During recent years, human breast milk has been documented as a potential source of bacteria for the newborn. Recently, we have reported the presence of fungi in breast milk from healthy mothers. It is well known that environmental and perinatal factors can affect milk bacteria; however, the impact on milk fungi is still unknown. The current report describes fungal communities (mycobiota) in breast milk samples across different geographic locations and the influence of the mode of delivery. We also provide novel insights on bacterium-fungus interactions, taking into account environmental and perinatal factors. We identified a core of four genera shared across locations, consisting of Malassezia, Davidiella, Sistotrema, and Penicillium, which have been reported to be present in the infant gut. Our data confirm the presence of fungi in breast milk across continents and support the potential role of breast milk in the initial seeding of fungal species in the infant gut.
KEYWORDS: fungi, Illumina sequencing, microbiota, mycobiota, breast milk
ABSTRACT
Recent studies report the presence of fungal species in breast milk of healthy mothers, suggesting a potential role in infant mycobiome development. In the present work, we aimed to determine whether the healthy human breast milk mycobiota is influenced by geographical location and mode of delivery, as well as to investigate its interaction with bacterial profiles in the same samples. A total of 80 mature breast milk samples from 4 different countries were analyzed by Illumina sequencing of the internal transcribed spacer 1 (ITS1) region, joining the 18S and 5.8S regions of the fungal rRNA region. Basidiomycota and Ascomycota were found to be the dominant phyla, with Malassezia and Davidiella being the most prevalent genera across countries. A core formed by Malassezia, Davidiella, Sistotrema, and Penicillium was shared in the milk samples from the different origins, although specific shifts in mycobiome composition were associated with geographic location and delivery mode. The presence of fungi in the breast milk samples was further confirmed by culture and isolate characterization, and fungal loads were estimated by quantitative PCR (qPCR) targeting the fungal ITS1 region. Cooccurrence network analysis of bacteria and fungi showed complex interactions that were influenced by geographical location, mode of delivery, maternal age, and pregestational body mass index. The presence of a breast milk mycobiome was confirmed in all samples analyzed, regardless of the geographic origin.
IMPORTANCE During recent years, human breast milk has been documented as a potential source of bacteria for the newborn. Recently, we have reported the presence of fungi in breast milk from healthy mothers. It is well known that environmental and perinatal factors can affect milk bacteria; however, the impact on milk fungi is still unknown. The current report describes fungal communities (mycobiota) in breast milk samples across different geographic locations and the influence of the mode of delivery. We also provide novel insights on bacterium-fungus interactions, taking into account environmental and perinatal factors. We identified a core of four genera shared across locations, consisting of Malassezia, Davidiella, Sistotrema, and Penicillium, which have been reported to be present in the infant gut. Our data confirm the presence of fungi in breast milk across continents and support the potential role of breast milk in the initial seeding of fungal species in the infant gut.
INTRODUCTION
Early human microbial gut colonization is an essential stepwise process with an impact on the immunological and metabolic programming of later health (1–3). Fungi residing in the human gut have been recognized as an important part of the gut microbiota, and although research on the field is scarce, the mycobiome could have important roles in human health status (4–8). Although information about fungal communities in the infant is generally lacking, there is evidence that fungal species (mainly yeast-like species) can be found in the gut early in life (9–11). A few reports have documented fungal transfer from mothers to infants, but little is known about how the mycobiome is shaped during this period (12–14). Recent prospective studies have revealed that altered gut mycobial patterns precede atopic wheeze and asthma development and have suggested fungal-bacterial interactions that would influence early-life patterns of microbial alpha diversity (15, 16).
Breast milk is an important source of bacteria to the infant, and together with oligosaccharides, contributes to the settlement of the gut microbiota characteristic of the healthy breast-fed child, with a strong impact on immune surveillance within the gastrointestinal environment and thereby also other membranes of the body (17–19). A recent study suggested the presence of a diversity of fungal species in human breast milk of healthy mothers, including Malassezia, Candida, and Saccharomyces as the most common genera, by means of high-throughput sequencing, microscopy, and other culture-independent techniques (20). Moreover, viable yeasts, predominantly Candida parapsilosis and Rhodotorula mucilaginosa species, were isolated and characterized. This finding provides a new angle to the infant mycobiome development and calls for further evaluation of the key determinants of their composition. Furthermore, complex interactions between bacteria and fungi have been reported in the human gut, oral cavity, skin, and vagina (16, 21–24), and, therefore, such are also likely to occur in breast milk.
In addition, accumulating evidence suggests that some environmental factors might influence breast milk composition (25–27). In particular, geographic location, delivery mode, maternal body mass index (BMI), and age have been suggested to have an impact on breast milk bacterial composition (28–35), although their potential impact on the milk’s fungal fraction is still to be elucidated.
In the present study, we characterized the breast milk mycobiota of healthy breast-feeding mothers from four different countries (Spain, Finland, South Africa, and China) in order to investigate the potential influence of geographic location and mode of delivery on its composition. Fungal loads in the samples were estimated, and cooccurrence networks between specific fungi and bacteria were analyzed for potential interactions depending on mode of delivery across the different countries.
RESULTS
Subject description.
The characteristics of the subjects participating in the study are listed in Table 1. The mean age of the mothers (n = 80) was 33.52 years (standard deviation [SD], ±4.87 years), with no statistical differences between countries. The mean pregestational BMI was 24.06 (SD, ±3.85), normal weight. Chinese mothers had significantly lower BMI, 21.71 (SD, ±1.97), considered normal weight. Differences in BMI between mothers delivering vaginally or by Caesarean section (C-section) were observed only in South African and Finnish women, where mothers delivering by C-section had higher BMIs, 26.67 ± 1.41 and 26.30 ± 2.57, respectively, although this difference was significant only in the South African group (P < 0.05).
TABLE 1.
Clinical characteristics of donors providing human milk samples for the studya
| Country | Delivery mode (no. of samples) | Age (yr) ± SD | P value | BMI ± SD | P value |
|---|---|---|---|---|---|
| Finland | C-section (10) | 35.20 ± 4.07 | 0.820 | 26.30 ± 2.57 | 0.185 |
| Vaginal (10) | 33.70 ± 6.02 | 22.65 ± 8.60 | |||
| Total (20) | 34.45 ± 5.06 | ns | 24.47 ± 6.46 | ns | |
| Spain | C-section (10) | 34.50 ± 2.59 | 0.288 | 24.34 ± 1.47 | 0.630 |
| Vaginal (10) | 32.20 ± 5.16 | 24.25 ± 1.43 | |||
| Total (20) | 33.35 ± 4.14 | ns | 24.30 ± 1.41 | ns | |
| South Africa | C-section (10) | 36.60 ± 6.08 | 0.944 | 26.67 ± 1.41 | 0.043 |
| Vaginal (10) | 31.50 ± 5.76 | 24.81 ± 2.67 | |||
| Total (20) | 34.05 ± 2.29 | ns | 25.75 ± 2.29 | ns | |
| China | C-section (10) | 32.60 ± 2.95 | 0.970 | 21.49 ± 2.29 | 0.449 |
| Vaginal (10) | 31.90 ± 4.25 | 21.92 ± 1.54 | |||
| Total (20) | 32.25 ± 3.58 | ns | 21.71 ± 1.97 | 0.004 | |
| All | C-section (40) | 34.72 ± 4.25 | 0.058 | 24.70 ± 2.83 | 0.072 |
| Vaginal (40) | 32.32 ± 5.20 | 23.41 ± 2.11 | |||
| Total (80) | 33.52 ± 4.87 | ns | 24.06 ± 3.85 | ns |
ns, not significant.
