Iron is a fundamental metal ion for living organisms as it facilitates various biological processes. The ferric uptake regulator (Fur) protein controls iron homeostasis in various bacterial species. It is believed that Fur’s iron-dependent regulatory action is sufficient for it to function as an iron sensor. However, we now establish that the bacterial pathogen Salmonella enables Fur to properly reflect changes in surrounding iron availability by fine-tuning its responsiveness to iron. This process requires a protein that hampers Fur DNA binding at low iron concentrations. In this way, Salmonella broadens the range of iron concentrations that Fur responds to. Our findings reveal a potentially widespread control mechanism of bacterial iron homeostasis.
KEYWORDS: EIIANtr, Salmonella enterica, nitrogen metabolic PTS
ABSTRACT
Iron is one of most abundant environmental metal ions but is highly limited in organisms. It is an important metal ion as it facilitates various biological processes, including catalysis of metabolic enzymes and DNA biogenesis. In bacteria, the ferric uptake regulator (Fur) protein controls iron uptake by regulating genes coding for iron transporters in response to iron concentration. This iron response is ascribed to Fur’s intrinsic affinity for iron because its binding to iron dictates its regulatory function. However, we now report that the pathogen Salmonella achieves a proper response of Fur to changes in environmental iron concentrations via EIIANtr (a nitrogen metabolic phosphotransferase system component). We establish that EIIANtr increases expression of iron transporter-coding genes under low-iron conditions (i.e., nanomolar ranges) in a Fur-dependent manner, which promotes Salmonella growth under such conditions. EIIANtr directly hampers Fur binding to DNA, thereby inducing expression of those genes. This regulation allows Salmonella to express Fur-regulated genes under low-iron conditions. Our findings reveal a potentially widespread control mechanism of bacterial iron uptake systems operating in response to iron availability.
IMPORTANCE Iron is a fundamental metal ion for living organisms as it facilitates various biological processes. The ferric uptake regulator (Fur) protein controls iron homeostasis in various bacterial species. It is believed that Fur’s iron-dependent regulatory action is sufficient for it to function as an iron sensor. However, we now establish that the bacterial pathogen Salmonella enables Fur to properly reflect changes in surrounding iron availability by fine-tuning its responsiveness to iron. This process requires a protein that hampers Fur DNA binding at low iron concentrations. In this way, Salmonella broadens the range of iron concentrations that Fur responds to. Our findings reveal a potentially widespread control mechanism of bacterial iron homeostasis.
INTRODUCTION
Iron is an abundant metal on Earth but is very limited in organisms due to its poor solubility (1). Because this metal is an essential cofactor for many biological processes, including reactions catalyzed by metabolic enzymes and DNA biogenesis (2), iron homeostasis is important for living organisms (1, 2). Not surprisingly, mammalian hosts and bacterial pathogens compete for this limiting metal ion during infection (3, 4). Therefore, it is important for bacteria to properly modulate their iron acquisition system in response to changes in iron availability, especially when the available iron concentration is low. Here, we report how a microorganism tunes the responsiveness of an iron sensor to properly control the iron uptake system.
The transcription factor ferric uptake regulator (Fur) functions as an iron sensor and plays a leading role in maintaining iron homeostasis in various bacterial species by controlling iron transporter-coding genes (5, 6). Fur represses expression of iron transporter genes under high-iron conditions where iron-bound Fur binds to its target promoters, thereby repressing gene transcription (5–7). It is reported that external iron concentrations below 5 to 10 µM cause dissociation of Fur from its targets, allowing expression of iron transporter-coding genes (5). Fur influences the expression of a variety of genes, including those that participate in iron acquisition and virulence (5, 8).
The ptsN gene encodes EIIANtr, a component of the nitrogen-metabolic phosphotransferase system (PTS) (9). This system lacks the membrane-bound complex that would normally control the activities of sugar PTSs in response to particular sugar availabilities (9). EIIANtr plays regulatory functions in various bacterial species by interacting with proteins involved in a variety of cellular processes. EIIANtr controls potassium uptake via TrkA and KdpD (10, 11), phosphate uptake via PhoR (12), virulence via SsrB (13), the stringent response via SpoT (14, 15), and amino sugar homeostasis via GlmS (16). The phosphorylation status of EIIANtr contributes some of those EIIANtr-mediated regulatory functions (10, 11, 15, 16). The phosphorylation of EIIANtr is known to be controlled by the extracellular abundance of nitrogen sources or by the cellular concentration of glutamine (15, 17).
The intracellular pathogen Salmonella enterica serovar Typhimurium is the etiologic agent of human gastroenteritis and murine typhoid fever. Salmonella resides inside macrophage phagosomes (18) where iron availability is limited due to the iron transporters being recruited by the mammalian host pumping out iron from those vesicles (19). However, the phagosome does not represent an iron-depleted condition given that iron-responding systems are activated in that compartment (20–22). Lack of the iron sensor Fur attenuates Salmonella virulence (23), suggesting that Salmonella must manage the low iron availability inside the phagosome and that the iron-sensing ability of Fur is critical for its virulence. Fur’s iron-sensing function is ascribed to its intrinsic affinity to iron (6, 24) given that iron binding dictates its DNA binding ability (6). The affinity of Fur for iron is in the low micromolar range (6, 25), which is believed sufficient for Fur to operate as an iron sensor.
Here, we now report that Fur’s response to iron requires EIIANtr in addition to its own ability to sense iron. EIIANtr tunes the responsiveness of Fur to iron by hampering Fur binding to DNA. This allows expression of the iron uptake system when surrounding iron concentrations drop to the nanomolar range (hereafter, low-iron conditions), which enables the intracellular pathogen Salmonella enterica to properly control its iron uptake system in response to iron availability.
