Abstract
One aspect of the function of the β-arrestins is to serve as scaffold or adapter molecules coupling G-protein coupled receptors (GPCRs) to signal transduction pathways distinct from traditional second messenger pathways. Here we report the identification of Dishevelled 1 and Dishevelled 2 (Dvl1 and Dvl2) as β-arrestin1 (βarr1) interacting proteins. Dvl proteins participate as key intermediates in signal transmission from the seven membrane-spanning Frizzled receptors leading to inhibition of glycogen synthase kinase-3β (GSK-3β), stabilization of β-catenin, and activation of the lymphoid enhancer factor (LEF) transcription factor. We find that phosphorylation of Dvl strongly enhances its interaction with βarr1, suggesting that regulation of Dvl phosphorylation and subsequent interaction with βarr1 may play a key role in the activation of the LEF transcription pathway. Because coexpression of the Dvl kinases, CK1ɛ and PAR-1, with Dvl synergistically activates LEF reporter gene activity, we reasoned that coexpression of βarr1 with Dvl might also affect LEF-dependent gene activation. Interestingly, whereas βarr1 or Dvl alone leads to low-level stimulation of LEF (2- to 5-fold), coexpression of βarr1 with either Dvl1 or Dvl2 leads to a synergistic activation of LEF (up to 16-fold). Additional experiments with LiCl as an inhibitor of GSK-3β kinase activity indicate that the step affected by βarr1 is upstream of GSK-3β and most likely at the level of Dvl. These results identify βarr1 as a regulator of Dvl-dependent LEF transcription and suggest that βarr1 might serve as an adapter molecule that can couple Frizzled receptors and perhaps other GPCRs to these important transcription pathways.
The arrestin proteins play critical roles in G protein-coupled receptor (GPCR) desensitization and internalization (1, 2). Phosphorylation of activated receptors by GPCR kinases facilitates binding of arrestin proteins that serve to physically uncouple activated receptors from bound G protein (3). This process, termed desensitization, effectively terminates many of the classical G protein signaling pathways. Two isoforms of β-arrestin originally discovered in the context of β2-adrenergic receptor desensitization [β-arrestin1 (βarr1) and β-arrestin2] have been identified (4, 5). In addition to their role as negative regulatory components in “conventional” GPCR signaling paradigms, it has recently become clear that the β-arrestins are multifunctional scaffold/adapter proteins (6).
One function of the β-arrestins as scaffold/adapter proteins is to facilitate the transmission of additional signals from activated GPCRs (6) [W.E.M. and R.J.L. (2001), http://www.stke.org/cgi/content/full/oc_sigtrans; 2001/69/pe1]. For example, the β-arrestins directly interact with several Src family kinases, including Src, Yes, and Hck (7–10). Agonist-activated GPCRs such as the β2-adrenergic and CXCR1 receptors recruit Src kinases to the receptor complexes via the β-arrestins. The recruitment of β-arrestin/Src complexes to the receptor is important for activation of mitogen-activated protein kinases, receptor internalization, and granule release (7, 8). β-Arrestin2 also has been found to interact with several components of classical mitogen-activated protein kinase signaling cascades. The β-arrestins interact with the c-Jun N-terminal kinase 3 (JNK3) and the apoptosis signal-regulating kinase 1 (ASK1) to enhance signal transmission to JNK3 (11, 12). Similarly, the β-arrestins interact with Raf and extracellular-regulated kinase 2 (Erk2) to facilitate GPCR-stimulated activation of extracellular-regulated kinase pathways (13, 14).
An important but still poorly characterized signaling pathway involves activation of seven membrane-spanning Frizzled receptors by Wnt glycoproteins (15, 16). This signaling pathway is conserved from flies to vertebrates and plays a key role in developmental and morphogenetic pathways. Binding of Wnt to Frizzled receptors initiates signal transmission leading to modulation of Dishevelled (Dvl) activity, inhibition of glycogen synthase kinase-3β (GSK-3β), and stabilization of β-catenin (15). In the absence of Wnt, β-catenin is phosphorylated by GSK-3β and targeted for degradation (17–19). Stabilized β-catenin accumulates in the cytosol and then translocates into the nucleus, where it interacts with the lymphoid enhancer factor (LEF)/T cell factor transcription factors to activate transcription of target genes (20). Interestingly, Dvl also activates the c-Jun amino-terminal kinase (JNK) through modulation of a signaling pathway independent of GSK-3β and β-catenin (21–23).
Dvl proteins consist of an amino-terminal DIX domain, a central PDZ domain, and a carboxyl-terminal DEP domain (24, 25). These conserved domains most likely mediate protein–protein interactions that are required for the ability of Dvl to stimulate LEF transcriptional activity. Stimulation of Frizzled receptors by Wnt proteins leads to hyperphosphorylation of Dvl, and recently several kinases, including protein kinase CKIɛ, protein kinase CKII, and PAR-1, have been shown to interact with Dvl and positively regulate LEF-dependent transcription (26–30).
Here we report that βarr1 interacts with Dvl proteins and provide evidence that βarr1 enhances Dvl-stimulated LEF transcriptional activity. These results delineate an unexpected signaling role for the β-arrestins and indicate that the utility of β-arrestin as a multifunctional adapter protein includes the modulation of signaling from “nonconventional” seven membrane-spanning receptors such as Frizzled.
Experimental Procedures
Cell Culture and Transfection.
HEK-293, NIH 3T3, and L cells were propagated according to instructions from the American Type Culture Collection. HEK-293 cells were transfected with either calcium phosphate (31) or FuGene6 according to the manufacturer's instructions (Roche Molecular Biochemicals). NIH 3T3 cells were transfected with Lipofectamine according to the manufacturer's instructions (Life Technologies, Rockville, MD). Cells were routinely harvested 48 h after transfection.
Yeast Two-Hybrid Screening.