Fungal cell detection in breast milk.
Eighty milk samples were analyzed by quantitative PCR (qPCR) targeting the ITS1–5.8S rRNA region. Results showed that 16 of 20 Spanish samples (80%; median value, 195,142 cells/ml), 9 of 20 Chinese samples (45%; median value, 170,732 cells/ml), 7 of 20 Finnish samples (35%; median value, 199,480 cells/ml), and 14 of 20 South African samples (70%; median value, 371,119 cells/ml) had detectable levels of fungi. No significant differences were observed between geographic locations or by mode of delivery (see Fig. S1 in the supplemental material).
The presence of fungal cells in the milk was further confirmed by culture in fungus-specific culture medium and identification of the isolates by 18S rRNA sequencing, as well as by microscopy after incubation of the milk samples with calcofluor white fungal stain. A summary of the results is available in Table S1 and Fig. S2 in the supplemental material.
Fungal composition of breast milk: impact of geographical area and perinatal factors.
After sequencing of the ITS1 fungal region, a mean of 107,765 taxonomically assigned, clean, filtered sequences per sample (SD, ±45,493), with an average length of 301 bp, was obtained. All breast milk samples contained fungal DNA, and they were dominated by two phyla: Basidiomycota (58.65%) and Ascomycota (41.03%). South African samples had significantly higher levels of Ascomycota and lower levels of Basidiomycota than the other countries (P < 0.05).
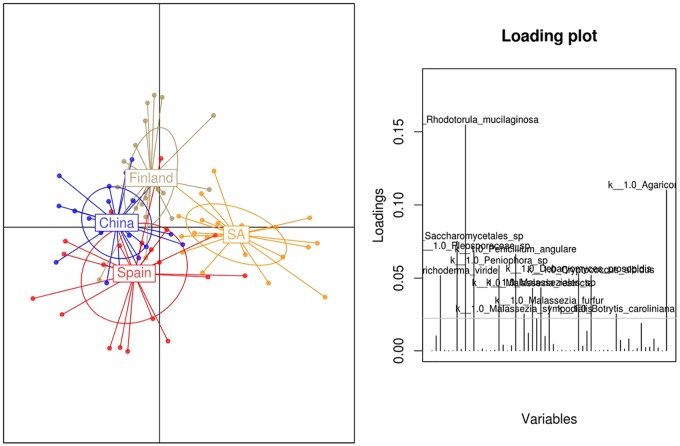
Discriminant analysis of principal components (DAPC), which transforms data using a principal-component analysis (PCA) and subsequently identifies clusters using discriminant analysis (DA), showed that South African samples clustered at a distance from the other countries, mainly due to the increased levels of Rhodotorula mucilaginosa (Fig. 1).
FIG 1.
Breast milk samples cluster separately according to fungal composition. DAPC analysis shows relationships in fungal composition among samples from different locations. A canonical loading plot shows differentially abundant bacterial OTUs in the groups. The individual peaks show the magnitude of the influence of each variable on the separation of the groups (threshold level = 0.05). Total number of samples, 80 (number per country, 20). SA, South Africa.
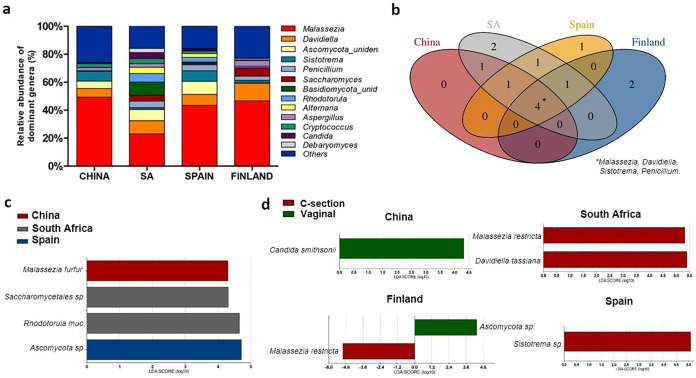
Taxonomic analysis at the genus level showed that breast milk samples were dominated by Malassezia (40.6% average abundance), followed by Davidiella (9.0%), which was prevalent regardless of the location or the donor’s type of delivery (Fig. 2a). The effects of country of origin and mode of delivery on breast milk fungal composition were analyzed and reflected that milk mycobiota differed significantly across geographic location (permutational multivariate analysis of variance [PERMANOVA], P = 0.005) and mode of delivery (PERMANOVA, P = 0.023). Redundant analysis (RDA) confirmed the effect of geographic location on breast milk fungal composition (P = 0.001), although that of mode of delivery did not reach statistical significance. The Kruskal-Wallis test was implemented to compare phylotypes at the genus level across samples. Results showed that Malassezia was statistically less abundant in South African samples (P < 0.05), and Penicillum and Rhodotorula abundances were lower in Chinese samples (P < 0.01), while Saccharomyces was more abundant in Spanish and Finnish samples (P < 0.01) than in samples from the rest of the locations. No statistically significant effect of maternal age, gestational BMI, or antibiotic intake during delivery was detected for breast milk microbial composition by using MaAsLin (multivariate analysis with linear model).
FIG 2.
Effect of geographical location and mode of delivery on fungal composition in breast milk samples. (a) Fungal relative abundances at the genus level across countries. Only genera present in more than 1% abundance in at least 20% of the samples are represented. (b) Shared phylotypes across countries at the genus level. *, core of four fungal genera shared across geographic locations. Venn diagram cutoff, 0.5. (c) Differentially abundant species in breast milk samples depending on geographic location, as inferred by the LefSe algorithm. The threshold for the logarithmic discriminant analysis (LDA) score was 2, with a P of <0.05. Number of samples per country, 20 (total number, 80). (d) Differentially abundant species in breast milk samples depending on delivery mode and geographic location, as inferred by the LefSe algorithm. The threshold for the LDA score was 2, with a P value of <0.05. Total number of samples, 80 (samples from vaginal deliveries, 40; samples from C-section deliveries, 40). SA, South Africa.
Despite the differences, a core of four genera shared across the four countries was identified, including Malassezia, Davidiella, Sistotrema, and Penicillium. Wallemia and Aspergillus were found only in samples from Finland, Botrytis and an unidentified Saccharomycetales were found only in South African samples, and an unidentified Malasseziales was found only in Spanish samples. Rhodotorula was present in samples from all countries except China (Fig. 2b).
Comparisons between samples were further analyzed at the species level. Linear discriminant analysis effect size (LefSe) results showed differentially abundant fungi between countries. Rhodotorula mucilaginosa and Saccharomycetales species were more abundant in South African samples, while Malassezia furfur was more prevalent in Chinese samples, and an Ascomycota sp. was more abundant in Spanish samples (Fig. 2c).
Taking into account mode of delivery, mycobiota compositions were different across the milk samples from different geographic origins. The Kruskal-Wallis test reflected that the occurrence of Cryptococcus was statistically significantly higher in milk samples of women delivering vaginally than in those who delivered by C-section (P = 0.028). At the species level, in Chinese breast milk samples, Candida smithsonii was significantly more abundant in samples of women with vaginal deliveries, Sistotrema species in samples from Spanish women with C-sections, Ascomycota species in samples from Finnish women with vaginal deliveries, Malasezzia restricta in samples from Finnish women with C-sections, and Malassezia restricta and Davidiella tassiana in samples from South African women with C-sections (LefSe analysis, P < 0.05) (Fig. 2d).
