Abstract
Objectives
Dental pulp tissue contains stem cells that can differentiate into multiple lineages under specific culture conditions; the origin of these dental pulp stem cells, however, is still unknown.
Materials and methods
Here we have utilized an α‐SMA‐GFP transgenic mouse model to characterize expression of a‐smooth muscle actin (SMA)‐GFP in subpassages of pulp‐tissue‐derived dental pulp cells, as perivascular cells express α‐SMA.
Results
During subculturing, percentages of cells expressing a‐SMA increased signi?cantly from passage 1 to 3. α‐SMA‐GFP‐positive cells expanded faster than α‐SMA‐GFP‐negative cells. The dental pulp cells at passage 3 were induced towards osteogenic, adipogenic or chondrogenic differentiation. All three differentiated cell lines expressed high levels of α‐SMA (mineralized nodules, lipid droplets and chondrocyte pellets). GFP expression colocalized with differentiated osteoblasts, adipocytes and chondrocytes. Co‐culturing the α‐SMA‐GFP‐positive cells with human endothelial cells promoted formation of tube‐like structures and robust vascular networks, in 3‐D culture.
Conclusions
Taken together, the a‐SMA‐GFP‐positive cells were shown to have multilieange differentiation ability and to promote vascularization in a co‐culture system with endothelial cells.
Introduction
Dental pulp is a highly vascularized tissue that contains dental pulp stem cells (DPSCs) 1. Since they were first described in the literature 2, DPSCs have been shown to have gene expression patterns that are similar to bone marrow stromal stem cells; these patterns involve expression of CD44, CD106, CD146, 3G5 and Stro‐1 3, 4. These cells also exhibit multipotentiality and can differentiate into odontoblasts, osteoblasts, adipocytes, chondrocytes and neural cells 5, 6. Previous research has attempted to isolate and characterize dental pulp progenitor/stem‐cell populations 7 and early studies suggested that they were derived from fibroblast‐like cells 8. Recent work on DPSCs has shown high expression of STRO‐1, a putative stem cell marker 9 and a mesenchymal stem cell progenitor population, based on STRO‐1, has been isolated from adult dental pulp and has shown multilineage potential 7, 10. The exact origin and location of the STRO‐1‐positive population remains unknown, although Shi et al. have suggested that DPSCs might reside in perivascular regions of dental pulp 9. Interestingly, Crisan et al. have reported that a subpopulation of human perivascular cells expressed both pericyte and mesenchymal stem‐cell (MSC) markers and that this isolated perivascular population was clonally multipotent in culture and was capable of expansion 11, 12. Taken together, these studies suggest that mesenchymal stem cells might be members of the pericyte family. However, no direct proof that confirms that dental pulp stem cells originate from dental perivascular cells/pericytes has been provided.
Alpha‐smooth muscle actin (α‐SMA) is expressed by perivascular cells/pericytes that reside around small blood vessels 13, 14. In developing teeth, this protein is expressed in the dental follicular area and outer dental epithelium; however, in developed teeth, it is found in the dental pulp and apical perivascular areas 15. In our study described here, we utilized an α‐SMA transgenic mouse model to study differentiation of dental pulp stem cells. This mouse model contains a green fluorescent protein (GFP) gene under the control of an α‐SMA promoter 14. Thus, α‐SMA expression can be detected by tracking GFP expression during cell proliferation and differentiation. Our research has found that a population of α‐SMA‐expressing cells acted as progenitors or DPSCs. We showed that α‐SMA‐positive cells cycled faster than α‐SMA‐negative cells and expression of α‐SMA was intense in all of the differentiated DPSC cultures (osteogenic, chondrogenic and adipogenic). These results provide evidence that perivascular cells and other cells may differentiate into DPSCs.
Materials and methods
Experimental mouse model
Dental pulp cultures from an α‐SMA‐GFP transgenic mouse model were used as the source of α‐SMA‐GFP‐positive cells. This mouse model has an α‐SMA promoter that drives expression of GFP; the model was developed by Dr Jen‐Yue Tsai et al. 16.