A rat βarr1 cDNA (containing a single point mutation converting Arg-161 to Gly) was cloned into the pAS2–1 yeast expression vector (CLONTECH). The pAS2–1(βarr1) plasmid was transformed into the PJ-69–4A yeast strain with a human heart cDNA library (CLONTECH) by standard yeast transformation protocols (32, 33). Rescued library plasmids from positive clones were sequenced with an Applied Biosystems DNA sequencer (Howard Hughes Nucleic Acid Facility, Duke University). The isolated plasmids were then cotransformed into yeast with either the pAS2–1 (βarr1) plasmid or pAS2–1, and the specificities of the interactions were confirmed by growth on −His and −Ade selective plates.
Plasmids.
Expression vectors for FLAG-βarr1, Myc-Dvl1, Myc-Dvl2, and MBP-Dvl1 have been described (9, 27, 34). pGL3-LEF and pCG-LEF[hemagglutinin (HA)] were kind gifts of Rudolf Grosschedl (University of Munich, Munich). An expression vector for HA-Dvl2 (180) was constructed by subcloning the Dvl2 insert from the yeast two-hybrid clone into a modified pcDNA3 vector containing an amino-terminal HA sequence.
Immunoprecipitations and Immunoblot Analysis.
Cell extracts were prepared by lysing cells in buffer A (20 mM Hepes/0.5% Nonidet P-40/250 mM NaCl/10% glycerol/2 mM EDTA/1 mM PMSF/2.5 μg/ml aprotinin/2.5 μg/ml leupeptin/100 μM sodium orthovanadate/50 mM sodium fluoride). FLAG-tagged βarr1 was immunopurified from clarified supernatant with the use of anti-FLAG affinity gel (Sigma). Myc-Dvl was immunopurified similarly, with the use of anti-Myc affinity gel (Covance, Princeton, NJ). Immunocomplexes were washed with buffer A, diluted in 2× sample buffer, and resolved by SDS/PAGE. Resolved proteins were transferred to nitrocellulose filters (Schleicher & Schuell), and nonspecific reactivity was blocked with Tris-buffered saline containing 0.1% Tween 20 and either 5% nonfat dried milk or 3% BSA. Antisera directed against FLAG, HA, and Myc (Santa Cruz Biotechnology) were used according to the manufacturer's specifications. Antiserum directed against Dvl2 has been described (24). Reactive proteins were detected by using the appropriate secondary antibodies in the enhanced chemiluminescence system (Amersham Pharmacia).
Dvl Phosphorylation and βarr1 Binding in Cells.
Wnt3A conditioned medium was prepared as described (35). NIH 3T3 cells expressing FLAG-tagged βarr1 were treated with Wnt3A-conditioned media overnight and lysed in buffer containing 50 mM Tris (pH 8.5), 1 mM EDTA with a 28-gauge needle. To dephosphorylate cell extracts, lysates were treated with 50 units of alkaline phosphatase (Roche Molecular Biochemicals) at 30°C for 30 min and diluted with an equal volume of buffer A. FLAG-tagged βarr1 was immunoprecipitated as described above. To dephosphorylate Dvl2 bound to βarr1, FLAG-βarr1 immunoprecipitates were washed three times with 50 mM Tris (pH 8.4), 1 mM EDTA and treated with 5 units of alkaline phosphatase as described above.
Dvl Phosphorylation and βarr1 Binding in Vitro.
Recombinant MBP-Dvl1 and His-βarr1 were purified as described (5, 36). MBP-Dvl1 was washed with protein kinase CKII buffer (20 mM Tris, pH 7.5/50 mM KCl/2 mM MgCl2) and phosphorylated with protein kinase CKII (New England Biolabs) and [γ-32P]ATP (60 μM; final concentration, 400 cpm/pmol) at 30°C for 30 min. Phosphorylated MBP-Dvl1 was visualized by exposure of dried gels to film. Stoichiometry of Dvl phosphorylation was determined by comparison of bands and standards with a PhosphorImager. For in vitro binding assays MBP-Dvl1 beads were incubated with 1 μg of His-βarr1, and bound βarr1 was analyzed by immunoblot as described above. Immunopurified Myc-Dvl2 from HEK-293 cells were washed with protein kinase CKII reaction buffer, phosphorylated with the addition of ATP, and used in binding assays with His-βarr1 as described above for MBP-Dvl1.
LEF-Luciferase Reporter Gene Assays.
HEK-293 cells were transfected with pGL3-LEF and pCG-LEF(HA) along with combinations of Myc-Dvl1, Myc-Dvl2, and FLAG-βarr1. Cells were harvested 48 h after transfection and lysed by repeated freeze/thaw in reporter lysis buffer (Promega). Luciferase activity was assayed in duplicate with the Luciferase Assay System according to the manufacturer's instructions (Promega).
Results
Identification of Dvl2 as a βarr1 Interacting Protein.
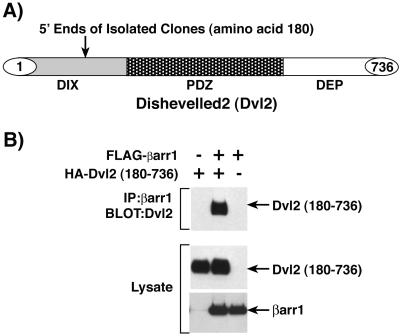
To search for novel βarr1 interacting proteins, we screened a human heart cDNA library with a βarr1-GAL4 DNA binding domain fusion protein as bait in the yeast two-hybrid system. We obtained two identical copies of a cDNA that encodes residues 180–736 of Dvl2 (Fig. 1A). Isolated Dvl2 clones were transformed into yeast and demonstrated specific interaction with the βarr1-GAL4 fusion protein when analyzed with the two-hybrid system (data not shown).
Figure 1.