Indices of alpha diversity and richness across the samples were similar, and no statistical differences were observed between geographic locations or delivery modes (Fig. S3).
Fungal and bacterial interactions: a network analysis.
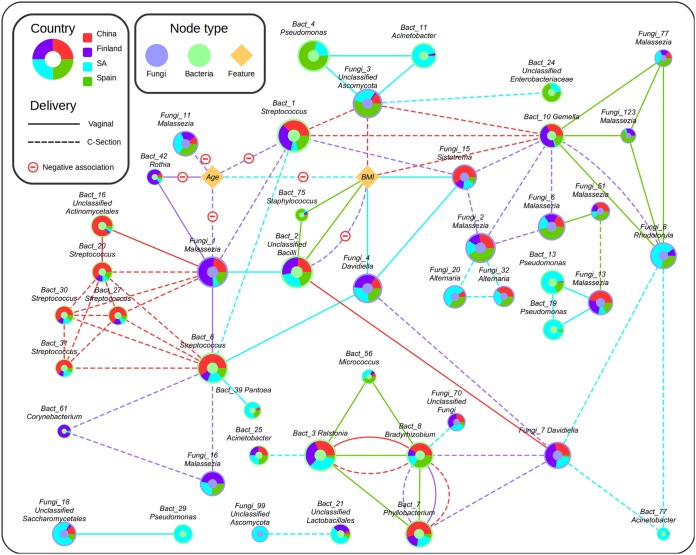
Network analyses of the bacteria and fungi present in the breast milk samples showed complex intra- and interdomain interactions, with different associations among organisms depending on the country of origin and delivery mode, some of which were also influenced by maternal features. For example, a Malassezia operational taxonomic unit (OTU) (Fungi_1) correlated positively with a Streptococcus (Bact_6) from vaginal delivery samples and with a Streptococcus (Bact_1) from C-section deliveries among Finnish samples, and the abundances were dependent on maternal age. The same Malassezia OTU correlated positively with several Streptococcus OTUs in samples from C-section deliveries from Chinese mothers and also positively with an unclassified member of Bacilli (Bact_2) from South African samples and vaginal deliveries. Significant influences of maternal age and BMI on specific bacterial and fungal organisms were also observed (Fig. 3). However, given that the density of fungal cells is at least 1 order of magnitude lower than that of bacteria, the influence of fungi on the breast milk ecosystem needs to be elucidated.
FIG 3.
Cooccurrence network of bacteria and fungi in breast milk samples depending on maternal features and delivery mode. Green nodes represent bacterial OTUs, blue nodes represent fungal OTUs, and yellow nodes represent features. Node size indicates OTU abundance. Pie chart colors represent the overall distribution of each OTU across country. Each link indicates a significant (P < 0.05) interaction between OTUs or features in samples from a given combination of country and delivery mode (vaginal, C-section). Link color denotes the country, and line type indicates delivery mode. SA, South Africa.
In order to study the diversity of the most common yeast in our samples, a phylogenetic tree of the most prevalent Malassezia OTUs detected in this work across geographic locations was determined, including known members of the Malassezia genus as a reference (Fig. S4). The tree shows a large diversity of Malassezia isolates with similarity to at least four known species, including OTUs which could potentially represent new species. With the exception of one OTU (Fungi 37, which was found to be uniquely present in China), all other sequences were found in all countries and appear to be therefore ubiquitous. In relation to mode of delivery, all the OTUs were present in breast milk from mothers with both delivery types.
DISCUSSION
Breast milk is a continuous source of microbes that are transmitted, together with many nutrients and protective compounds, to the infant gut during a critical period when the key regulatory systems of the body are immature (17, 18). Although bacteria inhabiting human breast milk have been extensively studied, the presence of fungi in the fluid had not been assessed until recently, when a diversity of fungal phylotypes in breast milk from healthy Spanish mothers was reported by our group (20). The mycobiome, the fungal fraction of the human microbiome, is present in lower abundances and has been much less explored than the bacterial fraction. However, its potential importance for human health and disease has stimulated an increased interest in this field (5–7, 10). In the infant, fungal species can be detected very early in life (10, 11, 13). However, the infant mycobiome is almost unexplored, and information about its development is scarce. To ascertain the presence of fungi in breast milk is difficult because of the possibility of contamination in samples with low microbial density, and therefore multiple approaches and strict negative controls are needed (15).
A recent study reported higher gut fungal diversities during the first months of life, which decreased over time, while the diversities of the bacterial fraction increased in reciprocal correlation, suggesting that potential interkingdom associations may drive microbial gut dynamics (36).
In the present study, we have confirmed the presence of diverse fungal communities in breast milk samples from Spain, Finland, China, and South Africa. Fungi were detected in all breast milk samples through massive DNA sequencing, with the two phyla Basidomycota and Ascomycota being the most prevalent and presenting reciprocal patterns of abundance in all countries except for South Africa, where Ascomycota levels were significantly higher, and Basidiomycota levels lower, than those of the other countries. At the genus level, Malassezia predominated in all countries, followed by Davidiella. In our previous work reporting the presence of fungi in breast milk, Malassezia also represented the most abundant genus (20). Other genera found in the current study, such as Alternaria, Rhodotorula, Saccharomyces, and Candida, were also found in the previous study.
Results yielded by qPCR showed that >70% of Spanish and South African samples, 45% of Chinese samples, and only 35% of Finish samples had detectable levels of fungal DNA. The median fungal load in all the samples was 2.5 × 105 cells/ml, in agreement with our previous results for Spanish samples.
Our findings reinforce the potential influence of environmental factors, in particular geographic location and delivery mode, on breast milk fungal composition. Samples from South Africa clustered at a distance from those from the other countries according to their fungal composition, because of the influence of the higher levels of Rhodotorula mucilaginosa in those samples (Fig. 1). Although differences among samples from different geographic locations were observed, a core constituted by four genera, Malassezia, Davidiella, Sistotrema, and Penicillium, was shared in all countries (Fig. 2b).
Breast milk mycobiota also differed depending on the mode of delivery (vaginal or C-section) across countries. Specific fungi, such as the genus Cryptococcus, appeared to be more prevalent among samples from mothers delivering vaginally, and specific shifts at species level were also observed within each country. No differences in fungal diversity or richness were observed in the present study. Previously, we identified changes in breast milk microbiota between locations, as well as in the milk metabolite profile (31, 35), using the same samples analyzed in this study.
Although the origin of breast milk fungi is unknown, most of the organisms detected in this study can be found in other human niches. Malassezia species are yeasts whose primary niche is the human body (and other animals). In healthy individuals, they are part of the normal microbiota, where they predominantly colonize the seborrheic parts of the skin (37), and are commonly found in infants (9, 38–40). Malassezia has also been detected in significant abundance in adult (11, 37, 41) and infant (42) fecal samples and therefore may play a role at the intestinal level; it has also been described as an oral commensal (43). Although Malassezia DNA has been detected in high proportions in breast milk before, no viable cells could be recovered by classic culture methods from breast milk, (20), and further efforts should be made to culture this organism, which has also been shown to be able to penetrate the cell and survive intracellularly.
