Abstract
Abstract. The Int‐6 gene is a site of mouse mammary tumour virus (MMTV) integration in murine tumours and INT6 protein has been identified independently as a subunit (eIF3e) of the eukaryotic translation initiation factor eIF3. In addition, the protein can interact with two other multi‐subunit complexes: the COP9 signalosome (CSN) and the proteasome. The role of INT6 in tumourigenesis is nonetheless currently unclear. Here, using immunofluorescence microscopy, we show that eIF3e/INT6 is localized in part to the nucleus, while other eIF3 components are cytoplasmic. Primary human fibroblasts, but not their transformed counterparts, showed reduced nuclear INT6 staining in some cells, and this reduction was maximal in early S phase. This variation in eIF3e/INT6 may indicate regulated shuttling between cellular compartments and would be consistent with the presence of a nuclear export signal as well as a nuclear localization signal in the protein sequence. Loss of regulation of eIF3e/INT6 redistribution may therefore be a significant feature of malignancy in human cells.
INTRODUCTION
The Int‐6 gene was originally identified as the site of mouse mammary tumour virus (MMTV) integration in two virally induced tumours and one pre‐neoplastic lesion (Marchetti et al. 1995). In each case, the MMTV integration occurred within an Int‐6 intron and in the opposite transcriptional orientation. The resulting Int‐6/MMTV hybrid mRNA has the potential to encode a C‐terminally truncated Int6 protein, which may act as a dominant negative oncoprotein. Nonetheless, no such protein has been reported in the MMTV‐induced tumours, such that the possibility of Int‐6 haploinsufficiency cannot be ruled out (Asano et al. 1997; M et al. 2001). Int‐6, which is expressed ubiquitously in adult mouse tissues and from day 8.5 of embryonic development, has been highly conserved throughout evolution, with related proteins found in fission yeast through to humans (Marchetti et al. 1995; Diella et al. 1997; Crane et al. 2000). The murine and human proteins are 100% identical, 48‐kDa proteins containing a bipartite nuclear localization sequence (NLS) and a putative N‐terminal nuclear export signal (NES). Potentially, therefore, INT6 could shuttle between the nucleus and the cytoplasm.
Human INT6 was identified independently as the eIF3e/p48 subunit of eukaryotic translation initiation factor 3 (eIF3) (Asano et al. 1997). The initiation of protein synthesis in eukaryotes is governed by at least 10 separable initiation factors and is primarily a cytoplasmic process. eIF3 is essentially responsible for binding the 40S ribosomal subunit and is the largest initiation factor, comprising 11 non‐identical subunits, eIF3a–eIF3k. At least five of these subunits have been implicated in human cancer; eIF3a, eIF3b, eIF3c and eIF3h are all over‐expressed in a variety of tumours. In contrast, the expression of eIF3e/INT6 is reduced in a proportion of mammary carcinomas and non‐small cell lung carcinomas (Watkins & Norbury 2002). Recent data have shown that eIF3e/INT6 can interact with p56, an interferon inducible protein and inhibitor of protein synthesis. This indicates that eIF3e may function as a negative regulator of translation initiation (Guo et al. 2000). Together with the original MMTV‐induced gene disruption seen in murine breast cancers, the evidence suggests that in mammals, INT6 could have some of the properties of a tumour suppressor gene, although it is frequently described as a proto‐oncogene.
As a component of a translation initiation factor, eIF3e/INT6 might be expected to be cytoplasmic, but the actual localization of the protein remains contentious. Murine Int‐6 was found to be perinuclear and associated with the Golgi apparatus (Diella et al. 1997), while in Drosophila the corresponding protein was cytoplasmic (Miyazaki et al. 1999). In human cells, however, endogenous INT6 was localized to PML bodies in the nucleus (Morris‐Desbois et al. 1999). Recent studies have shown that eIF3e/INT6 interacts not only with eIF3 but also with subunits of the COP9 signalosome, which is predominantly nuclear (Yahalom et al. 2001) and the 26S proteasome, which is distributed throughout mammalian cells (Hoareau Alves et al. 2002; Yen et al. 2003). These multi‐subunit complexes are involved in distinct aspects of the ubiquitin‐dependent proteolysis pathway. It is possible that eIF3e/INT6 has multiple roles and that its subcellular localization varies according to its function.
Here, we describe the characterization of an antibody that specifically detects human eIF3e/INT6 by immunofluorescence microscopy. Our data suggest that a major fraction of eIF3e/INT6 is nuclear and does not co‐localize with the bulk of eIF3. These findings provide new insight into the normal biological role of INT6, as well as giving an indication of how this function could be disrupted during tumourigenesis.