Identification of Dvl2 as a βarr1 interacting protein. (A) Yeast two-hybrid screening of a human heart cDNA library with βarr1 identified two independent clones of Dvl2. Both clones extend into the DIX domain and contain amino acids 180–736. The basic structure of Dvl2 and the position of the 5′ end of the identified clones are depicted. (B) HEK-293 cells were transfected with HA-Dvl2(180–736) and FLAG-βarr1, and cell extracts were immunoprecipitated with anti-FLAG antibody directed against βarr1. Immunoprecipitates were immunoblotted with anti-HA antibody to detect associated Dvl2(180–736) (Top). To ensure equivalent expression between transfections, whole-cell lysates were immunoblotted with anti-HA antibody to detect Dvl2(180–736) (Middle) and anti-FLAG antibody to detect βarr1 (Bottom). The results shown are representative of at least three independent experiments.
To determine whether βarr1 interacted with Dvl2 in cells, we constructed a HA-tagged Dvl2 (180) expression vector corresponding to the clones isolated in the yeast two-hybrid screen. HEK-293 cells were transfected with HA-tagged Dvl2 (180) and FLAG-tagged βarr1 together or individually. FLAG-βarr1 was immunoprecipitated, and immunoprecipitates were analyzed by immunoblot to determine whether βarr1 and Dvl2 (180) form a complex in cells (Fig. 1B Top). Dvl2 (180) immunoprecipitated with βarr1 only when both HA-Dvl2 (180) and FLAG-βarr1 were expressed together, indicating that these two proteins specifically interact in HEK-293 cells.
βarr1 Interacts with Both Dvl1 and Dvl2 in Cells.
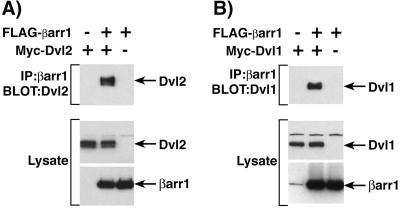
To further confirm the interaction between βarr1 and Dvl2, HEK-293 cells were transfected with full-length Myc-Dvl2 and FLAG-βarr1 together or individually. FLAG-βarr1 was immunoprecipated, and Myc-Dvl2 was present in the immunoprecipated complex only when both Myc-Dvl2 and FLAG-βarr1 were expressed (Fig. 2A Top). Moreover, Myc-Dvl1 was also found to interact with βarr1 in coimmunoprecipitation experiments (Fig. 2B Top). FLAG-βarr1 was immunoprecipated, and Myc-Dvl1 was present in the immunoprecipated complex only when both Myc-Dvl1 and FLAG-βarr1 were expressed.
Figure 2.
Interaction of βarr1 with Dvl1 and Dvl2. (A) HEK-293 cells were transfected with Myc-Dvl2 and FLAG-βarr1, and cell extracts were immunoprecipitated with anti-FLAG antibody directed against βarr1. (B) HEK-293 cells were transfected with Myc-Dvl1 and FLAG-βarr1, and cell extracts were immunoprecipitated with anti-FLAG antibody directed against βarr1. Immunoprecipitates were immunoblotted with anti-Myc antibody to detect associated Dvl proteins (Top). To ensure equivalent expression between transfections, whole-cell lysates were immunoblotted with anti-Myc antibody to detect Dvl (Middle) and anti-FLAG antibody to detect βarr1 (Bottom). The results shown are representative of at least three independent experiments.
Modulation of Dvl2 Interaction with βarr1 in Wnt3A-Stimulated Cells.
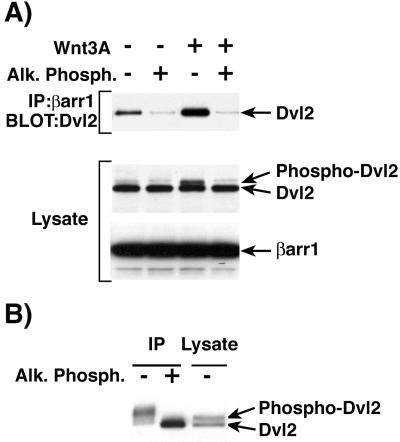
Wnt3A stimulation of endogenous Frizzled receptors in NIH 3T3 cells leads to Dvl phosphorylation (27). To examine the effect of Frizzled stimulation and Dvl2 phosphorylation on βarr1 binding, NIH 3T3 cells were transfected with FLAG-βarr1 for 36–48 h and then stimulated with Wnt3A-conditioned medium or control medium overnight. FLAG-βarr1 was immunoprecipitated from cell extracts treated with or without alkaline phosphatase. In cells stimulated with Wnt3A-conditioned medium there was a significant increase in the amount of endogenous Dvl2 immunoprecipitated with FLAG-βarr1 as compared with cells treated with control medium (Fig. 3A Top). Interestingly, alkaline phosphatase treatment of either unstimulated or Wnt3A-stimulated cell extracts dramatically reduced the binding of endogenous Dvl2 to βarr1. Similar results were observed in Wnt3A-treated HEK-293 cells (data not shown). Analysis of Dvl2 mobility by immunoblot with NIH 3T3 extracts from Wnt3A-stimulated cells demonstrated a large increase in the level of higher molecular weight Dvl2 that is markedly reduced by treatment with alkaline phosphatase (Fig. 3A Middle). These data confirm published results that indicate Dvl2 is phosphorylated upon Wnt3A stimulation and demonstrate that it is the phosphorylated form that preferentially interacts with βarr1 (27). The level of βarr1 in cell extracts treated with Wnt3A conditioned or control medium was the same, indicating that Wnt3A does not affect β-arrestin stability (Fig. 3A Bottom).
Figure 3.
