The poultry industry is faced with numerous challenges associated with infectious diseases and suboptimal performance of flocks. As microbiome research continues to grow, it is becoming clear that poultry health and production performance are partly influenced by nonpathogenic symbionts that occupy different habitats within the bird. This study has defined the baseline composition and overlaps between respiratory and gut bacteria in healthy, optimally performing chicken layers across all stages of the commercial farm sequence. Consequently, the study has set the groundwork for the development of interventions that seek to enhance production performance and to prevent and control infectious diseases through the modulation of gut and respiratory bacteria.
KEYWORDS: baseline microbiota, chicken gut microbiota, chicken respiratory microbiota, commercial chicken layers, microbiome
ABSTRACT
The digestive and respiratory tracts of chickens are colonized by bacteria that are believed to play important roles in the overall health and performance of the birds. Most of the current research on the commensal bacteria (microbiota) of chickens has focused on broilers and gut microbiota, and less attention has been given to layers and respiratory microbiota. This research bias has left significant gaps in our knowledge of the layer microbiome. This study was conducted to define the core microbiota colonizing the upper respiratory tract (URT) and lower intestinal tract (LIT) in commercial layers under field conditions. One hundred eighty-one chickens were sampled from a flock of >80,000 birds at nine times to collect samples for 16S rRNA gene-based bacterial metabarcoding. Generally, the body site and age/farm stage had very dominant effects on the quantity, taxonomic composition, and dynamics of core bacteria. Remarkably, ileal and URT microbiota were compositionally more related to each other than to that from the cecum. Unique taxa dominated in each body site yet some taxa overlapped between URT and LIT sites, demonstrating a common core. The overlapping bacteria also contained various levels of several genera with well-recognized avian pathogens. Our findings suggest that significant interaction exists between gut and respiratory microbiota, including potential pathogens, in all stages of the farm sequence. The baseline data generated in this study can be useful for the development of effective microbiome-based interventions to enhance production performance and to prevent and control disease in commercial chicken layers.
IMPORTANCE The poultry industry is faced with numerous challenges associated with infectious diseases and suboptimal performance of flocks. As microbiome research continues to grow, it is becoming clear that poultry health and production performance are partly influenced by nonpathogenic symbionts that occupy different habitats within the bird. This study has defined the baseline composition and overlaps between respiratory and gut bacteria in healthy, optimally performing chicken layers across all stages of the commercial farm sequence. Consequently, the study has set the groundwork for the development of interventions that seek to enhance production performance and to prevent and control infectious diseases through the modulation of gut and respiratory bacteria.
INTRODUCTION
The digestive and respiratory tracts of chickens are colonized by complex microbial communities, which are believed to play important roles in the overall health and performance of the birds (1–6). The bacterial composition of the chicken gut has been extensively studied, mainly because of its association with performance (6–10). Studies of commensal and pathogenic bacteria in the chicken gut have mainly focused on broilers (7, 11–14), likely due to their short life cycle and direct economic importance. Within the gut, the cecum has attracted the most attention because of its high microbial densities and health-related functions, such as urea recycling, carbohydrate fermentation, and water regulation (15). Most studies of the chicken gut microbiota have focused on one segment at a time; fewer studies have compared multiple segments from the same set of birds (15–18). On this basis, we previously undertook a large comprehensive study of 37 different commercial flocks of Cobb 500 broilers within a vertically integrated broiler system and defined the core microbiota in the ileum and cecum using 16S rRNA gene amplicon sequencing (7). However, broilers and layers are different in their compositional dynamics of microbiota, mainly due to differences in the length of life cycles, flock management protocols such as caged housing, and sex of birds. With that said, few studies of the layer’s gut have been published (14, 19–22), the majority of which explored the cecum of young layers aged 0 to 8 weeks under experimental conditions. The microbiota of young layers are likely not reflective of what is found in older birds. For example, the microbiota of older layers are constantly exposed to numerous stressors associated with management and physiological changes as the birds mature and enter the laying period. Additionally, data are not available for other gut segments such as the ileum, which harbors microbiota that can predict production performance, as we demonstrated in broilers (7).
Unlike the gut microbiota, the respiratory microbiota of both broilers and layers are understudied (7, 23–25). The respiratory microbiota of layers have been reported in only two studies, which showed variable results, possibly due to the effects of inadequate sample sizes and sampling time points, differences in sampling sites and techniques, layer breeds, and the use of management systems which are not widely applied in commercial settings (23, 25). In broilers, a 16S rRNA-based microbiota analysis revealed wide differences between tracheal and cecal microbiota in 42-day-old birds (24). In a previous paper, we defined the broiler trachea microbiota and found relationships to microbiota in the gut (cecum and ileum) and environment (litter) over several life cycles (7). While bacterial microbiota in the respiratory and gut ecosystems differed substantially in composition and dynamics across age groups, remarkable overlaps in taxonomic memberships were observed between these ecosystems (7). Some of the overlapping taxa were associated with depressed weight gain and have been consequently considered to be potential pathogens (7). Such comprehensive comparisons of gut and respiratory microbiota are lacking for layer chickens.
The current study was performed to comprehensively define the baseline upper respiratory tract (URT) and lower intestinal tract (LIT) bacterial microbiota in Hy-Line W-36 layers maintained in commercial settings under a standard management protocol (26). Potential interactions between URT and LIT microbiota were investigated throughout the lifetime of these layers, focusing on taxonomic compositions, dynamics of core bacteria as the chicken ages, and distribution and persistence of genera with known avian pathogens.
RESULTS
Flock performance.
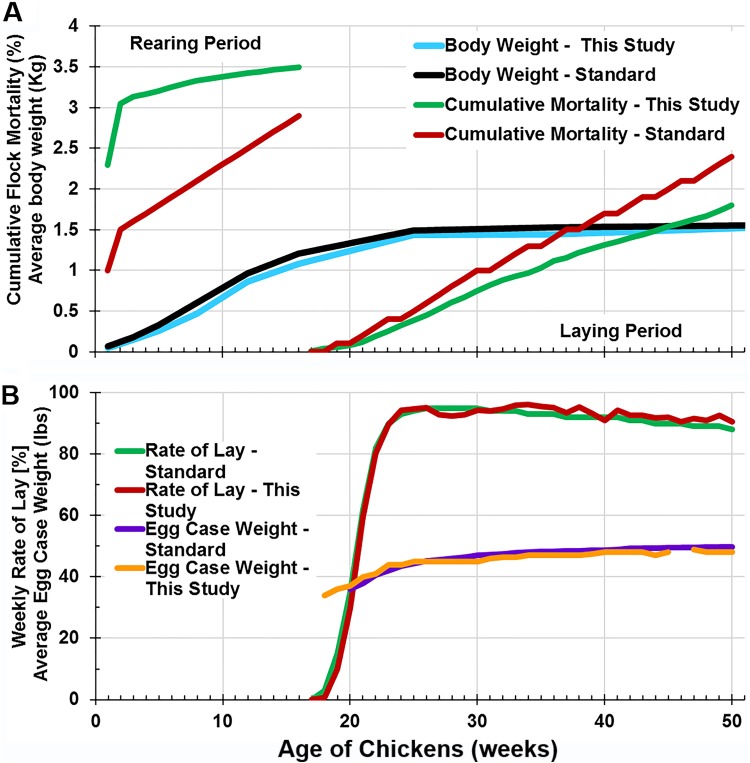
During acclimation at the first 2 weeks of the rearing period, flock mortality was higher than the average expectation for Hy-Line W-36 chickens (Fig. 1A). Thereafter, the rate of cumulative mortality was slightly lower than expected for the rest of the rearing period and during the laying period (Fig. 1A). Flock performance was also monitored in terms of weekly rate of lay and average egg case weight, and no major differences were observed between data generated in this study and the expected performance based on Hy-Line W-36 genetic potential (Fig. 1B). In addition, the average body weights of sampled birds were similar to the expected values throughout the study (Fig. 1A).
FIG 1.
Comparison of observed flock mortality and egg production with the expected performance based on the genetic potential of Hy-Line W-36 layers. (A) Cumulative flock mortality and average body weights (of sampled birds) during the rearing and laying periods. (B) Weekly rate of egg lay and egg case weights.
URT and LIT microbial communities are distinct, yet URT and ileum communities have some compositional similarities.
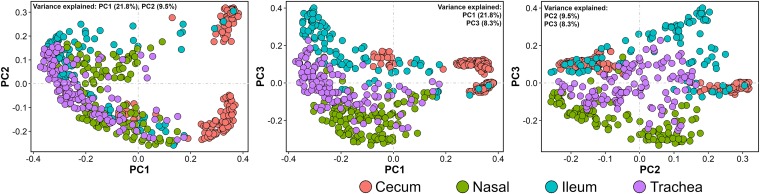
The compositions of URT (trachea and nasal cavity) and LIT (cecum and ileum) bacterial communities were explored through principal-coordinate (PCO) plots constructed using UniFrac distance metrics. As seen through three main planes of the microbial community space, the samples tended to cluster by the body site they were derived from (Fig. 2; see also Fig. S1 in the supplemental material). The analysis of similarity (ANOSIM) statistical test was performed using weighted UniFrac distance metrics to determine the extent of ecological differences observed on PCO plots. ANOSIM generates R values, which show the levels of variation between the communities being compared, with values closer to zero indicating no variation between communities and those close to +1 indicating high variation (27). Not surprisingly, and in accordance with the PCO plots (Fig. 2 and S1), microbiota from each body site were found to be compositionally distinct from those found in other body sites (ANOSIM, P < 0.001, R = 0.696). Variation was highest between cecum and nasal cavity (R = 0.877) and lowest between ileum and trachea (R = 0.400). Nevertheless, communities were less variable between respiratory sites (trachea versus nasal cavity, R = 0.522) than between gut sites (cecum versus ileum, R = 0.730). Furthermore, the community in the ileum was compositionally closer to URT communities (R = 0.400 for ileum versus trachea and R = 0.669 for ileum versus nasal cavity) than to the cecal community (R = 0.730 for ileum versus cecum).