Primary pulp tissue culture and dental pulp cell culture
Maxillary and mandibular molars from 6‐ to 8‐week‐old transgenic mice were dissected. Pulp tissue was isolated, minced and transferred to 100‐mm tissue culture dishes for 5 min, to allow cell attachment before addition of growth medium (αMEM containing 10% heat‐inactivated foetal calf serum) and the pulp tissue was cultured at 37 °C and 5% CO2. Cells emerging from the pulp were termed passage 0 cells (P0). When cultures reached 80% confluence, they were passaged using 0.25% trypsin‐EDTA, then subcultured (P1). These studies used cells up to passage 3 (P3).
FACS sorting of dental pulp‐derived α‐SMA‐GFP positive cells
The α‐SMA‐GFP‐positive population of dental pulp cells (P1–P3) was selectively purified based on intensity of GFP fluorescence, using a FACS Vantage Sorter. Sorted α‐SMA‐GFP‐positive cells were seen to be mainly perivascular cells, consisting of pericytes and smooth muscle cells.
Multilineage differentiation of α‐SMA‐GFP‐positive cells
Dental pulp cells at P3 were seeded in 24‐well culture plates at 1.0 × 105 density cells/well and were cultured in growth medium for 3 days. Culture medium was then exchanged for lineage‐specific induction medium to induce multilineage differentiation (osteogenesis: 50 μm ascorbate‐2‐phosphate, 10 mm β‐glycerophosphate, 0.01 μm 1,25‐dihydroxyvitamin D3; chondrogenesis: 10 ng/ml TGF‐β1, 100 nm dexamethasone, 6.25 μg/ml insulin, 50 nm ascorbate‐2‐phosphate, 110 mg/l sodium pyruvate; adipogenesis: 1 μm dexamethasone, 10 μm insulin, 200 μm indomethacin, 0.5 mm isobutyl‐methylxanthine). Osteogenic, chondrogenic and adipogenic differentiation cultures were stained with alizarin red, toluidine blue and oil red O, respectively, and then evaluated using fluorescence microscopy, as previously described 17.
RNA extraction and reverse transcription‐polymerase chain reaction
Total RNA was extracted from the specimens using the Qiagen RNeasy Mini Kit. Reverse transcription was carried out on 1 μg total RNA, using a murine leukaemia virus reverse transcriptase, according to the manufacturer's protocol. The resulting cDNA templates were used for RT‐PCR. PCR oligonucleotide primers were as follows:
OSX (F: 5′‐CACTCACACCCGGGAGAAGA‐3′; R: 5′‐GGTGGTCGCTTCGGGTAAA‐3′), ON (F: 5′‐AACGTCCTGGTCACCCTGTATG‐3′; R: 5′‐GATCTTCTTCACCCGCAGCTT‐3′), PPAR‐γ (F: 5′‐GACCACTCGCATTCCTTT‐3′; R: 5′‐CCACAGACTCGGCACTCA‐3′), LPL (F: 5′‐AGGGTGAGGAATCTAATG‐3′; R: 5′‐CAGGTGTTTCAACCGCTA‐3′), SOX9 (F: 5′‐GTTGATCTGAAGCGAGAGGG‐3′; R: 5′‐CATTGACGTCGAAGGTCTCA‐3′), Col‐1α1 (F: 5′‐AAGACCCAGACTGCCTCAAC‐3′; R: 5′‐TTGGCCCTAATTTTCCACTG‐3′) and GAPDH (F: 5′‐CTCACTGGCATGGCCTTCCG‐3′; R: 5′‐ACCACCCTGTTGCTGTAGCC‐3′).
Products were evaluated by electrophoresis on 2% agarose gels, stained with ethidium bromide and visualized with Quantity One software 18
Human umbilical vein endothelial cell preparation and labelling
Human umbilical vein endothelial cells (HUVECs) were labelled with DsRed, by transfection with a lentiviral vector as previously described 17. HUVECs were maintained on 0.1% gelatin‐coated plates using endothelial cell growth medium (EGM‐2).