Enhanced binding of βarr1 to Dvl2 in cells treated with Wnt3A. (A) NIH 3T3 cells were transfected with FLAG-βarr1 and treated with Wnt3A-conditioned medium. Cell extracts (± alk. phos. treatment) were immunoprecipitated with anti-FLAG antibody directed against βarr1. Immunoprecipitates were immunoblotted with anti-Dvl antibody to detect Dvl2 (Top). To ensure equivalent expression between transfections, whole-cell lysates were immunoblotted with anti-Dvl antibody to detect Dvl2 (Middle) and anti-FLAG antibody to detect βarr1 (Bottom). (B) NIH 3T3 cells transfected with FLAG-βarr1 and Myc-Dvl2 were immunoprecipitated with anti-FLAG antibody directed against βarr1. Immunoprecipitates (± alk. phos. treatment) were immunoblotted with anti-Myc antibody to detect Dvl2 (left two lanes). Whole-cell lysates were immunoblotted with anti-Myc antibody to demonstrate the electrophoretic mobility of phophorylated and unphosphorylated Dvl2 (right lane). The results shown are representative of at least three independent experiments. Alk. Phosph., alkaline phosphatase.
To obtain more evidence in support of the finding that βarr1 binds preferentially to phosphorylated Dvl2, NIH 3T3 cells were transfected with FLAG-βarr1 and Myc-Dvl2, FLAG-βarr1 was immunoprecipitated, and the immunocomplex was left untreated or was treated with alkaline phosphatase. The mobility of Dvl2 in βarr1 immunoprecipitates shifted to the lower molecular weight form after dephosphorylation by alkaline phosphatase, indicating that βarr1 binds preferentially to phosphorylated Dvl2 (Fig. 3B, left two lanes). Overexpression of Myc-Dvl2 generated two bands in the cell lysate (Fig. 3B, right lane). The higher molecular weight band is most likely the phosphorylated form of Dvl2 because it comigrated with the lower molecular weight band (unphosphorylated form) after the cell extracts were treated with alkaline phosphatase (data not shown). The Dvl2 in βarr1 immunoprecipitates appears to migrate even more slowly than the phosphorylated Dvl2 detectable in whole-cell lysates, which could indicate that βarr1 preferentially binds a less abundant, more highly phosphorylated form of Dvl2 (Fig. 3B, left lane).
In Vitro Interaction of βarr1 with Dvl Is Increased by Dvl Phosphorylation.
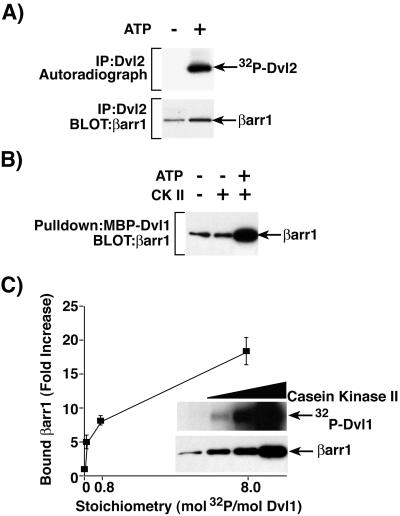
Several protein kinases (CKIɛ, CKII, and PAR-1) have been reported to bind and phosphorylate Dvl proteins (27, 29, 30). To more clearly elucidate the role of phosphorylation in Dvl binding to βarr1, HEK-293 cells were transfected with Myc-Dvl2 and the Dvl2 was immunoprecipitated with anti-Myc affinity beads. When immunoprecipitated Dvl2 was incubated with [γ-32P]ATP, the Dvl2 became phosphorylated, presumably by endogenous kinase(s) associated with Dvl2 (Fig. 4A Upper). The immunoprecipitated Myc-Dvl2 (unphosphorylated or phosphorylated in vitro) was incubated with 1 μg of recombinant His-tagged βarr1. Bound βarr1 was detected by immunoblot, and the results indicated that βarr1 interacts preferentially with the phosphorylated Dvl2 (Fig. 4A Lower).
Figure 4.
Enhanced binding of βarr1 to phosphorylated Dvl. (A) HEK-293 cells were transfected with Myc-Dvl2 and FLAG-βarr1, and cell extracts were immunoprecipitated with anti-Myc antibody directed against Dvl2. Immunoprecipitated Dvl2 was phosphorylated in the presence of [γ-32P]ATP by endogenous Dvl2-associated kinase(s). Immunoprecipitated Dvl2 was processed for autoradiography (Upper) or incubated with His-βarr1 for >3 h at 4°C. Washed Dvl2 immunoprecipitates were analyzed by immunoblot with anti-His antibody to detect associated βarr1 (Lower). (B) MBP-Dvl1 was phosphorylated by protein kinase CKII in the presence of ATP. Phosphorylated and unphosphorylated control MBP-Dvl1 was incubated with His-βarr1 for >3 h at 4°C, and associated βarr1 was detected by immunoblot with anti-His antibody directed against βarr1. (C) MBP-Dvl1 was phosphorylated to various stoichiometric levels by protein kinase CKII. Phosphorylated MBP-Dvl1 was then either eluted in sample buffer and processed for autoradiography (Inset Upper) or incubated with His-βarr1. MBP-Dvl1-associated βarr1 was detected by immunoblot with anti-His antibody directed against βarr1 (Inset Lower). The relative amounts of βarr1 bound to MBP-Dvl1 phosphorylated by protein kinase CKII were quantified as fold increases over that bound to unphosphorylated MBP-Dvl1. Results are the mean ± SEM of three independent experiments.
Recombinant MBP-Dvl1 was phosphorylated with protein kinase CKII to determine whether β-arrestin interacts preferentially with recombinant Dvl phosphorylated in vitro. Unphosphorylated (without the addition of ATP or without kinase and ATP) and in vitro phosphorylated MBP-Dvl1 were incubated with 1 μg of recombinant His-tagged βarr1. Phosphorylated MBP-Dvl1 bound significantly more His-tagged βarr1 (Fig. 4B). To determine whether the extent of phosphorylation of Dvl correlates with binding to βarr1, MBP-Dvl1 immobilized to amylose resin was phosphorylated in vitro by protein kinase CKII to a stoichiometry of 0.08, 0.8, and 8 mol phosphate/mol Dvl1 (Fig. 4C Inset Upper). The unphosphorylated and phosphorylated MBP-Dvl1 proteins were incubated with 1 μg of recombinant His-tagged βarr1. The incorporation of increasing amounts of phosphate into MBP-Dvl1 resulted in the appearance of increasing amounts of His-tagged βarr1 bound to MBP-Dvl1 (Fig. 4C Inset Lower). The relative amount of bound βarr1 bound to MBP-Dvl was quantified, and the data are presented in graphical form (Fig. 4C). The binding of βarr1 to MBP-Dvl1 increased 5.1- to 18.4-fold over that bound to unphosphorylated MBP-Dvl1 when MBP-Dvl1 was phosphorylated at stoichiometric levels of 0.08 to 8.