FIG 2.
Relationship between bacterial communities sampled from respiratory and gut sites. Principal coordinate (PCO) plots show sample distribution in microbial community space. Plots were drawn using an unweighted UniFrac distance matrix generated between samples with OTU communities rarefied to a depth of 5,000 sequences per sample. The percent variances explained by each principal coordinate are shown inside the plots. Nasal, nasal cavity.
Bacterial community compositions are affected by age.
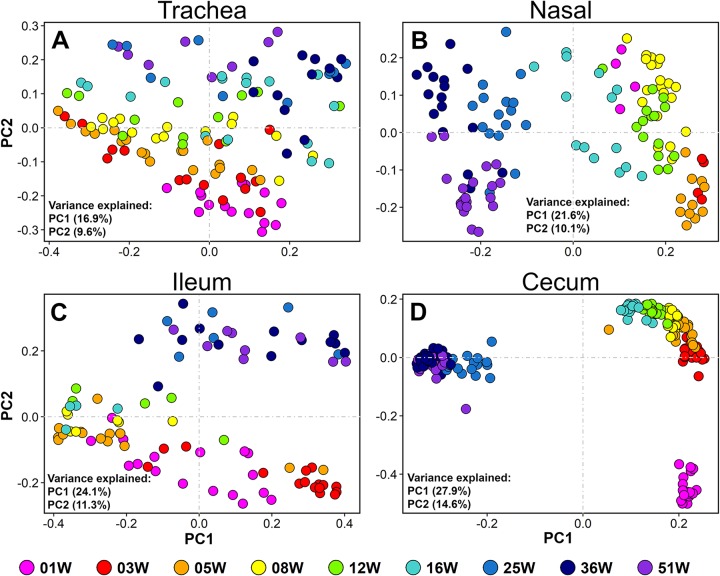
Patterns of bacterial community composition in each body site were examined by performing beta diversity analyses using bird age as the exploratory variable. Body site-specific PCO plots demonstrated that bird age had a tremendous influence on bacterial beta diversity (Fig. 3 and S2). Statistical comparisons using ANOSIM revealed that the effects of age were more prominent in the cecum (P = 0.0001, R = 0.834) and nasal cavity (P = 0.001, R = 0.580) and less prominent in ileum (P = 0.001, R = 0.363) and trachea (P = 0.001, R = 0.348). The tracheal community composition shifted very gradually as the chicken aged, without clear separation between brooding (0 to 5 weeks), growing (6 to 16 weeks), and laying (17 weeks onward) stages of the farm sequence (Fig. 3A and S2A). Nasal and ileal communities shifted gradually during the brooding and grow-out stages but more drastically after the birds transitioned to the laying stage (16 to 25 weeks) (Fig. 3B and C and S2B and C). Age had a dramatic effect on the cecal communities, in which samples formed three distinct clusters. The cecal community shifted drastically after the first week of life (1 to 3 weeks) and after transitioning to the laying stage (16 to 25 weeks) (Fig. 3D and S2D).
FIG 3.
Shifts of bacterial community structure with age. (A to D) Principal coordinate (PCO) plots. All four plots were drawn from the same unweighted UniFrac distance matrix. Nasal, nasal cavity.
URT and LIT possess a putative core microbiota.
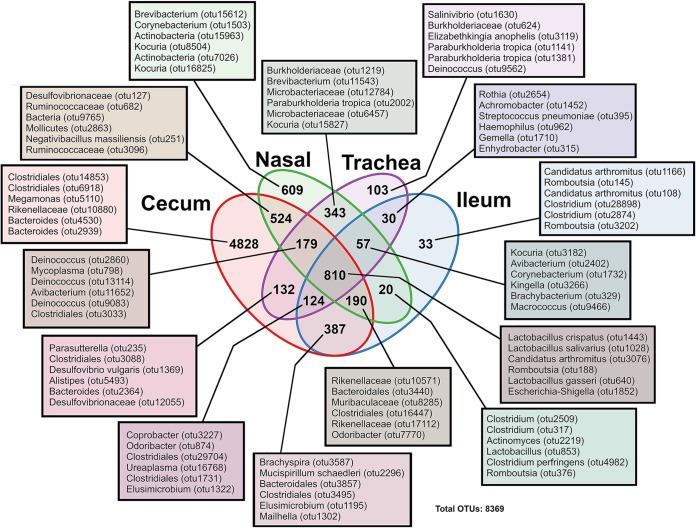
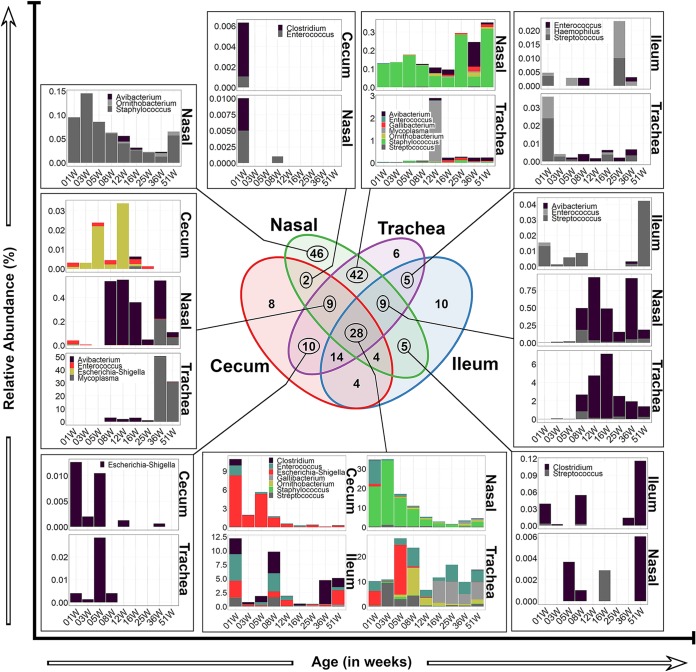
A total of 8,369 operational taxonomic units (OTUs) were clustered at 97% identity with the Uclust algorithm implemented in QIIME (28). Using the SILVA database (release 132 for QIIME), these OTUs were classified to all seven levels where possible. Since 16S rRNA gene-based classification below the genus level is generally considered not reliable, all OTUs referred to by species are named as approximations only. A Venn diagram was constructed to reveal bacterial OTUs that were unique or shared (core) between different body sites. Of 8,369 OTUs, 66.6% (5,573/8,369) were not present in more than one anatomical site (Fig. 4). These unique OTUs were distributed as follows: 86.6% (4,828/5,573) in cecum, 10.9% (609/5,573) in the nasal cavity, 1.8% (103/5,573) in the trachea, and 0.06% (33/5,573) in the ileum. The remaining OTUs (2,796) formed a putative core that was shared variably between sets of body sites. Of the 2,796 shared OTUs, 343 (12.2%) were uniquely shared between the URT sites (trachea versus nasal cavity), with the top 6 most abundant OTUs representing 3 orders (Burkholderiales, Enterobacterales, and Actinomycetales). The LIT had 387 OTUs (>13.8%) that were uniquely shared between cecum and ileum, with the top 6 most abundant OTUs being derived from 6 orders (Spirochaetales, Deferribacterales, Bacteroidales, Clostridiales, Elusimicrobiales, and Desulfovibrionales). Nonetheless, the majority of the shared OTUs (28.9%) were present in all body sites and were dominated by members belonging to 3 orders (Lactobacillales, Clostridiales, and Enterobacterales) (Fig. 4; see also Data Set S1).
FIG 4.
Venn diagram depicting the number of OTUs that were shared between body sites or unique to a given body site. The OTUs listed are the top 6 most abundant OTUs in that group; full lists of OTUs are provided in Data Set S1 in the supplemental material. The taxonomy given is of the lowest level assigned by SILVA database. Nasal, nasal cavity.
Dynamics of core OTUs across age groups vary depending on the body site.