MATERIALS AND METHODS
Antibodies and cell culture
The N‐terminal 20‐kDa portion of human INT6 was expressed as a glutathione S‐transferase fusion protein in Escherichia coli BL‐21. Gel‐purified fusion protein was used to raise a rabbit polyclonal antiserum (denoted CN25) following standard procedures (Crane et al. 2000). Antibodies were affinity purified against recombinant full‐length INT6, and were used at 1 µg/ml for immunoblotting and 2 µg/ml for immunofluorescence. Goat anti‐eIF3 antiserum was kindly donated by John Hershey and was used at 1/500 dilution for immunofluorescence. H1299 (human small cell lung carcinoma) and Cos7 (SV40‐transformed African green monkey kidney) cells were cultured in Dulbecco's modified Eagle's medium (DMEM) with 10% fetal calf serum (FCS) and 4 mm glutamine. HF19 and MRC5 (human primary lung fibroblasts) and MRC5vi (SV40‐transformed human fibroblasts) were grown in alpha‐MEM with 10% FCS and 4 mm glutamine. HT1080 (human fibrosarcoma) cells were grown in RPMI medium with 10% FCS and 4 mm glutamine. MCF‐10a (adherent human breast epithelium) were maintained in DMEM/Ham's F12 medium (50 : 50 ratio) with 10% FCS, 5 µg/ml hydrocortisone, 10 ng/ml EGF and 10 µg/ml insulin.
Immunofluorescence microscopy
Cells were grown on glass cover slips (Marienfield No. 1; 22 × 22 mm) and were fixed by immersion in ice‐cold 4% paraformaldehyde, 0.25 m Hepes (pH 7.4) for 30 min (4 °C). Cells were then washed twice in cold phosphate‐buffered saline (PBS) and permeabilized in PBS containing 0.5% Triton X‐100 for 30 min at 4 °C. Fixed cells were blocked by incubation in PBS, 0.1% Tween‐20, 10% FCS for 30 min at 37 °C. The coverslips were immersed in primary antibody diluted in the same blocking solution and incubated for 1 h at 37 °C. Cells were washed twice in PBS and incubated with the appropriate CY3‐conjugated secondary antibody (in PBS, 0.1% Tween‐20, 10% FCS) for 1 h at 37 °C. After further washing with PBS, cells were stained with Hoechst 33258 (1 µg/ml) in order to detect DNA, washed with distilled water, air dried in the dark for 15 min and then mounted on microscope slides (Gold Star) in 90% Glycerol, 50 mm Tris‐HCl pH 8.0, 1 mm p‐phenylene diamine. Images were acquired using a Zeiss Axioskop microscope equipped with a Planapochromat 100 × objective, an Axiocam cooled CCD camera and Axiovision software (Carl Zeiss Ltd, Welwyn Garden City, UK), and were assembled using Adobe Photoshop. For quantification of differential eIF3e/Int6 staining patterns from immunofluorescence data, 100 cells were counted and the nuclear localization pattern of eIF3e/Int6 was recorded. The cells were chosen from random fields of view and all counts were performed in triplicate for each slide.
Cell cycle synchronization
Duplicate samples of cells were grown in parallel on coverslips and in 100‐mm culture dishes. After 24 h, all cells were washed with medium containing 0.2% FCS and then incubated for a further 48 h in the same low serum medium. Medium containing 10% serum was then added back to the cells and incubation continued. Cells were harvested at intervals 16–24 h thereafter; coverslips were fixed and stained as described above; the corresponding 100‐mm culture dishes were simultaneously trypsinized and cells were prepared for analysis by propidium iodide staining and flow cytometry.
For arrest in early S phase, duplicate samples of cells were grown in parallel on coverslips and in 100‐mm culture dishes as above. At approximately 50% confluence, the cells were incubated in medium containing 5 µg/ml aphidicolin (Sigma, St Louis, MO, USA) for 16 h at 37 °C. Cells on coverslips were fixed and stained as described above. Cells in 100‐mm culture dishes were trypsinized and prepared for analysis by flow cytometry. In some experiments, a double block protocol was used: at 50% confluence, cells were incubated with 2 mm thymidine for 16 h. Cells were washed and allowed to grow in normal culture medium for 9 h before incubation with aphidicolin as described above.
Flow cytometry
Cells were harvested by trypsinization, collected by centrifugation and washed in 2 ml of ice‐cold PBS. Cells were fixed by re‐suspension in 2 ml ice‐cold 70% ethanol and incubation for 30 min at 4 °C. Fixed cells were washed with cold PBS, then re‐suspended in 2 ml PBS, 100 µg/ml Rnase A, 40 µg/ml propidium iodide and incubated in the dark for 15 min at room temperature (23 °C). Red fluorescence was measured for up to 10 000 cells using a Becton Dickinson FACScalibur cytometer and cell profiles were analysed using CellQuest software.
