (46).
Cdpi-P1-3 Proteomic Analysis
Top-down and bottom up proteomics strategies were used to determine the sequences and modifications of Cdpi-P1-3. For the bottom up analysis, the RP-HPLC fractions were reduced and alkylated with 10 mm DTT and 20 mm iodoacetamide. Centrifugal filters were used to eliminate excess alkylating agent. Proteins were resuspended in 25 mm ammonium bicarbonate (pH 7.8) containing 1 mm of CaCl2. Then, proteins were digested with a LysC/trypsin mix (Promega, Madison, WI) at an enzyme/protein ratio of 1:50 (w/w) at 37 °C for 18 h. After the digestion, each sample was resuspended in 0.1% (v/v) FA, and the protein digests were analyzed by an Orbitrap Fusion™ Lumos™ Tribrid™ Mass Spectrometer (Thermo Fisher, San Jose, CA) using a 75 × 25 mm, 2 μm, C18 column. Additionally, top-down proteomics was performed on the intact protein fractions, and the reduced/alkylated subunits using a Thermo Scientific 75 × 15 mm, 3 μm, C8 column. The analyses were performed using electron transfer dissociation (ETD). The parameters used for the MS acquisitions were: survey scan = 1 (μscan), mass range = 750–4,000 (m/z), maximum injection time = 50 ms, resolution = 240,000. The MS/MS acquisition parameters were: scans = 1 μscans, cycle time = 3 s, maximum injection time = 35 ms, isolation with = 0.7 Da, ETD activation time = 50 ms and dynamic exclusion was set to exclude selected precursor ions for 60 s after 10 fragmentation selections in 30 s.
Spectra were processed and analyzed with Proteome Discoverer™ Software (Version 2.0) using the SEQUEST algorithm (47, 48) using the Conus (taxon ID 6490) database found in UniprotKB. This database was created using the 2018 releases of the SwissProt, and TrEMBL databases from UniProt and adding the sequences of the precursors of Cdpi-P1-3 from the transcriptome resulting in 9677 sequences. For Proteome Discoverer, the search parameter settings were 10 ppm precursor mass tolerance, 0.2 Da product mass tolerance, unspecific enzyme with 2 missed cleavages and 0.1 FDR. Cysteine carbamidomethylation was selected as a fixed modification, and Pro hydroxylation, Glu carboxylation, N terminus cyclization (for Glu and Gln), and deamidation amidation were selected as variable modifications. The search parameters used for the peptide mapping analysis using the software Thermo BioPharma™ Finder™ were: 10–20 signal/noise threshold and 4 ppm mass tolerance for peak detection, 5 ppm with a minimum confidence of 0.80 and high protease specificity for trypsin for peptide identification and mass accuracy.
sPLA2 Assay for the Activity of Cdpi-P1-3
PLA2 activity was determined using sPLA2 assay kit (Cayman Chemical Company, Ann Arbor, MI) following the manufacturer's protocol. To achieve reproducible results, the amount of sPLA2s added to the well should cause an absorbance increase between 0.01 and 0.1/min. The samples were diluted to reach the enzymatic activity at this level. The assay buffer was 25 mm Tris-HCl, pH 7.5, containing 10 mm CaCl2, 100 mm KCl, and 0.3 mm Triton X-100. Assay buffer was used as a blank, and Apis mellifera venom PLA2 (1 μg/ml) was used as a positive control. We used 0.25 μl of injected venom from C. purpurascens diluted to a final volume of 100 μl and ∼1 μg/ml of Cdpi-P1. The optical density was measured every minute at λ = 405 nm using Thermomax microplate reader (Molecular devices, San Jose, CA). All tests were carried out by triplicate or more and mean values were calculated. Enzymatic activity was expressed as the increase in absorbance per minute. Specific activity was expressed as micromoles of fatty acid released per minute per milligram of protein.
Agar Plate Activity Assay
PLA2 activity was evaluated by indirect radial digestion in an agar plate according to the methods described by Gutierrez et al. (49). A solution of 25 ml of 1% (w/v) agarose dissolved in phosphate-buffered saline (PBS) pH 7.2 was melted in a boiling water bath and cooled to 50 °C. One ml of human whole blood (Bioreclamation IVT, Westbury, NY) was washed three times with (PBS) pH 7.2 using centrifugations at 1900 × g. Three hundred microliters of washed erythrocytes were combined with 300 μl of 1:3 egg yolk solution in PBS and 250 μl of 0.01 m CaCl2. The solution was added to the agar solution, stirred, and immediately poured into a 9 × 9-cm plastic Petri dish. Cylindrical holes of ∼2 mm in diameter were punched into the gel and emptied by suction. Samples were prepared in duplicates as follows: positive control containing 20 μg/ml of A. mellifera venom PLA2, 0.5 μl of C. purpurascens injected venom, and the Cdpi-P1 fraction (20 μg/ml) in a final volume of 10 μl in 0.2 mm sodium acetate pH 6.0, 0.15 mm NaCl. The holes were filled with a volume of 8 μl from each sample. Empty holes (negative controls) were filled with 8 μl of buffer. The plate was covered and incubated for 20 h at 37 °C.
Hemolytic Activity
The hemolytic effects of C. purpurascens injected venom on red blood cells (RBC) were evaluated using human blood (50). One ml of whole human blood (Bioreclamation IVT, Westbury, NY) was diluted in 10 ml PBS, pH 7.4, and then centrifuged at 1900 × g for 15 min at 4 °C. The RBCs were washed three consecutive times. The pellet rich in erythrocytes was resuspended in PBS to a final volume of 10 ml. A hundred microliters of the purified RBCs were combined with 2, 5, 10, 15, and 20 μl of injected venom. Final volume was adjusted to 300 μl with PBS. Positive control (maximum hemolysis) consisted of 200 μl of PBS in 1% (v/v) Triton X-100 and 100 μl RBC, whereas the negative control contained 200 μl of PBS and 100 μl RBC. The samples were incubated for 3 h at 37 °C in an incubator and then centrifuged at 1900 × g for 15 min at 4 °C. The absorbance of the supernatant was determined at λ = 540 nm. All samples were tested by triplicate. Percentage of hemolysis was calculated as follows:
Direct Assessment of the Activity by Mass Spectrometry
PLA2 activity was determined by mass spectrometry. For that purpose, A. mellifera venom PLA2 was used as a positive control in a final concentration of 10 μg/ml. Venom activity was determined using a volume of 3 μl of injected venom from C. purpurascens and the Cdpi-P1 fraction in a final concentration of ∼10 μg/ml. A stock solution of 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC) (Avanti Polar Lipids, Inc, Alabaster, AL) was prepared at 1 mg/ml diluted with methanol. Reaction was started by adding DOPC to a final concentration of 100 μg/ml. The final assay volume was 30 μl adjusted with the 50 mm sodium acetate (pH 7.4), 10 mm CaCl2, 100 mm KCl.
Calcium-dependent activity was determined using the conditions described above, using 25 mm ammonium acetate (pH 7.4) as buffer, 100 mm KCl, 2 mm EDTA. Control assays were A. mellifera venom PLA2, injected venom and the RP-HPLC fraction II without DOPC.
Samples were incubated for 3 h while gently agitated at 37 °C. Direct flow injection analysis (FIA) was performed on an Agilent 6530 Accurate Mass Q-TOF, calibrated with the mass range set to 3200 m/z and an extended dynamic range of 2 GHz. Analysis was performed in positive mode with the following settings: capillary voltage, 4000 V; nozzle voltage, 0 V; fragmentor voltage, 170 V; nebulizer pressure, 20 psi; drying gas, 11 L/min at 350 °C, and sheath gas, 12 L/min at 400 °C. The mass range was set from 100 to 1500 m/z, with a 1.0 spectra/s acquisition speed for the MS and MS/MS modes (51). The mobile phase used was 50% (v/v) HPLC H2O/50% (v/v) acetonitrile/0.1% (v/v) FA. The flow was 0.5 ml/min. The injection volume of sample was 10 μl.
Additional MS of venom sPLA2s activities were carried out using a Thermo Scientific Velos ProTM with a HESI probe. The analysis was performed by direct injection of 2 μl of sample and ran isocratically over 10 min in 90% (v/v) methanol/10% (v/v) acetonitrile in positive mode. The autosampler was set at 37 °C.
RESULTS
Fractionation of Cdpi-P1-3 and Characterization
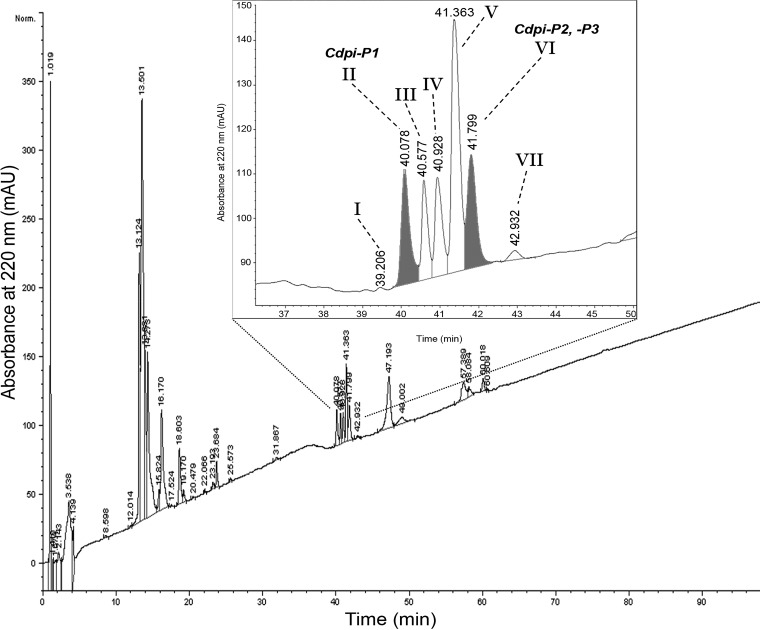
The RP-HPLC profile of the injected venom from C. purpurascens is shown in Fig. 1. The peaks eluted between 39 and 42 min (peaks I to VII) were collected and saved for further analysis.
