Abstract
Fibroblast growth factors (FGFs) are signalling peptides that control important cell processes such as proliferation, differentiation, migration, adhesion and survival. Through binding to different types of receptor on the cell surface, these peptides can have different effects on a target cell, the effect achieved depending on many features. Thus, each of the known FGFs elicits specific biological responses. FGF receptors (FGFR 1–5) initiate diverse intracellular pathways, which in turn lead to a variety of results. FGFs also bind the range of FGFRs with a series of affinities and each type of cells expresses FGFRs in different qualitative and quantitative patterns, which also affect responses. To summarize, cell response to binding of an FGF ligand depends on type of FGF, FGF receptor and target cell, all interacting in concert. This review aims to examine properties of the FGF family and its members receptors. It also aims to summarize features of intracellular signalling and highlight differential effects of the various FGFs in different circumstances.
Introduction
Fibroblast growth factors (FGFs) are secreted polypeptide ligands that can act in either paracrine or endocrine fashions to stimulate or uphold particular cell functions required for metabolism, tissue homeostasis and development. The mammalian FGF family consists of 23 related polypeptides. Eighteen of these have been grouped into five paracrine‐acting groups and one endocrine‐acting group, on the basis of amino acid sequence and structural analysis (reviewed in 1). These molecules have been studied for many years, but their functions are yet to be fully understood. The first FGFs were isolated from bovine brain tissue in the late 1970s 2, 3 and since then scientists have been working to understand their effects, as each member elicits different biological responses in different cells (reviewed in 4, 5, 6). It has been demonstrated that FGFs are involved in processes including cell proliferation, migration, differentiation, adhesion and survival. Targets of the FGFs are mainly two classes of receptor, the tyrosine kinase receptor family 7 and co‐receptors heparin sulphate proteoglycans 5. FGF–tyrosine kinase receptor interaction is not a straightforward process; different FGFs have different affinities for different receptors. Moreover, activation of multiple receptors affects subsequent downstream activity. Full understanding of fibroblast growth factor–receptor interactions however, will help comprehension of how any specific biological response is achieved. The variety of FGF affinities for the receptors, lead in turn to different effects on the cell 8.
Fibroblast growth factors and their receptors have important functions in development, metabolism, angiogenesis and tumourigenesis, and some also play an important role in embryogenesis, organ development and wound healing 1; thus it is essential to understand their wide‐ranging effects. In particular, the role of FGFs in eliciting PKC gamma, Ras‐MAP kinase (Ras‐MAPK) and Src‐mediated pathways (cf below) is crucial in tumourigenesis, and further understanding of this may be a further step in combatting cancer. Here, the members of the fibroblast growth factor family will be described with their receptors, and how interactions between them can lead to a plethora of cellular responses.
The FGFs
Fibroblast growth factors are secreted polypeptide ligands that bind to a variety of receptors located on the surface of target cells, of many tissues. Structure of FGFs has been revealed by high‐resolution X‐ray diffraction as well as by nuclear magnetic resonance imaging 9 and shows that the archetypal structure consists of 12 strands, linked together forming a 3‐fold symmetrical structure of beta sheets 10. All members of the FGF family have the same core of around 140 amino acids. Twenty‐two types of FGF have been recognized in humans, while FGF‐24 has been identified only in zebrafish embryos 11. FGF‐15 occurs in mice and is closely related to human FGF‐19 12 (Fig. 1).
Figure 1.
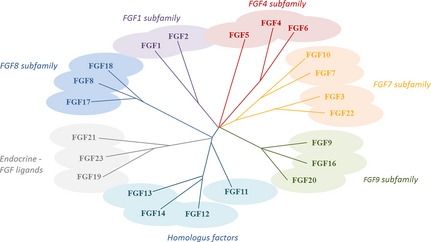
Fibroblast growth factor family tree. Modified after 14.
First FGFs to be discovered were FGF 1 and FGF 2, originally called acidic and basic FGFs (aFGF and bFGF) 2, 3, 13. These were found to have significant effects on cell migration, proliferation, differentiation and angiogenesis. Also, FGFs are sometimes referred to as heparin‐binding growth factors, as they have high affinity for heparin and heparan sulphate. Binding of FGFs to different glucosaminoglycans (such as heparan sulphate) makes FGFs resistant to degradation, hence they can remain as a reservoir in the extracellular matrix.
Fibroblast growth factors are commonly divided into subgroups 14 although members of each family have similar qualities. First, FGFs11–FGF15 are not always included in the FGF family; this is because unlike other FGFs, they do not have the ability to bind to and activate FGF receptors. Instead, they are called homologous factors, acknowledging that their genomic structure highly resembles FGFs. FGF1 and FGF2 (aFGF and bFGF) are members of the FGF1 subfamily. They were the first to be discovered, yet their physiological roles are still unclear. It is likely that they affect vascular tone or reduce blood pressure 15. However, it is known that FGF2 has angiogenic properties, promotes proliferation and migration, and inhibits apoptosis of endothelial cells 16. The FGF4 subfamily includes FGF4, FGF5 and FGF6. FGF4 is particularly important during organ development. It affects processes such as trophoblast proliferation as well as limb and heart valve development, while FGF5 is an important factor in hair growth cycle regulation 17. FGF3, FGF7, FGF10 and FGF22 are members of the FGF7 subfamily 14. FGF3 is crucial in development of inner ear structure, while FGF7 is central to kidney development; it is sometimes referred to as keratinocyte growth factor (KGF) 14. FGF22, FGF7 and FGF10 are presynaptic organizers involved in vesicle clustering and neurite branching 18. A further subfamily includes FGF8, FGF17 and FGF18. FGF8 is a significant player in limb, ear, eye and brain development and together with FGF17, FGF8 also produces effects on forebrain development 19; FGF18 is required for correct development of bone. The FGF9 subfamily consists of FGF9, FGF16 and FGF20. FGF9 upregulates proliferation of mesenchymal tissues, and initiates secretion of ligands for FGF3, FGF7, FGF10 and FGF22 subgroups. Accordingly, FGF9 knockout leads to reduced production of certain ligands and reduced mesenchymal–epithelial signalling 20.