Immunoblotting
To prepare whole cell protein extracts, cells were grown in 100‐mm culture dishes until 75% confluent. Cells were harvested and re‐suspended in 100 µl cold PBS followed by an equal volume of 2 × sodium dodecyl sulfate – polyacrylamide gel electrophoresis (SDS–PAGE) sample buffer (0.125 m Tris pH 6.8, 20% Glycerol, 4% SDS, 1%β‐mercaptoethanol, 0.04% bromophenol blue). Lysates were then heated at 98 °C for 5 min before loading onto SDS/polyacrylamide (10%) gels. eIF3e/INT6 was detected following semi‐dry blotting onto nitrocellulose membranes. Membranes were blocked with MPBST (PBS, 0.3% Tween‐20, 3% Marvel skimmed milk) for 30 min at room temperature and then incubated with the anti‐INT6 polyclonal antibody (1 µg/ml dilution in MPBST) followed by an HRP‐conjugated anti‐rabbit secondary antibody (1/1000 dilution). Protein bands were visualized using ECL chemiluminescence (Amersham Biosciences Little Chalfont, UK) according to the manufacturer's instructions.
Cell fractionation
To prepare nuclear and cytoplasmic protein fractions, cells were grown in three 100‐mm culture dishes to 75% confluence. Cells were trypsinized, collected by centrifugation and washed with 5 ml ice‐cold PBS. Cell pellets were re‐suspended in 500 µl TMN buffer: 10 mm Tris pH 7.5, 1.5 mm MgCl2, 10 mm NaCl, 1 mm NaF, 1 mm Na3VO4, 1 mm DTT plus EDTA‐free protease inhibitor cocktail (Roche, Lewes, UK). The suspension was incubated on ice for 15 min and cells were lysed in a 3‐ml dounce homogenizer (13 strokes). Homogenized cells were centrifuged at 1500 g for 5 min at 4 °C to pellet the nuclear fraction. The supernatant was retained as a cytoplasmic fraction. The nuclear pellet was re‐suspended in 500 µl TKM buffer: 50 mm Tris pH 7.5, 5 mm MgCl2, 25 mm KCl, 1 mm NaF, 1 mm Na3VO4, 1 mm DTT plus EDTA‐free protease inhibitor cocktail. Fractions were stored at −80 °C. Efficiency of fractionation was determined by immunoblotting using IHIC8 (rabbit polyclonal anti‐topoisomerase IIα serum at 1/500 dilution; kindly provided by Ian Hickson) and mouse anti‐β‐tubulin (Sigma) at 1/500 dilution.
Small interfering RNA (siRNA) duplex preparation
RNA oligonucleotides (Cruachem, Glasgow, UK) were designed corresponding to a 21‐base pair region of the INT6 gene, 219 base pairs downstream from the translation initiation codon:
INT6 sense: 5′‐GAACCACAGUGGUUGCACAUU; INT6 antisense: 5′‐UGUGCAACCACUGUGGUUCUU; reverse INT6 control sense: 5′‐UUACACGUUGGUGACACCAAG; reverse INT6 control antisense: 5′‐UUCUUGGUGUCACCAACGUGU.
These were dissolved at 50 µm in nuclease‐free water and stored at −80 °C. 50‐µm sense and antisense RNA oligonucleotides were denatured in siRNA annealing buffer (20 mm potassium acetate, 6 mm Hepes‐KOH pH 7.4, 0.4 mm magnesium acetate) for 1 min at 85 °C. The RNAs were allowed to anneal for 1 h at 37 °C and were diluted to a final concentration of 20 µm. Duplex formation was assessed by 5% agarose gel electrophoresis. INT6 and control siRNA duplex aliquots were stored at −20 °C.
RNA duplex transfection
Cells were grown to 20% confluence either on glass coverslips (for immunofluorescence) or in appropriate tissue culture plates. Cells were transfected with 200 nm siRNA duplexes in Optimem medium (Gibco, Rockville, MD, USA) + Oligofectamine lipid transfection reagent (Invitrogen, Renfrew, UK) according to the manufacturer's instructions and as previously described (Elbashir et al. 2001). Transfection was allowed to proceed for 4 h before addition of excess complete medium (+10% FCS). Transfected cells were incubated for 48–72 h before being harvested for immunofluorescence microscopy or immunoblotting.