Fig. 1.
Fractionation of the Cdpi-Ps. RP-HPLC of the injected venom of C. purpurascens. Five to ten shots of injected venom were applied to an analytical C8 column and eluted with a linear gradient of 1% (v/v) B/min over 100 min at a flow rate of 1 ml/min. The inset displays an amplification of the fractions that corresponds to the Cdpi-P1-3. Fraction II and VI which are shadowed corresponded to Cdpi-P1 and Cdpi-P2,-P3 respectively.
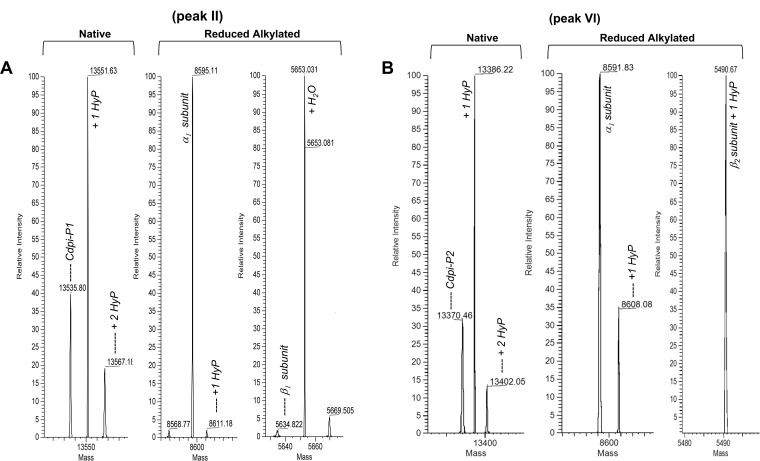
Preliminary MALDI-TOF mass spectrometry of the purified RP-HPLC fractions peaks designed as II and VI yielded an average molecular mass of 13,546 Da and 13,371 Da, respectively (data not shown). Additionally, the intact protein masses of the chromatographic fractions were determined by direct injection in the Orbitrap Fusion™ Lumos™ Tribrid™ Mass Spectrometer. The spectra deconvolution showed isotopically unresolved masses of 13,535.80 Da for the fraction peak II (Fig. 2A) and 13,370.46 Da for the fraction peak VI (Fig. 2B), which is within the range of the reported conodipines (31). The other chromatographic peaks (III to VI) contained different hydroxylation states of p21a, a conotoxin with a 9.3 kDa molecular mass (38).
Fig. 2.
Molecular mass determination of the Cdpi-P1-3 fractions. A, Fraction II corresponded to Cpdi-P1 with a deconvoluted molecular mass of 13,535.80 Da and two possible hydroxylations. After reduction and alkylation the obtained deconvoluted masses were 8,595.11 for the α-subunit and 5,634.82 Da for the β-subunit. B, Fraction VI corresponded to Cdpi-P2 with a deconvoluted molecular mass of 13,370.46 Da and two possible hydroxylations. After reduction and alkylation the obtained deconvoluted masses were 8,591.83 for the α-subunit and 5,490.67 Da for the β-subunit.
The two fractions containing the putative conodipines were reduced and alkylated yielding two peaks with molecular masses of 8595.11 Da and 5634.82 Da for fraction II (Fig. 2A) and 8591.83 Da and 5490.67 Da for fraction VI (Fig. 2B), which confirmed that these are heterodimeric proteins, which is characteristic of the previously reported conodipines. Notably, fraction II was more concentrated and of higher purity than fraction VI. Fraction VI major component was the nonhydroxylated state of p21a. For that reason, the conodipine in fraction II (Cdpi-P1) was selected for testing in the activity assayed.
Cdpi-P1 Partial Sequencing by Edman Degradation and Transcriptome Assembling
The reduced and alkylated subunits corresponding to the fraction peak II were separated by RP-HPLC and subjected to N terminus sequencing by Edman degradation. We were unable to sequence the β-subunit, presumably because it had a blocked N terminus (pyroglutamate) (Fig. 3); however, we obtained the first 34 residues N terminus of the α-subunit, yielding a KESCTKHSNGCSTPLRLPCQEYFROACDIHDNCY sequence (O = hydroxyproline). A preliminary BLAST search with the obtained N terminus sequence yield identities from 67% to 79% with the N terminus sequence of the conodipines reported from C. geographus (36), and an identity of 53% with the alpha chain of Cdpi-M from C. magus, the only conodipine isolated so far from Conus dissected venom (31). Sequence homology along with the molecular mass and the heterodimeric composition were strong indications of conodipines present in the injected venom of C. purpurascens.
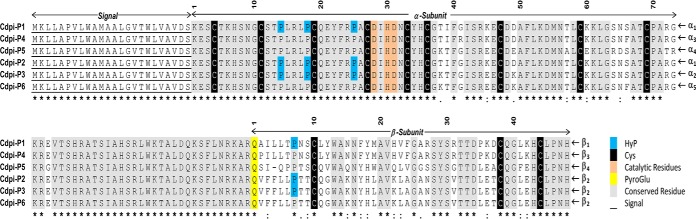
Fig. 3.
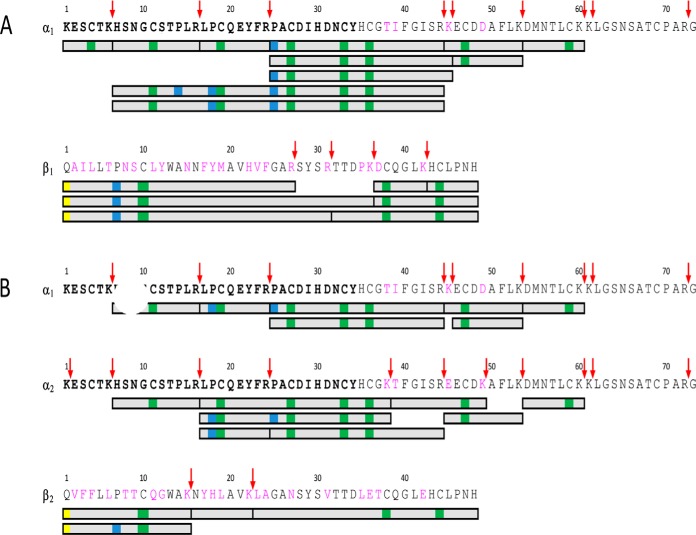
Sequence alignment of the Cdpi-Ps found in C. purpurascens transcriptome. Residues underlined indicate the signal region. The α-subunit and β-subunit are separated by an interchain peptide. Conserved residues are highlighted in gray, catalytic residues in orange, pyro-Gln residue in yellow and found Hyp in blue. Residue numbers corresponded to the mature protein subunits.
The partial N terminus sequence of the conodipine-P1 obtained by Edman degradation was used to search in the transcriptome of the venom duct of C. purpurascens. We found a set of six types of conodipines coded by the transcript: Cdpi-P1 to -P6 (Fig. 3) (GenBank accession MK493027-MK493032). The six conodipine precursor sequences have 178 residues, and the sequence found by Edman degradation of the first 34 amino acids was the same for all Cdpi-Ps. The predicted protein precursor sequence starts with a 24-residue signal peptide followed by the sequence that includes the α and β-subunits. Five different alphas (α1-α5) and four different betas (β1-β4) were found in the transcriptome. The combination of subunits α1β1 was assigned as Cdpi-P1, whereas the combination of α1β2, α2β2, α3β3, α4β4, and α5β2 were Cdpi-P2, -P3, P4, P5, and -P6 respectively (Fig. 3). The signal region and the cleavage sites for all six transcriptome-derived sequences were determined and their calculated molecular masses are shown in Table I.