βarr1 Enhances Dvl-Stimulated LEF Transcriptional Activity.
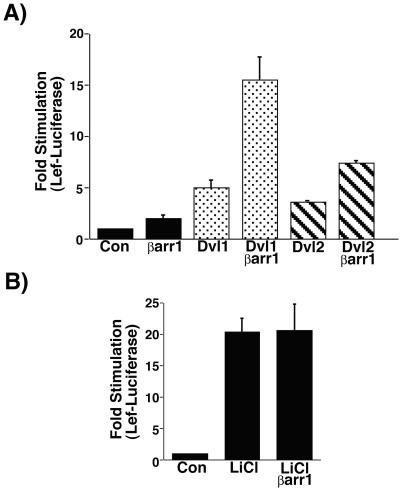
To determine whether βarr1 altered Dvl1-stimulated LEF transcriptional activity, we transfected βarr1, Dvl1, or both plasmids into HEK-293 cells with a LEF-luciferase reporter gene plasmid (Fig. 5A) (37, 38). Dvl1 alone stimulated LEF-Luciferase activity 5-fold over control samples, and βarr1 alone stimulated LEF-Lucifease activity 2-fold over control samples. Interestingly, the combination of βarr1 and Dvl1 stimulated LEF-Luciferase activity 16-fold over control samples. Similar results were obtained with Dvl2 except that Dvl2 itself is a weaker stimulator of LEF activity (Fig. 5A). Dvl2 alone stimulated LEF-Luciferase activity 3.6-fold over control samples, whereas the combination of βarr1 and Dvl2 stimulated LEF-Lucifease activity 7.4-fold over control samples. Relative Dvl levels were unaffected by transfection of βarr1 (data not shown). These results indicate that βarr1 synergistically enhances Dvl-stimulated LEF transcriptional activity.
Figure 5.
βarr1 enhances Dvl1- and Dvl2-stimulated LEF-mediated transcription. (A) HEK-293 cells were transfected with the reporter plasmid pGL3-LEF-FOS, HA-LEF, and combinations of Dvl1, Dvl2, and βarr1. Cell extracts were prepared, and LEF-dependent luciferase activity was quantified. Results are the mean ± SEM of 3–17 independent experiments. (B) HEK-293 cells transfected with the reporter plasmid, HA-LEF, and with or without βarr1 were treated for 16 h with 20 mM LiCl to measure the effect of βarr1 on LiCl-stimulated LEF-dependent transcriptional activity. Results are the mean ± SEM of six independent experiments.
Inhibition of GSK-3β is an important step in the signaling cascade leading to activation of LEF transcriptional activity. Because multiple lines of evidence indicate that Dvl proteins are upstream of GSK-3β, we hypothesized that if βarr1 was influencing LEF transcriptional activity through interaction with Dvl, then βarr1 would have no effect on LEF activity stimulated by direct inhibition of GSK-3β. LiCl binds to and inhibits GSK-3β activity, resulting in a strong stimulation of LEF transcriptional activity (39). We analyzed LiCl-stimulated LEF-Luciferase activity in HEK-293 cells transfected with control plasmids or βarr1 (Fig. 5B). LiCl stimulated LEF-Luciferase activity 20-fold over unstimulated cells, and this increase was unaffected by expression of β-arrestins. These results support our conclusion that βarr1 is acting upstream of GSK-3β and interacting with Dvl proteins to stimulate signal transduction, resulting in increased LEF transcriptional activity.
Discussion
The data presented here identify Dvl as a βarr1 interacting protein that couples the β-arrestins to a unappreciated signaling pathway. Dvl proteins were originally identified in Drosophila as proteins that regulate developmental processes, including the canonical Frizzled signaling pathway that controls embryonic segmentation and patterning as well as a separate pathway that controls planar cell polarity (40–42). More recently Dvl has been identified as a modulator of mammalian signaling involved in regulation of the LEF transcription factor as well as JNK activity (22). The LEF pathway appears to function both in mammalian systems and the canonical Frizzled pathway in Drosophila, whereas the JNK pathway appears to be present both in mammalian systems and the planar cell polarity pathway in Drosophila. The data presented here indicate that βarr1 directly interacts with Dvl and that phosphorylation of Dvl induced by activation of Frizzled receptors stimulates the interaction between βarr1 and Dvl. Additionally, βarr1 synergistically enhances Dvl-stimulated LEF activity. These results suggest that βarr1 plays an important role in the regulation of Dvl signaling.
Dvl appears to be a central regulatory protein in the modulation of all Frizzled receptor signaling pathways. Recently several proteins have been identified, such as Axin, Frat, Idax, and Nkd, which, like βarr1, bind to and/or regulate the activity of Dvl (34, 43–46). The Dvl proteins contain multiple domains normally found in other adapter proteins that are likely to mediate protein–protein interactions and regulate Dvl activity. For example, Nkd binds to Dvl and acts to promote signaling to the JNK pathway while antagonizing signaling to the LEF pathway (44). Nkd binds to the middle region of Dvl, which comprises the PDZ region. It is interesting that Nkd appears to act in opposition to βarr1 in that βarr1 stimulates the pathway leading to LEF activity. Also, in contrast to Nkd, we do not find that βarr1 has any effect on JNK signaling mediated by Dvl (data not shown).