Preparation of 3‐D collagen/fibronectin gels, and vascular network formation in vitro
Collagen/fibronectin gels were prepared as previously described, with minor modifications 19. Briefly, gel formulation consisted of 1.5 mg/ml collagen gel (PureCol, Fremont, CA,USA), 90 μg/ml human plasma fibronectin (Gibco/BRL, Gaithersburg, MD), 25 mm HEPES (Gibco) and 38.5% Complete EGM‐2 (Lonza, EGM‐2, Lonza, Walkersville, ML). HUVECs and α‐SMA‐GFP‐positive cells were mixed at a ratio of 4:1 and were suspended in gel matrix mixture at final cell concentration of 2 × 106 cells/ml. Cell suspensions were polymerized in a 48‐well plate (100 μl/well) for 30 min at 37 °C. Gels were then cultured in 250 μl EGM‐2 medium.
Statistics
Data points for each experimental group were expressed as mean (±SEM).
Results
α‐SMA‐GFP‐positive cells in dental pulp cell cultures
Cells emerging from the dental pulp had fibroblast‐like morphology (Fig. 1A). At passage 1, only a small fraction of total cells were GFP positive (Fig. 1A,a–c), but by the third passage (P3), GFP‐positive cells dominated the cultures (Fig. 1A,d–f). Different passages of dental pulp cells were analysed by FACS to determine percentages of GFP‐positive cells in the cultures. GFP‐positive cells accounted for 19 ± 4.32% (P0), 43 ± 5.45% (P1), 65 ± 6.37% (P2) and 88 ± 6.53% (P3) cells respectively (Fig. 1B).
Figure 1.
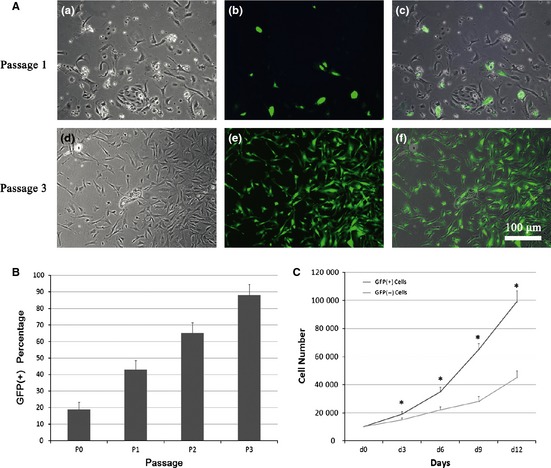
GFP expression during in vitro subculturing of cells derived from primary pulp tissue. (A) The percentage of α‐SMA‐GFP‐positive cells increased from passage 1 to passage 3. a, d: phase‐contrast images; b, e: fluorescence microscopy images; c, f: merged images (200×). (B) Quantification of GFP+ cells in the total cell population, in each passage. (C) Comparison of cell population growth between SMA+ and SMA− cells over 12 days.
To compare expansion of α‐SMA‐GFP‐positive cells with α‐SMA‐GFP‐negative cells, FACS‐sorted GFP‐positive and GFP‐negative cells were plated at identical densities on day 0, and total cell numbers were counted on days 3, 6, 9 and 12. At each time point, numbers of α‐SMA‐GFP positive cells were significantly higher than those of negative cells (Fig. 1C).
α‐SMA‐GFP expression in multilineage differentiation of dental pulp cells
Osteogenesis
DPSCs at P3 were cultured in osteogenic medium for up to 3 weeks (Fig. 2a). Mineralized nodules formed in the culture (Fig. 2b), as revealed by positive alizarin red‐S staining (Fig. 2d). Fluorescence microscopy indicated that all differentiated cells expressed GFP, and mineralized nodules exhibited intense GFP localization (Fig. 2b,c). Expression of osteogenic marker genes, osterix (OSX) and osteonectin (ON), was higher in cultures at days 7 and 14, as confirmed by osteogenic differentiation (Fig. 3). These results demonstrate that α‐SMA‐GFP‐positive cells retained their ability to express α‐SMA during osteogenic differentiation and that these α‐SMA cells then dominated the osteogenic cultures.