FGF19, FGF21 and FGF23 belong to a subfamily normally referred to as endocrine FGF ligands 21. One property that distinguishes these from other FGFs is that they need the presence of two types of klotho‐protein to form the FGF–receptor complex in tissue; this is a consequence of their low affinity to heparin sulphate. The two types of klotho protein (αKlotho and βKlotho) 22, 23, 24, 25, 26, 27, 28 are selectively used as co‐receptors by FGF19 subfamily members 21. FGF19 stimulates bile acid synthesis and initiates oxidation of fatty acids. FGF21 causes a fasting response by stimulating glucose uptake in adipocytes, thus reducing levels of glucose in the bloodstream. Injections of FGF21 to diabetic and obese mice leads to reduced blood concentration of insulin, glucagon, glucose and triglycerides. Continuous injection of FGF21 to obese mice reduces their body weight in the order of 20%. Finally, FGF23 is an important vitamin D regulator 21.
FGF receptors
The FGFs bind to two different classes of receptor and it is important to appreciate that many different ligands can activate the same receptor. FGFs bind simultaneously to low‐affinity, heparin sulphate proteoglycans 29 and high‐affinity FGF receptors. High‐affinity receptors consist of one extracellular component containing between one and three Ig‐SF domains, one transmembrane domain and one intracellular tyrosine kinase domain. High‐affinity receptors have one unique structure that distinguishes them from others namely an ‘acid box’ consisting of eight acidic residues located between the first and second Ig‐SF domains. First Ig‐SF domain and the acid box probably contribute to autoinhibition, while domains 2 and 3 are FGF ligand‐binding sites 6.
Five signalling FGF receptors (expressed as multiple splice variants) have been identified to date 30, 31, but it is believed that there are others hitherto undiscovered. The common name for these receptors is FGFR 1–5 and they are coded for by separate genes; differential splicing gives rise to multiple alternative forms of the receptors. For example, splicing of the gene that codes for Ig‐like domain 3 causes variants with different specificities for the binding site. Different isoforms of the receptors are expressed in different organs, for example, FGFR3IIIb is mainly located in epithelial tissues and IIIc forms are preferrentially expressed in tissues of mesenchymal origin. Both isoforms have separate ligands, which only bind to the specific receptor 30. This means that mesenchymal cells produce ligands that only activate IIIc receptor, achieving a paracrine signal 30.
Fibroblast growth factors receptors are unevenly distributed between body tissues and patterns in which they occur are specific to each tissue. Studies have shown that FGFR‐1 is expressed in skin, calvarial bones and growth plates and at high levels in the foetal brain 32, 33. FGFR‐2 is also found in the brain, growth plates and calvarial bone, but also in liver, lungs, intestine and kidneys 34, 35. FGFR‐3 also is expressed in brain, growth plates and calvarial bone as well as in lung, kidney and intestines 36, 37. FGFR‐4 can be found in lungs, kidney, liver, pancreas, intestine, foetal adrenals, spleen and striated muscle 38.
Low‐affinity FGF receptors are present on surfaces of most cells; they can also be called heparan sulphate proteoglycans (HSPGs). Heparan sulphate is a linear glycan, containing disaccharide unit repeats of glucuronic acid and N‐substituted glucosamine 39, 40. N‐sulphated molecules 5–10 disaccharide units in length with various modifications alternate along the glycan chain, with N‐acetylated stretches that are mainly unaltered 39, 40. These changes in N‐sulphated regions include C5 epimerization of glucuronic acid 3‐0‐ as well as 6‐0‐sulphation of N‐sulphated glucosamine 39, 40; FGFs mainly bind to these modified sulphated domains 40. Interestingly, alteration of heparan sulphate by sulphation and epimerization is controlled in a tissue‐specific fashion, contributing to a better finetuning of FGF activity.
Thus, HSPGs have two different very important functions. First, is that binding FGF to an HSPG protects the growth factor from degradation, thus it can act as an extracellular buffer. Secondly, HSPGs are involved in complex formation between FGFs and their FGFR. Binding FGFs to their respective receptors induces dimerization and formation of a ternary complex containing FGF, FGFR and heparan sulphate. However, the primary importance of HSPGs appear to be to maintain functional role of paracrine FGF activity, as they immobilize FGFs in the extracellular matrix neighbouring their site of secretion; this limits their activity on cells in the immediate proximity. Conversely, as endocrine FGFs have a poor affinity to HSPGs, they can easily enter the bloodstream and act at a distance from their site of synthesis. In contrast to paracrine FGFs that depend heavily on HSPGs, endocrine FGFs instead require Klotho co‐receptors to become biologically active 22, 23, 24, 25, 26, 27, 28.
FGF signalling
Signalling via activation of an FGFR requires receptor dimerization, a prerequisite for moving intracellular kinases closer to each other, initiating onset of different intracellular signalling pathways that lead to adjustment of gene expression 41. Formation of receptor dimers activates their intracellular tyrosine kinases, allowing them to transphosphorylate tyrosine residues on each dimer of the receptor 42. These residues can act as binding sites for signalling molecules containing Src homology‐2 or phosphotyrosine‐binding domains. Signalling molecules are often bound to different docking proteins and activated receptor kinases phosphorylate and activate their intracellular substrates. Most prominent of these are FGFR substrate 2a (FRS2a) 43 and phospholipase Cy1 (PLCy1) 44, 45. Upon activation, FRS2a initiates downstream signalling via one of two pathways – Ras‐MAPK or PI3K‐AKT 43, 44, 45 (Fig. 2).
Figure 2.

Overview of fibroblast growth factor ( FGF ) receptor signalling.