Table I. Calculated molecular masses for the different Cdpi-Ps.
| Monoisotopic (Da) | Average (Da) | Elemental composition | |
|---|---|---|---|
| Native Cdpi-P1 (α1β1) | 13,534.185 | 13,543.29 | C583H890N170O176S14 |
| Native Cdpi-P2 (α1β2) | 13,372.138 | 13,381.07 | C577H884N164O178S13 |
| Native Cdpi-P3 (α2β2) | 13,401.164 | 13,410.11 | C578H887N165O178S13 |
| Native Cdpi-P4 (α3β3) | 13,719.317 | 13,728.56 | C595H905N173O175S14 |
| Native Cdpi-P5 (α4β4) | 13,342.868 | 13,351.73 | C568H853N165O184S13 |
| Native Cdpi-P6 (α5β2) | 13,461.201 | 13,470.71 | C584H891N165O177S13 |
| Red/Alk α1-subunit | 8,596.872 | 8,602.65 | C357H564N111O117S10 |
| Red/Alk α2-subunit | 8,625.899 | 8,631.69 | C358H567N112O117S10 |
| Red/Alk α3-subunit | 8,755.988 | 8,761.88 | C367H577N114O116S10 |
| Red/Alk α4-subunit | 8,711.863 | 8,717.69 | C360H565N112O121S10 |
| Red/Alk α5-subunit | 8,685.935 | 8,691.78 | C364H571N112O116S10 |
| Red/Alk β1-subunit | 5,633.664 | 5,637.36 | C250H374N71O71S4 |
| Red/Alk β2-subunit | 5471.617 | 5474.14 | C244H368N65O73S3 |
| Red/Alk β3-subunit | 5659.681 | 5663.39 | C252H376N71O71S4 |
| Red/Alk β4-subunit | 5327.356 | 5330.75 | C232H336N65O75S3 |
Molecular masses were calculated using Thermo BioPharma™ Finder™ (Version 2.0) under Protein Sequence Editor, and includes the pyroglutamate, cysteine disulfide bonds or carbamidomethylation. Red/Alk = reduced and carbamidomethylated cysteine residues.
A BLAST search with the whole Cdpi-P1, -P2 and -P3 transcriptome sequences shows similarities with other conodipines and sPLA2s. The higher sequence identities ranged between 58 and 61% with the transcriptome-based conodipines from C. lenavati (Cdpi-L) (UniProtKB A0A0K8TUY3_9CAEN), C. tribblei (Cdpi-T) (UniProtKB A0A0C9RYL5_9CAEN), and C. geographus (Cdpi-G5, -G4 and -G3) (UniProtKB W4VS60_CONGE, W4VSC8_CONGE, and W4VSJ3_CONGE) (Fig. 4).
Fig. 4.
Sequence alignment of Cdpi-Ps with other cone snail conodipines. Cdpi-P1, Cdpi-P2 and Cdpi-P3 (GenBank Accession Number: MK493027-MK493032) aligned with the conodipines from C. geographus Cdpi-G3, -G4 and -G5 (UniProtKB - W4VSJ3, W4VSC8, and W4VS60 respectively), Cdpi-L from C. lenavati (UniProtKB - A0A0K8TUY3), Cdpi-T from Conus tribblei (UniProtKB - A0A0C9RYL5), and Cdpi-M from C. magus (UniProtKB - Q9TWL9 and Q9TWL8). Residues underlined indicate the signal region. Amino acid conservations are denoted by (*). Full stops (.) and colons (:) represent a low and high degree of similarity, respectively.
Cdpi-Ps Proteomics Sequence Analysis
RP-HPLC fractions I to VII were subjected to LysC/trypsin digestion and then analyzed by LC-MS/MS. The digested peptides sequences were obtained by peptide mapping analysis using the modified Conus database information. Only sequences with a confidence score of 80% or more were used. The MS/MS data of the peak II fraction provided 81% coverage for α1-subunit and 100% coverage for β1-subunit. This combination corresponded to Cdpi-P1 (α1β1) (Fig. 5A). It should be noted that there are distinctive residues that differentiate α1 from α2 and β1 from β2. Raw MS/MS spectra that distinguished Cdpi-P1 are shown on supplemental Fig. S1 and S2. These results agreed with the deconvoluted masses obtained for the native and reduced alkylated form of fraction II.
Fig. 5.
Determination of Cdpi-P1, -P2 and -P3 sequence by bottom up proteomics. Sequence coverage map was achieved after the trypsin/LysC digestion of the fraction II (A) and fraction VI (B). Peptide mapping analysis was performed by mass spectrometry. Gray bars indicate the identified digested peptides. Fragments sequences matches were determined using the conodipines sequences from the transcriptome. A, Fraction II sequence coverage corresponded to subunits α1 and β1, which match Cdpi-1. B, Fraction VI coverage corresponded to α1, α2, and β2-subunits, which subunits combination are a1β2 and α2β2 and match to Cdpi-P2 and Cdpi-P3 respectively. Covered fragments are in gray, Hyp in blue, carbamidomethyl cysteine in green and pyroglutamate in yellow. Cleavage residues for Trypsin/LysC are in magenta. Bold residues correspond to then Edman degradation obtained sequence. Some of the key MS/MS spectra are showing on the supplemental Figs. S1 to S4. For more details about the spectra and masses of the corresponding fragments, refer to the supplementary material.
We found isoforms of the conodipines formed by Pro hydroxylation (Hyp) on Pro14, Pro18, and Pro25 of the α1-subunit (Fig. 5A). Hyp was also found on Pro25 with the Edman degradation analysis and in the native and reduced alkylated deconvoluted masses were a +16 Da was observed (Fig. 2A). In the case of the Cdpi-P1 β1-subunit, we found evidence the presence of a pyroglutamate on the N terminus along with hydroxylation at Pro7 (Fig. 5A and supplemental Fig. S2).
For fraction VI, coverage of 64.4% was obtained for the α1-subunit and 100% coverage for β2-subunit, corresponding to Cdpi-P2 (α1β2) (Fig. 5B). We observed a 64.4% sequence match with the α2-subunit (Fig. 5B). Because only the β1-subunit was observed with coverage of 100%, we concluded that Cdpi-P2 (α1β2) and Cdpi-P3 (α2β2) co-eluted in fraction VI. However, Cdpi-P3 was less abundant than Cdpi-P2. It should be noted that the deconvolution of the native proteins gave a mass of 13,370 Da that corresponds to Cdpi-P2, and a mass of 13,402 Da, which corresponds to the Cdpi-P3 mass; however, it also matches with the molecular mass of Cdpi-P2 plus two Hyp's. Raw MS/MS spectra that distinguished Cdpi-P2 and Cdpi-P3 are shown on supplemental Fig. S3 and S4.
Reduced and alkylated samples of fraction peak II and VI were subjected to top-down proteomic analysis via ETD fragmentation. Fig. 6 shows the fragmentation maps of the two subunits α1 and β1 for fraction II and β2 for fraction VI. Each subunit showed a characteristic charge distribution, which was a key for subunit identification. Data on the matched fragments are included in the supplementary information (supplemental Fig. S5).
Fig. 6.
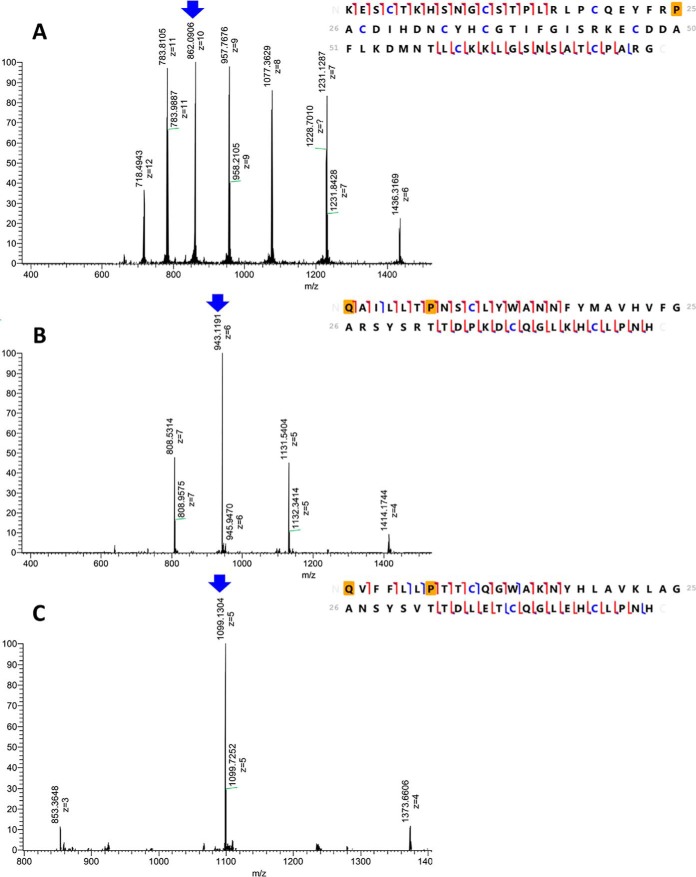
Top-down proteomics of the reduced and alkylated α1, α1 and β2-subunits of Cdpi-P. On the left side of the figure are the ESI-MS spectra of different charge distributions for each subunit. MS/MS by EThcD for these subunits were obtained for the peaks labeled with the blue arrow. Fragmentation maps, scoring and residue coverage were obtained using the software ProSight Lite (90). On the right side of the figure are the fragmentation maps for each subunit. The orange square represents the corresponded posttranslational modifications such as pyroglutamic acid and hydroxyproline. Carbamidomethylated cysteines are highlighted in blue. Red brackets are matched with c/z ions, whereas blue brackets are matched with b/y ions. The matching fragment list are shown in supplemental Fig. S5) A, ESI-MS spectra and fragmentation map of [M+10H]10+ = 862.1 corresponding to the α1-subunit in fraction II (42% sequence coverage). B, ESI-MS spectra and fragmentation map of [M+6H]6+ = 943.1 corresponding to the β1-subunit in fraction II (68% sequence coverage). C, ESI-MS spectra and fragmentation map of [M+5H]5+ = 1099.1 corresponding to the β2-subunit in fraction VI (64% sequence coverage).