Figure 2.
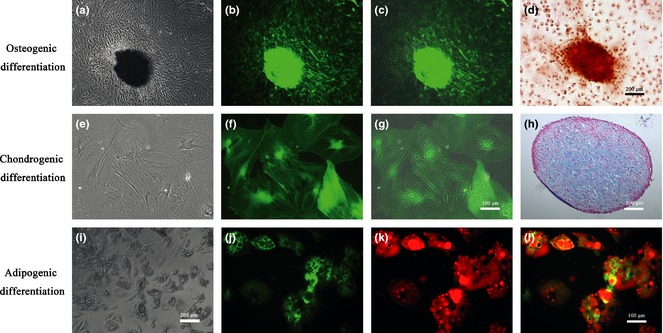
α‐ SMA ‐ GFP expression in multilineage differentiation of dental pulp stem cells ( DPSC s). Osteogenic differentiation of DPSCs. (a) morphology of DPSCs after culturing in osteogenic medium for 3 weeks. (b) Differentiated DPSCs expressed GFP after osteogenic induction. (c) Merged image shows that all osteogenically differentiated DPSCs expressed GFP. (d) Positive alizarin red‐S staining of mineralized nodules (100×). Chondrogenic differentiation of DPSCs. (e) After chondrogenic induction, cells changed morphology from fibroblast‐like to flat, multiangled shapes. (f) Chondrogenically differentiated cells expressed GFP. (g) Merged image shows that all chondrogenically differentiated DPSCs expressed GFP (200×). (h) Toluidine blue staining of proteoglycans in DPSC pellet, proving that cells differentiated into chondrocytes after 3 weeks (40×). Adipogenic differentiation of DPSCs. (i) DPSCs cultured in adipogenic induction medium for 2 weeks accumulated lipid droplets. (j) Lipid droplets are shown as black on green in fluorescence microscopic images; GFP‐positive staining remained during adipogenesis. (k) Positive oil red O staining for lipid droplets (in red on fluorescence microscope images). (l) Merged images show that differentiated DPSCs expressed GFP during adipogenesis (200×).
Figure 3.
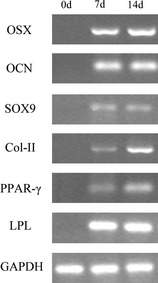
RT ‐ PCR analysis of gene expression during multilineage differentiation. Differentiated DPSC cultures at 7 and 14 days reveal upregulated expression of markers for osteogenic, chondrogenic and adipogenic lineages.
Chondrogenesis
DPSCs at P3 were cultured in chondrogenic medium for 7 days. Morphology of the cells in these cultures changed from fibroblast‐like in shape to flat, multi‐angled morphology (Fig. 2e), with intense GFP expression (Fig. 2f). The merged image confirms that all of chondrogenically differentiated cells expressed α‐SMA‐GFP (Fig. 2g). Cells were then pelleted and cultured as small masses for a further 3 weeks. Toluidine blue staining showed that cells had large nuclei with multiple nucleoli, similar to chondrocytes, and that most of the cells produced proteoglycans in their extracellular matrix (Fig. 2h). Expression of chondrogenic marker genes SOX9 and Col‐II also significantly increased during this differentiation period (Fig. 3). These results indicate that α‐SMA‐GFP‐positive cells expressed α‐SMA during chondrogenic differentiation.
Adipogenesis
DPSCs at P3 were cultured in adipogenic medium for 14 days, during which time, morphology of the cells transformed into a rounder shape with lipid‐filled vesicles (Fig. 2i). The lipids appeared as black areas under fluorescence microscopy, although GFP staining remained dominant in these cultures (Fig. 2j). Positive oil red O staining of lipid droplets (red under fluorescence microscopy) confirmed that the DPSCs had differentiated into adipocytes (Fig. 2k). Merged images show co‐localization of GFP expression and adipocyte staining (Fig. 2l). Expression of adipogenesis‐specific genes, PPAR‐γ and LPL, was significantly higher in these cultures on days 7 and 14 (Fig. 3). These results show that α‐SMA‐GFP‐positive cells expressed α‐SMA during adipogenesis.