Davidiella, the second most prevalent fungus found in the samples of this study, was detected in the only published study about the characterization of vaginal microbiota and mycobiota of asymptomatic women (44). In the same study, Candida was found to be the predominant genus. Therefore, these fungi may play an important role in the early colonization of vaginally born infants. In our previous study on breast milk fungi, Davidiella could not be detected (20), which could be associated with the differences on sequencing platforms and genes targeted in both studies, as has been previously shown (45, 46). In addition, Davidiella represents the sexual form of the Cladosporium genus (47). Fungi can have an asexual form (anamorph) and a sexual form (teleomorph) that may be classified into different genera. This sexual dimorphism can be a significant problem when classifying fungal sequences, and the use of different databases and/or sequencing of different genes can lead to conflicting classifications. In a study with pediatric inflammatory bowel disease (IBD) patients, Cladosporium cladosporiodes abundance decreased in patients with IBD, while Pichia jadinii and Candida parapsilosis increased, in comparison to controls (48).
Candida is probably the most ubiquitous genus of the human mycobiome. It is the major fungal genus detected in the adult oral cavity (49, 50) and has also been detected in the infant mouth, including several species as common inhabitants (C. parapsilosis, C. tropicalis, C. orthopsilosis, etc.) (9, 51, 52). Several Candida species are also commonly present in adult skin and fecal samples (7, 41) and in infant anal and fecal samples (9, 53). Although Candida can be responsible for vaginal infections (54), it is the most prevalent fungus in the vaginal mycobiome of healthy women (44). Transmission of Candida from mother to infant likely occurs, as the same fingerprinting of the DNA has shown identity between maternal Candida from vagina, rectum, oral cavity, and skin and infant Candida from oral cavity and rectum (14).
Other prevalent fungi detected in our samples are commonly found in several body niches. Saccharomyces is among the most abundant fungus in the gut (7, 41), and Saccharomyces cerevisiae has been reported to be highly prevalent and abundant in the infant oral and anal mycobiome during the first month of life (9). In a recent study, bacteria and fungi from fecal samples in children suffering atopic wheeze were analyzed, and Saccharomycetales taxa appeared to be decreased in the atopic wheeze group, while the species Pichia kudriavzevii was increased, compared to controls (16). Others, such as Penicillium or Aspergillus, can also be detected in fecal samples, and Debaromyces hansenii represents one of the main species present in the gut of breastfed infants (12). In the present study, we detected Debaromyces, although none of the sequences have been classified as D. hansenii. However, DNA from this species was previously detected in breast milk (20).
The study of interspecies interactions within a population is necessary to better understand the microbiota’s role. It is known that microorganisms can interact by competition and sometimes collaboration, thereby influencing microbiota composition and the host’s health. It has been demonstrated that cross talk between bacteria and fungi can exist, modulating host defense mechanisms, protecting against infections, or collaborating to cause them (55, 56). For example, synergies between oral Streptococcus oralis and C. albicans enhanced C. albicans invasion through the activation of host enzymes that cleave epithelial junction proteins (57). On the contrary, Streptococcus mutans showed the ability to modulate biofilm formation and to reduce C. albicans virulence in an animal model (58). Some vaginal isolates of Lactobacillus strains have shown antifungal activity in vitro against Candida spp., and probiotic Lactobacillus rhamnosus and Lactobacillus reuterii strains have shown in vitro efficacy against C. albicans responsible for vaginal infections (24). To understand microbial relationships, microbial network analyses are indispensable, allowing the identification and representation of the most influential members in a bacterial community and their interactions with other microorganisms (59). In a recent work, bacterial interactions in the colostrum and mature milk of Italian and Burundian mothers were analyzed and showed different bacterial networks among the two populations. The identified networks were complex and dynamic, changing from colostrum to mature milk (60). In the present study, we have analyzed cooccurrence relationships between bacteria and fungi in breast milk, observing a complex network of interactions between fungi and bacteria and within the same domain. Microbial interactions were influenced by delivery characteristics (mode of delivery and geographic location), and maternal features (maternal BMI and age) influenced the prevalence of particular microorganisms. Interesting positive correlations were observed between several Malassezia spp., the most prevalent fungi detected in breast milk by sequencing, and different streptococci, the latter representing one of the most common bacterial genera in breast milk (61). Interestingly, in our previous study, we observed a significant positive correlation between Malassezia and bacterial load (20), and further experimental research should analyze potential synergistic relationships between these genera.
Our data confirmed the presence of fungal DNA and fungal cells (including viable cells) in breast milk samples from healthy mothers from four different geographic locations, by using different approaches. This supports the existence of a “breast milk mycobiota” under healthy conditions. Differences in composition associated with mode of delivery and country of origin were observed. In addition, we observed some interdomain microbial interactions in breast milk that could lead to further in vitro studies. The presence of viable fungal cells suggests a potential influence of breast milk on the infant’s mycobiota development. However, data from the infant gut mycobiota is missing in the present study, and further studies should address the potential fungal transference from breast milk to the infant gut mycobiome. Although we tried to prevent the contamination of maternal skin mycobiota by cleaning the breast prior to sample collection (which has been previously shown to reduce bacteria in breast milk samples [62]), it should be taken into account that certain retrograde flux occurs during breastfeeding, and fungal species present in maternal skin and the infant’s mouth could be translocated to breast milk and vice versa (63). A greater understanding of the environmental influence on the bacterial and fungal communities and their metabolic functions is also needed.
MATERIALS AND METHODS
Subjects and sampling.
Breast milk samples at 1-month postpartum were obtained from 80 healthy, lactating women from 4 different geographical locations (20 in each location), including China (Beijing area), South Africa (Cape Town), Finland (southwestern area), and Spain (Valencia, Mediterranean area).
All mothers were practicing exclusive breastfeeding. Subjects were grouped according to mode of delivery: vaginal (n = 10 per country) and Caesarean section (C-section) (n = 10 per country). Maternal characteristics such as age, weight, and pregestational body mass index (BMI) were collected at the time of enrollment. All women who delivered via C-section received prophylactic antibiotics, except Finnish women, for whom no prophylaxis is routinely used per the hospital policy. All participants were given detailed oral and written information, and written informed consent was obtained for participation. The study protocol was approved by the ethics committees of the respective participating institutions: Bioethics Committee of CSIC and the Regional Ethics Committee for Biomedical Research (Spain), Ethics Committee, Hospital District of Southwest Finland (Finland), Medical Research Board of Peking University (China), and University of Cape Town, Human Research Ethics Committee (South Africa).
Before sample collection, nipples and mammary areolas were cleaned with soap and sterile water and soaked in chlorhexidine to reduce sampling of microorganisms residing on the skin. Milk samples were collected in a sterile tube manually, with the first drops discarded. All samples were frozen at −20°C until further processing. To avoid bias, samples were collected using the same standardized protocol in the four countries and were processed and analyzed in a single laboratory.
Microbial DNA extraction and sequencing.
Breast milk samples (1.5 ml) were centrifuged at 14,000 rpm for 20 min at 4°C to remove fat, and pellets were used for total DNA extraction, which involved mechanical and chemical cell lysis. Bead beating was carried out using FastPrep (FP120-230, Bio 101; ThermoSavant, Holbrook, NY, USA), and the InviMag stool DNA kit (Stratec Molecular, Berlin, Germany) was used with the King Fisher magnetic particle processor (Thermo Fisher Scientific Oy, Vantaa, Finland). The DNA extraction protocol was also used with water to serve as negative controls. Isolated DNA concentrations were measured using a Qubit 2.0 fluorometer (Life Technology, Carlsbad, CA, USA).
Primers targeting the highly variable fungal internal transcriber spacer ITS1 of the fungal 18S rRNA gene (forward, TAGAGGAAGTAAAAGTCGTAA; reverse, TTYRCTRCGTTCTTCATC) (64) with adaptors were used for sequencing on an Illumina MiSeq platform. Sequencing was carried out at the Foundation for the Promotion of Health and Biomedical Research, FISABIO (Valencia, Spain). No-template controls (NTCs) and negative controls during DNA extraction were included to rule out potential contaminations at the time of DNA extraction or sequencing.