Altogether, intact protein deconvolution, and top-down and bottom-up approaches showed that fraction II contained Cdpi-P1 and fraction VI contained Cdpi-P2 and Cdpi-P3 with the latter at a lower concentration. Given the purity and abundance of fraction II, Cdpi-P1 was selected for use in subsequent activity assays.
Agar Egg Yolk Plate Assay and Hemolytic Activity of Cdpi-P1 and Injected Venom
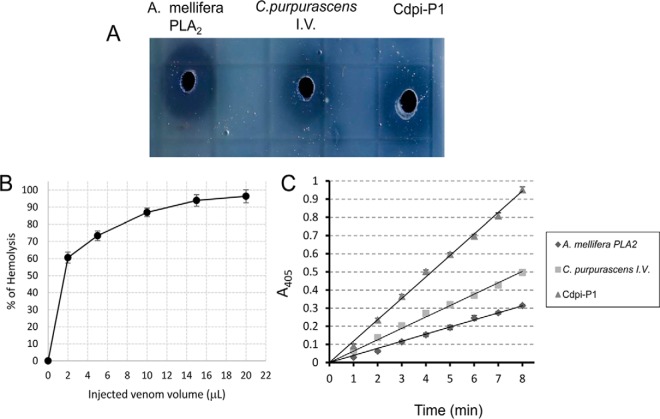
The activity of Cdpi-P1 and injected venom were determined by the formation of halos in agarose-erythrocyte egg yolk gels as a consequence of the enzymatic hydrolysis of lecithin to lysolecithin, and latter lysis of red blood cell membranes (Fig. 7A). All three, the positive control, A. mellifera PLA2, C. purpurascens-injected venom and Cpdi-P1 formed halos indicating the presence of PLA2 activity.
Fig. 7.
Cdpi-Ps activity. A, PLA2 activity was evaluated by the indirect radial hemolysis in agar plate method. PLA2 hydrolyzes the phospholipids of egg yolk, which are liberated into medium as fatty acids and glycerophospholipids. The fatty acids decrease the pH and produce the lysis of erythrocytes forming the halos. A. mellifera venom PLA2 was used as positive control. Halos were observed with C. purpurascens injected venom and the Cdpi-P1 fraction. B, Direct hemolytic activity of C. purpurascens injected venom toward human erythrocytes. Dose-response curve of the direct hemolytic activity of the injected venom shows that it can induce up to 95% hemolysis. C, Colorimetric sPLA2 assay. A. mellifera PLA2 (1 μg/ml) was used as positive control. For the assay, we tested 0.25 μl of injected venom from C. purpurascens diluted in a final volume of 100 μl and ∼1 μg/ml of Cdpi-P1. The line slope corresponded to the PLA2 activity. The resulted activity values for A. mellifera PLA2, the injected venom from C. purpurascens and Cdpi-P1 fraction were 0.088, 0.142, and 0.266 μmol/min respectively.
A dose-response curve showed the direct hemolytic activity of C. purpurascens venom is shown on Fig. 7B. These results confirmed that injected venom from C. purpurascens can induce hemolysis in human erythrocytes. Two microliters of injected venom can induce 60% of hemolysis while a plateau between 85 and 95% is reached above 10 μl of venom.
sPLA2 Assay for the Activity of Cdpi-P1 and Injected Venom
We determined PLA2 activities using a colorimetric assay based on the hydrolysis of a 1,2-dithio analog of diheptanoyl phosphatidylcholine that serves as a substrate for most sPLA2s. C. purpurascens injected venom and Cdpi-P1 were tested using the A. mellifera PLA2 as the positive control (Fig. 7C). We determined that C. purpurascens injected venom have an active PLA2, as the specific activity for Cdpi-P1 was 449.7 ± 7.9 μmol/min/mg (n = 5), compared with A. mellifera PLA2, which showed an 87.9 ± 2.1 μ/min/mg (n = 5) specific activity.
Direct Assessment of the Activity of Cdpi-Ps by Mass Spectrometry
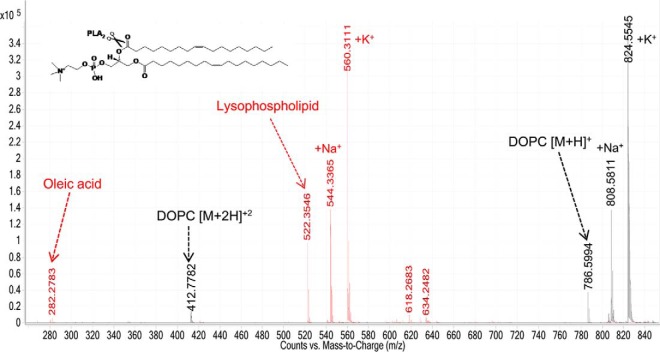
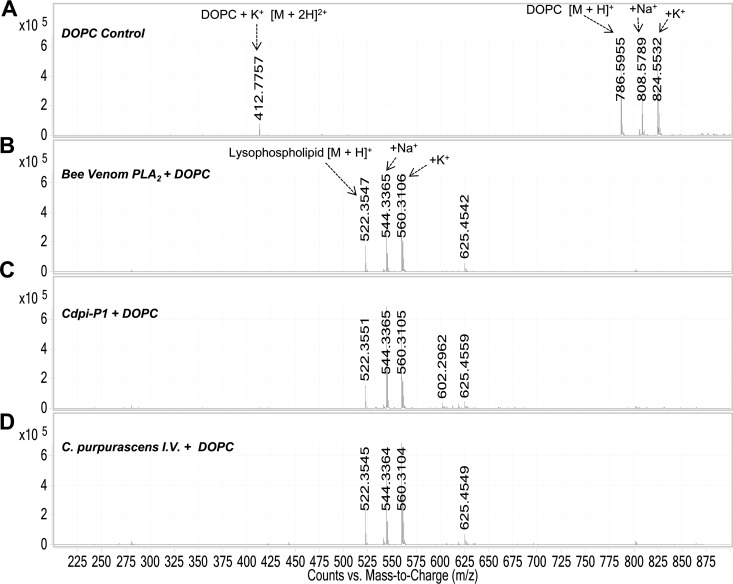
The direct measurement of venom PLA2 activity by LC-ESI-MS was carried out by monitoring by the formation of the lysophospholipid and fatty acid (products of the hydrolysis). We followed the formation of the hydrolysis products of DOPC after treatment with Cdpi-P1 (Fig. 8). The peak corresponding to DOPC showed a molecular mass of 786.59 Da. After Cdpi-P1 activity, a lysophospholipid with a mass of 522.35 Da was formed along with oleic acid with a mass of 282.28 Da. The same results were observed when A. mellifera PLA2 was used as the positive control and with the C. purpurascens-injected venom, where the DOPC peak disappeared after the treatment and corresponding lisophospholipid was formed (Fig. 9).
Fig. 8.
Direct assessment of the activity of Cdpi-P1 by mass spectrometry. Overlay of DOPC control spectra (black). Hydrolysis of DOPC by Cdpi-P1 (red). Cdpi-P1 hydrolyzes the phospholipid DOPC (786 Da) into the lipolysis products, the oleic acid (282 Da) and the lysophopholipid (522 Da). Sodium and potassium adducts could be observed.
Fig. 9.
Direct assessment of sPLA2s activities of by mass spectrometry. A, Control experiment with only DOPC. B, DOPC incubated with A. mellifera PLA2. C, DOPC incubated with Cdpi-P1 fraction D, DOPC incubated with C. purpurascens I.V. Notice that compared with the control, PLA2s hydrolyzes the DOPC (786 Da) to form the lysophospholipid (522 Da). Sodium and potassium adducts could be observed.
Other control experiments were performed using the venom from the snakes, Agkistrodon piscivorous piscivourous (APP) and Agkistrodon piscivorous locustoma (APL). Both snake venoms have been reported to have sPLA2s (52). Additionally, we tested the activity of the injected venoms from the fish-hunting cone snails, C. ermineus and C. striatus. All the above, except the C. striatus venom, formed the lysophospholipid (522 Da), indicating PLA2 activity (supplemental Fig. S6). MS analysis indicated the presence of molecular masses in the range of sPLA2 in all venoms except in the venom of C. striatus (data not shown).
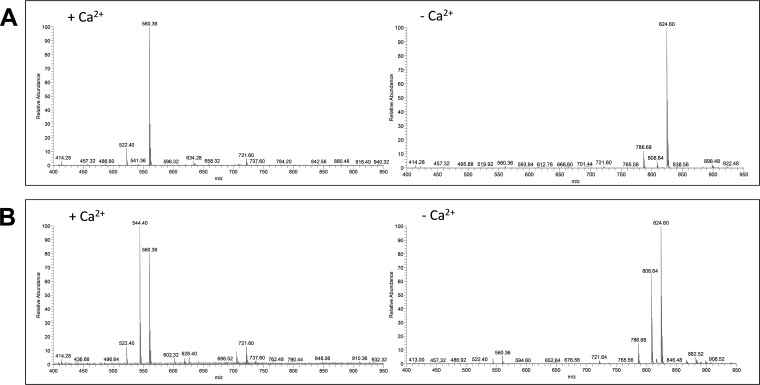
Because calcium plays an important role in the lipolytic activity of different venom sPLA2s (1, 31), we evaluated the activity of different venoms in absence of Ca2+ using a buffer with no calcium that contained EDTA (36). Fig. 10 shows that no lysophospholipid was formed in the absence of calcium for the A. mellifera PLA2 and the C. purpurascens-injected venom, indicating that in absence of calcium the PLA2 activity was abolished. The same results were obtained with C. ermineus, APP, and APL venom (data not shown).