Formation of vascular network within 3D collagen/fibronectin gels
Human umbilical vein endothelial cells were labelled with DsRed by transfection with a lentiviral vector (transfection efficiency ~90%) (Fig. 4A). To test the effect of α‐SMA‐GFP‐positive cells on formation of a vascular network by HUVECs, a co‐culture of α‐SMA‐GFP‐positive cells and DsRed‐ HUVECs was loaded into a 3‐D collagen‐fibronectin gel. In the presence of α‐SMA‐GFP‐positive cells, HUVECs began to elongate and interconnect by the day after plating (D1) (Fig. 4B,a–c). On day 7, α‐SMA‐GFP‐positive cells were condensed, and the emerging vascular networks were seen to primarily consist of HUVECs (Fig. 4B,e–g). On day 14, the α‐SMA‐GFP‐positive cells had proliferated significantly, and there was a robust, branched network of HUVECs visible within the gels (Fig. 4B,i–j). In control gels containing HUVECs only, HUVECs formed much shorter and smaller multi‐cellular cords which regressed and fell apart by day 7; no vascular network was found on either day 7 or day 14 (Fig. 4B,d,h,l).
Figure 4.
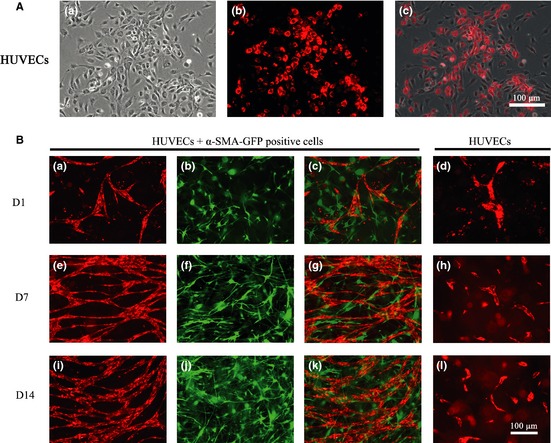
Formation of 3‐D vascular networks in vitro . (A) Human umbilical vein endothelial cells (HUVECs) labelled with DsRed with efficiency of over 90% (a–c) (200×). (B) Co‐culture of HUVECs with α‐SMA‐GFP‐positive cells. (a) HUVECs grew and interconnected with other cells by day 1; (b, c) α‐SMA‐GFP‐positive cells distributed themselves around the HUVECs; (e) vascular networks primarily consisted of HUVECs at day 7; (f, g) α‐SMA‐GFP‐positive cells become denser around HUVEC vascular networks; (i) By day 14, HUVECs formed a robust network; (j, k) α‐SMA‐GFP‐positive cells distributed themselves around the HUVEC network; (d, h, l) In the control group, HUVEC cultures without α‐SMA‐GFP‐positive cells proliferated slowly and were unable to form vascular networks by days 1, 7 and 14 (100×).