RESULTS
Immunofluorescence microscopy detects eIF3e/INT6 in the nucleus of mammalian cells
The anti‐eIF3e/INT6 antibody CN25 detected a protein of approximately 48 kDa in immunoblots of a number of mammalian whole cell extracts (Fig. 1a), in line with the previously described mobility of eIF3e/INT6 (Asano et al. 1997). In order to test the specificity of the antibody, transfection of small interfering RNA (siRNA) oligonucleotides (Elbashir et al. 2001) directed against INT6 was used. The abundance of the protein detected by the CN25 antibody on immunoblots was specifically reduced following transfection with INT6 siRNA, but not following transfection with a control RNA duplex in which the base sequence was reversed (Fig. 1b). The CN25 antibody was then used for immunofluorescence microscopy on H1299 cells. The antibody specifically recognized a nuclear antigen, the abundance of which was notably reduced in cells where eIF3e/INT6 levels had been decreased by INT6 siRNA transfection (Fig. 1c). We conclude that the protein recognized by CN25 in immunoblots and immunofluorescence is the authentic INT6 gene product. CN25 detected eIF3e/INT6 in a wide variety of mammalian cells. In each case the immunofluorescence signal was clearly localized to the nucleus. The localization of bulk eIF3 (detected with a polyclonal goat antibody raised against the whole eIF3 complex (Brown‐Leudi et al. 1982) in the same cell lines was by contrast clearly cytoplasmic (Fig. 2a). The peptide sequence of eIF3e/INT6 suggests that it contains both a nuclear localization signal and a nuclear export sequence. Thus, the protein might be capable of localizing to both the nucleus and the cytoplasm and potentially shuttling between the two compartments.
Figure 1.
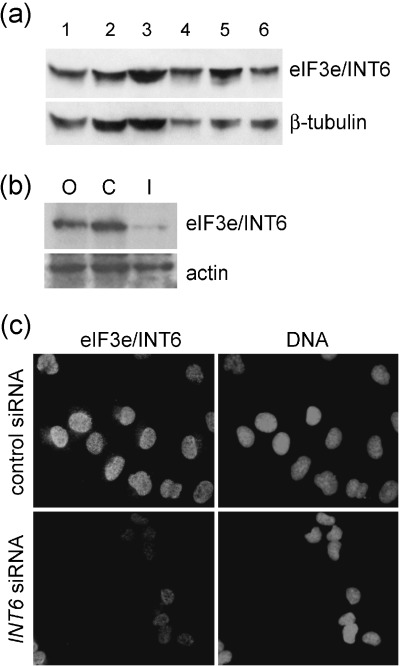
The CN25 antibody is specific for eIF3e/INT6. (a) Immunoblot analysis of mammalian cell line lysates using the CN25 anti‐INT6 antibody. The eIF3e/INT6 protein is detectable as a 48‐kDa band and the membrane was re‐probed with an antibody against β‐tubulin as a loading control. Lane 1, MRC5; 2, HF19; 3, MCF‐10a; 4, MRC5vi; 5, HT1080; 6, COS7. (b) CN25 immunoblotting of lysates of siRNA‐treated H1299 cells. Cells were transfected with O, Oligofectamine (lipid vector) alone; C, control (reversed sequence) oligonucleotide duplex; I, INT6 oligonucleotide duplex. The membrane was re‐probed with an antibody against actin as a loading control. (c) Immunofluorescence microscopy of siRNA‐treated H1299 cells stained with CN25. Identical exposure times were used for the detection of eIF3e/INT6 in cells transfected with control (reversed sequence) or INT6 siRNA duplexes. Hoechst 33258 fluorescence was used to reveal nuclear DNA in the same fields.
Figure 2.
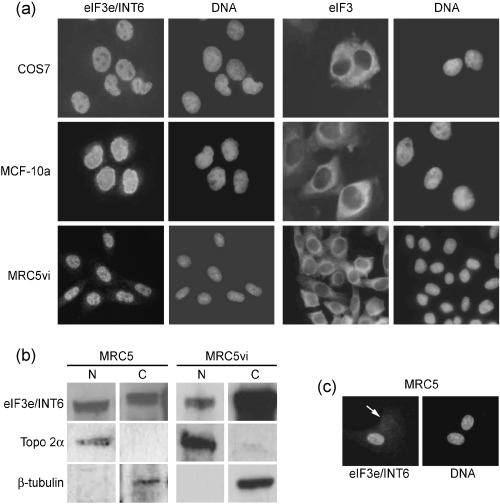
eIF3e/INT6 and its localization in mammalian cells. (a) Immunofluorescence microscopy with CN25 (anti‐INT6) and anti‐eIF3 antibodies on COS7, MCF‐10a and MRC5vi cells; DNA was stained with Hoechst 33258. (b) Immunoblotting with the CN25 antibody on nuclear and cytoplasmic fractions (N and C, respectively) of MRC5 and MRC5vi cells. Membranes were re‐probed with antibodies against β‐tubulin and topoisomerase 2α to confirm separation of nuclear and cytoplasmic fractions. (c) Immunofluorescence microscopy of MRC5 cells stained with CN25. Some cells showed a marked reduction in nuclear eIF3e/INT6 immunoreactivity (arrow).