Ras‐MAPK is one of the best studied signalling pathways. It involves docking protein FRS2α, which becomes activated by tyrosine residues on the activated FGF receptor. FRS2α is the core of a complex formed by adaptor Grb2, tyrosine phosphatase Shp2 and docking protein GAB1 46. To Grb2 binds guanine nucleotide exchange factor SOS, which in turn activates the Ras‐MAP kinase. The MAP kinases are regulatory proteins that affect different kinases and transcription factors and thereby regulate target genes. Effects gained by stimulating Ras‐MAP kinase are mainly mitogenic 6.
GAB1 also leads to activation of a further important pathway, by activation of PI‐3 kinase. P‐I‐3 activates PDK, which in turn activates ATK/PKB 43. Effects of ATK are anti‐apoptotic. Activated FGF receptors can also lead to hydrolysis of PtdIns and activation of PDK and ATK. FGFs can act on intracellular calcium levels through recruitment of Src homology‐2 domain of PLCγ to the receptor. Activation of PLCγ allows it to hydrolyse PIP2, which leads to formation of diaglycerol and IP3. Effect of diaglycerol and Ins P3 is release of calcium and activation of calcium‐dependent protein kinases, which affect cytoskeletal organization 41.
In addition, Src plays an unexpected role in receptor signalling. FGFR activation stimulates clathrin‐mediated endocytosis. FGF exposure increases number of CCPs, including those undergoing endocytosis, and this is mediated by Src and its phosphorylation target Eps8. Eps8 interacts with clathrin‐mediated endocytosis machinery and depletion of Eps8 inhibits FGFR signalling and immediate Erk signalling. Once internalized, FGFR passes through peripheral early endosomes on its way to recycling, via a Src‐ and Eps8‐dependent mechanism 45.
Diversity in cellular responses
Effects achieved by these signalling cascades are not shared through all cell types. There appears to be an apparently redundant number of receptors and ligands. Moreover, cross‐binding activity between ligands and receptors further complicates the picture and makes it difficult to use gene manipulation as a way forward. The MAPK pathway is always seen as a response to FGF ligand binding, while others, such as AKT activation, differ between cell types 47. Moreover, FGF signalling is affected by tissue‐specific HSPGs that can either amplify or block FGFR activation. Varying effects also seem to depend on cell state of differentiation, receptor phenotype and presence of other growth factors or cytokines 47.
A further explanation for the variety between cell responses is that intracellular signalling pathways are influenced by a number of regulators. Examples of these are the Sproty proteins, MAPK phosphatase 3 and SEF, which are inhibitory molecules that either bind to different molecules (Sproty protein) and inhibit them, or act as regulatory feedback [SEF has similar expression to FGF 48]. There are also excitatory molecules, which upregulate signalling pathways such as FLRT, a family of transmembrane proteins 14.
Finally, responses to FGFs are in some instances, concentration dependent. One study on lens epithelial cells has indicated that low concentrations (150 pg/ml) of FGF‐1 and FGF‐2 initiated proliferation; as higher concentrations (3 ng/ml) of FGF were added, the cells started to migrate. To achieve cell differentiation, an even higher concentration (40 ng/ml) of FGF was required 49. It also seems that time interval over which cells are exposed to FGFs matters. Proliferation and migration were achieved within 24 h, while their differentiation was seen after 4 days. It was also shown that proliferation and migration could occur simultaneously and higher concentration lead to more pronounced response.
Embryogenesis
Fibroblast growth factors and their receptors have long been implicated in control of embryogenesis 50. In the mouse, FGF‐4 is the first FGF to be expressed, from the 4‐cell stage and onwards 51; FGF‐4 knockout mice have been shown to suffer peri‐implantation lethality at stages E4–5. Development progresses normally up to the blastocyst stage, but they die immediately after implantation due to ICM (inner cell mass) defect formation 52. Moreover, along with FGF‐4 expression, the fifth cell division appears to be crucial for blastocyst formation 53. It is believed that FGF‐4 effects are mediated via FGFR2 as this receptor is the first to be expressed 54. Indeed, FGFR2 knockout mice suffer from early embryonic lethality comparable to FGF‐4 KO mice. In both cases, cause of death seems to be deficient formation and maintenance of the ICM 54.
Three receptors, FGFR1, 3 and 4, are all expressed in early development 54. Deletion of FGFR1 is lethal at stages E7.5–E9.5, as it prevents normal gastrulation and inhibits mesoderm/endoderm differentiation 55. FGFR 3 knockout mice survive, but display skeletal malformations, whereas FGFR4 knockout mice appear to have normal development 56.
Use of murine embryonic stem cells has shed further light on the role of FGFs in development. Homozygous FGF4 −/− ES cells proliferate normally when grown in culture. They maintain pluripotency, but their differentiated progeny have a much reduced lifespan. By blocking FGFR1 and FGFR3 in ES cells, it has been shown that these cells continue to proliferate and maintain their state of pluripotency, suggesting that this particular signalling mechanism does not initiate differentiation itself, but rather exerts a permissive effect 57. FGFR2, on the other hand, stimulates proliferation of ES cells from the 5th cell division to ICM formation, whereas FGFR1 and 3 studied in epiblast EC cells affected germ layer specification 58. Thus, it appears probable that FGF signalling in embryos has clear stage‐specific effects.