RESULTS
Expression of iron uptake genes requires EIIANtr under low-iron conditions.
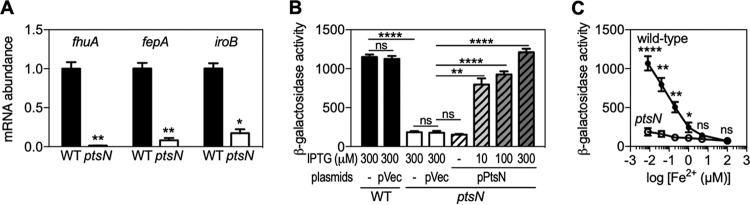
A recent proteomic study reported that the Escherichia coli ptsN mutant reduces the abundance of proteins involved in iron uptake system (26), which leads us to question whether EIIANtr has a role in iron response. To test this idea, we first examined expression of genes involved in iron importation (including fhuA, fepA, and iroB) in the wild-type strain and the ptsN mutant grown in acidified defined media to mimic the experience of Salmonella in acidic phagosomes, where iron uptake is important (18, 19). The wild-type strain displayed higher mRNA abundance of those genes than the ptsN mutant (Fig. 1A), indicating that EIIANtr is involved in controlling iron response genes. We further investigated the expression of the fhuA gene by measuring β-galactosidase activity produced by Salmonella strains with a pfhuA-lacZ fusion from its normal chromosomal location; the wild-type strain had ∼9-fold-higher activities than the ptsN mutant (Fig. 1B). Moreover, the defective fhuA expression of the ptsN mutant was due to a lack of EIIANtr protein because a plasmid expressing EIIANtr from a heterologous promoter restored fhuA expression to wild-type levels, but the empty vector did not (Fig. 1B).
FIG 1.
EIIANtr G expression of iron uptake genes under low-iron conditions. (A) mRNA abundance of fhuA, fepA, and iroB genes was determined in the wild-type (WT) and the ptsN mutant strains grown in acidified M9 medium (pH 5.8) to mid-log phase. (B and C) β-Galactosidase activities of Salmonella with a pfhuA-lacZ fusion in the normal chromosomal location and isogenic ptsN mutants with denoted plasmids (empty vector [pVec] or plasmid expressing EIIANtr from a heterologous promoter [pPtsN]) were determined. Bacteria were grown to mid-log phase in acidified M9 medium (pH 5.8) with 8 nM FeSO4 and IPTG at the denoted concentrations (B) or in acidified M9 medium (pH 5.8) with FeSO4 at the denoted concentrations (C). The means and standard deviations (SD) of results from at least three independent experiments are shown as follows: symbols or bars, mean values; error bars, SD. Two-tailed t tests were performed for comparisons between the wild-type strain and the ptsN mutant or between indicated strains, and statistical significance is indicated as follows: ns, not significant; *, P < 0.05; **, P < 0.01; ****, P < 0.0001.
We next investigated fhuA expression under conditions of various iron concentrations. Wild-type Salmonella induced an ∼18-fold increase in fhuA expression when iron concentrations dropped from 100 µM to 8 nM (Fig. 1C). Surprisingly, however, the ptsN mutant failed to do so under conditions of the same changes in iron concentrations even though it has the iron sensor Fur (Fig. 1C); the ptsN mutant displayed only an approximately 2-fold increase in fhuA expression (Fig. 1C). This EIIANtr-dependent regulation of fhuA expression was evident when iron concentrations were lower than 1 µM (Fig. 1C). These results suggest that Salmonella EIIANtr controls the iron uptake genes under low-iron conditions.
EIIANtr controls iron uptake gene expression in a Fur-dependent manner.
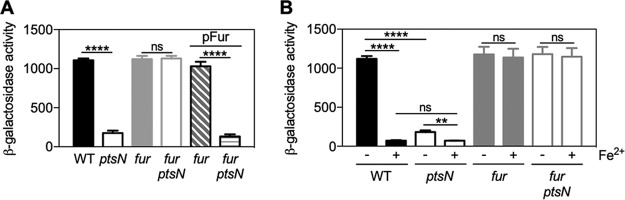
Given that expression of those iron uptake genes (including fhuA) is repressed by Fur (24, 27), we hypothesized that EIIANtr controls the iron uptake gene expression via Fur. Consistent with the previous notion that Fur represses its target genes under iron-replete conditions (5–7), a deletion of the fur gene did not alter fhuA expression under low-iron conditions. Lack of Fur abrogated the regulatory effects of EIIANtr on fhuA expression (Fig. 2A), indicating that Fur is necessary for EIIANtr-mediated regulation of fhuA expression. If EIIANtr regulates fhuA expression by altering Fur expression (i.e., if changes in Fur expression could in turn control expression of fhuA gene), heterologous EIIANtr-independent expression of Fur should abolish regulatory effects of EIIANtr on fhuA expression. However, lack of EIIANtr reduced fhuA transcription even when Fur was expressed from a heterologous promoter (Fig. 2A), indicating that EIIANtr likely controls fhuA expression independently of Fur expression and that Fur represses fhuA expression when EIIANtr is absent under low-iron conditions. Moreover, fur transcription occurred independently of EIIANtr (see Fig. S1A in the supplemental material), and Fur protein amounts were also comparable in the wild-type and the ptsN mutant strains (Fig. S1B). These findings suggest that the regulatory effect of EIIANtr occurs at posttranscriptional levels.
FIG 2.