Fungal load.
qPCR amplification and detection of the ITS1–5.8S rRNA conserved fungal region was performed as previously described (17), using the primers ITS1F (5′-TCCGTAGGTGAACCTGCGG) and 5.8 R (5′-CGCTGCGTTCTTCATCG). Each reaction mixture of 20 μl was composed of 10 μl of KAPA Sybr Fast qPCR master mix (KAPA Biosistems), 0.4 μl of each primer (10 μM concentration), and 2 μl of template DNA, with an annealing temperature of 61°C in a LightCycler 480 real-time PCR system (Roche Technologies). All amplifications were performed in duplicate, and a negative control was included in each reaction plate. Samples with threshold cycle (CT) values equal to or higher than the negative control were considered negative for fungal DNA.
Breast milk culture and identification of fungal colonies.
One-hundred-microliter volumes of selected breast milk samples were plated in four solid fungus-selective media: Sabouraud (Conda-Pronadisa) supplemented with chloramphenicol, 0.05 g/liter (Roche); Rose Bengal (Conda-Pronadisa) supplemented with chloramphenicol, 0.5 g/liter (Roche); YPD (40 g/liter dextrose, 40 g/liter peptone, 20 g/liter yeast extract, and 40 g/liter agar) supplemented with 25 μg/ml of streptomycin–25 U/ml of penicillin (Biowest); and YNB (Sigma) with 8% ethanol and 25 μg/ml of streptomycin–25 U/ml of penicillin (Biowest). All plates were incubated aerobically at 37°C, as previously described (17). DNA from the isolated colonies was extracted and amplified by PCR using primers targeting the 18S rRNA gene (forward, 5′-GTAGTCATATGCTTGTCTC; reverse, 5′-CCATTCCCCGTTACCCGTTG). PCR products were sequenced in an Applied Biosystems 3730/3730xl DNA analyzer at the University of Valencia (Spain), and isolates were identified by using the BLAST algorithm in the NCBI database, with a minimum of 98% sequence identity.
Microscopic analyses of fungi in milk.
In order to identify fungal cells in breast milk, samples were incubated with calcofluor white stain that dyes the cell walls of the fungi and yeasts. Samples were visualized with fluorescence microscopy using a Nikon Eclipse E90i microscope (Nikon Corporation) with a 100× objective. Image processing was performed using the NIS-Elements BR v3.22 software (Nikon).
Data analysis.
ITS1 reads were pair-end joined using the FLASH program (65), with default parameters applied. The resulting sequences were end-trimmed in 20-bp sliding windows with an average quality value of >30 and a length of >50 bp, using the Prinseq-lite program (66). Chimeric reads were eliminated using the UCHIME algorithm (67), resulting in a total of 9,797,578 reads. Taxonomy assignment of the remaining sequences was performed using the Ribosomal Database Project Classifier stand-alone tool (68) with the UNITE fungal ITS v7.2 trainset (69) and an 80% confidence threshold. Sequences were clustered into operational taxonomical units (OTUs) based on 99% identity, and representative OTU sequences were obtained using CD-hit software (70). OTU tables were rarefied to 9,200 sequences per sample to avoid variations in sequencing depth, and Shannon and Chao1 indexes were calculated using the “plyr” and “vegan” packages from R software (version 3.2.2) (71).
Statistical analysis.
Calypso software (version 8.2) (72) was used to obtain a Venn diagram for shared phylotypes, discriminant analysis of principal components (DAPC) was performed at the OTU level, using geographic location as a factor, and PERMANOVA and redundancy analysis (RDA) were applied to study the statistical effect of country and delivery mode on breast milk fungal composition. The Kruskal-Wallis test was implemented to study genus-level taxonomical differences between countries and delivery modes, using GraphPad PRISM 6 (GraphPad Software). The linear discriminant analysis effect size (LefSe) (73) algorithm was used to detect the most differentially abundant fungi between countries, and between vaginal and C-section deliveries in each country, at the species level. In order to control the potential effects of maternal age, maternal BMI predelivery, and antibiotic use at delivery, MaAsLin (multivariate analysis with linear model) (74), which finds associations between metadata and microbial abundances, was applied. Other statistical analysis and graphing were performed using GraphPad PRISM 6.
Analysis of bacterial and fungal cooccurence.
Sequences from the 16S rRNA gene of the same samples, from a report by Kumar et al. (31), were obtained from NCBI (SRA accession no. SRP082263 and submission ID SUB1772296). Quality filtering, chimera checking, and OTU clustering were done the same way as for the ITS1 reads.
RDP Classifier was used to taxonomically assign the bacterial (against the RDP 16S rRNA training set 16) (75) and fungal (against the UNITE v 07-04-2014 trainset) (70) representative OTU sequences. Samples with fewer than 1,500 sequences were excluded from the analysis.
For the bacterial data sets, OTUs with a higher relative abundance in any of the two controls than in the breast milk samples were treated as putative contaminants and discarded. This procedure could not be performed on the fungal data sets, since the sequencing of the two controls yielded too few reads. Nevertheless, the low fraction of reads assigned to putative contaminants in the bacterial data sets (2% on average) leads us to believe that the samples were essentially contamination-free. Both the bacterial and fungal OTU tables were rarefied to 1,500 sequences per sample. OTUs from both the bacterial and fungal data sets having an overall relative abundance higher than 1% of the total reads, or appearing in at least one sample with a relative abundance higher than 5%, were combined into a single table. Associations between pairs of bacterial and fungal OTUs were calculated using the maximal information coefficient, as implemented in MICtools (76). Pseudo P values were obtained by generating 200,000 null matrices and further transformed to Storey’s Q-values to correct for multiple hypothesis testing with the Benjamini-Hochberg method. Correlations with a false discovery rate lower than 0.01 were deemed significant. Further, we divided the samples into eight groups according to the combination of the four countries and two delivery modes. We used linear regression to calculate correlations between pairs of OTUs and factors (age, BMI) in a given group. For each group, only OTUs appearing in at least four samples and with a relative abundance higher than 2% in at least one sample were included. Correlations with a P value lower than 0.05 were deemed significant. Network analysis was performed on Cytoscape (77).
Phylogenetic relationships between Malassezia reads.
ITS sequences of the 20 most abundant OTUs assigned to the Malassezia genus by the RDP Classifier were combined with those of known Malassezia representatives from the UNITE v07-04-2014 database (69). A multiple sequence alignment was constructed with MAFFT v7.313 (78). Cryptococcus neoformans was selected as an outgroup, and its ITS sequence was added to the alignment using the “add” option from MAFFT. The resulting alignment was manually curated and further refined with MUSCLE v3.8.31 (79). Phylogenetic trees were inferred with RaxML v8 (80) and MrBayes v3.2 (81), using 1,000 replicates and 1,000,000 generations, respectively. TreeGraph2 (82) was used to combine and visualize the maximum likelihood and Bayesian inference trees.
Accession number(s).
All ITS1 sequences have been deposited in the European Nucleotide Archive (ENA) server under study ID PRJEB25581. Samples were deposited under accession numbers ERS2312706 to ERS2312785.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to all the participant families that provided biological samples for this study.