Teratocarcinoma
Different biological responses to FGF exposure can, in some instances, be concentration‐dependent, as for example, in the human teratocarcinoma cell line Tera 2 59, 60. These cells have been extensively used as a target line for FGFs as they express four FGF receptors (FGFR1‐4); the cells have been shown to respond differently to changes in concentration of FGFs. Cell population multiplication was stimulated by addition of 1–10 ng bFGF/ml, but bFGF‐effect was abrogated by addition of protamine sulphate. When high concentrations of bFGF were added, preferential effects on cell locomotion were observed. One hundred nanograms bFGF/ml stimulated cell movement, but only exerted a marginal effect on cell proliferationion 61. Moreover, when effects of four other members of the fibroblast growth factor family, FGF‐10, FGF‐16, FGF‐17 and FGF‐18, were examined, it was found that all four enhanced survival levels of Tera‐2 cells by counteracting apoptosis at concentrations between 1–10 ng/ml. When higher concentrations of any of the four FGFs were added, preferential effects on cell motility were observed 4. Greater difference were revealed when effects of FGF‐8, FGF‐9 and KGF on Tera 2 cells were examined. It was found that each of these factors promoted Tera 2 cell proliferation, albeit with different efficacy. Whereas dramatic effects on cell numbers were observed after addition of 1–10 μg FGF‐9/ml, a lower effect could be achieved by FGF‐8 or KGF. In contrast, KGF expressed the most potent effect on cell locomotion at higher concentration (100 μg/ml). Even though high concentrations of FGF‐8 and FGF‐9 stimulated cell movement, this effect was substantially lower than that of KGF 62. Likewise, FGF‐19, as well as FGF‐20, promoted Tera 2 cell proliferation. Whereas FGF‐20 promoted cell population expansion at low doses, FGF‐19 was required at high doses to achieve a comparable effect. Moreover, FGF‐19 did not significantly stimulate cell locomotion, while FGF‐20 promoted motility at high doses 63. In one recent study, it was demonstrated that FGF23 acted in a similar way to FGF19, whereas FGF24 was virtually without effect on proliferation or expansion of Tera 2 cell numbers 64.
Oligodendrocytes
A number of studies has shown that activation of MAPK leads to proliferation of oligodendrocyte precursors and endothelial cells. Inhibition of PLCγ, on the other hand, did not affect oligodendrocytes 5. Whereas ligand activation of the MAPK pathway has been shown to stimulate proliferation in other cell types, investigations of chondrocytes has shown that MAPK activation, unlike PLCγ and PI‐3, leads to interruption of the cell cycle. Effects of FGF2 on development of oligodendrocytes have been studied and it has been found that FGF2 induces different, stage‐specific responses in the cells 65, 66. One experiment on oligodendrocytes has shown that FGF‐2 induced proliferation in the cells while FGFs‐8, ‐9 and ‐17 had no effect, no matter how high the concentration was or how long the duration. This difference in response is due to FGF‐2 rapidly activating the MAPK pathway, while FGF‐8, ‐9 and ‐17 had much weaker and slower effects on it. Consequences of FGF‐2 on differentiation have also been closely studied 66 the conclusions being that FGF‐2 inhibits differentiation of oligodendrocyte progenitors. However, further studies showed that FGF‐9, unlike FGF‐2, did not block oligodendrocyte differentiation.
Effects of FGFs on mature oligodendrocytes have also been examined: FGF‐2 induces multiple responses in mature cell line such as elongation, inhibition of myelin protein synthesis and re‐entering the cell cycle. These were studied after FGF‐8, ‐9, ‐17 and ‐18 addition, when it was shown that only FGF‐2 had any cell cycle effect. FGFs‐9 and ‐18 provided the same results as FGF‐2 on oligodendrocyte differentiation, but treatment with FGF‐9 and ‐18 did not result in loss of myelin‐like membranes, as observed in FGF‐2 treated cells. FGF‐8 and FGF‐17 did not increase cell size, which suggests that these cells' response distinguished between different FGFs 66.
Expression patterns of the receptors may also play a certain role in response of FGF signalling. It has been confirmed that cells express different FGFRs during different developmental phases. FGFR expression pattern changes dramatically as cells differentiate. An example of this is FGFR expression in oligodendrocytes during differentiation where FGFR1 is expressed throughout development; while FGFR2 is more prominent during terminal differentiation, FGFR3 is downregulated at the end of oligodendrocyte differentiation 65, 66.
It is also possible that different FGF‐FGFR interactions lead to different responses. This theory has been tested by adding FGFR inhibitors to oligodendrocytes during different phases of development. Results indicated that during the progenitor phase, activation of FGFR‐1 only (by FGF‐2) 67, 68 induced proliferation, while inhibition of proliferation required FGF‐8, FGF‐17 or FGF‐18, bound to FGFR‐3. When the same experiment was performed on differentiated oligodendrocytes it revealed that activation of FGFR‐1 was required for cells to re‐enter the cell cycle, not FGFR‐3. Cell elongation required activation of FGFR‐2 by FGF‐2, FGF‐9 and FGF‐18 67, 68 (Fig. 3).
Figure 3.
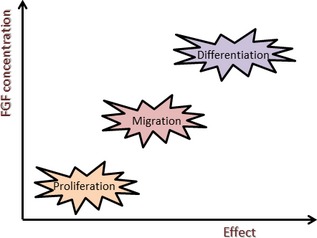
Relationship between concentration and biological effects.
Concluding remarks
Fibroblast growth factors play a pivotal role in regulation of key developmental processes. There is mounting evidence for importance of correct spatial and temporal regulation of expression of FGFs and their receptors. This review has highlighted differential effects of the 23 hitherto discovered mammalian members of the FGF family. These ligands interact with a family of tyrosine kinase receptors that can elicit a variety of biological responses. We conclude that different FGFs do not necessarily have the same effect on one type of cell, as different FGFs exert different responses. Moreover, a variety of intracellular pathways is activated to differing extents depending on which ligand initiates activation.
One particular type of FGF can also give rise to varied responses at different stages of development. The picture is further complicated by different cellular expression patterns of tyrosine kinase receptors during different phases of development. Finally, response to FGF activation may depend on availability of substrates and other intracellular regulators. A key question is then how these different responses are generated and more specifically how very similar elicitations can lead to changes in secondary patterns of gene expression, which direct intracellular signal transduction to generate any particular cell response.