Fig. 10.
Calcium dependence activity. A, DOPC incubated with A. mellifera PLA2 in presence of calcium (left) and in presence of EDTA (right). B, DOPC incubated with C. purpurascens injected venom in presence of calcium (left) and in presence of EDTA (right). Notice that in presence of EDTA the sPLA2 activity is abolished.
DISCUSION
Here we describe the isolation and characterization of three conodipines (Cdpi-P1, -P2, and -P3) found in the injected venom of C. purpurascens. These are the first conodipines described in injected venom, the actual concoction deliver to the prey. Other Cdpis have been found in the dissected venom of C. magus and of C. geographus (31, 36); however, because no conodipines had been found in the injected venom, it was suggested that these enzymes were not involved in the envenomation process (32). The isolation of Cdpi-Ps clearly show that these enzymes are part of the envenomation strategy of certain cone snail species. Conodipine sequences have been obtained from the transcriptome of several cone snail species, which might be an indication of their prevalence in cone snail venom and even a key component for envenomation (34, 53–55).
Conodipines are heterodimers, however, their alpha and beta subunits are encoded by a single precursor with a signal peptide sequence and a peptide linker between the two subunits. Like other venom PLA2s, conodipines have high intra and interspecies sequence variability as shown in the phylogenetic tree of known conodipines (supplemental Fig. S7). Different isoforms were reported within the same species (54). In our case, six conodipine isoforms were found in the transcriptome, with higher sequence variability on the beta subunit. All Cdpi-Ps precursors have the same signal and remarkable similarity in their subunits linker. The catalytic His-Asp dyad is conserved as in the other conodipines precursors reported.
The complete sequences of Cdpi-P1, -P2, and -P3 from C. purpurascens-injected venom were obtained by a combination of proteomics (MS/MS) and transcriptomic (RNAseq) data analysis. The mature Cdpi-Ps contains nine cysteines on the α-subunit and three on the β-subunit. Using homology modeling (supplemental Fig. S8), we surmised that the third cysteine (Cys19) in the alpha subunit is the one involved in intermolecular disulfide bonding. Our proteomic analysis shows that these conodipines are hydroxylated at Pro in both the alpha and beta subunits and several hydroxylation-related isoforms were observed. This differential and selective hydroxylation has been reported in conotoxins and it is found in the venom of C. purpurascens (37, 38, 56, 57). The importance of Hyp modification in conopeptides is still unclear, but it is possible they could contribute to the formation of hydrogen bonds and facilitate the binding strength and selectivity toward a target (58). For the Cdpi-Ps, there is the possibility that the Hyp might be involved in enzyme dimerization and stabilization. In addition to Hyp modifications, we found that all Cdpi-Ps contain N-terminal pyro-Glu/Gln residues. This moiety is a major modification in secretory proteins, and it is thought to assist in receptor-ligand interactions and in the stabilization of the N terminus against degradation (59).
Venom sPLA2s have been reported as either as monomeric or multimeric complexes with at least one subunit catalytically active. The mechanism of action of multimeric PLA2s is more complex when compared with their monomeric counterparts. One of the first examples of a heterodimeric PLA2 is the snake β-bungarotoxin (60); heterodimeric sPLA2s have been isolated from scorpion venom such as Imperatoxin (61). Other examples include Heteromtoxin (62) and Hemilipin (63), which have a large PLA2 active subunit, and a small subunit linked by a disulfide bond. The function of the small subunit is unknown, although they have been categorized as members of the Kunitz protease inhibitor superfamily (64). Likewise, the catalytic residues of the conodipines are conserved and located on the alpha subunit, which is larger than the beta. However, there is a high sequence variability on the beta subunit, and they have no sequence relationship with the Kunitz protease inhibitor superfamily.
Among all sPLA2 isoforms, the amino acid residues involved in the Ca2+ binding and catalysis are highly conserved. The His/Asp dyad and a Tyr are also highly conserved in the active site of these enzymes (65, 66). Conodipines have the conserved His/Asp dyad that corresponds to the residues His-30 and Asp-31. Additionally, Asp-28 has been suggested as an active residue because it may possibly form hydrogen bonds with His-30 (9, 31). A study with species of the phylum Cnidarian, Mollusca, and Turnicata found a conserved catalytic site with the sequence X-Cys-Asp-X-His-Asp-X-Cys-Tyr-X-Cys (67), which matches with the domain observed in all Cdpi-Ps and other Cdpis. The precise role of these residues and the other conserved amino acids has not been determined. Although the His/Asp dyad has been a hallmark of these enzymes, other group of PLA2 enzymes with a His/Lys dyad has been described in snake venom (68–74). No His/Lys dyad conodipines have been reported so far. In addition to the calcium binding dyad, other venom PLA2s have an extra calcium binding motif of Trp/Tyr-Cys-Gly-X-Gly. It is notable that this motif is not present in any conodipines, which also underwrites conodipines as unique group of sPLA2.
We assessed the activity of Cdpi-P1-3 by colorimetry and an agarose-erythrocyte egg yolk gel assay. We found enzymatic activity for the venom fraction containing Cdpi-P1 and for the C. purpurascens injected venom. A direct assessment of Cdpi-P1 activity was performed by MS indicating that is a fast and reliable method for PLA2s activity studies. We evaluated the DOPC lipolysis in presence of Cdpi-P1 and the injected venom of C. purpurascens, C. ermineus and Agkistrodon piscivorous. We found that the activity of Cdpi-Ps is Ca2+-dependent. The use of MS to follow the enzymatic activity of PLA2s has been reported, as venom toxins have been tested in cultured neurons and in cardiolipin (75, 76). It has been shown that MS can be used as a screening method with high sensitivity and specificity. These MS-based techniques not only allow the characterization of enzymes, but also enable studies on the lipid composition of membranes. The use of MS to determine the activity of venom enzymes by MS is promising for a fast and reliable method for enzyme kinetics. We recently reported that other venom enzyme's activities, such as hyaluronidases, could be follow by MS (40).
Venom sPLA2s could be potent hemolytic agents (77–81); i.e. C. loroisii venom produced significant alterations in the hematological parameters, including the reduction of the red blood cells and induced edema (82). Our results show that the injected venom of C. purpurascens exerts a notable hemolytic effect. This may be because of Cdpi-P isoforms and/or also because of other active enzymes as proteases present in the venom (83, 84). The damage of the red cell membrane by the action of venom molecular components could be a direct or a multistep process, where not only does the sPLA2s participate in the hemolysis process, but also other enzymes.
Conodipines, similar to other peptides and proteins present in venom, may be naturally tailored by cone snails to efficiently adapt to a variety of different preys (85). We not only found different conodipines in C. purpurascens, but we also found that some individuals of this species kept in our lab did not express measurable quantities of conodipines and no PLA2 activity was detected in their injected venom. It would be important to determine the factors that conditioned the selective expression of these venom enzymes by some specimens. Furthermore, that we found PLA2 activity in the venom of C. ermineus, but not in C. striatus, which demonstrate that not all fish-hunting cone snails express these enzymes. Additionally, it should be noted that conodipines are not only restricted to fish-hunting species, as conodipines have been found on the closely related worm-hunting species, C. tribblei and C. lenavati. This demonstrates that conodipines are found in cone snails of their prey preference.
It is unclear how conodipines have evolved and adapted within this genus. The intraspecific and interspecies variability and adaptations shown so far indicates a biochemical conundrum that requires further study. However, it has been recently proposed that group IX PLA2s, belonging to the phylum Mollusca, Cnidarian and Turnicata, share a common evolutionary origin with group XIV PLA2s of bacteria and fungi. Furthermore, there are indications that group IX PLA2s go through species-specific duplication and that evolutionary pressures in different lineages promoted ancestral molecule modifications (67).
Venom PLA2s may have medical and therapeutic applications, such as prevention of prion peptide-induced cell death (84), viral inhibition (85, 86), hepatoxicity protection (86), inhibition of cancer cell proliferation (88), neuroinflammatory modulation (87), and mediation of antigen delivery (88). Given the large intraspecific and interspecies structural variability shown by venom PLA2s along with their associated activity, it seems that their use can be optimized for specific applications and the discovery and testing new PLA2s will be important for a variety of uses in research, medicine and industry.
Disclaimer
Certain commercial equipment, instruments, or materials are identified in this paper in order to specify the experimental procedure adequately. Such identification is not intended to imply recommendation or endorsement by the National Institute of Standards and Technology, nor is it intended to imply that the materials or equipment identified are necessarily the best available for the purpose.