As INT6 was identified as a component of eIF3 by biochemical fractionation of cytoplasmic extracts (Asano et al. 1997), it would be surprising if eIF3e/INT6 were solely nuclear. Cytoplasmic eIF3e/INT6 might be masked from immunofluorescent detection by the CN25 antibody, for example, as a result of interaction with the eIF3 complex. Indeed, immunoblotting analysis of nuclear and cytoplasmic fractions from fibroblasts showed that eIF3e/INT6 was clearly detectable in both fractions (Fig. 2b). It therefore seems that, while the immunofluorescence signal detected with the CN25 antibody represents genuine nuclear eIF3e/INT6, another fraction of the protein is cytoplasmic and not detected by immunofluorescence under the conditions used here. In a minor, but significant, percentage of MRC5 primary fibroblasts the INT6 signal was not strongly nuclear, but instead was diffusely distributed at a lower level throughout the cell (Fig. 2c). This diffuse staining was not seen in the SV40‐transformed MRC5 cell line, MRC5vi.
Nuclear eIF3e/INT6 is reduced in cycling MRC5 cells
To determine whether the diffuse eIF3e/INT6 staining pattern was related to a particular stage of the cell cycle, MRC5 cells were rendered quiescent by serum deprivation and then induced to re‐enter the cell cycle semi‐synchronously by the addition of serum. Flow cytometry of propidium iodide‐stained cells showed that serum withdrawal arrested the cells with a 2C DNA content. By 16 h after serum re‐feeding, cells were predominantly in G1 and early S‐phase; by 22 h, most cells were in late S‐phase and G2/M, and by 24 h some cells had passed through mitosis and re‐entered G1 (Fig. 3a). Immunofluoresence microscopy using the CN25 antibody showed that the eIF3e/INT6 signal was strongly nuclear in quiescent cells. The diffuse staining pattern was not tightly restricted to a particular time point in this experiment, but this pattern only emerged in cycling populations (Fig. 3b) and was most frequently seen during G1 or early S phase.
Figure 3.
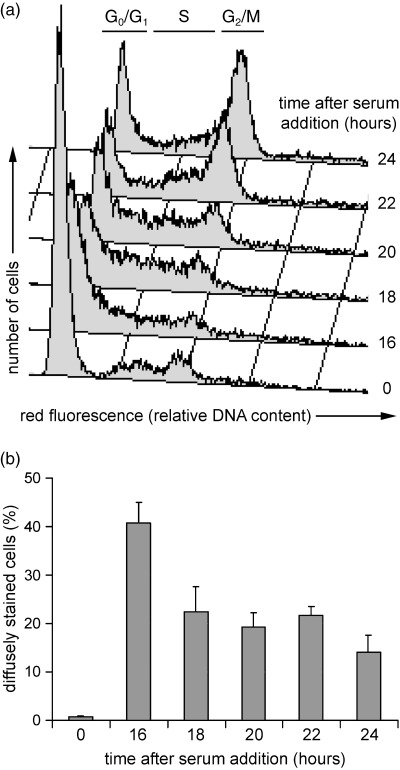
Reduction of nuclear eIF3e/INT6 in cycling MRC5 cells. (a) Flow cytometric analysis of propidium iodide‐stained MRC5 cells synchronized by serum deprivation (time 0) and re‐feeding. The regions of the histogram occupied by cells in G0/G1, S and G2/M phases are indicated. (b) Quantification of the percentages of MRC5 cells (immunostained with CN25 anti‐INT6 antibody) with diffuse eIF3e/INT6 staining at the indicated times after serum re‐feeding.
Differential localization of eIF3e/INT6 does not occur in transformed MRC5 cells
As the previous experiment suggested that the diffuse eIF3e/INT6 staining was most prominent in cells in G1 or S phase, we next examined cells treated with aphidicolin, a DNA polymerase α inhibitor. Flow cytometry of aphidicolin‐treated MRC5 and MRC5vi cells showed that they were successfully arrested in early S phase (MRC5vi required double blocking with excess thymidine in order to achieve full arrest; data not shown). Immunofluorescence microscopy of the samples using CN25 revealed that the proportion of diffusely stained MRC5 cells increased after aphidicolin treatment (Fig. 4), in line with the results of the serum re‐feeding experiments. In the transformed MRC5vi cells, the pattern of eIF3e/INT6 immunofluorescence revealed by CN25 staining was constitutively nuclear, such that very few diffusely stained cells could be detected in either the S phase arrested or the untreated populations.
Figure 4.