Perhaps further insight into the mechanism by which βarr1 affects Dvl function may come from studies aimed at identifying kinases that phosphorylate Dvl and regulate its function. Members of the casein kinase family (protein kinase CKIɛ and protein kinase CKII) as well as the PAR-1 kinase have been shown to phosphorylate Dvl and positively influence the ability of Dvl to stimulate LEF reporter gene activity (28–30). Inhibition of these kinases with pharmacological inhibitors and/or antisense oligonucleotides destabilizes β-catenin, suggesting a functional influence of Dvl phosphorylation on stimulation of LEF activity. Overexpression of Dvl alone leads to low-level LEF activity, and concomitant expression of CKIɛ or PAR-1 with Dvl synergistically activates LEF-dependent transcription activity (29, 47). The enhanced Dvl activity on LEF induced by protein kinases CK1ɛ and PAR-1 is strikingly similar to what we have observed with overexpression of βarr1. Moreover, we have shown that phosphorylation of Dvl promotes the assembly of the β-arrestin/Dvl complex. Phosphorylation of Dvl may lead to the recruitment of proteins like βarr1 that stimulate some function of Dvl in mediating signal transmission to GSK-3β. It is likely that the precise nature of the components in the Dvl complex determines whether the signal proceeds to LEF or to JNK. It is still unknown precisely which residues are phosphorylated by the various Dvl kinases or what phosphorylation sites lead to enhanced βarr1 binding. Our in vitro studies, however, indicate that βarr1 binding is dramatically increased when the stoichiometry of Dvl1 phosphorylation by protein kinase CKII reaches 1 mol/mol, but we do not know the identity of the endogenous kinase(s) responsible for the stimulation of β-arrestin/Dvl interaction.
Additional evidence that βarr1 functions at the level of Dvl and not further downstream comes from experiments that use LiCl to stimulate LEF activity. LiCl directly binds to GSK-3β and inhibits activity leading to induction of reporter gene activity (39). In this experimental system, βarr1 does not affect LEF activity, suggesting that the step at which βarr1 functions is upstream of GSK-3β and most likely at the level of Dvl. We have not observed β-catenin stabilization by either Dvl or β-arrestin/Dvl in our experiments, although we do see strong stimulation of LEF activity (Fig. 5 and data not shown). However, several groups have reported an increase in cytoplasmic β-catenin induced by strong transient overexpression of Dvl (27, 34). These results suggest that in our cells there is a small but undetectable change in the levels of cytoplasmic β-catenin or that activation of Dvl leads to induction of LEF through a mechanism independent of β-catenin stabilization. Interestingly, a recent report indicates that only a small pool of β-catenin is competent to interact with LEF and suggests that mechanisms distinct from accumulation of β-catenin are also important in transcriptional regulation (48).
Stimulation of GPCR phosphorylation on Ser/Thr residues by GPCR kinases leads to a marked increase in the affinity of the β-arrestins for activated receptors (3). Although this theme is consistent for most if not all GPCRs, no cytoplasmic β-arrestin interacting proteins have been shown to undergo a phosphorylation event that promotes interaction with β-arrestin. Thus, Dvl represents a β-arrestin interacting protein whose interaction is shown to be modulated by phosphorylation, similar to the situation with membrane-bound GPCRs. It will be interesting to determine whether any other cytoplasmic targets of β-arrestin follow this paradigm. As phosphorylation of Dvl is induced by agonist activation of Frizzled, the binding of βarr1 to Dvl is likely to play an important role in the regulation of this pathway.
Although much of the focus on the activation of LEF transcriptional activity has centered on the signaling pathway from Frizzled receptors through Dvl, several recent reports suggest that other GPCR and G protein signaling events can modulate LEF activity (49–51). For example, stimulation of the FPb prostanoid receptor with prostaglandin F2α leads to activation of LEF-dependent transcription (50). Perhaps interaction of βarr1 with Dvl provides a direct means by which GPCRs communicate with this pathway. Under some circumstances Dvl proteins are recruited to the plasma membrane in response to agonist stimulation (52). However, it remains unclear whether membrane recruitment of Dvl is required for activation of LEF transcription pathways. Constitutively active mutants of Gα12 and Gα13 bind to E-cadherin, resulting in release of cadherin-bound β-catenin that stimulates LEF-dependent gene expression (49). Because FP prostanoid receptors activate Gα12 and Gα13, it seems plausible that the interaction of certain G proteins with E-cadherin could be an important factor in the stimulation of LEF activity (53). However, although both FPa and FPb prostanoid receptors activate Gα12 and Gα13 proteins, only the FPb prostanoid receptor stimulated LEF activity, implicating other regulators of GPCR signaling in the activation of LEF activity.
Although GPCR signaling cascades are involved in many aspects of cell biology, the connections between GPCRs and LEF-dependent transcription pathways are just beginning to be understood. Studies aimed at identifying novel β-arrestin interacting proteins indicate that the β-arrestin/Dvl interface may be a key link in the connection between these two pathways. Further studies aimed at understanding the mechanism by which β-arrestin affects Dvl function will be crucial not only for understanding how GPCR signaling components activate LEF transcription activity, but also for understanding how Dvl proteins receive and transmit signals through this important pathway.
Acknowledgments
We thank Stephane A. Laporte for critical reading of the manuscript; Shinji Takada for Wnt3A cells; Donna Addison, Mary Holben, and Julie Turnbough for excellent secretarial assistance; and Sabrina T. Exum, W. Carl Stone, and Grace Irons for technical assistance. This work was supported in part by National Institutes of Health Grant HL16037. R.J.L. is an Investigator of the Howard Hughes Medical Institute.