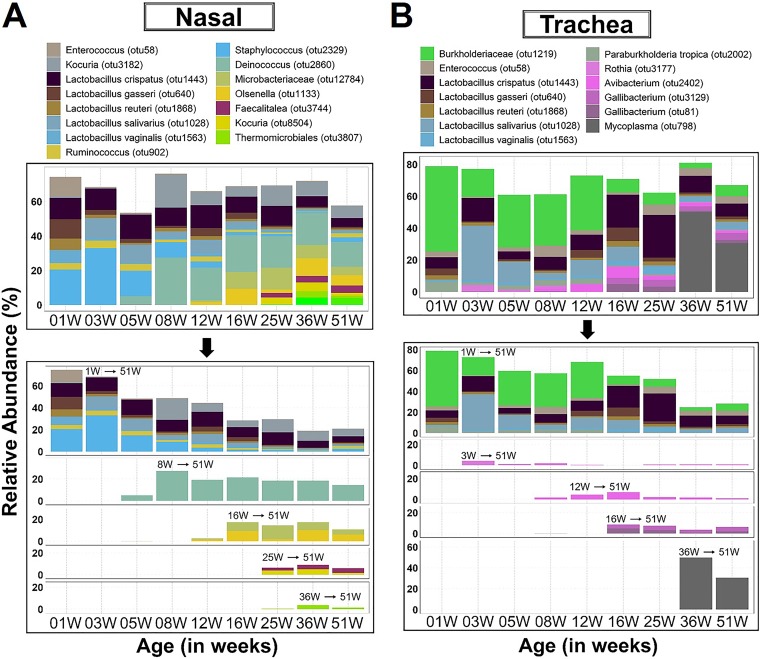
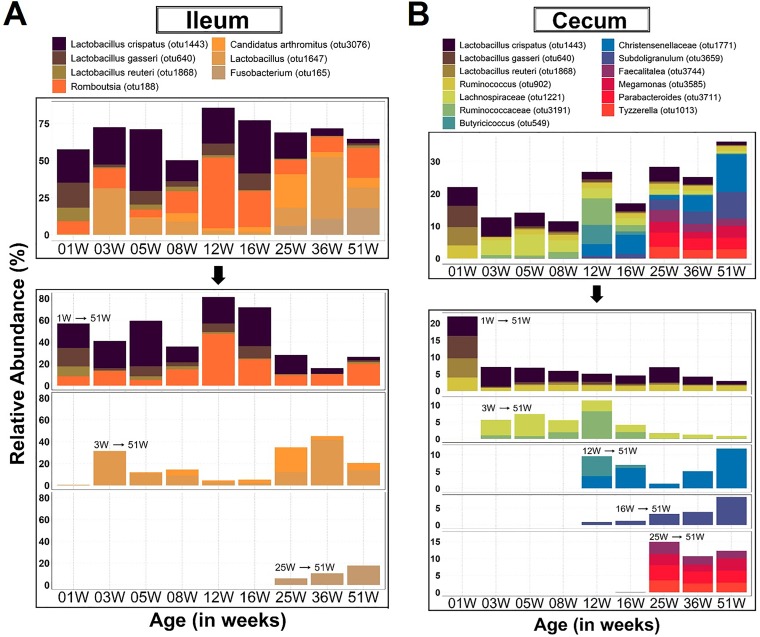
Core microbiota in a body site were defined as those bacteria that occurred in ≥75% of the birds sampled from a given age group and had a mean relative abundance of ≥3.5% in at least one age group during the study timeline. These parameters are within the range of values used to validly define core in other studies (29) and further reduce the core to a manageable few prevalent OTUs. As such, in most body sites, the core microbiota occupied a majority of the total available niche space. The core microbiota observed during the first week of life consistently occupied more than 50% of the total niche space in the ileum, nasal cavity, and trachea but barely surpassed 30% of the space in the cecum (Fig. 5 and 6). After the initial colonization of the core in the cecum, the relative abundance of those bacteria dropped off substantially and continued to decline gradually with age while still maintaining a substantial presence. In contrast, the initial colonizers of the ileum, nasal cavity, and trachea maintained a substantial presence for the duration of the brood and grow-out periods, but many noticeably declined during the laying period (25 weeks and older) (Fig. 5 and 6).
FIG 5.
Dynamics of core respiratory microbiota across age groups. OTUs were identified as core based on ≥75% occurrence in chickens sampled within a given age, 100% occurrence across all subsequent ages, and at least 3.5% relative abundance in one of the ages. 1W→51W, etc. within the chart indicates the time point at which an OTU emerged as core and its persistence over time. Note that some OTUs emerged earlier and persisted below core abundance levels but became core at the indicated time points. Nasal, nasal cavity.
FIG 6.
Dynamics of core gut microbiota across age groups. OTUs were identified as core based on ≥75% occurrence in chickens sampled within a given age, 100% occurrence across all subsequent ages, and at least 3.5% relative abundance in one of the ages. 1W→51W, etc. within the chart indicates the time point at which an OTU emerged as core and its persistence over time. Note that some OTUs emerged earlier and persisted below core abundance levels but became core at the indicated time points.
OTUs representing three species of Lactobacillus (L. reuteri, L. gasseri, and L. crispatus) were initial colonizers commonly found in URT and LIT cores for the entire duration of the study. No other core OTUs were shared between the LIT sites. Ruminococcus and Faecalitalea were common to the cores of the nasal cavity and cecum during the rearing (brood and grow-out) and layer stages, respectively (Fig. 5A and 6B). Lactobacillus vaginalis, Lactobacillus salivarius, and Enterococcus were shared between the cores of the URT (Fig. 5A and B).
Except for Faecalitalea in nasal cavity and cecum, no other overlap of late-emerging core OTUs was observed between the body sites. The late colonizers (OTUs that became core after the first week of life) were distributed as follows: Deinococcus, Olsenella, Microbacteriaceae, Faecalitalea, Thermomicrobiales, and Bacteroides caecigallinarum in nasal cavity; Rothia, Avibacterium, Gallibacterium, and Mycoplasma in trachea; Lactobacillus aviarius, “Candidatus Arthromitus,” and Fusobacterium in ileum; and Ruminococcaceae, Christensenellaceae, Butyricicoccus, Subdoligranulum, Tyzzerella, Lachnospiraceae, Parabacteroides, Megamonas, and Faecalitalea in cecum. The dynamics and relative abundances of the emerging late core OTUs differed depending on the body site. However, the general trend was a decline of the total abundance of the initial core over time, but the total abundance was restored by the newly emerging core OTUs (Fig. 5 and 6). Interestingly, three genera with potential to cause disease in chickens (Avibacterium, Gallibacterium, and Mycoplasma) were identified as late core OTUs in trachea. Especially interesting is Mycoplasma, which emerged in the middle of the laying stage and persisted in a high relative abundance until the end of study timeline (Fig. 5 and 6).
Ecological relationships between body sites were also visualized further through a heat map of all core OTUs. The OTU phylotypes clustered as expected based on known genetic relationships between the represented bacteria. However, major ecological relationships were revealed by the hierarchical clustering of the samples in that the respiratory sites were more related to each other than to either of the gut sites. Yet, the cecum was compositionally distinct from the other three body sites. Subtle ecological relationships were observed in each body site with regard to the three stages of the layer farm sequence, but the samples smoothly grouped according to consecutive age groups (see Fig. S3).
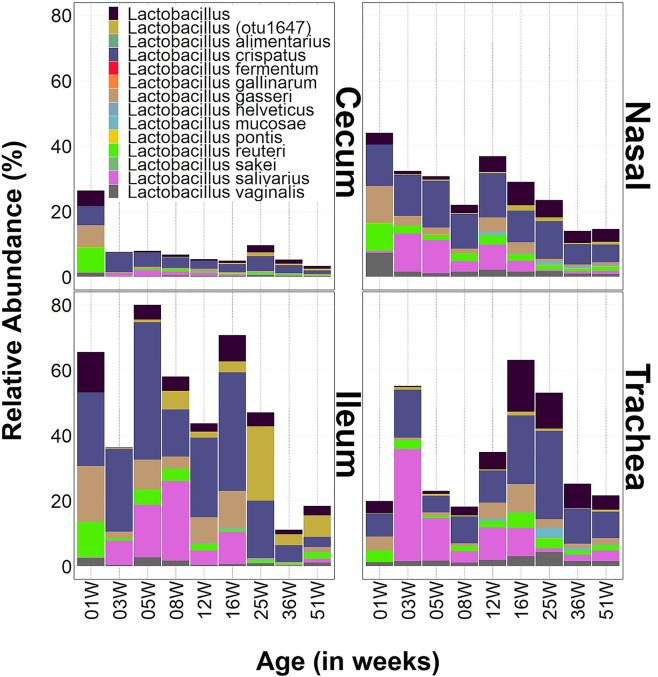
Lactobacillus was ubiquitously present in all body sites, but it occupied significant proportions of the niche spaces in the ileum, nasal cavity, and trachea.
As mentioned previously, the genus Lactobacillus was commonly found in all body sites sampled in this study. Of those OTUs classified to species level by the SILVA database, five species appear to maintain an identifiable presence in each of the sites: L. crispatus, L. gasseri, L. reuteri, L. salivarius, and L. vaginalis. Of these, L. salivarius emerged to prominence after the initial colonization (week 3) and was common in the gut sites until week 25 and common in the respiratory sites through week 51 (although at a reduced abundance after week 16) (Fig. 7). Lactobacillus (otu1647) was also included for comparison, given its prominent presence in the ileal core. This OTU was identified by the NCBI 16S rRNA sequence database as Lactobacillus aviarius (Fig. 7), which we previously found to exist at high levels in broiler ileum (7).
FIG 7.
Ecology of Lactobacillus in URT and LIT sites. Bar charts show changes in relative abundances of genus-level bacterial taxa (vertical axis) with chicken layer age (horizontal axis). 01W, 03W, etc. indicate ages of birds in weeks. Nasal, nasal cavity.
Many of the other OTUs identified to the species level were shared among the body sites but at a very low relative abundance. Lactobacillus mucosae was one such species, which had OTUs shared among the four body sites, but its distribution among the sites was sporadic. It was prominent only in the respiratory system at weeks 12, 25, and 36. Another species was Lactobacillus pontis, which was more commonly distributed in the nasal cavity than in other sites. Lactobacillus sakei and Lactobacillus fermentum were composed of one OTU each, and both were found ubiquitously (Fig. 7).