One obvious result is that even though FGF and FGFR families are large, there does not appear to be any functional redundancy in the system. This issue has been addressed in studies where one FGF has been deleted in mouse knockout experiments and other FGFs have been increased to compensate for the lacking factor. A good example is knockout mice lacking FGF10 that are characterized by mammary gland hypoplasia, salivary gland aplasia and pulmonary agenesis 69, 70, 71. In contrast, knockout of the closely related FGF7 fails in kidney development – a phenotype quite distinct from FGF10 knockout mice 72. Even though FGF7 and FGF10 are branching morphogens 73, structurally very similar, it seems clear that they cannot substitute for each other. Moreover, FGF7 and FGF22 are both involved in development of presynaptic terminals of hippocampal pyramidal neurons. However, these two ligands affect formation of different synapses, and it has been conclusively demonstrated that they cannot replace each other 74.
Vast amounts of genetic data on mammalian development have pointed at the importance of a finely orchestrated role for the FGF family in normal development. FGFs are part of an extended gene family including TGFbeta/BMPs, Hedgehog, Notch and Wnt 41. Albeit structurally different, they all combine their efforts to steer undifferentiated cells toward lineage determination, proliferation, locomotion and differentiation. This concept also suggests that there is a role for crosstalk between activation of FGF‐receptors and other signalling pathways. There is mounting evidence that FGF‐activation may activate or repress other signalling pathways such as TGFbeta/BMP, IGF, IHH/PTHIH and Notch 5. However, the best‐characterized developmental crosstalk is that between FGF and Wnt. In such distinct areas as trachea development in Drosophila, mesoderm induction in Xenopus, and CNS, kidney and tooth development in lower mammals 75 crosstalk between FGFs and Wnt can lead to convergence or divergence of signalling routes activated by each pathway. Moreover, it has been suggested that activation of one signal can confer competence of another 76.
However, still puzzling questions remain to be explored. Interplay between endocrine and paracrine FGFs is an area that has recently attracted some attention. Some clues have emerged from studies of adipose tissue where postprandial induction of FGF1 and FGF21 link back to peroxisome proliferator‐activated receptor gamma (PPAR gamma) 77, 78. FGF1 is an adipose tissue morphogen acting via PPAR gamma, a role that is believed to safeguard nutrient supply 79, 80. This process also requires FGF21 for a successful outcome 81. FGF21, however, stimulates PPAR gamma in such a way that this could be seen as a positive feedback loop 82. Hence, nuclear receptors could be seen as one way of integrating endocrine FGF and paracrine FGF action.
Much attention has been given to the role of FGFs in tumourigenesis and development 83. Recently, the role of FGFs in ageing has attracted some promising attention. A series of studies using FGF23 knockout mice has pointed at accelerated ageing and reduced endogenous expression of other FGFs. Moreover, ability to respond to FGFs appears to be obscured in such mice. This has been the basis for testing FGFs on a therapeutic scale for such age‐related diseases as osteoarthritis, cardiovascular disease and type II diabetes. 14.
In conclusion, the FGF signalling systems must be tightly regulated and integrated to uphold metabolism, ensure tissue repair and avoid dysregulation of development 84. Even minor mutations or changes in gene activation can eventually cause tissue damage or malignant disease 5. However, on the bright side, the FGF–FGFR system still remains a promising area for future drug development.
References
- 1. Itoh N, Ornitz DM (2011) Fibroblast growth factors: from molecular evolution to roles in development, metabolism and disease. J. Biochem. 149, 121–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gospodarowicz D, Bialecki T, Greenburg G (1978) Purification of the fibroblast growth factor activity from bovine brain. J. Biol. Chem. 253, 3736–3743. [PubMed] [Google Scholar]
- 3. Böhlen P, Esch F, Baird A, Gospodarowicz D (1985) Acidic fibroblast growth factor from bovine brain: amino terminal sequence and comparison with basic FGF. EMBO J. 4, 1951–1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Granerus M, Engström W (2000) Dual effects of four members of the fibroblast growth factor member family on multiplication and motility in human teratocarcinoma cells in vitro. Anticancer Res. 20, 3527–3532. [PubMed] [Google Scholar]
- 5. Dailey L, Ambrosetti D, Mansukhani A, Basilico C (2005) Mechanisms underlying differential responses to FGF signaling. Cytokine Growth Factor Rev. 16, 233–247. [DOI] [PubMed] [Google Scholar]
- 6. Wesche J, Haglund K, Haugsten M (2011) Fibroblast growth factors and their receptors in cancer. Biochem. J. 437, 199–213. [DOI] [PubMed] [Google Scholar]
- 7. Fanti WJ, Johnson DE, Williams LT (1993) Signalling by receptor tyrosine kinases. Ann. Rev. Biochem. 62, 453–481. [DOI] [PubMed] [Google Scholar]