EIIANtr-mediated regulation of fhuA requires Fur. β-Galactosidase activities of Salmonella with a pfhuA-lacZ fusion in the normal chromosomal location and isogenic strains deleted for ptsN, fur, and fur ptsN genes with or without a plasmid expressing Fur from a heterologous promoter (pFur) were determined. Bacteria were grown to mid-log phase in acidified M9 medium (pH 5.8) (A) or the same medium with (+) or without (−) 100 µM FeSO4 (B). The means and standard deviations (SD) of results from at least three independent experiments are shown as follows: bars, mean values; error bars, SD. Two-tailed t tests were performed for comparisons between indicated strains, and statistical significance is indicated as follows: ns, not significant; **, P < 0.01; ****, P < 0.0001.
Given that iron renders Fur’s regulatory function (6) and that Fur repressed fhuA expression in the ptsN mutant under low-iron conditions (Fig. 2A), we next investigated the effects of exogenous iron on EIIANtr- and Fur-mediated regulation of fhuA. The addition of iron (100 μM of FeSO4) greatly reduced fhuA expression levels in the wild-type strain (Fig. 2B). Under this condition, the ptsN mutant showed fhuA expression comparable to that exhibited by the wild-type strain (Fig. 2B). Moreover, lack of Fur resulted in high levels of fhuA expression independently of both EIIANtr and iron levels (Fig. 2B). Taken together, these findings indicate that EIIANtr promotes expression of iron uptake genes (including fhuA) via Fur under low-iron conditions.
EIIANtr interacts with Fur.
Since EIIANtr controls its targets via protein-protein interaction (10, 11, 13–16), we wondered whether it interacts with Fur. To examine interactions of EIIANtr and Fur, a bacterial two-hybrid assay was used in which β-galactosidase levels are dependent on the proximity of fused proteins to fragments (i.e., T25 and T18) of the Bordetella pertussis adenylate cyclase in an E. coli strain lacking its own adenylate cyclase (28). Coexpression of T25-EIIANtr and T-18-Fur produced ∼33-fold-higher levels of β-galactosidase activity than the strain expressing T25-EIIANtr and the T-18 fragment or empty vectors (Fig. 3A). However, β-galactosidase activities from the strain expressing T25-EIIANtr and T-18-Fur were lower than seen with expression of positive-control plasmids. Given that some EIIANtr-mediated regulation of biological functions is dependent on its phosphorylation status (10, 11, 15, 16), we wondered if phosphorylation is necessary for the interaction of EIIANtr with Fur. The unphosphorylatable EIIANtr variant (H73A) gave expression levels of β-galactosidase activities similar to those seen with wild-type EIIANtr (Fig. 3A), indicating that the phosphorylation status of EIIANtr is not critical for the interaction with Fur. This is similar to results revealing other roles of EIIANtr occurring independently of its phosphorylation status (12–14, 16, 29). Taken together, these results indicate that EIIANtr interacts with Fur.
FIG 3.
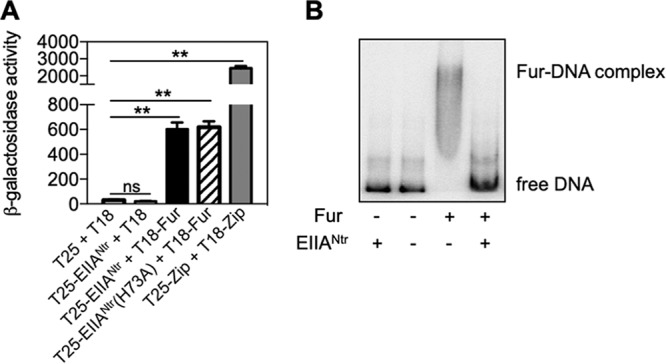
EIIANtr inhibits Fur binding to DNA. (A) β-Galactosidase activities were determined from cya E. coli mutant strains harboring the denoted plasmid combinations grown in acidified M9 medium containing 0.5 mM IPTG. The means and standard deviations (SD) of results from at least three independent experiments are shown as follows: bars, mean values; error bars, SD. Two-tailed t tests were performed for comparisons between indicated strains, and statistical significance is indicated as follows: ns, not significant; **, P < 0.01. (B) In vitro binding of Fur to the fhuA promoter with or without EIIANtr. The fhuA promoter DNA (80 fmol) was incubated with Fur (2 µM) and EIIANtr (10 µM) proteins. Data are representative of results from at least three independent experiments.
EIIANtr controls iron uptake gene expression by hampering Fur binding to DNA.
Given that EIIANtr interacts with Fur (Fig. 3A), it is possible that EIIANtr controls fhuA expression by interfering with Fur binding to DNA. To test this, a gel shift assay was conducted using purified Fur and EIIANtr proteins with the fhuA promoter DNA. Purified Fur bound to the fhuA promoter DNA, which formed a complex in vitro (Fig. 3B). Addition of EIIANtr to this reaction hampered Fur binding to the fhuA promoter (Fig. 3B). However, EIIANtr alone did not bind to the fhuA promoter (Fig. 3B), indicating that prevention of Fur binding to DNA by EIIANtr was not due to competition between EIIANtr and Fur for binding the promoter. These results suggest that EIIANtr promotes iron uptake gene expression by relieving Fur-mediated repression via inhibition of Fur binding to its target promoter regions under low-iron conditions.
Control of iron uptake gene expression by EIIANtr under low-iron conditions ensures Salmonella growth.