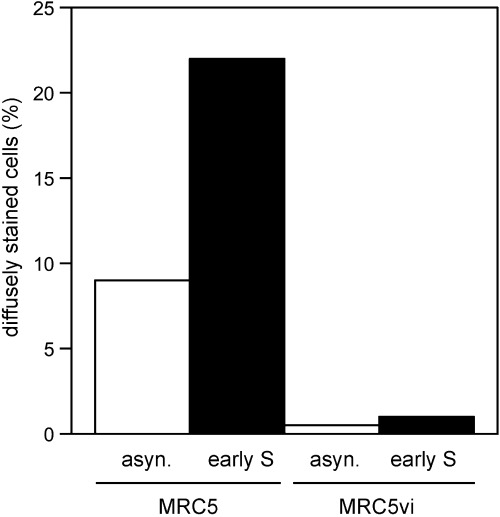
CN25 (anti‐INT6) immunostaining of asynchronous (asyn.) MRC5 and MRC5vi cells and cells arrested in early S phase, as indicated. The percentages of cells exhibiting diffuse eIF3e/INT6 staining are shown.
Differential localization of eIF3e/INT6 also occurs in other fibroblast populations
To determine whether the diffuse CN25 staining pattern was specific to MRC5 or a more general property of primary fibroblasts, we examined the eIF3e/INT6 localization in HF19 fibroblasts. In these cells, the pattern of CN25 immunofluorescence was predominantly nuclear but diffuse staining was also visible (Fig. 5a). Serum starvation of HF19 cells demonstrated that eIF3e/INT6 was constitutively nuclear in quiescent cells. After 20 h of serum stimulation, the diffuse CN25 staining pattern was again detected in the cycling cells (Fig. 5b and c). HT1080 fibrosarcoma cells and HF19 cells were also treated with aphidicolin. Flow cytometry showed that aphidicolin treatment successfully arrested both cell populations in early S phase (data not shown). CN25 immunofluorescence revealed that the proportion of diffusely stained HF19 cells was increased by aphidicolin treatment (Fig. 5c). Very few diffusely stained HT1080 cells could be detected and the eIF3e/INT6 staining pattern was not affected by the presence of aphidicolin. The results obtained with HF19 and HT1080 cells, therefore, closely reflect those obtained with MRC5 and MRC5vi fibroblasts.
Figure 5.
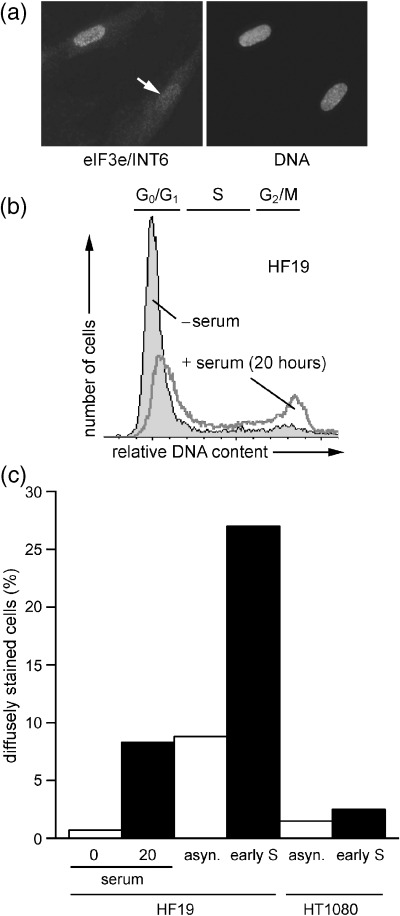
eIF3e/INT6 localization in HF19 and HT1080 cells. (a) Immunofluorescence microscopy of HF19 cells stained with CN25 (anti‐INT6) antibody; DNA was stained with Hoechst 33258. Some cells showed a marked reduction in nuclear eIF3e/INT6 immunoreactivity (arrow). (b) Flow cytometric analysis of propidium iodide‐stained HF19 cells synchronized by serum deprivation (− serum) and re‐feeding (20 h). The regions of the histogram occupied by cells in G0/G1, S and G2/M phases are indicated. (c) Percentages of HF19 and HT1080 cells with diffuse eIF3e/INT6 staining following CN25 (anti‐INT6) immunofluorescence microscopy. Cells were synchronized by serum deprivation (0) or 20 h serum re‐feeding (20), asynchronous (asyn.) or arrested in early S phase, as indicated.