DATA AVAILABILITY
This article contains supplemental Fig. S1–S8, supplemental Table S1 and other data files. Raw and processed bottom up proteomics data has been deposited to MassIVE (https://massive.ucsd.edu/ProteoSAFe/static/massive.jsp) project ID MSV000083408. Exported results can also be found in supplemental excel files Pur_4L_II_TD_Results and Pur_4L_VI_TD_Results.
Supplementary Material
Footnotes
* This work was partially supported by the Florida Sea Grant Program (R/LR-MB-28).
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- PLA2
- phospholipase A2
- sPLA2
- secreted PLA2
- cPLA2
- cytosolic PLA2
- iPLA2
- Ca2+-independent PLA2
- LpPLA2
- lipoprotein associated PLA2
- AdPLA2
- adipose PLA2
- LPPLA2s
- lysosomal PLA2
- Cdpi
- conodipines
- RP-HPLC
- reversed phased HPLC
- Tris
- tris(hydroxymethyl)aminomethane
- BLAST
- basic local alignment search tool
- FA
- formic acid
- ETD
- electron transfer dissociation
- FDR
- false discovery rate
- PBS
- phosphate-buffered saline
- RBC
- red blood cells
- DOPC
- 1,2-dioleoyl-sn-glycero-3-phosphocholine
- FIA
- flow injection analysis.
REFERENCES
- 1. Burke J. E., and Dennis E. A. (2009) Phospholipase A2 structure/function, mechanism, and signaling. J. Lipid Res. 50, S237–S242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kini R. M. (2003) Excitement ahead: structure, function and mechanism of snake venom phospholipase A2 enzymes. Toxicon 42, 827–840 [DOI] [PubMed] [Google Scholar]
- 3. Funk C. D. (2001) Prostaglandins and leukotrienes: advances in eicosanoid biology. Science 294, 1871–1875 [DOI] [PubMed] [Google Scholar]
- 4. Rivera R., and Chun J. (2008) Biological effects of lysophospholipids. Rev. Physiol. Biochem. Pharmacol. 160, 25–46 [DOI] [PubMed] [Google Scholar]
- 5. Dennis E. A., Cao J., Hsu Y. H., Magrioti V., and Kokotos G. (2011) Phospholipase A2 enzymes: physical structure, biological function, disease implication, chemical inhibition, and therapeutic intervention. Chem. Rev. 111, 6130–6185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. de Paula R. C., Castro H. C., Rodrigues C. R., Melo P. A., and Fuly A. L. (2009) Structural and pharmacological features of phospholipases A2 from snake venoms. Protein Pept. Lett. 16, 899–907 [DOI] [PubMed] [Google Scholar]
- 7. Hariprasad G., Srinivasan A., and Singh R. (2013) Structural and phylogenetic basis for the classification of group III phospholipase A2. J. Mol. Model. 19, 3779–3791 [DOI] [PubMed] [Google Scholar]
- 8. Schaloske R. H., and Dennis E. A. (2006) The phospholipase A2 superfamily and its group numbering system. Biochim. Biophys. Acta 1761, 1246–1259 [DOI] [PubMed] [Google Scholar]
- 9. Six D. A., and Dennis E. A. (2000) The expanding superfamily of phospholipase A(2) enzymes: classification and characterization. Biochim. Biophys. Acta 1488, 1–19 [DOI] [PubMed] [Google Scholar]
- 10. Simakov O., Marletaz F., Cho S. J., Edsinger-Gonzales E., Havlak P., Hellsten U., Kuo D. H., Larsson T., Lv J., Arendt D., Savage R., Osoegawa K., de Jong P., Grimwood J., Chapman J. A., Shapiro H., Aerts A., Otillar R. P., Terry A. Y., Boore J. L., Grigoriev I. V., Lindberg D. R., Seaver E. C., Weisblat D. A., Putnam N. H., and Rokhsar D. S. (2013) Insights into bilaterian evolution from three spiralian genomes. Nature 493, 526–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Teixeira C. F., Landucci E. C., Antunes E., Chacur M., and Cury Y. (2003) Inflammatory effects of snake venom myotoxic phospholipases A2. Toxicon 42, 947–962 [DOI] [PubMed] [Google Scholar]
- 12. Gutierrez J. M., and Lomonte B. (1995) Phospholipase A2 myotoxins from Bothrops snake venoms. Toxicon 33, 1405–1424 [DOI] [PubMed] [Google Scholar]
- 13. Harris J. B., and Cullen M. J. (1990) Muscle necrosis caused by snake venoms and toxins. Electron Microsc. Rev. 3, 183–211 [DOI] [PubMed] [Google Scholar]
- 14. Lambeau G., and Gelb M. H. (2008) Biochemistry and physiology of mammalian secreted phospholipases A2. Annu. Rev. Biochem. 77, 495–520 [DOI] [PubMed] [Google Scholar]
- 15. Kini R. M., and Chan Y. M. (1999) Accelerated evolution and molecular surface of venom phospholipase A2 enzymes. J. Mol. Evol. 48, 125–132 [DOI] [PubMed] [Google Scholar]
- 16. Ibrahim S. A. (1967) Phospholipase A in scorpion venoms. Toxicon 5, 59–60 [DOI] [PubMed] [Google Scholar]
- 17. Kuchler K., Gmachl M., Sippl M. J., and Kreil G. (1989) Analysis of the cDNA for phospholipase A2 from honeybee venom glands. The deduced amino acid sequence reveals homology to the corresponding vertebrate enzymes. Eur. J. Biochem. 184, 249–254 [DOI] [PubMed] [Google Scholar]
- 18. Garcia-Arredondo A., Rojas-Molina A., Ibarra-Alvarado C., Lazcano-Perez F., Arreguin-Espinosa R., and Sanchez-Rodriguez J. (2016) Composition and biological activities of the aqueous extracts of three scleractinian corals from the Mexican Caribbean: Pseudodiploria strigosa, Porites astreoides and Siderastrea siderea. J. Venom Anim. Toxins Incl. Trop. Dis. 22, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Huang T. F., and Chiang H. S. (1994) Effect on human platelet aggregation of phospholipase A2 purified from Heloderma horridum (beaded lizard) venom. Biochim. Biophys. Acta 1211, 61–68 [DOI] [PubMed] [Google Scholar]
- 20. Shiomi K. A., Kazama A., Shimakura K., and Nagashima Y. (1998) Purification and properties of phospholipases A2 from the crown-of-thorns starfish (Acanthaster planci) venom. Toxicon 36, 589–599 [DOI] [PubMed] [Google Scholar]
- 21. Ota E., Nagai H., Nagashima Y., and Shiomi K. (2006) Molecular cloning of two toxic phospholipases A2 from the crown-of-thorns starfish Acanthaster planci venom. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 143, 54–60 [DOI] [PubMed] [Google Scholar]
- 22. Kishimura H., Ojima T., Hayashi K., and Nishita K. (2000) cDNA cloning and sequencing of phospholipase A2 from the pyloric ceca of the starfish Asterina pectinifera. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 126, 579–586 [DOI] [PubMed] [Google Scholar]
- 23. Nevalainen T. J., Quinn R. J., and Hooper J. N. (2004) Phospholipase A2 in porifera. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 137, 413–420 [DOI] [PubMed] [Google Scholar]
- 24. Nevalainen T. J., Peuravuori H. J., Quinn R. J., Llewellyn L. E., Benzie J. A., Fenner P. J., and Winkel K. D. (2004) Phospholipase A2 in cnidaria. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 139, 731–735 [DOI] [PubMed] [Google Scholar]
- 25. Razpotnik A., Krizaj I., Sribar J., Kordis D., Macek P., Frangez R., Kem W. R., and Turk T. (2010) A new phospholipase A2 isolated from the sea anemone Urticina crassicornis - its primary structure and phylogenetic classification. FEBS J. 277, 2641–2653 [DOI] [PubMed] [Google Scholar]
- 26. Heo Y., Kwon Y. C., Shin K., Yoon W. D., Han C. H., Yum S., and Kim E. (2016) cDNA and gene structures of two phospholipase A2 isoforms, acidic PLA2 PA4 and PLA2 PA3A/PA3B/PA5, in Nemopilema nomurai jellyfish venom. Toxicon 122, 160–166 [DOI] [PubMed] [Google Scholar]
- 27. Memar B., Jamili S., Shahbazzadeh D., and Pooshang Bagheri K. (2016) Description of histopathological changes induced by the venom of the Persian Gulf Lionfish (Pterois russelli) in a mouse model of multiorgan toxicity. Toxicon 122, 94–102 [DOI] [PubMed] [Google Scholar]
- 28. Han S. K., Yoon E. T., Scott D. L., Sigler P. B., and Cho W. (1997) Structural aspects of interfacial adsorption. A crystallographic and site-directed mutagenesis study of the phospholipase A2 from the venom of Agkistrodon piscivorus piscivorus. J. Biol. Chem. 272, 3573–3582 [PubMed] [Google Scholar]
- 29. Harris J. B., and Scott-Davey T. (2013) Secreted phospholipases A2 of snake venoms: effects on the peripheral neuromuscular system with comments on the role of phospholipases A2 in disorders of the CNS and their uses in industry. Toxins 5, 2533–2571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kang T. S., Georgieva D., Genov N., Murakami M. T., Sinha M., Kumar R. P., Kaur P., Kumar S., Dey S., Sharma S., Vrielink A., Betzel C., Takeda S., Arni R. K., Singh T. P., and Kini R. M. (2011) Enzymatic toxins from snake venom: structural characterization and mechanism of catalysis. FEBS J. 278, 4544–4576 [DOI] [PubMed] [Google Scholar]