Abbreviations
- GPCR
G protein-coupled receptor
- βarr1
β-arrestin1
- JNK
c-Jun N-terminal kinase
- GSK-3β
glycogen synthase kinase-3β
- Dvl
Dishevelled
- LEF
lymphoid enhancer factor
- HA
hemagglutinin
References
- 1.Lefkowitz R J. J Biol Chem. 1998;273:18677–18680. doi: 10.1074/jbc.273.30.18677. [DOI] [PubMed] [Google Scholar]
- 2.Pierce K L, Luttrell L M, Lefkowitz R J. Oncogene. 2001;20:1532–1539. doi: 10.1038/sj.onc.1204184. [DOI] [PubMed] [Google Scholar]
- 3.Pitcher J A, Freedman N J, Lefkowitz R J. Annu Rev Biochem. 1998;67:653–692. doi: 10.1146/annurev.biochem.67.1.653. [DOI] [PubMed] [Google Scholar]
- 4.Lohse M J, Benovic J L, Codina J, Caron M G, Lefkowitz R J. Science. 1990;248:1547–1550. doi: 10.1126/science.2163110. [DOI] [PubMed] [Google Scholar]
- 5.Attramadal H, Arriza J L, Aoki C, Dawson T M, Codina J, Kwatra M M, Snyder S H, Caron M G, Lefkowitz R J. J Biol Chem. 1992;267:17882–17890. [PubMed] [Google Scholar]
- 6.Miller W E, Lefkowitz R J. Curr Opin Cell Biol. 2001;13:139–145. doi: 10.1016/s0955-0674(00)00190-3. [DOI] [PubMed] [Google Scholar]
- 7.Luttrell L M, Ferguson S S, Daaka Y, Miller W E, Maudsley S, Della Rocca G J, Lin F, Kawakatsu H, Owada K, Luttrell D K, et al. Science. 1999;283:655–661. doi: 10.1126/science.283.5402.655. [DOI] [PubMed] [Google Scholar]
- 8.Barlic J, Andrews J D, Kelvin A A, Bosinger S E, DeVries M E, Xu L, Dobransky T, Feldman R D, Ferguson S S, Kelvin D J. Nat Immunol. 2000;1:227–233. doi: 10.1038/79767. [DOI] [PubMed] [Google Scholar]
- 9.Miller W E, Maudsley S, Ahn S, Khan K D, Luttrell L M, Lefkowitz R J. J Biol Chem. 2000;275:11312–11319. doi: 10.1074/jbc.275.15.11312. [DOI] [PubMed] [Google Scholar]
- 10.Imamura, T., Huang, J., Dalle, S., Ugi, S., Usui, I., Luttrell, L. M., Miller, W. E., Lefkowitz, R. J. & Olefsky, J. M. (2001) J. Biol. Chem., in press. [DOI] [PubMed]
- 11.McDonald P H, Chow C W, Miller W E, Laporte S A, Field M E, Lin F T, Davis R J, Lefkowitz R J. Science. 2000;290:1574–1577. doi: 10.1126/science.290.5496.1574. [DOI] [PubMed] [Google Scholar]
- 12.Miller W E, McDonald P H, Cai S F, Field M E, Davis R J, Lefkowitz R J. J Biol Chem. 2001;276:27770–27777. doi: 10.1074/jbc.M102264200. [DOI] [PubMed] [Google Scholar]
- 13.DeFea K A, Zalevsky J, Thoma M S, Dery O, Mullins R D, Bunnett N W. J Cell Biol. 2000;148:1267–1281. doi: 10.1083/jcb.148.6.1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luttrell L M, Roudabush F L, Choy E W, Miller W E, Field M E, Pierce K L, Lefkowitz R J. Proc Natl Acad Sci USA. 2001;98:2449–2454. doi: 10.1073/pnas.041604898. . (First Published February 20, 2001; 10.1073/pnas.041604898) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wodarz A, Nusse R. Annu Rev Cell Dev Biol. 1998;14:59–88. doi: 10.1146/annurev.cellbio.14.1.59. [DOI] [PubMed] [Google Scholar]
- 16.Peifer M, Polakis P. Science. 2000;287:1606–1609. doi: 10.1126/science.287.5458.1606. [DOI] [PubMed] [Google Scholar]
- 17.Ikeda S, Kishida S, Yamamoto H, Murai H, Koyama S, Kikuchi A. EMBO J. 1998;17:1371–1384. doi: 10.1093/emboj/17.5.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kitagawa M, Hatakeyama S, Shirane M, Matsumoto M, Ishida N, Hattori K, Nakamichi I, Kikuchi A, Nakayama K. EMBO J. 1999;18:2401–2410. doi: 10.1093/emboj/18.9.2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu C, Kato Y, Zhang Z, Do V M, Yankner B A, He X. Proc Natl Acad Sci USA. 1999;96:6273–6278. doi: 10.1073/pnas.96.11.6273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sharpe C, Lawrence N, Martinez Arias A. BioEssays. 2001;23:311–318. doi: 10.1002/bies.1045. [DOI] [PubMed] [Google Scholar]
- 21.Boutros M, Paricio N, Strutt D I, Mlodzik M. Cell. 1998;94:109–118. doi: 10.1016/s0092-8674(00)81226-x. [DOI] [PubMed] [Google Scholar]
- 22.Li L, Yuan H, Xie W, Mao J, Caruso A M, McMahon A, Sussman D J, Wu D. J Biol Chem. 1999;274:129–134. doi: 10.1074/jbc.274.1.129. [DOI] [PubMed] [Google Scholar]
- 23.Moriguchi T, Kawachi K, Kamakura S, Masuyama N, Yamanaka H, Matsumoto K, Kikuchi A, Nishida E. J Biol Chem. 1999;274:30957–30962. doi: 10.1074/jbc.274.43.30957. [DOI] [PubMed] [Google Scholar]
- 24.Semenov M V, Snyder M. Genomics. 1997;42:302–310. doi: 10.1006/geno.1997.4713. [DOI] [PubMed] [Google Scholar]
- 25.Sussman D J, Klingensmith J, Salinas P, Adams P S, Nusse R, Perrimon N. Dev Biol. 1994;166:73–86. doi: 10.1006/dbio.1994.1297. [DOI] [PubMed] [Google Scholar]