DISCUSSION
Although human eIF3e/INT6 was described as a subunit of a translation initiation factor, its physiological function remains unclear. eIF3e/INT6 has since been found to interact with a variety of proteins including the interferon‐inducible p56 (Guo & Sen 2000; Guo et al. 2000), the HTLV‐1 Tax protein (Desbois et al. 1996), subunits of the COP9 signalosome, a regulator of transcription factors and the cell cycle (Yahalom et al. 2001), and the 26S proteasome (Hoareau Alves et al. 2002; Yen et al. 2003). The study of Guo et al. suggests that eIF3e/INT6 may act to inhibit eIF3 activity and thus translation initiation (Guo et al. 2000). The regulation of translation is already strongly linked to malignancy. A critical threshold level of protein synthesis must be exceeded in order for a cell to pass the restriction point and thus become committed to entering S phase (Brooks 1977; Zetterberg et al. 1995). Furthermore, the over‐expression of a number of translation initiation factors has been detected in human tumours and transformed cell lines (Watkins & Norbury 2002). In contrast, reduced expression of eIF3e/INT6 has been demonstrated in approximately 40% of mammary carcinomas and 30% of non‐small cell lung carcinomas (Marchetti et al. 2001). This could relate to a role as a negative regulator of eIF3 and is consistent with the Int‐6 gene disruption by MMTV proviral sequences seen in mouse mammary tumours (Marchetti et al. 1995; Diella et al. 1997). It seems likely, however, that eIF3e/INT6 has additional roles unrelated to the initiation of translation.
Using immunofluorescence microscopy, we found that a fraction of eIF3e/INT6 localized to the nucleus where it might perform such an alternative function (1, 2). Immunoblotting analysis with the same antibody detected the protein in both the nuclear and cytoplasmic fractions of cell extracts (Fig. 2b). We infer that eIF3e/INT6 is present in both the nucleus and the cytoplasm, but that the epitope recognized by CN25 is masked when eIF3e/INT6 is cytoplasmic. Redistribution of eIF3e/INT6 from the nucleus to the cytoplasm would therefore have been seen only as a decrease in nuclear staining under these conditions. The reasons underlying the discrepancy between our data and those published previously are unclear, but presumably involve differences in antibody specificity and/or sample preparation. We consider it unlikely that intrinsic differences between cell lines are the underlying cause, as a similar pattern of nuclear staining was seen using the CN25 antibody in a variety of cell lines in this study. The specific down‐regulation of immunoreactivity by siRNA transfection gives us confidence that the nuclear protein detected in our experiments is indeed eIF3e/INT6, but this type of control was not performed in any of the earlier studies, as the technique was developed only recently.
In proliferating diploid fibroblasts, nuclear eIF3e/INT6 was substantially reduced in approximately 10–20% of cells. Synchronization experiments showed that this percentage increased to as much as 40% in early S phase (Fig. 3b), implying either a greater requirement for eIF3e/INT6 in the cytoplasm or the need for its exclusion from the nucleus at this stage in the cell cycle. The loss of eIF3e/INT6 from the nucleus could be interpreted as movement of the protein into the cytoplasm to interact in a regulatory capacity with a large protein complex (such as eIF3 or the proteasome) or to prevent its interaction with a nuclear protein complex such as the COP9 signalosome. Proteolytic degradation of eIF3e/INT6 could also account for the observed reduction in nuclear staining. Specific eIF3e/INT6 degradation cannot be discounted, as the diffusely stained cells were always in the minority, making it difficult to assess concomitant changes in overall eIF3/INT6 protein levels. The cell cycle transition most sensitive to partial inhibition of protein synthesis is the restriction point in G1 (Zetterberg et al. 1995; Pyronnet & Sonenberg 2001). By the time fibroblasts enter S phase, the requirement for high levels of protein synthesis has passed. This suggests that the apparent redistribution of eIF3e/INT6 in early S phase is unlikely to be linked to an alteration in global protein synthesis. It is possible that the disappearance of eIF3e/INT6 from the nucleus reflects a more subtle, and hitherto unexpected, alteration in the pattern of translation in early S phase. Alternatively, this change in eIF3e/INT6 localization might involve a function of eIF3e/INT6 that is unrelated to translation. It is currently not clear why only a subpopulation of the fibroblasts exhibited the diffuse pattern of CN25 staining. This could indicate that the disappearance of eIF3e/INT6 from the nucleus occurs in every cell but is very transient, such that the proportion of cells affected at any given instant is comparatively low, even in synchronized populations. Alternatively, the cells exhibiting loss of nuclear eIF3e/INT6 may be a specific subset in a particular physiological state, for example those experiencing stochastic low‐level DNA damage or oxidative stress. Interestingly, the early S phase redistribution of eIF3e/INT6 that was seen in normal diploid MRC5 and HF19 fibroblasts was not apparent in SV40‐immortalized MRC5vi or HT1080 fibrosarcoma cells (4, 5). Loss of regulation of eIF3e/INT6 redistribution may therefore be a significant and widespread feature of tumourigenesis in fibroblasts, and conceivably other cell types. Clarification of this point will require further studies.