- 31. McIntosh J. M., Ghomashchi F., Gelb M. H., Dooley D. J., Stoehr S. J., Giordani A. B., Naisbitt S. R., and Olivera B. M. (1995) Conodipine-M, a novel phospholipase A2 isolated from the venom of the marine snail Conus magus. J. Biol. Chem. 270, 3518–3526 [DOI] [PubMed] [Google Scholar]
- 32. Leonardi A., Biass D., Kordis D., Stocklin R., Favreau P., and Krizaj I. (2012) Conus consors snail venom proteomics proposes functions, pathways, and novel families involved in its venomic system. J. Proteome Res. 11, 5046–5058 [DOI] [PubMed] [Google Scholar]
- 33. Robinson S. D., Safavi-Hemami H., McIntosh L. D., Purcell A. W., Norton R. S., and Papenfuss A. T. (2014) Diversity of conotoxin gene superfamilies in the venomous snail, Conus victoriae. PLoS ONE 9, e87648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Barghi N., Concepcion G. P., Olivera B. M., and Lluisma A. O. (2015) High conopeptide diversity in Conus tribblei revealed through analysis of venom duct transcriptome using two high-throughput sequencing platforms. Mar. Biotechnol. 17, 81–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Abalde S., Tenorio M. J., Afonso C. M. L., and Zardoya R. (2018) Conotoxin diversity in Chelyconus ermineus (Born, 1778) and the convergent origin of piscivory in the Atlantic and Indo-Pacific cones. Genome Biol Evol 10, 2643–2662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Safavi-Hemami H., Hu H., Gorasia D. G., Bandyopadhyay P. K., Veith P. D., Young N. D., Reynolds E. C., Yandell M., Olivera B. M., and Purcell A. W. (2014) Combined proteomic and transcriptomic interrogation of the venom gland of Conus geographus uncovers novel components and functional compartmentalization. Mol. Cell. Proteomics 13, 938–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hopkins C., Grilley M., Miller C., Shon K.-J., Cruz L. J., Gray W. R., Dykert J., Rivier J., Yoshikami D., and Olivera B. M. (1995) A new family of Conus peptides targeted to the nicotinic acetylcholine receptor. J. Biol. Chem. 270, 22361–22367 [DOI] [PubMed] [Google Scholar]
- 38. Möller C., and Marí F. (2011) 9.3 KDa components of the injected venom of Conus purpurascens define a new five-disulfide conotoxin framework. Peptide Sci. 96, 158–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gasteiger E., Gattiker A., Hoogland C., Ivanyi I., Appel R. D., and Bairoch A. (2003) ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 31, 3784–3788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Moller C., Clark E., Safavi-Hemami H., DeCaprio A., and Mari F. (2017) Isolation and characterization of Conohyal-P1, a hyaluronidase from the injected venom of Conus purpurascens. J. Proteomics 164, 73–84 [DOI] [PubMed] [Google Scholar]
- 41. Haas B. J., Papanicolaou A., Yassour M., Grabherr M., Blood P. D., Bowden J., Couger M. B., Eccles D., Li B., Lieber M., MacManes M. D., Ott M., Orvis J., Pochet N., Strozzi F., Weeks N., Westerman R., William T., Dewey C. N., Henschel R., LeDuc R. D., Friedman N., and Regev A. (2013) De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 8, 1494–1512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Schaffer A. A., Aravind L., Madden T. L., Shavirin S., Spouge J. L., Wolf Y. I., Koonin E. V., and Altschul S. F. (2001) Improving the accuracy of PSI-BLAST protein database searches with composition-based statistics and other refinements. Nucleic Acids Res. 29, 2994–3005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Petersen T. N., Brunak S., von Heijne G., and Nielsen H. (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8, 785–786 [DOI] [PubMed] [Google Scholar]
- 44. Nielsen M., Lundegaard C., Lund O., and Kesmir C. (2005) The role of the proteasome in generating cytotoxic T-cell epitopes: insights obtained from improved predictions of proteasomal cleavage. Immunogenetics 57, 33–41 [DOI] [PubMed] [Google Scholar]
- 45. Duckert P., Brunak S., and Blom N. (2004) Prediction of proprotein convertase cleavage sites. Protein Eng. Des. Sel. 17, 107–112 [DOI] [PubMed] [Google Scholar]
- 46. Sievers F., Wilm A., Dineen D., Gibson T. J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Soding J., Thompson J. D., and Higgins D. G. (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tabb D. L. (2015) The SEQUEST family tree. J. Am. Soc. Mass. Spectrom. 26, 1814–1819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yates J. R. 3rd, Eng J. K., McCormack A. L., and Schieltz D. (1995) Method to correlate tandem mass spectra of modified peptides to amino acid sequences in the protein database. Anal. Chem. 67, 1426–1436 [DOI] [PubMed] [Google Scholar]
- 49. Gutierrez J. M., Avila C., Rojas E., and Cerdas L. (1988) An alternative in vitro method for testing the potency of the polyvalent antivenom produced in Costa Rica. Toxicon 26, 411–413 [DOI] [PubMed] [Google Scholar]
- 50. Accary C., Hraoui-Bloquet S., Hamze M., Mallem Y., El Omar F., Sabatier J. M., Desfontis J. C., and Fajloun Z. (2014) Protein content analysis and antimicrobial activity of the crude venom of Montivipera bornmuelleri; a viper from Lebanon. Infect. Disord. Drug Targets 14, 49–55 [DOI] [PubMed] [Google Scholar]
- 51. Kang Y. P., Lee W. J., Hong J. Y., Lee S. B., Park J. H., Kim D., Park S., Park C. S., Park S. W., and Kwon S. W. (2014) Novel approach for analysis of bronchoalveolar lavage fluid (BALF) using HPLC-QTOF-MS-based lipidomics: lipid levels in asthmatics and corticosteroid-treated asthmatic patients. J. Proteome Res. 13, 3919–3929 [DOI] [PubMed] [Google Scholar]
- 52. Jia Y., Ermolinsky B., Garza A., and Provenzano D. (2017) Phospholipase A2 in the venom of three cottonmouth snakes. Toxicon 135, 84–92 [DOI] [PubMed] [Google Scholar]
- 53. Robinson S. D., Li Q., Lu A., Bandyopadhyay P. K., Yandell M., Olivera B. M., and Safavi-Hemami H. (2017) The Venom Repertoire of Conus gloriamaris (Chemnitz, 1777), the Glory of the Sea. Mar. Drugs 15, pii: E145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Aman J. W., Imperial J. S., Ueberheide B., Zhang M. M., Aguilar M., Taylor D., Watkins M., Yoshikami D., Showers-Corneli P., Safavi-Hemami H., Biggs J., Teichert R. W., and Olivera B. M. (2015) Insights into the origins of fish hunting in venomous cone snails from studies of Conus tessulatus. Proc. Natl. Acad. Sci. U.S.A. 112, 5087–5092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Baruscotti M., Westenbroek R., Catterall W. A., DiFrancesco D., and Robinson R. B. (1997) The newborn rabbit sino-atrial node expresses a neuronal type I-like Na+ channel. J. Physiol. 498, 641–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Cruz L. J., Gray W. R., Olivera B. M., Zeikus R. D., Kerr L., Yoshikami D., and Moczydlowski E. (1985) Conus geographus toxins that discriminate between neuronal and muscle sodium channels. J. Biol. Chem. 260, 9280–9288 [PubMed] [Google Scholar]
- 57. Lopez-Vera E., Walewska A., Skalicky J. J., Olivera B. M., and Bulaj G. (2008) Role of hydroxyprolines in the in vitro oxidative folding and biological activity of conotoxins. Biochemistry 47, 1741–1751 [DOI] [PubMed] [Google Scholar]
- 58. Franco A., Pisarewicz K., Moller C., Mora D., Fields G. B., and Marí F. (2006) Hyperhydroxylation: a new strategy for neuronal targeting by venomous marine molluscs. Molluscs, pp. 83–103, Springer; [DOI] [PubMed] [Google Scholar]
- 59. Schilling S., Wasternack C., and Demuth H. U. (2008) Glutaminyl cyclases from animals and plants: a case of functionally convergent protein evolution. Biol. Chem. 389, 983–991 [DOI] [PubMed] [Google Scholar]
- 60. Chang C. C., and Lee C. Y. (1963) Isolation of neurotoxins from the venom of Bungarus Multicinctus and their modes of neuromuscular blocking action. Arch. Int. Pharmacodyn. Ther. 144, 241–257 [PubMed] [Google Scholar]
- 61. Zamudio F. Z., Gurrola G. B., Arevalo C., Sreekumar R., Walker J. W., Valdivia H. H., and Possani L. D. (1997) Primary structure and synthesis of Imperatoxin A (IpTx(a)), a peptide activator of Ca2+ release channels/ryanodine receptors. FEBS Lett. 405, 385–389 [DOI] [PubMed] [Google Scholar]
- 62. Incamnoi P., Patramanon R., Thammasirirak S., Chaveerach A., Uawonggul N., Sukprasert S., Rungsa P., Daduang J., and Daduang S. (2013) Heteromtoxin (HmTx), a novel heterodimeric phospholipase A(2) from Heterometrus laoticus scorpion venom. Toxicon 61, 62–71 [DOI] [PubMed] [Google Scholar]