Given that EIIANtr is required for expression of iron uptake genes under low-iron conditions (Fig. 1C; see also Fig. 2B) and that EIIANtr hampers Fur binding to DNA by interacting with Fur (Fig. 3), we wondered if excess amounts of iron might alter interaction between EIIANtr and Fur. To test this idea, the bacterial two-hybrid assay was done with a supply of 100 µM iron. Although coexpression of T25-EIIANtr and T-18-Fur produced ∼33-fold-higher levels of β-galactosidase activity than were seen with the control strains (Fig. 3A), the same strain showed basal levels of β-galactosidase activity in the presence of excess iron (Fig. S2A). Furthermore, the addition of iron to the mixture of purified Fur with or without EIIANtr and the target promoter DNA abolished EIIANtr effects on Fur binding to target DNA (Fig. S2B). This is consistent with the finding that regulatory effects of EIIANtr were not observed under conditions of high iron concentrations (Fig. 1C; see also Fig. 2B). These results further support the notion that EIIANtr plays a critical role in controlling iron uptake gene expression under low-iron conditions, such as inside the host phagosome.
We then wondered whether EIIANtr-mediated regulation of iron uptake genes could provide any advantage with respect to Salmonella physiology. Given that iron supports bacterial growth (30, 31), we examined iron-dependent growth phenotypes of the Salmonella wild-type strain and the ptsN mutant using iron concentrations differentially activating iron uptake genes in those two strains (Fig. 1C). As expected, supply of low concentrations of iron (250 nM FeSO4) increased growth of wild-type Salmonella by >80% (Fig. 4). However, it resulted in an increase of only <30% for the Salmonella ptsN mutant (Fig. 4). These results suggest that the EIIANtr-mediated activation of iron uptake genes promotes Salmonella growth under low-iron conditions.
FIG 4.
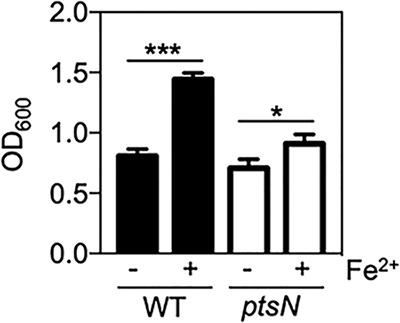
EIIANtr promotes Salmonella growth under low-iron conditions. Bacterial growth was determined for the Salmonella wild-type strain and the ptsN mutant in acidified M9 medium with (+) or without (−) 0.25 µM FeSO4. The optical density at 600 nm (OD600) of bacteria was measured. The means and standard deviations (SD) from three independent experiments are shown as follows: bars, mean values; error bars, SD. Two-tailed t tests were performed for comparisons between indicated strains, and statistical significance is indicated as follows: *, P < 0.05; ***, P < 0.001.
DISCUSSION
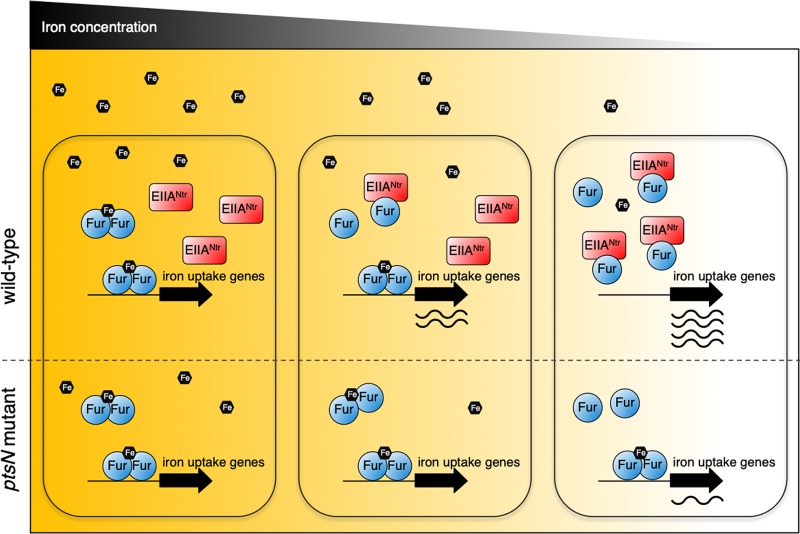
Signal-sensing regulatory systems respond to environmental changes. The output of such systems is dependent on the sensitivity of the system to environmental signals. How does an organism tune the sensitivity of certain sensory systems? Here, we have established that EIIANtr allows the iron sensor Fur to appropriately respond to environmental iron concentrations. Our findings suggest that Salmonella fine-tunes the responsiveness of Fur to iron via EIIANtr (Fig. 5), thereby properly controlling the iron uptake system and potentially other biological functions (5, 8, 27). This provides a new insight into the mechanism of Fur’s iron sensing. Given that EIIANtr and Fur proteins coexist in >600 different bacterial organisms (see Table S1 in the supplemental material), the EIIANtr-mediated tuning of Fur’s iron response is likely a widespread mechanism in other bacteria.
FIG 5.
EIIANtr enables Salmonella to properly control iron uptake gene expression in response to iron availability by modulating Fur. A model depicting iron-dependent activation of iron uptake gene expression by Fur and EIIANtr is depicted. Transcription of iron uptake genes (including fhuA) is repressed by Fur at high iron concentrations. When iron concentrations decrease, iron-unbound Fur levels increase, but only a low level of expression of those genes is seen in the ptsN mutant. In the wild-type strain, EIIANtr facilitates expression of iron uptake genes by hampering binding of Fur to its target promoters. This enables Salmonella to properly control the iron uptake system under low-iron conditions.
As described above, it has been believed that Fur is sufficient to operate as an iron sensor by itself because Fur binding to iron dictates its DNA binding ability (5, 6). We have now established that Fur actually requires an additional protein factor, EIIANtr, to properly respond to changes of iron concentrations (Fig. 1; see also Fig. 5) in addition to its own ability to sense iron. In the absence of EIIANtr, the high affinity of Fur for iron (6, 25) favors formation of the iron-bound form and repression of iron uptake genes even under conditions of low iron concentrations (Fig. 1A). When iron concentrations drop to below certain levels, iron-unbound Fur levels increase due to less availability of iron. Under this condition, EIIANtr promotes iron uptake gene expression (Fig. 1A), possibly because Fur has a chance of interacting with EIIANtr than iron. Alternatively or in addition, EIIANtr binding to Fur may block Fur’s binding to iron.