Discussion
Dental pulp stem cells have been shown to exhibit multipotency and can differentiate into several cell types, including odontoblasts, osteoblasts, chondrocytes, adipocytes and neural‐like cells. They have high proliferation rates in monolayer culture conditions in vitro. Based on this, dental pulp tissue may represent a promising reservoir of stem cells for tooth tissue engineering and clinical applications 2, 3, 18. However, the exact origin and precise anatomical location of these dental pulp stem cells are still not clear 7. Previous studies have found that STRO‐1‐positive DPSCs express the vascular antigen CD146 and the pericyte marker 3G5, suggesting that these cells may reside in perivascular niches 9. Crisan et al. have reported that blood vessel walls contain a reserve of progenitor cells that may originate as mesenchymal stem cells and other related adult stem cell types. Although that study did not include dental pulp in its tissue samples, their results suggested that both foetal and adult mesenchymal stem cells are derived from the pericyte lineage 12. Our recent study demonstrates that mechanical stress can significantly regulate osteogenic and odontogenic differentiation of dental pulp stem cells and additionally, dental pulp stem cells express typical pericyte characteristics, such as α‐SMA and PDGF‐Rβ, suggesting that dental pulp stem cells may derive from perivascular cells 20. During normal physical metabolism or tissue repair, these progenitor cells could differentiate into dental pulp stem cells, then further differentiate into odontoblasts or other cell types 21. In this work, we have shown that dental pulp multilineage cells originate from perivascular cells. As perivascular cells express the cytoskeletal protein α‐smooth muscle actin, we used an α‐SMA‐GFP transgenic mouse model to define a perivascular‐derived progenitor population derived from dental pulp 22. We showed that these cells actively differentiate into multiple lineages and persist during differentiation.
Using the same mouse model, Kalajzic et al. have reported that α‐SMA‐GFP‐positive cells were associated with the microvasculature within dental pulp, alveolar crest and the rich vascular area of periodontal ligaments, in adult mice 15. They found that α‐SMA‐GFP‐positive cells from periodontal ligaments and dental follicles proliferate faster than α‐SMA‐GFP‐negative cells 15. Consistent with their results, we found that α‐SMA‐GFP‐positive cells from dental pulp expanded faster than their α‐SMA‐GFP‐negative counterparts. This explains the significant increase in GFP‐positive cells between P0 and P3 [from 19% (P0) to 88% (P3)]. Thus, after subculturing cells derived from pulp explants, DPSCs used in the present study contained a higher number of GFP positive cells compared to those of other studies, that used primary tissue digestion. α‐SMA is not only specific to pericytes, but is also present in vascular smooth muscle cells of larger blood vessels. Therefore, the α‐SMA‐GFP‐positive cells in our study may represent both pericytes and smooth muscle cells. Based on previous work linking mesenchymal cells and pericytes 11, it is likely that most of the subcultured GFP‐positive cells had originated from pericytes. It will be interesting in future studies to assess this hypothesis by staining for pericyte surface marker, 3G5.
Vascularization of tissue constructs remains a major challenge for stem cell‐based tissue regeneration, as established blood vessel networks are essential for providing nutrients and oxygen for tissue regeneration. Vascularization occurs by differentiation, migration and connection of endothelial progenitor cells in response to stimuli, such as cell–cell contacts and growth factor presence 23. Perivascular cells, especially pericytes, play an important role in vascularization. During vascularization, these cells are first recruited to the relevant site; they proliferate and then contact and communicate with endothelial cells 12, 24. The following mechanisms are essential for successful vascularization: endothelial cells contact one another and interact to form networks 12, 25, 26, growth factors are synthesized 27, 28 and a mural wall of new blood vessels is formed 12, 26. 3‐D collagen gel models have previously been created to study vascularization in vitro 29. Our previous work successfully showed that bone‐marrow‐derived pericytes can stimulate endothelial cells to form tube‐like structures and to subsequently create robust vascular networks in a 3‐D collagen–fibronectin scaffold 17. In the present study, we have demonstrated that α‐SMA‐GFP‐expressing cells from dental pulp also promote vascularization. The next challenges are to use the same model to complete vascularization in vivo and to implant a pre‐vascularized structure to determine stability of vascularized structures, and their integration with the host vascular network.
In summary, this study has proven that dental pulp stem cells may be located in perivascular regions, as pericytes. α‐SMA‐GFP‐positive cells proliferated rapidly and dominated DPSC cultures by passage 3 and these cells were able to differentiate into multiple lineages; they maintained their expression of α‐SMA‐GFP during differentiation and co‐localized with mineralized nodules, chondrocytes and lipid droplets. α‐SMA‐GFP‐positive cells also promoted vascularization in a co‐culture system with endothelial cells.