- 63. Jridi I., Catacchio I., Majdoub H., Shahbazeddah D., El Ayeb M., Frassanito M. A., Ribatti D., Vacca A., and Borchani L. (2015) Hemilipin, a novel Hemiscorpius lepturus venom heterodimeric phospholipase A2, which inhibits angiogenesis in vitro and in vivo. Toxicon 105, 34–44 [DOI] [PubMed] [Google Scholar]
- 64. Kwong P. D., McDonald N. Q., Sigler P. B., and Hendrickson W. A. (1995) Structure of beta 2-bungarotoxin: potassium channel binding by Kunitz modules and targeted phospholipase action. Structure 3, 1109–1119 [DOI] [PubMed] [Google Scholar]
- 65. Arni R. K., and Ward R. J. (1996) Phospholipase A2 - a structural review. Toxicon 34, 827–841 [DOI] [PubMed] [Google Scholar]
- 66. Murakami M. T., and Arni R. K. (2003) A structure based model for liposome disruption and the role of catalytic activity in myotoxic phospholipase A2s. Toxicon 42, 903–913 [DOI] [PubMed] [Google Scholar]
- 67. Nevalainen T. J., Morgado I., and Cardoso J. C. (2013) Identification of novel phospholipase A2 group IX members in metazoans. Biochimie 95, 1534–1543 [DOI] [PubMed] [Google Scholar]
- 68. Scott D. L., Achari A., Vidal J. C., and Sigler P. B. (1992) Crystallographic and biochemical studies of the (inactive) Lys-49 phospholipase A2 from the venom of Agkistridon piscivorus piscivorus. J. Biol. Chem. 267, 22645–22657 [PubMed] [Google Scholar]
- 69. Ward R. J., Chioato L., de Oliveira A. H., Ruller R., and Sa J. M. (2002) Active-site mutagenesis of a Lys49-phospholipase A2: biological and membrane-disrupting activities in the absence of catalysis. Biochem. J. 362, 89–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Lomonte B., Angulo Y., and Calderon L. (2003) An overview of lysine-49 phospholipase A2 myotoxins from crotalid snake venoms and their structural determinants of myotoxic action. Toxicon 42, 885–901 [DOI] [PubMed] [Google Scholar]
- 71. dos Santos J. I., Soares A. M., and Fontes M. R. (2009) Comparative structural studies on Lys49-phospholipases A(2) from Bothrops genus reveal their myotoxic site. J Struct Biol 167, 106–116 [DOI] [PubMed] [Google Scholar]
- 72. Lomonte B., and Rangel J. (2012) Snake venom Lys49 myotoxins: From phospholipases A(2) to non-enzymatic membrane disruptors. Toxicon 60, 520–530 [DOI] [PubMed] [Google Scholar]
- 73. Cintra-Francischinelli M., Pizzo P., Rodrigues-Simioni L., Ponce-Soto L. A., Rossetto O., Lomonte B., Gutierrez J. M., Pozzan T., and Montecucco C. (2009) Calcium imaging of muscle cells treated with snake myotoxins reveals toxin synergism and presence of acceptors. Cell. Mol. Life Sci. 66, 1718–1728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Mora-Obando D., Fernandez J., Montecucco C., Gutierrez J. M., and Lomonte B. (2014) Synergism between basic Asp49 and Lys49 phospholipase A2 myotoxins of viperid snake venom in vitro and in vivo. PLoS ONE 9, e109846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Paoli M., Rigoni M., Koster G., Rossetto O., Montecucco C., and Postle A. D. (2009) Mass spectrometry analysis of the phospholipase A(2) activity of snake pre-synaptic neurotoxins in cultured neurons. J. Neurochem. 111, 737–744 [DOI] [PubMed] [Google Scholar]
- 76. Hsu Y. H., Dumlao D. S., Cao J., and Dennis E. A. (2013) Assessing phospholipase A2 activity toward cardiolipin by mass spectrometry. PLoS ONE 8, e59267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Tuichibaev M. U., Akhmedova N. U., and Muksimov F. A. (1988) Hemolytic effect of phospholipase A2 and orientotoxin from venom of the great hornet, Vespa orientalis. Biokhimiia 53, 434–443 [PubMed] [Google Scholar]
- 78. Vogel C. W., Pluckthun A., Muller-Eberhard H. J., and Dennis E. A. (1981) Hemolytic assay for venom phospholipase A2. Anal. Biochem. 118, 262–268 [DOI] [PubMed] [Google Scholar]
- 79. Jiang M. S., Fletcher J. E., and Smith L. A. (1989) Factors influencing the hemolysis of human erythrocytes by cardiotoxins from Naja naja kaouthia and Naja naja atra venoms and a phospholipase A2 with cardiotoxin-like activities from Bungarus fasciatus venom. Toxicon 27, 247–257 [DOI] [PubMed] [Google Scholar]
- 80. Rudenko S. V., and Semenchenko A. (1995) Change in erythrocyte volume and spectrum of membrane proteins induced by melittin, phospholipase A2 and bee venom. Biokhimiia 60, 734–745 [PubMed] [Google Scholar]
- 81. Arce-Bejarano R., Lomonte B., and Gutierrez J. M. (2014) Intravascular hemolysis induced by the venom of the Eastern coral snake, Micrurus fulvius, in a mouse model: identification of directly hemolytic phospholipases A2. Toxicon 90, 26–35 [DOI] [PubMed] [Google Scholar]
- 82. Saminathan R., Babuji S., Sethupathy S., Viswanathan P., Balasubramanian T., and Gopalakrishanakone P. (2006) Clinico-toxinological characterization of the acute effects of the venom of the marine snails, Conus loroisii. Acta Trop. 97, 75–87 [DOI] [PubMed] [Google Scholar]
- 83. Möller C., Vanderweit N., Bubis J., and Marí F. (2013) Comparative analysis of proteases in the injected and dissected venom of cone snail species. Toxicon 65, 59–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Safavi-Hemami H., Moller C., Mari F., and Purcell A. W. (2013) High molecular weight components of the injected venom of fish-hunting cone snails target the vascular system. J. Proteomics 91, 97–105 [DOI] [PubMed] [Google Scholar]
- 85. Calvete J. J., Sanz L., Angulo Y., Lomonte B., and Gutierrez J. M. (2009) Venoms, venomics, antivenomics. FEBS Lett. 583, 1736–1743 [DOI] [PubMed] [Google Scholar]
- 86. Kim H., Keum D. J., Kwak J., Chung H. S., and Bae H. (2014) Bee venom phospholipase A2 protects against acetaminophen-induced acute liver injury by modulating regulatory T cells and IL-10 in mice. PLoS ONE 9, e114726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Chung E. S., Lee G., Lee C., Ye M., Chung H. S., Kim H., Bae S. J., Hwang D. S., and Bae H. (2015) Bee venom phospholipase A2, a novel Foxp3+ regulatory T cell inducer, protects dopaminergic neurons by modulating neuroinflammatory responses in a mouse model of Parkinson's disease. J. Immunol. 195, 4853–4860 [DOI] [PubMed] [Google Scholar]
- 88. Almunia C., Bretaudeau M., Held G., Babon A., Marchetti C., Castelli F. A., Menez A., Maillere B., and Gillet D. (2013) Bee venom phospholipase A2, a good “chauffeur” for delivering tumor antigen to the MHC I and MHC II peptide-loading compartments of the dendritic cells: the case of NY-ESO-1. PLoS ONE 8, e67645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Schneider T. D., and Stephens R. M. (1990) Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 18, 6097–6100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Fellers R. T., Greer J. B., Early B. P., Yu X., LeDuc R. D., Kelleher N. L., and Thomas P. M. (2015) ProSight Lite: graphical software to analyze top-down mass spectrometry data. Proteomics 15, 1235–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. The UniProt, C. (2017) UniProt: the universal protein knowledgebase. Nucleic Acids Res. 45, D158–D169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Edgar R. C. (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Arnold K., Bordoli L., Kopp J., and Schwede T. (2006) The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22, 195–201 [DOI] [PubMed] [Google Scholar]
- 94. Cavazzini D., Meschi F., Corsini R., Bolchi A., Rossi G. L., Einsle O., and Ottonello S. (2013) Autoproteolytic Activation of a Symbiosis-regulated Truffle Phospholipase A2. J. Biol. Chem. 288, 1533–1547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Sugiyama M., Ohtani K., Izuhara M., Koike T., Suzuki K., Imamura S., and Misaki H. (2002) A novel prokaryotic phospholipase A2. Characterization, gene cloning, and solution structure. J. Biol. Chem. 277, 20051–20058 [DOI] [PubMed] [Google Scholar]
- 96. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., and Ferrin T. E. (2004) UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This article contains supplemental Fig. S1–S8, supplemental Table S1 and other data files. Raw and processed bottom up proteomics data has been deposited to MassIVE (https://massive.ucsd.edu/ProteoSAFe/static/massive.jsp) project ID MSV000083408. Exported results can also be found in supplemental excel files Pur_4L_II_TD_Results and Pur_4L_VI_TD_Results.