A.B.-A. and M.C.C. acknowledge the European Research Council (ERC) under the European Union's Horizon 2020 research and innovation program (ERC Starting Grant project no. 639226). F.P.-S. was supported by Ministerio de Economía y Competitividad (MINECO) grant no. CTM2016-80095-C2-1-R (NOVAMAR). The Chinese group acknowledges support from Key Projects of Beijing Science and Technology (D141100004814002) and the Natural Scientific Foundation of Beijing (Z140001).
The authors declare that they have no conflicts of interest.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.02994-18.
REFERENCES
- 1.Chervonsky AV. 2010. Influence of microbial environment on autoimmunity. Nat Immunol 11:28–35. doi: 10.1038/ni.1801. [DOI] [PubMed] [Google Scholar]
- 2.Houghteling PD, Walker WA. 2015. Why is initial bacterial colonization of the intestine important to infantsʼ and childrenʼs health? J Pediatr Gastroenterol Nutr 60:294–307. doi: 10.1097/MPG.0000000000000597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tamburini S, Shen N, Wu HC, Clemente JC. 2016. The microbiome in early life: implications for health outcomes. Nat Med 22:713–722. doi: 10.1038/nm.4142. [DOI] [PubMed] [Google Scholar]
- 4.Cui L, Morris A, Ghedin E. 2013. The human mycobiome in health and disease. Genome Med 5:63. doi: 10.1186/gm467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sokol H, Leducq V, Aschard H, Pham H-P, Jegou S, Landman C, Cohen D, Liguori G, Bourrier A, Nion-Larmurier I, Cosnes J, Seksik P, Langella P, Skurnik D, Richard ML, Beaugerie L. 2016. Fungal microbiota dysbiosis in IBD. Gut 66:1039–1048. doi: 10.1136/gutjnl-2015-310746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hatoum R, Labrie S, Fliss I. 2012. Antimicrobial and probiotic properties of yeasts: from fundamental to novel applications. Front Microbiol 3:421. doi: 10.3389/fmicb.2012.00421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Underhill DM, Iliev ID. 2014. The mycobiota: interactions between commensal fungi and the host immune system. Nat Rev Immunol 14:405–416. doi: 10.1038/nri3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Laforest-Lapointe I, Arrieta M-C. 2018. Microbial eukaryotes: a missing link in gut microbiome studies. mSystems 3:e00201-17. doi: 10.1128/mSystems.00201-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ward TL, Dominguez-Bello MG, Heisel T, Al-Ghalith G, Knights D, Gale CA. 2018. Development of the human mycobiome over the first month of life and across body sites. mSystems 3:e00140-17. doi: 10.1128/mSystems.00140-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ward TL, Knights D, Gale CA. 2017. Infant fungal communities: current knowledge and research opportunities. BMC Med 15:30. doi: 10.1186/s12916-017-0802-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seddik HA, Ceugniez A, Bendali F, Cudennec B, Drider D. 2016. Yeasts isolated from Algerian infants’s feces revealed a burden of Candida albicans species, non-albicans Candida species and Saccharomyces cerevisiae. Arch Microbiol 198:71–81. doi: 10.1007/s00203-015-1152-x. [DOI] [PubMed] [Google Scholar]
- 12.Schei K, Avershina E, Øien T, Rudi K, Follestad T, Salamati S, Ødegård RA. 2017. Early gut mycobiota and mother-offspring transfer. Microbiome 5:107. doi: 10.1186/s40168-017-0319-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Drell T, Štšepetova J, Simm J, Rull K, Aleksejeva A, Antson A, Tillmann V, Metsis M, Sepp E, Salumets A, Mändar R. 2017. The influence of different maternal microbial communities on the development of infant gut and oral microbiota. Sci Rep 7:9940. doi: 10.1038/s41598-017-09278-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bliss JM, Basavegowda KP, Watson WJ, Sheikh AU, Ryan RM. 2008. Vertical and horizontal transmission of Candida albicans in very low birth weight infants using DNA fingerprinting techniques. Pediatr Infect Dis J 27:231–235. doi: 10.1097/INF.0b013e31815bb69d. [DOI] [PubMed] [Google Scholar]
- 15.Salter SJ, Cox MJ, Turek EM, Calus ST, Cookson WO, Moffatt MF, Turner P, Parkhill J, Loman NJ, Walker AW. 2014. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol 12:87. doi: 10.1186/s12915-014-0087-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arrieta M-C, Arévalo A, Stiemsma L, Dimitriu P, Chico ME, Loor S, Vaca M, Boutin RCT, Morien E, Jin M, Turvey SE, Walter J, Parfrey LW, Cooper PJ, Finlay B. 2017. Associations between infant fungal and bacterial dysbiosis and childhood atopic wheeze in a nonindustrialized setting. J Allergy Clin Immunol 142:424–434. doi: 10.1016/j.jaci.2017.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jost T, Lacroix C, Braegger CP, Rochat F, Chassard C. 2014. Vertical mother-neonate transfer of maternal gut bacteria via breastfeeding. Environ Microbiol 16:2891–2904. doi: 10.1111/1462-2920.12238. [DOI] [PubMed] [Google Scholar]
- 18.Allan Walker W, Shuba Iyengar R. 2015. Breast milk, microbiota, and intestinal immune homeostasis. Pediatr Res 77:220–228. doi: 10.1038/pr.2014.160. [DOI] [PubMed] [Google Scholar]
- 19.Boix-Amorós A, Collado MC, Mira A. 2016. Relationship between milk microbiota, bacterial load, macronutrients, and human cells during lactation. Front Microbiol 7:492. doi: 10.3389/fmicb.2016.00492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boix-Amorós A, Martinez-Costa C, Querol A, Collado MC, Mira A. 2017. Multiple approaches detect the presence of fungi in human breastmilk samples from healthy mothers. Sci Rep 7:13016. doi: 10.1038/s41598-017-13270-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoffmann C, Dollive S, Grunberg S, Chen J, Li H, Wu GD, Lewis JD, Bushman FD. 2013. Archaea and fungi of the human gut microbiome: correlations with diet and bacterial residents. PLoS One 8:e66019. doi: 10.1371/journal.pone.0066019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peleg AY, Hogan DA, Mylonakis E. 2010. Medically important bacterial-fungal interactions. Nat Rev Microbiol 8:340–349. doi: 10.1038/nrmicro2313. [DOI] [PubMed] [Google Scholar]
- 23.Mason KL, Erb Downward JR, Mason KD, Falkowski NR, Eaton KA, Kao JY, Young VB, Huffnagle GB. 2012. Candida albicans and bacterial microbiota interactions in the cecum during recolonization following broad-spectrum antibiotic therapy. Infect Immun 80:3371–3380. doi: 10.1128/IAI.00449-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parolin C, Marangoni A, Laghi L, Foschi C, Ñahui Palomino RA, Calonghi N, Cevenini R, Vitali B. 2015. Isolation of vaginal lactobacilli and characterization of anti-Candida activity. PLoS One 10:e0131220. doi: 10.1371/journal.pone.0131220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sundekilde U, Downey E, O’Mahony J, O’Shea C-A, Ryan C, Kelly A, Bertram H. 2016. The effect of gestational and lactational age on the human milk metabolome. Nutrients 8:304. doi: 10.3390/nu8050304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McGuire MK, Meehan CL, McGuire MA, Williams JE, Foster J, Sellen DW, Kamau-Mbuthia EW, Kamundia EW, Mbugua S, Moore SE, Prentice AM, Kvist LJ, Otoo GE, Brooker SL, Price WJ, Shafii B, Placek C, Lackey KA, Robertson B, Manzano S, Ruíz L, Rodríguez JM, Pareja RG, Bode L. 2017. What’s normal? Oligosaccharide concentrations and profiles in milk produced by healthy women vary geographically. Am J Clin Nutr 105:1086–1100. doi: 10.3945/ajcn.116.139980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gay M, Koleva P, Slupsky C, Toit E, Eggesbo M, Johnson C, Wegienka G, Shimojo N, Campbell D, Prescott S, Munblit D, Geddes D, Kozyrskyj A, InVIVO LactoActive Study Investigators. 2018. Worldwide variation in human milk metabolome: indicators of breast physiology and maternal lifestyle? Nutrients 10:1151. doi: 10.3390/nu10091151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cabrera-Rubio R, Collado MC, Laitinen K, Salminen S, Isolauri E, Mira A. 2012. The human milk microbiome changes over lactation and is shaped by maternal weight and mode of delivery. Am J Clin Nutr 96:544–551. doi: 10.3945/ajcn.112.037382. [DOI] [PubMed] [Google Scholar]