- 8. Olsen SK, Ibrahami OA, Raucci A, Zhang F, Eliseenkova AV, Yayon A et al (2004) Insights into the molecular basis for fibroblast growth factor receptor autoinhibition and ligand binding promiscuity. Proc. Natl. Acad. Sci. USA 27, 935–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Arunkumar AI, Srisailam T, Krishnaswamy T, Kumar S, Kathir KM, Chi Y et al (2002) Structure and stability if an acidic fibroblast growth factor from Notophthalamus viridescens . J. Biol. Chem. 277, 46424–46432. [DOI] [PubMed] [Google Scholar]
- 10. Zhu X, Komiya H, Chirino A, Faham S, Fox GM, Arakawa T et al (1991) Three‐dimensional structures of acidic and basic fibroblast growth factors. Science 251, 90–93. [DOI] [PubMed] [Google Scholar]
- 11. Fisher S, Draper BW, Neumann CJ (2003) The zebrafish fgf24 mutant identifies an additional level of Fgf signaling involved in vertebrate forelimb initiation. Development 130, 3515–3524. [DOI] [PubMed] [Google Scholar]
- 12. Nishimura T, Utsunomiya M, Hoshikawa M, Ohuchi H, Itoh N (1999) Structure and expression of a novel human FGF, FGF‐19, expressed in the fetal brain. Biochem. Biohys. Acta 1444, 148–151. [DOI] [PubMed] [Google Scholar]
- 13. Fitzgerald KA, O'neill LA, Geraring AJ, Callard RE (2001). The Cytokine Factsbook and Webfacts, 2nd edn Burlington, ON: Elsevier Science. [Google Scholar]
- 14. Beenken A, Mohammadi M (2009) The FGF family: biology, pathophysiology and therapy. Nat. Rev. Drug Discov. 8, 235–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cuevas P, Carceller F, Ortega S, Nieto I, Giménez‐Gallego G (1991) Hypotensive activity of fibroblast growth factor. Science 254, 1208–1210. [DOI] [PubMed] [Google Scholar]
- 16. Zhou M, Sutliff RL, Paul RJ, Lorenz JN, Hoying JB, Haudenschild CC et al (1998) Fibroblast growth factor 2 control of vascular tone. Nat. Med. 4, 201–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pethö‐Schramm A, Müller HJ, Paus R (1996) FGF5 and the murine hair cycle. Arch. Dermatol. Res. 288, 264–266. [DOI] [PubMed] [Google Scholar]
- 18. Umemori H, Linhoff MW, Ornitz DM, Sanes JR (2004) FGF22 and its close relatives are presynaptic organizing molecules in the mammalian brain. Cell Press 118, 257–270. [DOI] [PubMed] [Google Scholar]
- 19. Bernhard R, von Bohlen OH (2003) Fibroblast growth factors and their receptors in the central nervous system. Cell Tissue Res. 313, 139–157. [DOI] [PubMed] [Google Scholar]
- 20. Colvin JS, White AC, Pratt SJ, Ornitz DM (2001) Lung hypoplasia and neonatal death in FGF9‐null mice identify gene as an essential regulator of lung mesenchyme. Development 128, 2095–2106. [DOI] [PubMed] [Google Scholar]
- 21. Wu A, Coulter S, Liddle C, Wong A, Eastham‐Anderson J, French DM et al (2011) FGF19 regulates cell proliferation, glucose and bile acid metabolism via FGFR4‐dependent and independent pathways. PLoS One 000, 0017868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Harmer NJ (2006) Insights into the role of heparan sulphate in fibroblast growth factor signalling. Biochem. Soc. Trans. 34, 442–445. [DOI] [PubMed] [Google Scholar]
- 23. Mohammadi M, Olsen SK, Ibrahimi OA (2005) Structural basis for fibroblast growth factor activation. Cytokine Growth Factor Rev. 16, 107–137. [DOI] [PubMed] [Google Scholar]
- 24. Sleeman M, Fraser J, McDonald M, Yuan S, Whte D, Grandison P et al (2001) Identification of a new fibroblast growth factor receptor, FGFR5. Gene 271, 171–182. [DOI] [PubMed] [Google Scholar]
- 25. Zhang H, Dessimoz J, Beyer TA, Krampert M, Williams LT, Werner S et al (2004) FGF receptor 1‐IIIb is dispensible for skin morphogenesis and wound healing. Eur. J. Cell Biol. 83, 3–11. [DOI] [PubMed] [Google Scholar]
- 26. Beer HD, Vindevoghe L, Gait MJ, Revest JM, Duan DR, Mason I et al (2000) FGF receptor 1‐IIIb is a naturally occurring functional receptor for FGFs that is preferentially expressed in the skin and the brain. J. Biol. Chem. 275, 16091–16097. [DOI] [PubMed] [Google Scholar]
- 27. Ledoux D, Mereau A, Pieri I, Barritault D, Courty J (1991) High affinity receptors to acidic and basic FGF are detected mainly in adult membrane preparations but not in liver, intestine, lung or stomach. Growth Factors 5, 221–231. [DOI] [PubMed] [Google Scholar]
- 28. Steiling H, Werner S (2003) Fibroblast growth factors; key players in epithelial morphohgenesis, repair and cytoprotection. Curr. Opin. Biotechnol. 14, 533–537. [DOI] [PubMed] [Google Scholar]
- 29. Scotet E, Houssaint E (1995) The choice between alternative IIIb and IIIc exons of the FGFR‐3 gene is not strictly tissue specific. Biochim. Biophys. Acta 1264, 238–242. [DOI] [PubMed] [Google Scholar]
- 30. Claus P, Grothe C (2001) Molecular cloning and developmental expression of rat FGF receptor 3. Histochem. Cell Biol. 115, 147–155. [DOI] [PubMed] [Google Scholar]
- 31. Stark KL, McMahon JA McMahon AP (1991) FGFR‐4, a new member of the FGF receptor family expressed in the definitive endoderm and skeletal muscle lineages of the mouse. Development 113, 641–651. [DOI] [PubMed] [Google Scholar]
- 32. Asada M, Shinomiya M, Suzuki M, Honda E, Sugimoto R, Iketika M et al (2008) Glycosaminoglycan affinity of the complete fibroblast growth factor family. Biochim. Biophys. Acta 1790, 40–48. [DOI] [PubMed] [Google Scholar]