Acknowledgements
This work was funded by the Natural Science Foundation of China (81071273, 31170929), the Foundation for Authors of National Excellent Doctoral Dissertation (FANEDD 200977) and the Program for New Century Excellent Talents (NCET‐08‐0373), Funding for Distinguished Young Scientists in Sichuan (2010JQ0066).
References
- 1. Sloan AJ, Waddington RJ (2009) Dental pulp stem cells: what, where, how? Int. J. Paediatr. Dent. 19, 61–70. [DOI] [PubMed] [Google Scholar]
- 2. Gronthos S, Mankani M, Brahim J, Robey PG, Shi S (2000) Postnatal human dental pulp stem cells (DPSCs) in vitro and in vivo. Proc. Natl. Acad. Sci. USA 97, 13625–13630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gronthos S, Brahim J, Li W, Fisher LW, Cherman N, Boyde A et al (2002) Stem cell properties of human dental pulp stem cells. J. Dent. Res. 81, 531–535. [DOI] [PubMed] [Google Scholar]
- 4. Shi S, Robey PG, Gronthos S (2001) Comparison of human dental pulp and bone marrow stromal stem cells by cDNA microarray analysis. Bone 29, 532–539. [DOI] [PubMed] [Google Scholar]
- 5. Arthur A, Shi S, Zannettino AC, Fujii N, Gronthos S, Koblar SA (2009) Implanted adult human dental pulp stem cells induce endogenous axon guidance. Stem Cells 27, 2229–2237. [DOI] [PubMed] [Google Scholar]
- 6. Iohara K, Zheng L, Ito M, Tomokiyo A, Matsushita K, Nakashima M (2006) Side population cells isolated from porcine dental pulp tissue with self‐renewal and multipotency for dentinogenesis, chondrogenesis, adipogenesis, and neurogenesis. Stem Cells 24, 2493–2503. [DOI] [PubMed] [Google Scholar]
- 7. Shi S, Bartold PM, Miura M, Seo BM, Robey PG, Gronthos S (2005) The efficacy of mesenchymal stem cells to regenerate and repair dental structures. Orthod. Craniofac. Res. 8, 191–199. [DOI] [PubMed] [Google Scholar]
- 8. Fitzgerald M, Chiego DJ Jr, Heys DR (1990) Autoradiographic analysis of odontoblast replacement following pulp exposure in primate teeth. Arch. Oral Biol. 35, 707–715. [DOI] [PubMed] [Google Scholar]
- 9. Shi S, Gronthos S (2003) Perivascular niche of postnatal mesenchymal stem cells in human bone marrow and dental pulp. J. Bone Miner. Res. 18, 696–704. [DOI] [PubMed] [Google Scholar]
- 10. Yang X, Zhang W, van den Dolder J, Walboomers XF, Bian Z, Fan M et al (2007) Multilineage potential of STRO‐1+ rat dental pulp cells in vitro . J. Tissue Eng. Regen. Med. 1, 128–135. [DOI] [PubMed] [Google Scholar]
- 11. Caplan AI (2008) All MSCs are pericytes? Cell Stem Cell 3, 229–230. [DOI] [PubMed] [Google Scholar]
- 12. Crisan M, Yap S, Casteilla L, Chen CW, Corselli M, Park TS et al (2008) A perivascular origin for mesenchymal stem cells in multiple human organs. Cell Stem Cell 3, 301–313. [DOI] [PubMed] [Google Scholar]
- 13. Wang J, Niu W, Nikiforov Y, Naito S, Chernausek S, Witte D et al (1997) Targeted overexpression of IGF‐I evokes distinct patterns of organ remodeling in smooth muscle cell tissue beds of transgenic mice. J. Clin. Invest. 100, 1425–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yokota T, Kawakami Y, Nagai Y, Ma JX, Tsai JY, Kincade PW et al (2006) Bone marrow lacks a transplantable progenitor for smooth muscle type alpha‐actin‐expressing cells. Stem Cells 24, 13–22. [DOI] [PubMed] [Google Scholar]