- 29.Khodayar-Pardo P, Mira-Pascual L, Collado MC, Martínez-Costa C. 2014. Impact of lactation stage, gestational age and mode of delivery on breast milk microbiota. J Perinatol 34:599–605. doi: 10.1038/jp.2014.47. [DOI] [PubMed] [Google Scholar]
- 30.Cabrera-Rubio R, Mira-Pascual L, Mira A, Collado MC. 2016. Impact of mode of delivery on the milk microbiota composition of healthy women. J Dev Orig Health Dis 7:54–60. doi: 10.1017/S2040174415001397. [DOI] [PubMed] [Google Scholar]
- 31.Kumar H, Du Toit E, Kulkarni A, Aakko J, Linderborg KM, Zhang Y, Nicol MP, Isolauri E, Yang B, Collado MC, Salminen S. 2016. Distinct patterns in human milk microbiota and fatty acid profiles across specific geographic locations. Front Microbiol 7:1619. doi: 10.3389/fmicb.2016.01619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li S-W, Watanabe K, Hsu C-C, Chao S-H, Yang Z-H, Lin Y-J, Chen C-C, Cao Y-M, Huang H-C, Chang C-H, Tsai Y-C. 2017. Bacterial composition and diversity in breast milk samples from mothers living in Taiwan and mainland China. Front Microbiol 8:965. doi: 10.3389/fmicb.2017.00965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hoashi M, Meche L, Mahal LK, Bakacs E, Nardella D, Naftolin F, Bar-Yam N, Dominguez-Bello MG. 2016. Human milk bacterial and glycosylation patterns differ by delivery mode. Reprod Sci 23:902–907. doi: 10.1177/1933719115623645. [DOI] [PubMed] [Google Scholar]
- 34.Toscano M, De Grandi R, Peroni DG, Grossi E, Facchin V, Comberiati P, Drago L. 2017. Impact of delivery mode on the colostrum microbiota composition. BMC Microbiol 17:205. doi: 10.1186/s12866-017-1109-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gómez-Gallego C, Morales JM, Monleón D, Du Toit E, Kumar H, Linderborg KM, Zhang Y, Yang B, Isolauri E, Salminen S, Collado MC. 2018. Human breast milk NMR metabolomic profile across specific geographical locations and its association with the milk microbiota. Nutrients 10:E1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fujimura KE, Sitarik AR, Havstad S, Lin DL, Levan S, Fadrosh D, Panzer AR, LaMere B, Rackaityte E, Lukacs NW, Wegienka G, Boushey HA, Ownby DR, Zoratti EM, Levin AM, Johnson CC, Lynch SV. 2016. Neonatal gut microbiota associates with childhood multisensitized atopy and T cell differentiation. Nat Med 22:1187–1191. doi: 10.1038/nm.4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Findley K, Oh J, Yang J, Conlan S, Deming C, Meyer JA, Schoenfeld D, Nomicos E, Park M, Becker J, Benjamin B, Blakesley R, Bouffard G, Brooks S, Coleman H, Dekhtyar M, Gregory M, Guan X, Gupta J, Han J, Hargrove A, Ho S, Johnson T, Legaspi R, Lovett S, Maduro Q, Masiello C, Maskeri B, McDowell J, Montemayor C, Mullikin J, Park M, Riebow N, Schandler K, Schmidt B, Sison C, Stantripop M, Thomas J, Thomas P, Vemulapalli M, Young A, Kong HH, Segre JA. 2013. Topographic diversity of fungal and bacterial communities in human skin. Nature 498:367–370. doi: 10.1038/nature12171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suhr MJ, Banjara N, Hallen-Adams HE. 2016. Sequence-based methods for detecting and evaluating the human gut mycobiome. Lett Appl Microbiol 62:209–215. doi: 10.1111/lam.12539. [DOI] [PubMed] [Google Scholar]
- 39.Gouba N, Raoult D, Drancourt M. 2013. Plant and fungal diversity in gut microbiota as Revealed by molecular and culture investigations. PLoS One 8:e59474. doi: 10.1371/journal.pone.0059474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jo J-H, Deming C, Kennedy EA, Conlan S, Polley EC, Ng W-I, Segre JA, Kong HH, Kong HH. 2016. Diverse human skin fungal communities in children converge in adulthood. J Invest Dermatol 136:2356–2363. doi: 10.1016/j.jid.2016.05.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nash AK, Auchtung TA, Wong MC, Smith DP, Gesell JR, Ross MC, Stewart CJ, Metcalf GA, Muzny DM, Gibbs RA, Ajami NJ, Petrosino JF. 2017. The gut mycobiome of the Human Microbiome Project healthy cohort. Microbiome 5:153. doi: 10.1186/s40168-017-0373-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Strati F, Di Paola M, Stefanini I, Albanese D, Rizzetto L, Lionetti P, Calabrò A, Jousson O, Donati C, Cavalieri D, De Filippo C. 2016. Age and gender affect the composition of fungal population of the human gastrointestinal tract. Front Microbiol 7:1227. doi: 10.3389/fmicb.2016.01227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dupuy AK, David MS, Li L, Heider TN, Peterson JD, Montano EA, Dongari-Bagtzoglou A, Diaz PI, Strausbaugh LD. 2014. Redefining the human oral mycobiome with improved practices in amplicon-based taxonomy: discovery of Malassezia as a prominent commensal. PLoS One 9:e90899. doi: 10.1371/journal.pone.0090899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Drell T, Lillsaar T, Tummeleht L, Simm J, Aaspõllu A, Väin E, Saarma I, Salumets A, Donders GGG, Metsis M. 2013. Characterization of the vaginal micro- and mycobiome in asymptomatic reproductive-age Estonian women. PLoS One 8:e54379. doi: 10.1371/journal.pone.0054379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Clooney AG, Fouhy F, Sleator RD, O’ Driscoll A, Stanton C, Cotter PD, Claesson MJ. 2016. Comparing apples and oranges?: next generation sequencing and its impact on microbiome analysis. PLoS One 11:e0148028. doi: 10.1371/journal.pone.0148028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Allali I, Arnold JW, Roach J, Cadenas MB, Butz N, Hassan HM, Koci M, Ballou A, Mendoza M, Ali R, Azcarate-Peril MA. 2017. A comparison of sequencing platforms and bioinformatics pipelines for compositional analysis of the gut microbiome. BMC Microbiol 17:194. doi: 10.1186/s12866-017-1101-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schubert K, Groenewald JZ, Braun U, Dijksterhuis J, Starink M, Hill CF, Zalar P, de Hoog GS, Crous PW. 2007. Biodiversity in the Cladosporium herbarum complex (Davidiellaceae, Capnodiales), with standardisation of methods for Cladosporium taxonomy and diagnostics. Stud Mycol 58:105–156. doi: 10.3114/sim.2007.58.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chehoud C, Albenberg LG, Judge C, Hoffmann C, Grunberg S, Bittinger K, Baldassano RN, Lewis JD, Bushman FD, Wu GD. 2015. Fungal signature in the gut microbiota of pediatric patients with inflammatory bowel disease. Inflamm Bowel Dis 21:1948–1956. doi: 10.1097/MIB.0000000000000454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ghannoum MA, Mukherjee PK. 2013. The human mycobiome and its impact on health and disease. Curr Fungal Infect Rep 7:345–350. doi: 10.1007/s12281-013-0162-x. [DOI] [Google Scholar]