The presence of lactobacilli in the cecum was suppressed after the initial colonization during the first week of life from approximately 25% to below 10% for the remainder of study timeline. In contrast, lactobacilli variably occupied significant proportions of the niche spaces in the ileum, nasal cavity, and trachea. In the ileum and nasal cavity, these bacteria variably occupied 30% to 80% and 20% to 40% of the total niche space, respectively, until week 36, when their levels dropped. In the trachea, lactobacilli consistently occupied approximately 20% of total niche space except at 3, 12, 16, and 25 weeks, where their levels jumped to approximately 60% (Fig. 7).
Genera with known poultry pathogens were shared between URT and LIT ecosystems.
Bacteria associated with several poultry diseases belong to several genera, including, but not limited to, Clostridium (30), Staphylococcus, Streptococcus, Enterococcus (31), Avibacterium, Haemophilus, Gallibacterium, Ornithobacterium (32, 33), Mycoplasma (34), and Escherichia-Shigella (35). Figure 8 and S4 illustrate that OTUs belonging to these genera were shared between the URT and LIT sites and display their dynamics across age groups. The 28 OTUs shared among the four sites were subsumed in seven genera: Clostridium, Enterococcus, Escherichia-Shigella, Gallibacterium, Ornithobacterium, Staphylococcus, and Streptococcus. They accounted for large proportions of the niche spaces available in those sites (up to 11% in cecum, 35% in nasal cavity, 12% in ileum, and 25% in trachea). In the cecum and nasal cavity, the initial proportions of these OTUs were high but declined with age. However, their proportions in the ileum and trachea were much more variable throughout life. Escherichia-Shigella OTUs dominated in the cecum, while Staphylococcus OTUs were dominant in the nasal cavity. In the ileum, the dominant OTUs were split between Clostridium, Enterococcus, Escherichia-Shigella, and Streptococcus at various points of life. In the trachea, the relative abundances of Enterococcus, Escherichia-Shigella, Streptococcus, Ornithobacterium, and Gallibacterium moved to prominence during different points in life (Fig. 8).
FIG 8.
Age- and farm stage-related changes in the compositions of ten genera with known pathogens of poultry. Bar charts show changes in relative abundances of genus-level bacterial taxa (vertical axis) with chicken layer age (horizontal axis). 01W, 03W, etc. indicate ages of birds in weeks. The missing bar charts are shown in Fig. S4 in the supplemental material. Nasal, nasal cavity.
Other OTUs were variously shared among the body sites in different combinations. Examples include some Ornithobacterium OTUs that were either unique to nasal cavity, shared between nasal cavity and trachea, or shared among all four sites. Clostridium OTUs were common in LIT sites and nasal cavity but not shared between the LIT and the trachea unless they were shared between all four sites. One Mycoplasma OTU identified as Mycoplasma gallisepticum by blasting the NCBI 16S database was shared between nasal cavity and trachea but was most common in the trachea at 12 weeks. The other Mycoplasma OTU (identified as M. synoviae by the NCBI 16S database) was shared between the nasal cavity and trachea in weeks 36 and 51 but was also present in the cecum at week 16. Enterococcus OTUs were shared variably between gut and respiratory sites. Streptococcus OTUs were shared variably between the ileum and respiratory sites, whereas only OTUs shared among all four sites were ever found in the cecum. Staphylococcus OTUs were found and shared between the respiratory sites except those that were shared among the four sites. Some Escherichia-Shigella OTUs were shared uniquely between the cecum and URT sites, while others were present in all four sites. For Avibacterium-Haemophilus OTUs to be found in LIT sites, they had to be present in the nasal cavity or trachea (Fig. 8).
Four bacterial poultry pathogens were reliably identified to the species level using additional methods. The presence of Mycoplasma synoviae was confirmed by searching the NCBI 16S rRNA database (confirmed at 100% identity) and serological evidence of IgG antibodies (see Fig. S5). The other 3 pathogens were confirmed through species-specific PCR and Sanger sequencing of the 16S rRNA gene and cpa5 gene fragments for Ornithobacterium rhinotracheale and Clostridium perfringens, respectively (36, 37) and iroN, hylF, ompT, iss, and iutA genes for avian pathogenic Escherichia coli (38). In addition to bacterial pathogens, antibody evidence indicated that the flock was exposed to avian reovirus (Fig. S5). Although the pathogenicity of the reovirus indicated by these antibodies was not determined, reoviruses of unknown pathogenicity are detected in almost all commercial poultry farms (39, 40).
DISCUSSION
The bacterial microbiota of the chicken gut has been studied extensively, mainly because of its association with weight gain in broilers (7, 8, 10, 41, 42) and egg production in layers (2–4, 43). However, chicken respiratory bacterial microbiota studies are scarce (7, 23–25), and chicken layers have been reported in only two studies (23, 25). The two studies on layers have provided much-needed data on respiratory microbiota, but they were limited in terms of sample sizes, sampling time points, and use of management systems that are not widely applied in commercial settings (23, 25). In the present study, we report the dynamics of the URT microbiota of healthy layers raised in commercial farm settings according to a standard management protocol (26). Furthermore, a side-by-side comparison of the URT and LIT microbiota demonstrates that the developing URT and LIT microbiota are distinct, yet they have an overlapping core that changes over time. This strongly indicates the existence of microbiota interactions between the gut and respiratory systems, possibly through aerosolization of fecal bacteria. Remarkably, microbiota in the ileum and URT were compositionally more related to each other than to that in the cecum, suggesting the existence of some environmental similarities between ileum and respiratory ecosystems, despite anatomical and physiological differences.
The body site and bird’s age play prominent roles in the development of bacterial microbiota in different breeds of layer and broiler chickens (7, 19, 23). It is therefore not surprising that a substantial amount of variation in microbially diverse populations across samples was attributable to body site and bird’s age (Fig. 2 and 3; see also Fig. S1 and S2 in the supplemental material). Interestingly, the layer stage separated noticeably from the previous stages in all four sites, more so in cecum than in other body sites (Fig. 3 and S2). Factors associated with flock transfer to the laying house at 16 weeks of age and nutritional change from developer to layer diets (26) are deemed likely to have contributed to the remarkable shift in bacterial beta diversity observed in the laying stage. The rapid community change could also be due to BMD50 (bacitracin methylene disalicylate) antibiotic supplementation during the first 4 weeks of the laying stage (17 to 22 weeks of age) to prevent necrotic enteritis caused or complicated by Gram-positive bacteria such as Clostridium spp. (see Fig. S6). It is unclear which of the three factors (new environment, nutritional change, or antibiotic treatment) had the greatest systemic effect on core composition. Nonetheless, pairwise comparison of bacterial community variation between different body sites indicated that, while URT communities were distantly related to the cecal community, they were relatively close to the ileal community. Interestingly, the least variation was observed between trachea and ileal communities (ANOSIM, R = 0.4) despite the anatomical disparities between these body sites. The ileum thus appears to have played a pivotal role in the development of microbiota communities in both URT and LIT ecosystems and could be a target for microbiota-based performance enhancement intervention strategies to modulate microbiota in both URT and LIT.
A side-by-side comparison between OTUs inhabiting the URT and LIT ecosystems has allowed the core microbiota to be defined based on their overlaps between body sites, emergence and persistence as the chicken aged and passed through the farm sequence stages, relative abundance in individual birds, and distribution between birds. Using this definition, we have illuminated previously unappreciated striking similarities between URT and ileal cores with regard to the amount of the ecological niche they occupied (≥50% at all stages) and persistence over the layer’s lifetime (Fig. 5 and 6A). The URT and ileal core bacteria acquired during the first week of age were present at high levels (>20%) during the rearing period (brood and grow-out stages) and persisted at detectable levels during the laying stage (Fig. 5 and 6A). The effects of the initial colonizers on the production performance of layers could not be inferred from this study. However, several broiler studies have shown that bacteria acquired immediately after hatch are crucial for optimal performance (21, 44–47). Such studies are yet to be performed in layer breeds, although the modulation of gut microbiota with direct fed microbials (probiotics) in older birds (7 to 59 weeks) was associated with improved egg quality and laying performance (2–5).
Host-related factors such as age, sex, and breed as well as environmental factors such as biosecurity level, housing, litter, feed access, and climate are implicated in the establishment of chicken microbiota after hatching (19). Therefore, the core microbiota reported here for Hy-Line W-36 layers may not generally apply to other layer breeds or management settings. Glendinning et al. (23) have recently reported the URT microbiota of Novogen Brown layers sampled at 2 days, 3 weeks, and 30 months of age in the United Kingdom. Nasal and buccal bacterial communities commonly contained high levels of Staphylococcus, Enterobacteriaceae, unclassified Lactobacillus, and Lactobacillus reuteri (23). Other dominant URT microbiota included Faecalibacterium prausnitzii and Staphylococcus equorum in the nasal cavity, and L. vaginalis and L. salivarius in the buccal cavity (23). A somewhat similar set of taxa were observed in the Hy-Line W-36 layer URT cores (Fig. 5), with the exception of Enterobacteriaceae and Faecalibacterium, which existed at levels below the core microbiota threshold defined in this study. However, high levels of previously unreported taxa such as Deinococcus and Burkholderiaceae were present in the Hy-Line W-36 URT cores (Fig. 5), which may reflect the effects of layer breed, flock management, or sampling techniques between the two studies. The subtle variations between the study on Novogen Brown layers and the data presented here underscore the need to provide comprehensive metadata, including management practice, sampling, and analysis techniques to enable reasonable evaluation of the effects of core bacterial taxa on the performance of different layer breeds.