- 15. San Miguel SM, Fatahi MR, Li H, Igwe JC, Aguila HL, Kalajzic I (2010) Defining a visual marker of osteoprogenitor cells within the periodontium. J. Periodontal Res. 45, 60–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Maeda S, Sutliff RL, Qian J, Lorenz JN, Wang J, Tang H et al (1999) Targeted overexpression of parathyroid hormone‐related protein (PTHrP) to vascular smooth muscle in transgenic mice lowers blood pressure and alters vascular contractility. Endocrinology 140, 1815–1825. [DOI] [PubMed] [Google Scholar]
- 17. Cai X, Lin Y, Friedrich CC, Neville C, Pomerantseva I, Sundback CA et al (2009) Bone marrow derived pluripotent cells are pericytes which contribute to vascularization. Stem Cell Rev. 5, 437–445. [DOI] [PubMed] [Google Scholar]
- 18. Grottkau BE, Purudappa PP, Lin YF (2010) Multilineage differentiation of dental pulp stem cells from green fluorescent protein transgenic mice. Int. J. Oral Sci. 2, 21–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Verseijden F, Jahr H, Posthumus‐van Sluijs SJ, Ten Hagen TL, Hovius SE, Seynhaeve AL et al (2009) Angiogenic capacity of human adipose‐derived stromal cells during adipogenic differentiation: an in vitro study. Tissue Eng. A 15, 445–452. [DOI] [PubMed] [Google Scholar]
- 20. Cai X, Zhang Y, Yang M, Gong P, Grottkau BE, Lin Y (2011) Uniaxial cyclic tensile stretch inhibits osteogenic and odontogenic differentiation of human dental pulp stem cells. J. Tissue Eng. Regen. Med. 5, 347–353. [DOI] [PubMed] [Google Scholar]
- 21. Carlile MJ, Sturrock MG, Chisholm DM, Ogden GR, Schor AM (2000) The presence of pericytes and transitional cells in the vasculature of the human dental pulp: an ultrastructural study. Histochem. J. 32, 239–245. [DOI] [PubMed] [Google Scholar]
- 22. Farrington‐Rock C, Crofts NJ, Doherty MJ, Ashton BA, Griffin‐Jones C, Canfield AE (2004) Chondrogenic and adipogenic potential of microvascular pericytes. Circulation 110, 2226–2232. [DOI] [PubMed] [Google Scholar]
- 23. Mathieu S, El‐Battari A, Dejou J, About I (2005) Role of injured endothelial cells in the recruitment of human pulp cells. Arch. Oral Biol. 50, 109–113. [DOI] [PubMed] [Google Scholar]
- 24. von Tell D, Armulik A, Betsholtz C (2006) Pericytes and vascular stability. Exp. Cell Res. 312, 623–629. [DOI] [PubMed] [Google Scholar]
- 25. Caplan AI (2000) Tissue engineering designs for the future: new logics, old molecules. Tissue Eng. 6, 1–8. [DOI] [PubMed] [Google Scholar]
- 26. Rouwkema J, de Boer J, Van Blitterswijk CA (2006) Endothelial cells assemble into a 3‐dimensional prevascular network in a bone tissue engineering construct. Tissue Eng. 12, 2685–2693. [DOI] [PubMed] [Google Scholar]
- 27. Caplan AI (2009) Why are MSCs therapeutic? New data: new insight. J. Pathol. 217, 318–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Haynesworth SE, Baber MA, Caplan AI (1996) Cytokine expression by human marrow‐derived mesenchymal progenitor cells in vitro: effects of dexamethasone and IL‐1 alpha. J. Cell. Physiol. 166, 585–592. [DOI] [PubMed] [Google Scholar]
- 29. Akita M, Murata E, Merker HJ, Kaneko K (1997) Formation of new capillary‐like tubes in a three‐dimensional in vitro model (aorta/collagen gel). Ann. Anat. 179, 137–147. [DOI] [PubMed] [Google Scholar]