- 33. Gallagher JT (2001) Heparan sulphate: growth control with a restricted sequence menu. J. Clin. Inv. 108, 357–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kurusu H, Choi M, Ogawa Y, Dickson AS, Goetz R, Elisenkova AV et al (2007) Tissue specific expression of betaKlotho and fibroblast growth factor receptor isoforms determines metabolic activity of FGF19 and FGF21. J. Biol. Chem. 282, 26687–26695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kurusu H, Ogawa Y, Miyoshi M, Yamamoto M, Nandi A, Rosenblatt KP et al (2006) Regulation of fibroblast growth factor 23 signalling by Klotho. J. Biol. Chem. 281, 6120–6123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ogawa Y, Kurusu H, Yamamoto M, Nandi A, Rosenblatt KP, Goetz R et al (2007) Beta Kloyho is required for metabolic activity of fibroblast growth factor 21. Proc. Natl. Acad. Sci. USA 104, 7432–7437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tomiyama K, Maeda R, Urakawa I, Yamazaki Y, Tanaka T, Ito S et al (2010) relevant use of Klotho in FGF19 subfamily signaling system in vivo. Proc. Natl. Acad. Sci. USA 107, 1666–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Urakawa I, Yamasaki Y, Shimada T, Ijima K, Hasegawa H, Okawa K et al (2006) Klotho converts canonical FGF receptor into a specific receptor for FGF23. Nature 444, 770–774. [DOI] [PubMed] [Google Scholar]
- 39. Yie J, Wang W, Deng L, Tam LT, Stevens J, Chen MM et al (2012) Understanding the physical interactions in the FGF21/FGFR/beta klotho complex: structural requirements and implications in FGF21 signalling. Chem. Biol. Drug Des. 79, 398–410. [DOI] [PubMed] [Google Scholar]
- 40. Yang C, Jin C, Li X, Wang F, McKeehan WL, Luo Y (2012) Differential specificity of endocrine FGF19 and FGF21 to FGFR1 and FGFR4 with KLB. PLoS One 7, e33870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Schlessinger J (2000) Cell signaling by receptor tyrosine kinases. Cell 103, 211–225. [DOI] [PubMed] [Google Scholar]
- 42. Furdui CM, Lew ED, Schlessinger J, Anderson KS (2006) Autophosphorylation of FGFR1 kinase is mediated by a sequential and precisely ordered reaction. Mol. Cell 21, 711–717. [DOI] [PubMed] [Google Scholar]
- 43. Gotoh N (2008) Regulation of growth factor signaling by FRS2 family docking/scaffold adaptor proteins. Cancer Sci. 99, 1319–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Carpenter G, Ji Q (1999) Phospholipase C gamma as a signal transducing element. Exp. Cell Res. 253, 15–24. [DOI] [PubMed] [Google Scholar]
- 45. Turner N, Grose R (2010) Fibroblast growth factor signaling: from development to cancer. Nat. Rev. Cancer 10, 116–129. [DOI] [PubMed] [Google Scholar]
- 46. Eswarakumar VP, Lax I, Schlessinger J (2005) Cellular signaling by fibroblast growth factor receptors. Cytokine Growth Factor Rev. 16, 139–149. [DOI] [PubMed] [Google Scholar]
- 47. Katoh M, Nakagama H (2013) FGF receptors. Cancer biology and treatment. Med. Res. Rev. doi: 10.1002/med.21288. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 48. Tsang M, Friesel R, Kudoh T, Dawid IB (2002) Identification of Sef, a novel modulator of FGF signaling. Nat. Cell Biol. 4, 165–169. [DOI] [PubMed] [Google Scholar]
- 49. McAvoy JV, Chamberlain CG (1989) Fibroblast growth factor (FGF) induces different responses in lens epithelial cells depending on its concentration. Development 107, 221–228. [DOI] [PubMed] [Google Scholar]
- 50. Böttcher RT, Niehrs C (2005) Fibroblast growth factor signaling during early vertebrate development. Endocr. Rev. 26, 63–77. [DOI] [PubMed] [Google Scholar]
- 51. Rappolee D, Basilico C, Patel Y, Werb Z (1994) Expression and function of FGF‐4 in periimplantation development development in mouse embryos. Development 120, 2259–2269. [DOI] [PubMed] [Google Scholar]
- 52. Feldman B, Poueymirou W, Papaioannou VE, deChiara TM, Goldfarb M (1995) Requirement of FGF‐4 for postimplantation mouse development. Science 267, 246–249. [DOI] [PubMed] [Google Scholar]
- 53. Chai N, Patel Y, Jacobson K, McMahon J, McMahon A, Rappolee D (1998) FGF is an essential regulator of the fifth cell division in preimplantation mouse embryos. Dev. Biol. 198, 105–115. [DOI] [PubMed] [Google Scholar]
- 54. Rappolee DA, Patel Y, Jocobson K (1998) Expression of fibroblast growth factors in periimplantation mouse embryos. Mol. Reprod. Dev. 51, 254–264. [DOI] [PubMed] [Google Scholar]
- 55. Arman E, Haffner–Krausz R, Chen Y, Heath JK, Lonai P (1998) Targeted disruption of fibroblast growth factor receptor 2 suggests a role for FGF signaling in pregastrulation mammalian development. Proc. Natl. Acad. Sci. USA 95, 5082–5087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Deng CX, Wynshaw‐Boris A, Shen MM, Daugherty C, Ornitz DM, Leder P (1994) Murine FGFR1 is required for early postimplantation growth and axial organization. Genes Dev. 8, 3045–3057. [DOI] [PubMed] [Google Scholar]
- 57. Kunath T, Saba‐El‐Leil MK, Almoussailleakh M, Wray J, Meloche S, Smith A (2007) FGF stimulation of the Erk1/2 cascade triggers transition of pluripotent embryonic stem cells from self renewal to lineage commitment. Development 134, 2895–2902. [DOI] [PubMed] [Google Scholar]
- 58. Lanner F, Rossant J (2010) The role of FGF/Erk signaling in pluripotent cells. Development 137, 3351–3360. [DOI] [PubMed] [Google Scholar]
- 59. Thompson S, Stern PL, Webb M, Walsh FS, Engström W, Evans EP et al (1984) Cloned human teratoma cells differentiate into neuron like cells and other cell types in retinoic acid. J. Cell Sci. 72, 37–64. [DOI] [PubMed] [Google Scholar]
- 60. Engström W (1986) Differential effects of epidermal growth factor on cell locomotion and cell proliferation in a cloned human embryonal carcinoma derived cell line in vitro. J. Cell Sci. 86, 47–55. [DOI] [PubMed] [Google Scholar]