The cecal microbiota characterized in different breeds of layers under different management conditions are similar at higher levels of taxonomic classification (family, order, and phylum) (19–22). The high-resolution core cecal microbiota data presented here forms an excellent basis for comparison with future studies, especially for Hy-Line W-36 layers. Our efforts to find ileal microbiota reports on chicken layers were not successful, which led us to believe that these microbiota had not been characterized before the present study was conducted. The ileum core occupied a larger ecological niche space (≥50% at all stages) than the cecum core (∼10% to 35%). However, the ileal core had comparatively fewer members (Fig. 6). We speculate that the ileal core is better at competitive exclusion of other bacteria, including potential pathogens, than the cecal core. In addition, the presence of three species of Lactobacillus (L. reuteri, L. gasseri, and L. crispatus) in the cores of all four sites studied emphasizes the importance of taking a more holistic approach in studying poultry microbiota.
A notable result from this study was that the ecologies of Lactobacillus were more similar between ileum and the URT than between cecum and other body sites. The effects of Lactobacillus and other microbiota could not be deduced, because body weights were very even between birds of the same age and eggs laid by individual birds were not tracked due to the placement of multiple birds in each cage. With that said, we and others have previously found consistent negative correlations between genus Lactobacillus abundance and Cobb 500 broiler performance (7, 48). In Dekalb XL single comb white leghorn layers, directly fed Lactobacillus (unspecified species) was reported to improve the feed conversion of growing birds (7 to 19 weeks old) and, paradoxically, worsen the feed conversion of laying birds (20 to 59 weeks old) (2). Further investigation is needed to determine whether these negative associations are causal or coincidental. Nonetheless, performance-enhancing benefits of several Lactobacillus species have been demonstrated in chickens (49–52). Notably, species such as L. crispatus and L. salivarius, which had concordant temporal dynamics and abundance in the URT and ileum (Fig. 5 and 6), can competitively exclude the colonization of some intestinal pathogens (51, 53, 54).
Interactions between URT and LIT microbiota were evident because of the several overlapping bacterial taxa observed at core or subcore levels. Such interactions can promote the enrichment and transfer of pathogenic bacteria between the URT and LIT as indicated by the cooccurrence of several putative pathogenic genera in these body sites. For examples, among the potential pathogens commonly found in URT and LIT ecosystems, Avibacterium, Ornithobacterium, and Mycoplasma were dominant in URT, and Staphylococcus was dominant in the nasal cavity, while Clostridium and Escherichia-Shigella were mainly found in the gut. M. synoviae, O. rhinotracheale, Clostridium perfringens, and avian pathogenic E. coli were identified to species thorough Sanger sequencing of non-16S marker genes and/or serologically. In addition, Campylobacter coli, a known human pathogen (55), was identified from cecal samples by sequencing a fragment of the ceuE gene (56). It was not possible to determine which OTUs corresponded with these pathogens, because numerous OTUs identified to the same genus by the SILVA database were often present in the same samples and those present rarely found matches of >99% identity within the NCBI 16S database. The persistence of bacterial pathogens at subclinical levels is common in commercial chicken flocks (7, 15, 24, 57); this deserves further study, since these pathogens have the potential to exacerbate the effects of superinfecting transkingdom pathogens such as respiratory and enteric viruses (58–60). Furthermore, nonbacterial domains (archaea, protists, fungi, and viruses) should be considered if the roles of microbiota in chicken health and performance were to be fully understood.
This study reveals the comparative dynamics of URT and LIT microbiota of chicken layers during the rearing (brooding and grow-out) and laying phases in commercial farm settings. Comprehensive metadata were collected throughout the study to check the immune status, clinical health, and production performance of the birds. Still, several inherent limitations of uncontrolled field conditions made it impossible to make meaningful correlations between production performance and antibody titers induced by vaccination or reovirus infection. Likewise, the effects of live vaccines on microbiota were confounded by overlapping vaccination timelines and could not be elucidated in this study. However, under experimental conditions, live vaccines have been associated with significant changes in microbiota compositions (61, 62). Such changes may have long-term effects on immunity to unrelated pathogens and production performance (6, 63). Understanding how live vaccines affect microbiota, egg production, and susceptibility to nontarget pathogens will be an important step toward improving management practices and maximizing productivity. Other limitations that need to be addressed in future studies include, but are not limited to, a side-by-side comparison of breeds and management systems as well as sampling across different seasons and flock cycles (7, 18).
Conclusion.
The baseline microbiota of chicken layers across all stages of the commercial farm sequence was until now largely undefined, and the relationships between respiratory and gut microbiota are unknown. The high-resolution data generated in this study can be used as a baseline for comparison with other breeds of layers. It has demonstrated that overlap, and possibly interaction, between gut and respiratory microbiota, including pathogens, is common in all stages of the commercial farm sequence. Therefore, these data can be used as a starting point for the development of effective microbiota-based interventions to enhance production performance and to prevent and control disease in commercial chicken layers.
MATERIALS AND METHODS
Experimental design.
A flock of >80,000 Hy-Line W-36 commercial layers was prospectively sampled for 1 year (November 2015 to November 2016) during the brood (≤5 weeks or younger), grow-out (6 to 16 weeks), and laying (≥17 weeks) stages of the commercial farm sequence. A total of 181 birds were sampled at 9 time points as follows: 20 birds at 1, 3, 8, and 51 weeks of age (WOA), 21 birds at 5 WOA, 16 birds at 12, 16, and 25 WOA, and 32 birds at 36 WOA. A detailed sampling timeline is shown in Fig. S6 in the supplemental material. The birds were transported from the commercial farm to a necropsy facility at the Ohio Agricultural Research and Development Center (OARDC) for euthanasia and tissue collection (within 3 h of sampling). Relevant flock metadata, including vaccination history, antibiotic treatment, nutritional changes, body weights, livability, and egg production, were collected to support interpretation of the data reported herein and to support future meta-analysis of similarly conducted studies. Flock management, including bird placement in cages, performance monitoring, vaccination, and nutritional changes at different stages/phases of the farm sequence, were followed generally as described in the Hy-Line W-36 commercial chicken management guide (26). The flock was vaccinated against Newcastle disease virus (NDV), infectious bronchitis virus (IBV), infectious bursal disease virus (IBDV), avian encephalomyelitis virus (AEV), fowl pox, and Mycoplasma gallisepticum as recommended in the management guide (Fig. S5) (26). In addition to the recommended vaccines, the flock was vaccinated against infectious laryngotracheitis virus and Salmonella enterica serovar Typhimurium (Fig. S5).
Euthanasia and tissue collection.
At each sampling age, blood was taken from live birds via the brachial wing vein before euthanasia to measure serum antibody responses against selected vaccines and to gather antibody evidence of exposure to Mycoplasma synoviae and reovirus of unknown pathogenicity that is ubiquitously present in commercial farms (Fig. S5) (2, 3). The birds were humanely euthanized in accordance with protocol number 2015A00000056-R1 approved by The Ohio State University Institutional Animal Care and Use Committee (IACUC). This protocol complies with the U.S. Animal Welfare Act, Guide for Care and Use of Laboratory Animals, and Public Health Service Policy on Humane Care and Use of Laboratory Animals. The Ohio State University is accredited by the Association for the Assessment and Accreditation of Laboratory Animal Care International (AAALAC). In brief, the animals were exposed to carbon dioxide (CO2) in a euthanasia chamber (1 to 3 birds). The CO2 flow was maintained at 10% to 30% displacement of chamber volume/minute until euthanasia was complete. Euthanasia was confirmed by the absence of breathing and lack of a heartbeat. After euthanasia, bird weight and tissue samples were collected.
Sample processing and sequencing.
Tracheas were aseptically excised in a biosafety cabinet and washed by flushing through 2 to 5 ml of sterile phosphate-buffered saline (PBS) several times using a pipette. Afterwards, the outer nares were thoroughly wiped with a cotton ball saturated with 100% ethanol. Approximately 100 to 500 μl of sterile PBS was drawn with a pipette. The PBS was flushed and retrieved through the nares (above the fold at the opening of the nostril); this process was repeated several times for both nares. On average, approximatyely 0.5 to 2 ml of nasal wash was collected from each bird depending on the sampling age. The samples were kept on ice until further processing. Bacteria in trachea and nasal washes were pelleted by centrifugation (10,000 × g) at 4°C for 10 min. The supernatants were discarded, and the bacterial pellets were resuspended in 200 µl of PBS and stored at −80°C until use for DNA extraction. DNA was extracted from the bacterial pellets using Qiagen DNeasy Blood and Tissue kit (Qiagen Sciences Inc., Germantown, MD) according to the manufacturer’s protocol except that the volume of buffers AW1 and AW2 was increased from 500 µl to 730 µl.
Full ileum (from the ileocecal junction to the Meckel's diverticulum) and cecum (both pouches) were excised, placed in sterile tubes, and snap-frozen in liquid nitrogen. They were then stored in a −80°C freezer until the time of homogenization. Frozen ilea and ceca (including tissues and digesta) were homogenized by grinding with a mortar and pestle to achieve a powdery consistence. Homogenization was performed in a biosafety cabinet, and liquid nitrogen was used to keep the samples frozen while being homogenized. Portions of the tissue homogenates (0.3 g) were placed inside sterile 2.0-ml screw-cap tubes for DNA extraction. DNA was extracted from the tissue homogenates using a DNeasy PowerSoil kit (Qiagen Sciences Inc., Germantown, MD) according to the manufacturer’s protocol with a few modifications in vortexing and centrifugation times in some steps as follows: step 4, vortex for 20 min; step 5, centrifuge for 2 min; steps 8 and 11, centrifuge for 2 min.