Bacteria have perhaps chosen to evolve by tuning the activity of Fur through the use of auxiliary protein instead of altering the affinity of Fur for iron, which might be feasible with respect to adjusting Fur-mediated iron metabolism in various bacterial species, depending on their habitats. As a result of tuning of Fur’s activity via EIIANtr, Fur not only attains better resolution in reflecting iron availability but also could potentially assimilate signals sensed and/or processed via EIIANtr. The human pathogen Acinetobacter baumannii controls iron metabolism by altering the activity of Fur via BlsA (32), which is a photoreceptor that responds to light and temperature signals (33). Thus, BlsA allows A. baumannii to integrate light and temperature signals into iron uptake systems. Our findings suggest that EIIANtr may enable Salmonella to control iron metabolism in response to environmental changes potentially impacting EIIANtr, such as changes in nitrogen or amino sugar sources (16, 17) or other potential signals changing cellular amounts of EIIANtr.
In addition to Fur, there is a membrane-bound iron sensor, PmrB, that is a constituent of the PmrA/PmrB two-component regulatory system (34). Interestingly, the Fur and PmrA iron-activated regulators negatively control Salmonella pathogenicity island 2 (SPI-2) gene expression (20, 21). As PmrB and Fur sense extracytoplasmic iron and cytoplasmic iron, respectively (5, 35), Salmonella controls SPI-2 gene expression in response to both extracytoplasmic and cytoplasmic iron levels via those iron sensors. As EIIANtr hampers Fur binding to DNA (Fig. 3B), it probably favors induction of SPI-2 gene expression by relieving Fur-mediated negative regulation. Paradoxically, EIIANtr reduces the transcription of SPI-2 genes by interfering with the binding of the major regulator of SPI-2, SsrB, to SPI-2 gene promoters (13). By impeding both Fur and SsrB, Salmonella probably achieves appropriate expression of virulence genes under low-iron conditions inside the host (19, 36).
We have shown that EIIANtr plays an important role in controlling iron uptake systems under low-iron conditions. This ability to cope with low iron availability is probably critical for Salmonella during infection given that mammalian hosts utilize a strategy to withhold iron (37), because an increase of iron availability results in enhanced growth and/or virulence of many bacterial pathogens in vitro and in vivo (30, 31) and iron depletion reduces intracellular bacterial growth (38). As NRAMP1 protein removes iron from macrophage phagosomes (19), an Nramp1−/− mouse lacking this iron transporter is highly sensitive to Salmonella infection (39). Moreover, lack of Fur highly attenuates Salmonella virulence in Nramp1+/+ mice whereas this defect in virulence is reduced in Nramp1−/− mice (23). We previously reported that EIIANtr promotes virulence by preventing hyperactivation of Salmonella pathogenicity island 2 (SPI-2) gene expression (13). Our findings reported here suggest that the attenuated virulence of the ptsN mutant Salmonella in mice (13) might be due to both ectopic expression of SPI-2 genes and failure to import sufficient iron inside the host.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The S. enterica serovar Typhimurium strains used in this study were derived from strain SL1344. The strains and plasmids used in this study are listed in Table 1. Phage P22-mediated transduction was performed as described previously (40). All Salmonella strains were grown aerobically at 30 or 37°C in LB or M9 minimal medium at pH 5.8 supplemented with 0.5% Casamino Acids. Antibiotics were used at the following concentrations: ampicillin, 50 μg/ml; chloramphenicol, 25 μg/ml; kanamycin (Km), 50 μg/ml; and streptomycin, 50 μg/ml. Primers used for the construction of bacterial strains and plasmids, reverse transcription-quantitative PCR (qRT-PCR), and electrophoretic mobility shift assay (EMSA) are listed in Table 2.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Reference or source |
|---|---|---|
| Strains | ||
| Salmonella enterica SL1344 | Wild type, Strr | 46 |
| Salmonella enterica SR3203 | ΔptsN | 13 |
| Salmonella enterica SR4101 | pfur-lacZ (Kmr) | This work |
| Salmonella enterica SR4102 | pfur-lacZ (Kmr) ΔptsN | This work |
| Salmonella enterica SR4103 | fur-FLAG | This work |
| Salmonella enterica SR4104 | fur-FLAG ΔptsN | This work |
| Salmonella enterica SR4125 | pfhuA-lacZ (Kmr) | This work |
| Salmonella enterica SR4125 | pfhuA-lacZ (Kmr) ΔptsN | This work |
| Salmonella enterica SR4131 | pfhuA-lacZ (Kmr) Δfur | This work |
| Salmonella enterica SR4132 | pfhuA-lacZ (Kmr) Δfur ΔptsN | This work |
| Escherichia coli DH5α | F– supE44 ΔlacU169 (φ80 lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | 47 |
| Escherichia coli BTH101 | F– cya-99 araD139 galE15 galK16 rpsL1 (Strr) hsdR2 mcrA1 mcrB1 | 28 |
| Plasmids | ||
| pCP20 | reppSC101ts Apr Cmr FLP+ cI857+ | 41 |
| pCE70 | repR6K Kmr Cmr FRT tnpR lacZY | 43 |
| pKD3 | repR6K Apr FRT Cmr FRT | 41 |
| pKD13 | repR6K Apr FRT Kmr FRT | 41 |
| pKD46 | reppSC101ts Apr paraBAD γ β exo | 41 |
| pUHE21-2lacIq | reppMB1 Apr lacIq | 48 |
| pFur | reppMB1 Apr lacIq fur | 21 |
| pJJ37 | reppMB1 Apr lacIq ptsN-His6 | 13 |
| pJJ48 | reppMB1 Apr lacIq fur-His6 | This work |
| pKT25 | Kmr repp15A | 44 |
| pUT18 | Apr reppMB1 | 44 |
| pT25-ptsN | Kmr repp15A ptsN | 13 |
| pT25-ptsN(H73A) | Kmr repp15A ptsN(H73A) | 12 |
| pT25-zip | Kmr repp15A zip | 44 |
| pT18-fur | Apr reppMB1 fur | This work |
| pT18-zip | Apr reppMB1 zip | 44 |
Apr, ampicillin resistance; Cmr, chloramphenicol resistance; Kmr, kanamycin resistance; Strr, streptomycin resistance.