- 50.Kraneveld EA, Buijs MJ, Bonder MJ, Visser M, Keijser BJF, Crielaard W, Zaura E. 2012. The relation between oral Candida load and bacterial microbiome profiles in Dutch older adults. PLoS One 7:e42770. doi: 10.1371/journal.pone.0042770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stecksén-Blicks C, Granström E, Silfverdal SA, West CE. 2015. Prevalence of oral Candida in the first year of life. Mycoses 58:550–556. doi: 10.1111/myc.12355. [DOI] [PubMed] [Google Scholar]
- 52.Kleinegger CL, Lockhart SR, Vargas K, Soll DR. 1996. Frequency, intensity, species, and strains of oral Candida vary as a function of host age. J Clin Microbiol 34:2246–2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.LaTuga MS, Ellis JC, Cotton CM, Goldberg RN, Wynn JL, Jackson RB, Seed PC. 2011. Beyond bacteria: a study of the enteric microbial consortium in extremely low birth weight infants. PLoS One 6:e27858. doi: 10.1371/journal.pone.0027858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Trama JP, Mordechai E, Adelson ME. 2005. Detection and identification of Candida species associated with Candida vaginitis by real-time PCR and pyrosequencing. Mol Cell Probes 19:145–152. doi: 10.1016/j.mcp.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 55.Ten Oever J, Netea MG. 2014. The bacteriome-mycobiome interaction and antifungal host defense. Eur J Immunol 44:3182–3191. doi: 10.1002/eji.201344405. [DOI] [PubMed] [Google Scholar]
- 56.Sam QH, Chang MW, Chai LYA. 2017. The fungal mycobiome and its interaction with gut bacteria in the host. Int J Mol Sci 18:E330. doi: 10.3390/ijms18020330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xu H, Sobue T, Bertolini M, Thompson A, Dongari-Bagtzoglou A. 2016. Streptococcus oralis and Candida albicans synergistically activate μ-calpain to degrade E-cadherin from oral epithelial junctions. J Infect Dis 214:925–934. doi: 10.1093/infdis/jiw201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Barbosa JO, Rossoni RD, Vilela SFG, de Alvarenga JA, Velloso MDS, Prata MCDA, Jorge AOC, Junqueira JC. 2016. Streptococcus mutans can modulate biofilm formation and attenuate the virulence of Candida albicans. PLoS One 11:e0150457. doi: 10.1371/journal.pone.0150457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Layeghifard M, Hwang DM, Guttman DS. 2017. Disentangling interactions in the microbiome: a network perspective. Trends Microbiol 25:217–228. doi: 10.1016/j.tim.2016.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Drago L, Toscano M, De Grandi R, Grossi E, Padovani EM, Peroni DG. 2017. Microbiota network and mathematic microbe mutualism in colostrum and mature milk collected in two different geographic areas: Italy versus Burundi. ISME J 11:875–884. doi: 10.1038/ismej.2016.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jost T, Lacroix C, Braegger C, Chassard C. 2013. Assessment of bacterial diversity in breast milk using culture-dependent and culture-independent approaches. Br J Nutr 110:1253–1262. doi: 10.1017/S0007114513000597. [DOI] [PubMed] [Google Scholar]
- 62.Sakwinska O, Moine D, Delley M, Combremont S, Rezzonico E, Descombes P, Vinyes-Pares G, Zhang Y, Wang P, Thakkar SK. 2016. Microbiota in breast milk of Chinese lactating mothers. PLoS One 11:e0160856. doi: 10.1371/journal.pone.0160856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ramsay DT, Kent JC, Owens RA, Hartmann PE. 2004. Ultrasound imaging of milk ejection in the breast of lactating women. Pediatrics 113:361–367. doi: 10.1542/peds.113.2.361. [DOI] [PubMed] [Google Scholar]
- 64.Toju H, Tanabe AS, Yamamoto S, Sato H. 2012. High-coverage ITS primers for the DNA-based identification of Ascomycetes and Basidiomycetes in environmental samples. PLoS One 7:e40863. doi: 10.1371/journal.pone.0040863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Magoc T, Salzberg SL. 2011. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27:2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schmieder R, Edwards R. 2011. Quality control and preprocessing of metagenomic datasets. Bioinformatics 27:863–864. doi: 10.1093/bioinformatics/btr026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. 2011. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang Q, Garrity GM, Tiedje JM, Cole JR. 2007. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kõljalg U, Nilsson RH, Abarenkov K, Tedersoo L, Taylor AFS, Bahram M, Bates ST, Bruns TD, Bengtsson-Palme J, Callaghan TM, Douglas B, Drenkhan T, Eberhardt U, Dueñas M, Grebenc T, Griffith GW, Hartmann M, Kirk PM, Kohout P, Larsson E, Lindahl BD, Lücking R, Martín MP, Matheny PB, Nguyen NH, Niskanen T, Oja J, Peay KG, Peintner U, Peterson M, Põldmaa K, Saag L, Saar I, Schüßler A, Scott JA, Senés C, Smith ME, Suija A, Taylor DL, Telleria MT, Weiss M, Larsson K-H. 2013. Towards a unified paradigm for sequence-based identification of fungi. Mol Ecol 22:5271–5277. doi: 10.1111/mec.12481. [DOI] [PubMed] [Google Scholar]
- 70.Li W, Godzik A. 2006. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 71.R Development Core Team. 2011. A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: http://www.R-project.org. [Google Scholar]
- 72.Zakrzewski M, Proietti C, Ellis JJ, Hasan S, Brion M-J, Berger B, Krause L. 2016. Calypso: a user-friendly web-server for mining and visualizing microbiome–environment interactions. Bioinformatics 33:btw725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C. 2011. Metagenomic biomarker discovery and explanation. Genome Biol 12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Morgan XC, Tickle TL, Sokol H, Gevers D, Devaney KL, Ward DV, Reyes JA, Shah SA, LeLeiko N, Snapper SB, Bousvaros A, Korzenik J, Sands BE, Xavier RJ, Huttenhower C. 2012. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol 13:R79. doi: 10.1186/gb-2012-13-9-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cole JR, Wang Q, Fish JA, Chai B, McGarrell DM, Sun Y, Brown CT, Porras-Alfaro A, Kuske CR, Tiedje JM. 2014. Ribosomal Database Project: data and tools for high throughput rRNA analysis. Nucleic Acids Res 42:D633–D642. doi: 10.1093/nar/gkt1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Albanese D, Riccadonna S, Donati C, Franceschi P. 2017. A practical tool for maximal information coefficient analysis. https://github.com/minepy/mictools. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol 61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Stöver BC, Müller KF. 2010. TreeGraph 2: Combining and visualizing evidence from different phylogenetic analyses. BMC Bioinformatics 11:7. doi: 10.1186/1471-2105-11-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.