DNA was quantified using a NanoDrop 2000c spectrophotometer (Thermo Fisher Scientific, Waltham, MA). The DNA samples (containing 1 to 100 ng/µl; volume, 20 µl/sample) were sequenced at the University of Minnesota Genomics Center (Minneapolis, MN). Amplification of the 16S rRNA gene was performed using KAPA HiFidelity Hot Start polymerase (Kapa Biosystems, Inc., Wilmington, MA) for two rounds of PCR. For the first round, the 515F (5′-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGGTGCCAGCMGCCGCGGTAA-3′) and 806R (5′-GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGGACTACHVGGGTWTCTAAT-3′) Nextera primers (64) (Integrated DNA Technologies, Coralville, IA) were used to amplify the V4 hypervariable region using the following cycling parameters: one cycle of 95°C for 5 min, followed by 25 cycles of 98°C for 20 s, 55°C for 15 s, and 72°C for 1 min. The products were then diluted 1:100, and 5 µl was used in a second round of PCR using forward (5′-AATGATACGGCGACCACCGAGATCTACAC[i5]TCGTCGGCAGCGTC-3′) and reverse (5′-CAAGCAGAAGACGGCATACGAGAT[i7]GTCTCGTGGGCTCGG-3′) indexing primers (Integrated DNA Technologies). The second PCR used the following cycling parameters: 1 cycle at 95°C for 5 min, followed by 10 cycles of 98°C for 20 s, 55°C for 15 s, and 72°C for 1 min. Pooled size-selected samples were denatured with NaOH, diluted to 8 pM in Illumina’s HT1 buffer, spiked with 20% PhiX, and heat denatured at 96°C for 2 min immediately prior to loading. A MiSeq 600 (2 × 300 bp) cycle v3 kit (Illumina, San Diego, CA) was used to sequence the samples. This protocol represents an optimization of the Earth Microbiome Project protocol (65), reported elsewhere (66).
High-throughput data processing.
Sequence reads were sorted by barcode to generate fastq files for each sample. Proximal and distal primers and adapters were trimmed from the reads using BBDuk, and paired-end reads were merged using BBMerge, both from the BBTools (v35) suite of DNA/RNA analysis software provided freely by the Joint Genome Institute (67). 16S rRNA amplicon primers were removed using Cutadapt package (v1.4.2) (68). All sequences were joined in QIIME (v1.9.0) (28) to generate a fasta file. Sequences of lengths less than 245 and greater than 260 bp were filtered using Mothur (v1.35) (69). Chimeras were removed in Mothur using Uchime (v4.2.40) (70). Open reference OTU picking was performed using Uclust in QIIME (v1.9.0), with the clustering radius set at 97% similarity. The SILVA (release 132) reference database (71) was used for taxonomic classification. After filtering the operational taxonomic units (OTUs) not classified as bacteria and those classified as Cyanobacteria (which likely represented chloroplast DNA from plant material in feed) and the removal of OTUs with counts less than 10 and rarefaction to 5,000 reads per sample in QIIME (v1.9.0), 8,369 OTUs were retained. All further analyses were conducted in R (72) using the packages GUniFrac (73), biomformat (74), dplyr (75), vegan (76), pairwiseAdonis (77), ape (78), dendextend (79), phytools (80), VennDiagram (81), and ggplot2 (82).
DNA sequence analysis.
Alpha and beta diversity were assessed in R using the vegan package. An unweighted UniFrac distance matrix was generated for analyses of beta diversity, including PCO analysis. Statistical comparisons of Shannon’s diversity index, species richness, evenness, and dissimilarity dispersion per age group were conducted using a pairwise Wilcoxon rank sum test with Bonferroni’s correction. Multivariate analysis of variance for comparisons between groups was performed using permutational multivariate analysis of variance (PERMANOVA) in R package pairwiseAdonis. ANOSIM comparisons were performed in QIIME. The selection of core microbiota was performed independently for each body site, using a serial filtering scheme. The first filter pulled OTUs that were present in more than 75% of samples within an age group. The second filter chose OTUs that were common among the filtered lists of all age groups. The third filter chose only those OTUs that were present at a mean relative abundance of more than 3.5% in at least one age group.
Data availability.
Raw data files and metadata are publicly available in the NCBI BioSample database under accession numbers SAMN10590833 through SAMN10591480 (https://www.ncbi.nlm.nih.gov/Traces/study/?acc=PRJNA509940). Raw data files are also available through Sequence Read Archive (SRA) (https://www.ncbi.nlm.nih.gov/sra/PRJNA509940). Pipe scripts for sequence processing and generation of OTU table are publicly available at https://github.com/kjmtaylor22/cwl-cats/blob/master/CWL85_Suppl%20Text%20File%201_QiimePipeline.txt. R scripts are publicly available at https://github.com/kjmtaylor22/cwl-cats.
Supplementary Material
ACKNOWLEDGMENTS
We thank the management team of the commercial farm participating in this study for providing chickens and giving us access to all flock treatment and production performance data related to this study. Sequence processing and analysis were performed using the resources of the Molecular & Cellular Imaging Center, Ohio Agricultural Research and Development Center, The Ohio State University.
This project was supported by Agriculture and Food Research Initiative competitive grant number 2015-68004-23131 from the USDA National Institute of Food and Agriculture (to C.-W.L. and T.J.J.).
C.-W.L. and T.J.J. developed the broad concept for the study. C.-W.L., J.M.N., and M.C.A. designed and managed the study. J.M.N., M.C.A., H.J., M.E., M.K.C., and C.-W.L. collected the samples. J.M.N., M.C.A., and A.G. performed sample processing and DNA extraction. B.P.W. and T.J.J. managed the DNA sequencing. K.J.M.T., J.M.N., M.C.A., and S.W. performed bioinformatics analyses. J.M.N., K.J.M.T., and M.C.A. wrote the manuscript with input from all authors. C.-W.L. provided reagents and sampling resources. All authors read and approved the final manuscript.
We declare no competing interests.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.03137-18.
REFERENCES
- 1.Clavijo V, Flórez MJV. 2018. The gastrointestinal microbiome and its association with the control of pathogens in broiler chicken production: a review. Poult Sci 97:1006–1021. doi: 10.3382/ps/pex359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nahashon SN, Nakaue HS, Mirosh LW. 1996. Performance of single comb white leghorn fed a diet supplemented with a live microbial during the growth and egg laying phases. Anim Feed Sci Technol 57:25–38. doi: 10.1016/0377-8401(95)00852-7. [DOI] [Google Scholar]
- 3.Inatomi T. 2016. Laying performance, immunity and digestive health of layer chickens fed diets containing a combination of three probiotics. Sci Postprint 1:e00058. [Google Scholar]
- 4.Lei K, Li YL, Yu DY, Rajput IR, Li WF. 2013. Influence of dietary inclusion of Bacillus licheniformis on laying performance, egg quality, antioxidant enzyme activities, and intestinal barrier function of laying hens. Poult Sci 92:2389–2395. doi: 10.3382/ps.2012-02686. [DOI] [PubMed] [Google Scholar]
- 5.Kurtoglu V, Kurtoglu F, Seker E, Coskun B, Balevi T, Polat ES. 2004. Effect of probiotic supplementation on laying hen diets on yield performance and serum and egg yolk cholesterol. Food Addit Contam 21:817–823. doi: 10.1080/02652030310001639530. [DOI] [PubMed] [Google Scholar]
- 6.Simon K, Verwoolde MB, Zhang J, Smidt H, de Vries Reilingh G, Kemp B, Lammers A. 2016. Long-term effects of early life microbiota disturbance on adaptive immunity in laying hens. Poult Sci 95:1543–1554. doi: 10.3382/ps/pew088. [DOI] [PubMed] [Google Scholar]
- 7.Johnson TJ, Youmans BP, Noll S, Cardona C, Evans NP, Karnezos TP, Ngunjiri JM, Abundo MC, Lee CW. 2018. A consistent and predictable commercial broiler chicken bacterial microbiota in antibiotic-free production displays strong correlations with performance. Appl Environ Microbiol 84:e00362-18. doi: 10.1128/AEM.00362-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Torok VA, Hughes RJ, Mikkelsen LL, Perez-Maldonado R, Balding K, MacAlpine R, Percy NJ, Ophel-Keller K. 2011. Identification and characterization of potential performance-related gut microbiotas in broiler chickens across various feeding trials. Appl Environ Microbiol 77:5868–5878. doi: 10.1128/AEM.00165-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nava GM, Bielke LR, Callaway TR, Castañeda MP. 2005. Probiotic alternatives to reduce gastrointestinal infections: the poultry experience. Anim Health Res Rev 6:105–118. doi: 10.1079/AHR2005103. [DOI] [PubMed] [Google Scholar]