TABLE 2.
Primers used in this study
| Primer | Sequence (5′–3′) |
|---|---|
| Strain construction | |
| fur-Red-F | CGC TTC CTC GTT TAA AAA TTC TGG AAG TTC TTC AGG AAC CTG TAG GCT GGA GCT GCT TCG |
| fur-Red-F | TCG TGA TGA TGC TGT TGC GTC AGT TCA AAA ACG GAT TTA CAT TCC GGG GAT CCG TCG ACC |
| fur-lacZ-F | GCG ACT GCC GCG AAG ACG AGC ACG CGC ACG ATG ACG CGA CTA AAT AAG TGC CCG TCG TTT TAC AAC GTC G |
| fur-lacZ-R | CAA CAT CAA GCG GCA GGA AAG AGG AGG ATA TAA AAA AGC CAA CCG GGC GGC GTG TAG GCT GGA GCT GCT TC |
| fhuA-Red-F | ATC GTT TAC GTT ATC ATT CAC TTT CAT CAG AGA TAT ACC ATG TAG GCT GGA GCT GCT TCG |
| fhuA-Red-R | CCT GCG CTA ATG GGT TGG TTG GAT CGG CGG TCA GGT TAT TAT TCC GGG GAT CCG TCG ACC |
| Plasmid construction | |
| pHis6x-Fur-F | CAT GTT CTG AAT TCA AAT TAT GCA TCA CCA TCA CCA TCA CGC AAT GAC TGA CAA CAA TAC CGC ATT AAA |
| pHis6x-Fur-R | ACC GGG CGG TTG GAT CCT CGA AAG ATT T |
| pUT18C-Fur-F | TTA GCA ACA GGA GGA TCC CCG CAT GAC T |
| qRT-PCR | |
| fhuA-qRT-F | GTT CAA CCG AAA GAA GAA ACC ATT A |
| fhuA-qRT-R | GTT TTT TCG ATA GGT GTA TCA GTT TTG |
| fepA-qRT-F | AGA AGA TTC ATT CCC TGA CCT TAC TG |
| fepA-qRT-R | TAT CGG TTT TGT CTT CCG CCA TCA |
| iroB-qRT-F | ATG CGT ATT CTG TTT GTC GGT CCA |
| iroB-qRT-R | CAG TAC TTC ATG GCC ATT AAC ACG A |
| rrs-qRT-F | CCA CAA AAC TTA TGG ATT TAT GCG T |
| rrs-qRT-R | TTT ACG CCC AGT AAT TCC GAT T |
| EMSA | |
| Fur-EMSA-F1 | CTC GAC GAC ATC CTC AAC GCC TAA TCT |
| Fur-EMSA-R1 | AAA CGA GGA AGC GTT ACT TTC AGG CCA G |
Construction of bacterial strains.
The method of Datsenko and Wanner (41) was used for chromosomal gene deletion and epitope tagging.
For construction of the fur deletion strain, the kanamycin resistance (Kmr) cassette from plasmid pKD13 was amplified using primers fur-RED-F and fur-RED-R. The resulting PCR products were introduced into the SL1344 strain containing plasmid pKD46, followed by selection for Δfur::kan transformants. The Kmr cassette was removed using plasmid pCP20 (41).
A Salmonella strain expressing the Fur protein with a FLAG tag at the C terminus in the normal fur chromosomal location was constructed. The Kmr cassette from plasmid pKD13 was amplified using primers Fur-FLAG-F and Fur-FLAG-R, and the PCR products were introduced into the SL1344 strain harboring plasmid pKD46. The Kmr cassette was removed using plasmid pCP20.
A strain carrying a lacZ fusion to the fhuA gene was constructed as described previously (42). The Kmr cassette from plasmid pKD13 was amplified using primers fhuA-RED-F and fhuA-RED-R. The resulting PCR products were introduced into the SL1344 strain harboring plasmid pKD46, and the Kmr cassette was removed using plasmid pCP20. Finally, the lacZY genes were introduced into the flippase recognition target (FRT) site using plasmid pCE70 (43).
A strain carrying a lacZ fusion to the fur gene was constructed as described previously (42). The Kmr cassette from plasmid pKD13 was amplified using primers fur-lacZ-F/fur-lacZ-R. The resulting PCR products were introduced into the SL1344 strain harboring plasmid pKD46, and the Kmr cassette was removed using plasmid pCP20. Finally, the lacZY genes were introduced into the FRT site using plasmid pCE70 (43).
Plasmid construction.
A plasmid expressing Fur protein with a His6 tag at N terminus from the lac promoter was constructed as follows: the fur gene was amplified from wild-type Salmonella (SL1344) using primers pHis6x-Fur-F/pHis6x-Fur-R, and the PCR fragments were introduced between the EcoRI and BamHI sites of the pUHE21-2lacIq plasmid vector.