ACKNOWLEDGEMENTS
We thank Ian Hickson, Simon Morley and John Hershey for providing antibodies, and Phill North, Sue Houlbrook and Sally Davies for cell stocks and invaluable cell culture advice. This work was supported by Cancer Research UK and the Association for International Cancer Research.
REFERENCES
- Asano K, Merrick WC, Hershey JW (1997) The translation initiation factor eIF3‐p48 subunit is encoded by int‐6, a site of frequent integration by the mouse mammary tumour virus genome. J. Biol. Chem. 272, 23477–23480. [DOI] [PubMed] [Google Scholar]
- Brooks RF (1977) Continuous protein synthesis is required to maintain the probability of entry into S phase. Cell 12, 311–317. [DOI] [PubMed] [Google Scholar]
- Brown‐Luedi ML, Meyer LJ, Milburn SC, Yau PM, Corbett S, Hershey JW (1982) Protein synthesis initiation factors from human HeLa cells and rabbit reticulocytes are similar: comparison of protein structure, activities, and immunochemical properties. Biochemistry 21, 4202–4206. [DOI] [PubMed] [Google Scholar]
- Crane R, Craig R, Murray R, Dunand‐Sauthier I, Humphrey T, Norbury C (2000) A fission yeast homolog of Int‐6, the mammalian oncoprotein and eIF3 subunit, induces drug resistance when overexpressed. Mol. Biol. Cell 11, 3993–4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desbois C, Rousset R, Bantignies F, Jalinot P (1996) Exclusion of Int‐6 from PML nuclear bodies by binding to the HTLV‐I Tax oncoprotein. Science 273, 951–953. [DOI] [PubMed] [Google Scholar]
- Diella F, Levi G, Callahan R (1997) Characterization of the INT6 mammary tumour gene product. DNA Cell Biol. 16, 839–847. [DOI] [PubMed] [Google Scholar]
- Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T (2001) Duplexes of 21‐nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411, 494–498. [DOI] [PubMed] [Google Scholar]
- Guo J, Sen GC (2000) Characterization of the interaction between the interferon‐induced protein P56 and the Int6 protein encoded by a locus of insertion of the mouse mammary tumour virus. J. Virol. 74, 1892–1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Hui DJ, Merrick WC, Sen GC (2000) A new pathway of translational regulation mediated by eukaryotic initiation factor 3. EMBO J. 19, 6891–6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoareau Alves K, Bochard V, Rety S, Jalinot P (2002) Association of the mammalian proto‐oncoprotein Int‐6 with the three protein complexes eIF3, COP9 signalosome and 26S proteasome. FEBS Lett. 527, 15–21. [DOI] [PubMed] [Google Scholar]
- Marchetti A, Buttitta F, Miyazaki S, Gallahan D, Smith GH, Callahan R (1995) Int‐6, a highly conserved, widely expressed gene, is mutated by mouse mammary tumour virus in mammary preneoplasia. J. Virol. 69, 1932–1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchetti A, Buttitta F, Pellegrini S, Bertacca G, Callahan R (2001) Reduced expression of INT‐6/eIF3‐p48 in human tumours. Int. J. Oncol. 18, 175–179. [DOI] [PubMed] [Google Scholar]
- Miyazaki S, Rasmussen S, Imatani A, Diella F, Sullivan DT, Callahan R (1999) Characterization of the Drosophila ortholog of mouse eIF‐3p48/INT‐6. Gene 233, 241–247. [DOI] [PubMed] [Google Scholar]
- Morris‐Desbois C, Bochard V, Reynaud C, Jalinot P (1999) Interaction between the Ret finger protein and the Int‐6 gene product and co‐localisation into nuclear bodies. J. Cell Sci. 112, 3331–3342. [DOI] [PubMed] [Google Scholar]
- Pyronnet S, Sonenberg N (2001) Cell‐cycle‐dependent translational control. Curr. Opin. Genet. Dev. 11, 13–18. [DOI] [PubMed] [Google Scholar]
- Watkins SJ, Norbury CJ (2002) Translation initiation and its deregulation during tumourigenesis. Br. J. Cancer 86, 1023–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yahalom A, Kim TH, Winter E, Karniol B, Von Arnim AG, Chamovitz DA (2001) Arabidopsis eIF3e (INT‐6) associates with both eIF3c and the COP9 signalosome subunit CSN7. J. Biol. Chem. 276, 334–340. [DOI] [PubMed] [Google Scholar]
- Yen HC, Gordon C, Chang EC (2003) Schizosaccharomyces pombe Int6 and Ras homologs regulate cell division and mitotic fidelity via the proteasome. Cell 112, 207–217. [DOI] [PubMed] [Google Scholar]
- Zetterberg A, Larsson O, Wiman KG (1995) What is the restriction point? Curr. Opin. Cell Biol. 7, 835–842. [DOI] [PubMed] [Google Scholar]