- 10.Stanley D, Denman SE, Hughes RJ, Geier MS, Crowley TM, Chen H, Haring VR, Moore RJ. 2012. Intestinal microbiota associated with differential feed conversion efficiency in chickens. Appl Microbiol Biotechnol 96:1361–1369. doi: 10.1007/s00253-011-3847-5. [DOI] [PubMed] [Google Scholar]
- 11.Pedroso AA, Batal AB, Lee MD. 2016. Effect of in ovo administration of an adult-derived microbiota on establishment of the intestinal microbiome in chickens. Am J Vet Res 77:514–526. doi: 10.2460/ajvr.77.5.514. [DOI] [PubMed] [Google Scholar]
- 12.Mohd Shaufi MA, Sieo CC, Chong CW, Gan HM, Ho YW. 2015. Deciphering chicken gut microbial dynamics based on high-throughput 16S rRNA metagenomics analyses. Gut Pathog 7:4. doi: 10.1186/s13099-015-0051-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Danzeisen JL, Kim HB, Isaacson RE, Tu ZJ, Johnson TJ. 2011. Modulations of the chicken cecal microbiome and metagenome in response to anticoccidial and growth promoter treatment. PLoS One 6:e27949. doi: 10.1371/journal.pone.0027949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Han Z, Willer T, Pielsticker C, Gerzova L, Rychlik I, Rautenschlein S. 2016. Differences in host breed and diet influence colonization by. Gut Pathog 8:56. doi: 10.1186/s13099-016-0133-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oakley BB, Lillehoj HS, Kogut MH, Kim WK, Maurer JJ, Pedroso A, Lee MD, Collett SR, Johnson TJ, Cox NA. 2014. The chicken gastrointestinal microbiome. FEMS Microbiol Lett 360:100–112. doi: 10.1111/1574-6968.12608. [DOI] [PubMed] [Google Scholar]
- 16.Stanley D, Hughes RJ, Moore RJ. 2014. Microbiota of the chicken gastrointestinal tract: influence on health, productivity and disease. Appl Microbiol Biotechnol 98:4301–4310. doi: 10.1007/s00253-014-5646-2. [DOI] [PubMed] [Google Scholar]
- 17.Ranjitkar S, Lawley B, Tannock G, Engberg RM. 2016. Bacterial succession in the broiler gastrointestinal tract. Appl Environ Microbiol 82:2399–2410. doi: 10.1128/AEM.02549-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oakley BB, Vasconcelos EJR, Diniz PPVP, Calloway KN, Richardson E, Meinersmann RJ, Cox NA, Berrang ME. 2018. The cecal microbiome of commercial broiler chickens varies significantly by season. Poult Sci 97:3635–3644. doi: 10.3382/ps/pey214. [DOI] [PubMed] [Google Scholar]
- 19.Kers JG, Velkers FC, Fischer EAJ, Hermes GDA, Stegeman JA, Smidt H. 2018. Host and environmental factors affecting the intestinal microbiota in chickens. Front Microbiol 9:235. doi: 10.3389/fmicb.2018.00235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Videnska P, Sedlar K, Lukac M, Faldynova M, Gerzova L, Cejkova D, Sisak F, Rychlik I. 2014. Succession and replacement of bacterial populations in the caecum of egg laying hens over their whole life. PLoS One 9:e115142. doi: 10.1371/journal.pone.0115142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ballou AL, Ali RA, Mendoza MA, Ellis JC, Hassan HM, Croom WJ, Koci MD. 2016. Development of the chick microbiome: how early exposure influences future microbial diversity. Front Vet Sci 3:2. doi: 10.3389/fvets.2016.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Polansky O, Sekelova Z, Faldynova M, Sebkova A, Sisak F, Rychlik I. 2015. Important metabolic pathways and biological processes expressed by chicken cecal microbiota. Appl Environ Microbiol 82:1569–1576. doi: 10.1128/AEM.03473-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Glendinning L, McLachlan G, Vervelde L. 2017. Age-related differences in the respiratory microbiota of chickens. PLoS One 12:e0188455. doi: 10.1371/journal.pone.0188455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sohail MU, Hume ME, Byrd JA, Nisbet DJ, Shabbir MZ, Ijaz A, Rehman H. 2015. Molecular analysis of the caecal and tracheal microbiome of heat-stressed broilers supplemented with prebiotic and probiotic. Avian Pathol 44:67–74. doi: 10.1080/03079457.2015.1004622. [DOI] [PubMed] [Google Scholar]
- 25.Shabbir MZ, Malys T, Ivanov YV, Park J, Shabbir MA, Rabbani M, Yaqub T, Harvill ET. 2015. Microbial communities present in the lower respiratory tract of clinically healthy birds in Pakistan. Poult Sci 94:612–620. doi: 10.3382/ps/pev010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hy-Line-International. 2016. Hy-Line W-36 commercial layers management guide. Hy-Line International, Des Moines, IA. [Google Scholar]
- 27.Chapman MG, Underwood AJ. 1999. Ecological patterns in multivariate assemblages: information and interpretation of negative values in ANOSIM tests. Mar Ecol Prog Ser 180:257–265. doi: 10.3354/meps180257. [DOI] [Google Scholar]
- 28.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Astudillo-García C, Bell JJ, Webster NS, Glasl B, Jompa J, Montoya JM, Taylor MW. 2017. Evaluating the core microbiota in complex communities: a systematic investigation. Environ Microbiol 19:1450–1462. doi: 10.1111/1462-2920.13647. [DOI] [PubMed] [Google Scholar]
- 30.Logue CM. 2013. Clostridial diseases, p 943–970. In Swayne DE. (ed), Diseases of poultry. John Wiley & Sons, Ames, IA. [Google Scholar]
- 31.Logue CM, Andreasen CB, Thayer SG, Waltman WD, Bricker JM, Saif YM, Hampson DJ, Fulton RM, Sanchez S, Abdul-Aziz T, Barnes HJ. 2013. Other bacterial diseases, p 971–1053. In Swayne DE. (ed), Diseases of poultry. John Wiley & Sons, Ames, IA. [Google Scholar]
- 32.Glisson JR, Hofacre CL, Christensen J, Ruiz JA, Sandhu TS, Chin RP, van Empel PCM, Hafez HM, Jackwood MW, Saif YM. 2013. Pasteurellosis and other respiratory bacterial infections, p 807–873. In Swayne DE. (ed), Diseases of poultry. Wiley & Sons, Ames, IA. [Google Scholar]
- 33.Blackwell PJ, Soriano-Vargas E. 2013. Infectious coryza and related bacterial infections, p 859–873. In Swayne DE. (ed), Diseases of poultry. John Wiley & Sons, Ames, IA. [Google Scholar]
- 34.Ferguson-Noel N. 2013. Mycoplasmosis, p. 875–941. In Swayne DE. (ed), Diseases of poultry. John Wiley & Sons, Ames, IA. [Google Scholar]
- 35.Nolan LK, Barnes HJ, Vaillancourt J-P, Abdul-Aziz T, Logue CM. 2013. Colibacillosis, p 751–805. In Swayne DE. (ed), Diseases of poultry. Wiley & Sons, Ames, IA. [Google Scholar]
- 36.Abdelwhab EM, Lüschow D, Hafeza HM. 2013. Development of real-time polymerase chain reaction assay for detection of Ornithobacterium rhinotracheale in poultry. Avian Dis 57:663–666. doi: 10.1637/10517-022213-ResNoteR. [DOI] [PubMed] [Google Scholar]
- 37.Baums CG, Schotte U, Amtsberg G, Goethe R. 2004. Diagnostic multiplex PCR for toxin genotyping of Clostridium perfringens isolates. Vet Microbiol 100:11–16. doi: 10.1016/S0378-1135(03)00126-3. [DOI] [PubMed] [Google Scholar]
- 38.Johnson TJ, Wannemuehler Y, Doetkott C, Johnson SJ, Rosenberger SC, Nolan LK. 2008. Identification of minimal predictors of avian pathogenic Escherichia coli virulence for use as a rapid diagnostic tool. J Clin Microbiol 46:3987–3996. doi: 10.1128/JCM.00816-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jones RC. 2000. Reovirus Infections. Rev Sci Tech 19:614–625. doi: 10.20506/rst.19.2.1237. [DOI] [PubMed] [Google Scholar]
- 40.Pantin-Jackwood MJ, Day JM, Jackwood MW, Spackman E. 2008. Enteric viruses detected by molecular methods in commercial chicken and turkey flocks in the United States between 2005 and 2006. Avian Dis 52:235–244. doi: 10.1637/8174-111507-Reg.1. [DOI] [PubMed] [Google Scholar]
- 41.Mookiah S, Sieo CC, Ramasamy K, Abdullah N, Ho YW. 2014. Effects of dietary prebiotics, probiotic and synbiotics on performance, caecal bacterial populations and caecal fermentation concentrations of broiler chickens. J Sci Food Agric 94:341–348. doi: 10.1002/jsfa.6365. [DOI] [PubMed] [Google Scholar]
- 42.Bae Y, Koo B, Lee S, Mo J, Oh K, Mo I. 2017. Bacterial diversity and its relationship to growth performance of broilers. Korean J Vet Res 57:159–167. [Google Scholar]
- 43.Yan W, Sun C, Yuan J, Yang N. 2017. Gut metagenomic analysis reveals prominent roles of Lactobacillus and cecal microbiota in chicken feed efficiency. Sci Rep 7:45308. doi: 10.1038/srep45308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baldwin S, Hughes RJ, Hao Van TT, Moore RJ, Stanley D. 2018. At-hatch administration of probiotic to chickens can introduce beneficial changes in gut microbiota. PLoS One 13:e0194825. doi: 10.1371/journal.pone.0194825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yeo J, Kim KI. 1997. Effect of feeding diets containing an antibiotic, a probiotic, or yucca extract on growth and intestinal urease activity in broiler chicks. Poult Sci 76:381–385. doi: 10.1093/ps/76.2.381. [DOI] [PubMed] [Google Scholar]
- 46.Zulkifli I, Abdulllah N, Azrin NM, Ho YW. 2000. Growth performance and immune response of two commercial broiler strains fed diets containing Lactobacillus cultures and oxytetracycline under heat stress conditions. Br Poult Sci 41:593–597. doi: 10.1080/713654979. [DOI] [PubMed] [Google Scholar]