A plasmid expressing T18-Fur fusion protein was constructed as follows: the fur gene was amplified from wild-type Salmonella (SL1344) using primers pUT18C-Fur-F/pHis6x-Fur-R and then introduced between the BamHI and EcoRI sites of pUT18C (44).
β-Galactosidase assay.
β-Galactosidase assays were carried out with at least three biological replicates with technical duplicates, and the activity was determined as described previously (45).
RNA isolation and reverse transcriptase-quantitative PCR (qRT-PCR).
Salmonella strains were grown as described above, and total RNA was isolated using an RNeasy minikit (Qiagen). After DNase treatment of the isolated RNA, cDNA was synthesized using Omni Transcript reverse transcription reagents (Qiagen) and random hexamers (Invitrogen). Quantification of the cDNA was carried out using 2× iQ SYBR green Supermix (Bio-Rad), and real-time amplification of the PCR products was performed using an iCycler iQ real-time detection system (Bio-Rad). The primers used for detection of the gene transcripts are listed in Table 2. Data were normalized to 16S rRNA expression levels.
Western blotting.
Salmonella strains encoding the Fur-FLAG protein from the normal chromosomal location were grown under the indicated conditions. Bacteria were collected by centrifugation, and cell lysates were prepared using B-PER solution (Pierce). Proteins from cell lysates were resolved by 12% SDS-PAGE, and the Fur and DnaK proteins were detected using anti-FLAG (Sigma; 1:2,000) and anti-DnaK (Abcam; 1:5,000) antibodies. The blots were developed using anti-mouse IgG horseradish peroxidase-linked antibody (GE Healthcare; 1:5,000) with an ECL detection system (Amersham Biosciences).
Bacterial two-hybrid assay.
E. coli BTH101 (which lacks the cya gene) was used for this assay. Derivatives of plasmids pUT18 and pKT25 were introduced into BTH101 cells as indicated. These cells were grown overnight in M9 minimal media containing ampicillin (100 µg/ml) and kanamycin (50 µg/ml). They were transferred to 1 ml of the same fresh medium containing 0.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside) at a dilution of 1:100 and grown for 8 h with shaking at 30°C. β-Galactosidase activities were determined as described above.
Protein purification.
His6-tagged Fur protein was expressed in E. coli strain BL21(DE3) as follows: bacterial cells were grown in LB at 37°C until the optical density at 600 nm (OD600) reached 0.5, and expression of those proteins was induced by addition of IPTG (0.5 M) followed by growth at 30°C for 5 h. Cells were harvested, washed, and suspended in buffer A (20 mM Tris [pH 8.0], 150 mM NaCl, 20 mM imidazole). The cells were then disrupted by sonication, and cell debris was removed by centrifugation at 20,000 × g at 4°C for 30 min. The supernatant was applied to a 1.5-ml nickel-nitrilotriacetic acid (Ni-NTA) agarose column equilibrated in buffer A, washed with 25 column volumes of the same buffer, and eluted using a gradient of buffer A and buffer B (20 mM Tris [pH 8.0], 150 mM NaCl, 250 mM imidazole). The fractions were then collected and analyzed by SDS/PAGE, and selected fractions were dialyzed against buffer C (20 mM Tris [pH 8.0], 150 mM NaCl, 10% glycerol).
Electrophoretic mobility shift assay (EMSA).
EMSAs were performed to determine binding of Fur to DNA in vitro. DNA fragments corresponding to the fhuA promoter was amplified by PCR using 32P-labeled primers EMSA-fhuA-F1/EMSA-fhuA-R1 with wild-type Salmonella chromosomal DNA as a template. The promoter DNA was purified from agarose gels using a gel extraction kit (Qiagen). The labeled DNA probe (16 fmol) was incubated with the His6-Fur protein in the presence or absence of EIIANtr-His6 at room temperature for 20 min in 20 μl of binding buffer (10 mM Tris [pH 7.5], 50 mM KCl, 5 mM MgCl2, 0.5 mM dithiothreitol [DTT], 2.5% glycerol) containing 50 ng/μl poly(dI-dC). The reaction mixtures were resolved by 6% PAGE, and the radiolabeled DNA fragments were visualized using a BAS2500 system (Fuji Film).
Protein co-occurrence.
Co-occurrence of EIIANtr and Fur proteins across sequenced organisms was analyzed using STRING software version 11.0.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dongwoo Shin for providing a plasmid, pFur.
This research was supported by a grant (14162MFDS972) from the Ministry of Food and Drug Safety of Korea in 2018. This work was also supported by the BK21 Plus Program of the Department of Agricultural Biotechnology, Seoul National University, Seoul, South Korea.
We declare that we have no conflict of interest.
J.C. and S.R. designed the research; J.C. performed the experiments; J.C. and S.R. analyzed the data; and J.C. and S.R. wrote the paper.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.03026-18.