- 61. Schofield PN, Granerus M, Lee A, Ekström TJ, Engström W (1992) Concentration dependent modulation of basic fibroblast growth factor action on multiplication and locomotion of teratocarcinoma cells. FEBS Lett. 298, 154–158. [DOI] [PubMed] [Google Scholar]
- 62. Granerus M, Engström W (2003) Effects of FGF8, FGF9 and keratinocyte growth factor on multiplication and locomotion in human teratocarcinoma cells in vitro. Anticancer Res. 23, 1313–1316. [PubMed] [Google Scholar]
- 63. Granerus M, Engström W (1996) Growth factors and apoptosis. Cell Prolif. 29, 309–314. [DOI] [PubMed] [Google Scholar]
- 64. Laestander C, Engström W (2013) Effects of fibroblast growth factors 23 and 24 on cell multiplication and locomotion in a human embryonal carcinoma cell line (Tera 2) in vitro. Cell Prolif. 46, 495. [Google Scholar]
- 65. Bansal R (2002) Fibroblast growth factors and their receptors in oligodendrocyte development: implications for demyelinisation and remyelinisation. Dev. Neurosci. 46, 24–35. [DOI] [PubMed] [Google Scholar]
- 66. Fortin D, Rom E, Sun H, Yayon A, Bansal R (2005) Distinct fibroblast growth factor/FGF receptor signaling pairs initiate diverse cellular responses in the oligodendrocyte lineage. J. Neurosci. 25, 7470–7479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Frederick TJ, Wood TL (2004) IGF‐1 and FGF‐2 coordinately enhance cyclin D1 and cyclin E‐cdk2 association and activity to promote G1 progression in oligodendrocyte progenitor cells. Mol. Cell. Neurosci. 25, 480–492. [DOI] [PubMed] [Google Scholar]
- 68. Bansal R, Magge S, Winkler S (2003) Specific inhibitor of FGF receptor signaling: FGF‐2 mediated effects on proliferation, differentiation and MAPK activation are inhibited by PD173074 in oligodendrocyte cells. J. Neurosci. Res. 74, 486–493. [DOI] [PubMed] [Google Scholar]
- 69. Mailleux AA, Spencer‐Dene B, Dillon C, Ndiaye D, Savona‐Baron C, Itoh N et al (2002) Role of FGF10‐FGFR2b signaling during mammary gland development in the mouse embryo. Development 129, 53–60. [DOI] [PubMed] [Google Scholar]
- 70. Parsa S, Ramasamy SK, de Langhe S, Gupte VV, Haigh JJ, Medina D et al (2013) Terminal end bud 7 maintenance in mammary gland is dependent upon FGFR2b signaling. Dev. Biol 317, 121–131. [DOI] [PubMed] [Google Scholar]
- 71. Pond AC, Bin X, Batts T, Roarty K, Hilsenbeck S, Rosen JM (2013) Fibroblast growth factor receptor signaling is essential for normal mammary gland development and stem cell function. Stem Cells 31, 178–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Quio J (1999) FGF7 modulates ureteric bud growth and nephron number in the developing kidney. Development 126, 547–554. [DOI] [PubMed] [Google Scholar]
- 73. Sekine K, Ohuchi H, Fujiwara M, Yoshizawa T, Sato T, Yagishita N et al (1999) FGF10 is essential for limb and lung formation. Nat. Genet. 21, 138–141. [DOI] [PubMed] [Google Scholar]
- 74. Terauchi A, Johnson‐Venkatesh EM, Toth AB, Javed D, Sutton MA, Umemori H (2010) Distinct FGFs promote differentiation of excitatory and inhibitory synapses. Nature 465, 783–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Moon RT, Brown JD, Torres M (1997) WNTs modulate cell fate and behavior during vertebrate development. Trends Genet. 13, 157–162. [DOI] [PubMed] [Google Scholar]
- 76. Trueb B, Amann R, Gerber SD (2013) Role of FGFRL1 and other FGF signaling proteins in early kidney development. Cell. Mol. Life Sci. 70, 2505–2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Jonker JW, Suh M, Atkins AR, Ahmadjan M, Li P, Whyte J et al (2012) A PPAR gamma‐FGF1 axis is required for adaptive adipose remodeling and metabolic homeostasis. Nature 485, 391–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Dutchak PA, Katafuchi T, Bookout AL, Choi JH, Yu RT, Mangelsdorf DJ et al (2012) Fibroblast growth factor 21 regulates PPAR gamma activity and the antidiabetic actions of thiazolidinediones. Cell 148, 556–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Tontonoz P, Spiegelman BM (2008) Fat and beyond: the diverse biology of PPAR gamma. Ann. Rev. Biochem. 77, 289–312. [DOI] [PubMed] [Google Scholar]
- 80. Potthoff MJ, Kliewer SA, Mangelsdorf DJ (2012) Endocrine fibroblast growth factors 15/19 and 21: from feast to famine. Genes Dev. 26, 312–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Hotta Y, Nakamura H, Konishi M, Murata Y, Takagi H, Matsumura S et al (2009) Fibroblast growth factor 21 regulates lipolysis in white adipose tissue but is not required for ketogenesis and triglyceride clearance in liver. Endocrinology 150, 4625–4633. [DOI] [PubMed] [Google Scholar]
- 82. Ming AY, Yoo E, Vorontsov EN, Altamentova SM, Kilkenny DM, Rocheleau JV (2012) Dynamics and distribution of KlothoB (KLB) and fibroblast growth factor receptor 1 (FGFR1) in living cells reveal the fibroblast growth factor 21 (FGF21) induced receptor complex. J. Biol. Chem. 2878, 19997–20006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Salottin J, Dias MH, Koga MM, Armelin HA (2013) Fibroblast growth factor 2 causes G2/M cell cycle arrest in Ras driven tumour cells through a src‐dependent pathway. PLoS One 8, e72582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Auciello G, Cunningham DL, Tatar T, Heath JK, Rappoport JZ (2013) Regulation of fibroblast growth factor receptor signaling and trafficking by src and Eps8. J. Cell Sci. 126, 613–624. [DOI] [PMC free article] [PubMed] [Google Scholar]