- 47.Bai SP, Wu AM, Ding XM, Lei Y, Bai J, Zhang KY, Chio JS. 2013. Effects of probiotic-supplemented diets on growth performance and intestinal immune characteristics of broiler chickens. Poult Sci 92:663–670. doi: 10.3382/ps.2012-02813. [DOI] [PubMed] [Google Scholar]
- 48.Stanley D, Hughes RJ, Geier MS, Moore RJ. 2016. Bacteria within the gastrointestinal tract microbiota correlated with improved growth and feed conversion: challenges presented for the identification of performance enhancing probiotic bacteria. Front Microbiol 7:187. doi: 10.3389/fmicb.2016.00187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.De Cesare A, Sirri F, Manfreda G, Moniaci P, Giardini A, Zampiga M, Meluzzi A. 2017. Effect of dietary supplementation with Lactobacillus acidophilus D2/CSL (CECT 4529) on caecum microbioma and productive performance in broiler chickens. PLoS One 12:e0176309. doi: 10.1371/journal.pone.0176309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Forte C, Manuali E, Abbate Y, Papa P, Vieceli L, Tentellini M, Trabalza-Marinucci M, Moscati L. 2018. Dietary Lactobacillus acidophilus positively influences growth performance, gut morphology, and gut microbiology in rurally reared chickens. Poult Sci 97:930–936. doi: 10.3382/ps/pex396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kizerwetter-Swida M, Binek M. 2009. Protective effect of potentially probiotic Lactobacillus strain on infection with pathogenic bacteria in chickens. Pol J Vet Sci 12:15–20. [PubMed] [Google Scholar]
- 52.Lan PT, Binh LT, Benno Y. 2003. Impact of two probiotic Lactobacillus strains feeding on fecal lactobacilli and weight gains in chicken. J Gen Appl Microbiol 49:29–36. doi: 10.2323/jgam.49.29. [DOI] [PubMed] [Google Scholar]
- 53.Pascual M, Hugas M, Badiola JI, Monfort JM, Garriga M. 1999. Lactobacillus salivarius CTC2197 prevents Salmonella enteritidis colonization in chickens. Appl Environ Microbiol 65:4981–4986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Neal-McKinney JM, Lu X, Duong T, Larson CL, Call DR, Shah DH, Konkel ME. 2012. Production of organic acids by probiotic lactobacilli can be used to reduce pathogen load in poultry. PLoS One 7:e43928. doi: 10.1371/journal.pone.0043928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ketley JM. 1997. Pathogenesis of enteric infection by Campylobacter. Microbiology 143:5–21. doi: 10.1099/00221287-143-1-5. [DOI] [PubMed] [Google Scholar]
- 56.Denis M, Soumet C, Rivoal K, Ermel G, Blivet D, Salvat G, Colin P. 1999. Development of a m-PCR assay for simultaneous identification of Campylobacter jejuni and C. coli. Lett Appl Microbiol 29:406–410. doi: 10.1046/j.1472-765X.1999.00658.x. [DOI] [PubMed] [Google Scholar]
- 57.De Boeck C, Kalmar I, Dumont A, Vanrompay D. 2015. Longitudinal monitoring for respiratory pathogens in broiler chickens reveals co-infection of Chlamydia psittaci and Ornithobacterium rhinotracheale. J Med Microbiol 64:565–574. doi: 10.1099/jmm.0.000047. [DOI] [PubMed] [Google Scholar]
- 58.Pan Q, Liu A, Zhang F, Ling Y, Ou C, Hou N, He C. 2012. Co-infection of broilers with Ornithobacterium rhinotracheale and H9N2 avian influenza virus. BMC Vet Res 8:104. doi: 10.1186/1746-6148-8-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Roussan DA, Haddad R, Khawaldeh G. 2008. Molecular survey of avian respiratory pathogens in commercial broiler chicken flocks with respiratory diseases in Jordan. Poult Sci 87:444–448. doi: 10.3382/ps.2007-00415. [DOI] [PubMed] [Google Scholar]
- 60.Stipkovits L, Egyed L, Palfi V, Beres A, Pitlik E, Somogyi M, Szathmary S, Denes B. 2012. Effect of low-pathogenicity influenza virus H3N8 infection on Mycoplasma gallisepticum infection of chickens. Avian Pathol 41:51–57. doi: 10.1080/03079457.2011.635635. [DOI] [PubMed] [Google Scholar]
- 61.Park SH, Kim SA, Rubinelli PM, Roto SM, Ricke SC. 2017. Microbial compositional changes in broiler chicken cecal contents from birds challenged with different Salmonella vaccine candidate strains. Vaccine 35:3204–3208. doi: 10.1016/j.vaccine.2017.04.073. [DOI] [PubMed] [Google Scholar]
- 62.Tarabichi Y, Li K, Hu S, Nguyen C, Wang X, Elashoff D, Saira K, Frank B, Bihan M, Ghedin E, Methé BA, Deng JC. 2015. The administration of intranasal live attenuated influenza vaccine induces changes in the nasal microbiota and nasal epithelium gene expression profiles. Microbiome 3:74. doi: 10.1186/s40168-015-0133-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Siegerstetter SC, Schmitz-Esser S, Magowan E, Wetzels SU, Zebeli Q, Lawlor PG, O'Connell NE, Metzler-Zebeli BU. 2017. Intestinal microbiota profiles associated with low and high residual feed intake in chickens across two geographical locations. PLoS One 12:e0187766. doi: 10.1371/journal.pone.0187766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer M, Gormley N, Gilbert JA, Smith G, Knight R. 2012. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J 6:1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gilbert JA, Meyer F. 2012. Modeling the earth microbiome. Microbe Wash DC 7:64–69. doi: 10.1128/microbe.7.64.1. [DOI] [Google Scholar]
- 66.Gohl DM, Vangay P, Garbe J, MacLean A, Hauge A, Becker A, Gould TJ, Clayton JB, Johnson TJ, Hunter R, Knights D, Beckman KB. 2016. Systematic improvement of amplicon marker gene methods for increased accuracy in microbiome studies. Nat Biotechnol 34:942–949. doi: 10.1038/nbt.3601. [DOI] [PubMed] [Google Scholar]
- 67.Bushnell B, Rood J, Singer E. 2017. BBMerge—accurate paired shotgun read merging via overlap. PLoS One 12:e0185056. doi: 10.1371/journal.pone.0185056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 17:10–12. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 69.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. 2011. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glockner FO. 2013. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.R Core Team. 2018. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 73.Chen J, Bittinger K, Charlson ES, Hoffmann C, Lewis J, Wu GD, Collman RG, Bushman FD, Li H. 2012. Associating microbiome composition with environmental covariates using generalized UniFrac distances. Bioinformatics 28:2106–2113. doi: 10.1093/bioinformatics/bts342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.McMurdie P, Paulson J. 2015. biomformat: an interface package for the BIOM file format. https://www.bioconductor.org/packages/release/bioc/html/biomformat.html.
- 75.Wickham H, Francois R, Henry L, Müller K. 2016. dplyr: a grammar of data manipulation. R package version 0.5. 0 R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 76.Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O’hara R, Simpson GL, Solymos P, Stevens MHH, Wagner H. 2013. Package ‘vegan’. Community Ecology Package, version 2 R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 77.Arbizu M. 2017. Pairwiseadonis: pairwise multilevel comparison using adonis. R package version 0.0 1 R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 78.Paradis E, Claude J, Strimmer K. 2004. APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20:289–290. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- 79.Galili T. 2015. Dendextend: extending R’s dendrogram functionality. R package version 0.18 R Foundation for Statistical Computing, Vienna, Austria: https://cran.r-project.org/web/packages/dendextend/index.html. [Google Scholar]
- 80.Revell LJ. 2012. phytools: an R package for phylogenetic comparative biology (and other things). Methods Ecol Evol 3:217–223. doi: 10.1111/j.2041-210X.2011.00169.x. [DOI] [Google Scholar]
- 81.Chen H, Boutros PC. 2011. VennDiagram: a package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinformatics 12:35. doi: 10.1186/1471-2105-12-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wickham H. 2016. ggplot2: elegant graphics for data analysis. Springer-Verlag, New York, NY. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw data files and metadata are publicly available in the NCBI BioSample database under accession numbers SAMN10590833 through SAMN10591480 (https://www.ncbi.nlm.nih.gov/Traces/study/?acc=PRJNA509940). Raw data files are also available through Sequence Read Archive (SRA) (https://www.ncbi.nlm.nih.gov/sra/PRJNA509940). Pipe scripts for sequence processing and generation of OTU table are publicly available at https://github.com/kjmtaylor22/cwl-cats/blob/master/CWL85_Suppl%20Text%20File%201_QiimePipeline.txt. R scripts are publicly available at https://github.com/kjmtaylor22/cwl-cats.