- 26.Willert K, Brink M, Wodarz A, Varmus H, Nusse R. EMBO J. 1997;16:3089–3096. doi: 10.1093/emboj/16.11.3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee J S, Ishimoto A, Yanagawa S. J Biol Chem. 1999;274:21464–21470. doi: 10.1074/jbc.274.30.21464. [DOI] [PubMed] [Google Scholar]
- 28.Song D H, Sussman D J, Seldin D C. J Biol Chem. 2000;275:23790–23797. doi: 10.1074/jbc.M909107199. [DOI] [PubMed] [Google Scholar]
- 29.Sun T Q, Lu B, Feng J J, Reinhard C, Jan Y N, Fantl W J, Williams L T. Nat Cell Biol. 2001;3:628–636. doi: 10.1038/35083016. [DOI] [PubMed] [Google Scholar]
- 30.Peters J M, McKay R M, McKay J P, Graff J M. Nature (London) 1999;401:345–350. doi: 10.1038/43830. [DOI] [PubMed] [Google Scholar]
- 31.Cullen B R. Methods Enzymol. 1987;152:684–704. doi: 10.1016/0076-6879(87)52074-2. [DOI] [PubMed] [Google Scholar]
- 32.Gietz R D, Schiestl R H, Willems A R, Woods R A. Yeast. 1995;11:355–360. doi: 10.1002/yea.320110408. [DOI] [PubMed] [Google Scholar]
- 33.James P, Halladay J, Craig E A. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kishida S, Yamamoto H, Hino S, Ikeda S, Kishida M, Kikuchi A. Mol Cell Biol. 1999;19:4414–4422. doi: 10.1128/mcb.19.6.4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shibamoto S, Higano K, Takada R, Ito F, Takeichi M, Takada S. Genes Cells. 1998;3:659–670. doi: 10.1046/j.1365-2443.1998.00221.x. [DOI] [PubMed] [Google Scholar]
- 36.Chen W, Komuniecki P R, Komuniecki R. Biochem J. 1999;339:103–109. [PMC free article] [PubMed] [Google Scholar]
- 37.Hsu S C, Galceran J, Grosschedl R. Mol Cell Biol. 1998;18:4807–4818. doi: 10.1128/mcb.18.8.4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sakanaka C, Weiss J B, Williams L T. Proc Natl Acad Sci USA. 1998;95:3020–3023. doi: 10.1073/pnas.95.6.3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stambolic V, Ruel L, Woodgett J R. Curr Biol. 1996;6:1664–1668. doi: 10.1016/s0960-9822(02)70790-2. [DOI] [PubMed] [Google Scholar]
- 40.Perrimon N, Mahowald A P. Dev Biol. 1987;119:587–600. doi: 10.1016/0012-1606(87)90061-3. [DOI] [PubMed] [Google Scholar]
- 41.Klingensmith J, Nusse R, Perrimon N. Genes Dev. 1994;8:118–130. doi: 10.1101/gad.8.1.118. [DOI] [PubMed] [Google Scholar]
- 42.Theisen H, Purcell J, Bennett M, Kansagara D, Syed A, Marsh J L. Development (Cambridge, UK) 1994;120:347–360. doi: 10.1242/dev.120.2.347. [DOI] [PubMed] [Google Scholar]
- 43.Li L, Yuan H, Weaver C D, Mao J, Farr G H, 3rd, Sussman D J, Jonkers J, Kimelman D, Wu D. EMBO J. 1999;18:4233–4240. doi: 10.1093/emboj/18.15.4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yan D, Wallingford J B, Sun T Q, Nelson A M, Sakanaka C, Reinhard C, Harland R M, Fantl W J, Williams L T. Proc Natl Acad Sci USA. 2001;98:3802–3807. doi: 10.1073/pnas.071041898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hino S, Kishida S, Michiue T, Fukui A, Sakamoto I, Takada S, Asashima M, Kikuchi A. Mol Cell Biol. 2001;21:330–342. doi: 10.1128/MCB.21.1.330-342.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fearnhead N S, Britton M P, Bodmer W F. Hum Mol Genet. 2001;10:721–733. doi: 10.1093/hmg/10.7.721. [DOI] [PubMed] [Google Scholar]
- 47.Kishida M, Hino Si S, Michiue T, Yamamoto H, Kishida S, Fukui A, Asashima M, Kikuchi A. J Biol Chem. 2001;276:33147–33155. doi: 10.1074/jbc.M103555200. [DOI] [PubMed] [Google Scholar]
- 48.Gottardi C J, Wong E, Gumbiner B M. J Cell Biol. 2001;153:1049–1060. doi: 10.1083/jcb.153.5.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Meigs T E, Fields T A, McKee D D, Casey P J. Proc Natl Acad Sci USA. 2001;98:519–524. doi: 10.1073/pnas.021350998. . (First Published January 2, 2001; 10.1073/pnas.021350998) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fujino H, Regan J W. J Biol Chem. 2001;276:12489–12492. doi: 10.1074/jbc.C100039200. [DOI] [PubMed] [Google Scholar]
- 51.Liu T, DeCostanzo A J, Liu X, Wang H, Hallagan S, Moon R T, Malbon C C. Science. 2001;292:1718–1722. doi: 10.1126/science.1060100. [DOI] [PubMed] [Google Scholar]
- 52.Yanagawa S, van Leeuwen F, Wodarz A, Klingensmith J, Nusse R. Genes Dev. 1995;9:1087–1097. doi: 10.1101/gad.9.9.1087. [DOI] [PubMed] [Google Scholar]
- 53.Pierce K L, Fujino H, Srinivasan D, Regan J W. J Biol Chem. 1999;274:35944–35949. doi: 10.1074/jbc.274.50.35944. [DOI] [PubMed] [Google Scholar]