REFERENCES
- 1.Neilands JB. 1981. Iron absorption and transport in microorganisms. Annu Rev Nutr 1:27–46. doi: 10.1146/annurev.nu.01.070181.000331. [DOI] [PubMed] [Google Scholar]
- 2.Boccio JR, Iyengar V. 2003. Iron deficiency: causes, consequences, and strategies to overcome this nutritional problem. Biol Trace Elem Res 94:1–32. doi: 10.1385/BTER:94:1:1. [DOI] [PubMed] [Google Scholar]
- 3.Sheldon JR, Laakso HA, Heinrichs DE. 18 March 2016. Iron acquisition strategies of bacterial pathogens. Microbiol Spectr 4. doi: 10.1128/microbiolspec.VMBF-0010-2015. [DOI] [PubMed] [Google Scholar]
- 4.Nairz M, Dichtl S, Schroll A, Haschka D, Tymoszuk P, Theurl I, Weiss G. 2018. Iron and innate antimicrobial immunity—depriving the pathogen, defending the host. J Trace Elem Med Biol 48:118–133. doi: 10.1016/j.jtemb.2018.03.007. [DOI] [PubMed] [Google Scholar]
- 5.Andrews SC, Robinson AK, Rodriguez QF. 2003. Bacterial iron homeostasis. FEMS Microbiol Rev 27:215–237. doi: 10.1016/S0168-6445(03)00055-X. [DOI] [PubMed] [Google Scholar]
- 6.Bagg A, Neilands JB. 1987. Ferric uptake regulation protein acts as a repressor, employing iron(II) as a cofactor to bind the operator of an iron transport operon in Escherichia coli. Biochemistry 26:5471–5477. doi: 10.1021/bi00391a039. [DOI] [PubMed] [Google Scholar]
- 7.D'Autreaux B, Pecqueur L, Gonzalez de Peredo A, Diederix RE, Caux-Thang C, Tabet L, Bersch B, Forest E, Michaud-Soret I. 2007. Reversible redox- and zinc-dependent dimerization of the Escherichia coli Fur protein. Biochemistry 46:1329–1342. doi: 10.1021/bi061636r. [DOI] [PubMed] [Google Scholar]
- 8.Carpenter BM, Whitmire JM, Merrell DS. 2009. This is not your mother's repressor: the complex role of Fur in pathogenesis. Infect Immun 77:2590–2601. doi: 10.1128/IAI.00116-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Powell BS, Court DL, Inada T, Nakamura Y, Michotey V, Cui X, Reizer A, Saier MH Jr, Reizer J. 1995. Novel proteins of the phosphotransferase system encoded within the rpoN operon of Escherichia coli. Enzyme IIANtr affects growth on organic nitrogen and the conditional lethality of an erats mutant. J Biol Chem 270:4822–4839. doi: 10.1074/jbc.270.9.4822. [DOI] [PubMed] [Google Scholar]
- 10.Lee CR, Cho SH, Yoon MJ, Peterkofsky A, Seok YJ. 2007. Escherichia coli enzyme IIANtr regulates the K+ transporter TrkA. Proc Natl Acad Sci U S A 104:4124–4129. doi: 10.1073/pnas.0609897104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luttmann D, Heermann R, Zimmer B, Hillmann A, Rampp IS, Jung K, Gorke B. 2009. Stimulation of the potassium sensor KdpD kinase activity by interaction with the phosphotransferase protein IIANtr in Escherichia coli. Mol Microbiol 72:978–994. doi: 10.1111/j.1365-2958.2009.06704.x. [DOI] [PubMed] [Google Scholar]
- 12.Luttmann D, Gopel Y, Gorke B. 2012. The phosphotransferase protein IIANtr modulates the phosphate starvation response through interaction with histidine kinase PhoR in Escherichia coli. Mol Microbiol 86:96–110. doi: 10.1111/j.1365-2958.2012.08176.x. [DOI] [PubMed] [Google Scholar]
- 13.Choi J, Shin D, Yoon H, Kim J, Lee CR, Kim M, Seok YJ, Ryu S. 2010. Salmonella pathogenicity island 2 expression negatively controlled by EIIANtr-SsrB interaction is required for Salmonella virulence. Proc Natl Acad Sci U S A 107:20506–20511. doi: 10.1073/pnas.1000759107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karstens K, Zschiedrich CP, Bowien B, Stulke J, Gorke B. 2014. Phosphotransferase protein EIIANtr interacts with SpoT, a key enzyme of the stringent response, in Ralstonia eutropha H16. Microbiology 160:711–722. doi: 10.1099/mic.0.075226-0. [DOI] [PubMed] [Google Scholar]
- 15.Ronneau S, Petit K, De Bolle X, Hallez R. 2016. Phosphotransferase-dependent accumulation of (p)ppGpp in response to glutamine deprivation in Caulobacter crescentus. Nat Commun 7:11423. doi: 10.1038/ncomms11423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yoo W, Yoon H, Seok YJ, Lee CR, Lee HH, Ryu S. 2016. Fine-tuning of amino sugar homeostasis by IIANtr in Salmonella Typhimurium. Sci Rep 6:33055. doi: 10.1038/srep33055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee CR, Park YH, Kim M, Kim YR, Park S, Peterkofsky A, Seok YJ. 2013. Reciprocal regulation of the autophosphorylation of enzyme INtr by glutamine and alpha-ketoglutarate in Escherichia coli. Mol Microbiol 88:473–485. doi: 10.1111/mmi.12196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rathman M, Sjaastad MD, Falkow S. 1996. Acidification of phagosomes containing Salmonella typhimurium in murine macrophages. Infect Immun 64:2765–2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cassat JE, Skaar EP. 2013. Iron in infection and immunity. Cell Host Microbe 13:509–519. doi: 10.1016/j.chom.2013.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Choi J, Groisman EA. 2013. The lipopolysaccharide modification regulator PmrA limits Salmonella virulence by repressing the type three-secretion system Spi/Ssa. Proc Natl Acad Sci U S A 110:9499–9504. doi: 10.1073/pnas.1303420110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Choi E, Kim H, Lee H, Nam D, Choi J, Shin D. 2014. The iron-sensing Fur regulator controls expression timing and levels of Salmonella pathogenicity island 2 genes in the course of environmental acidification. Infect Immun 82:2203–2210. doi: 10.1128/IAI.01625-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Srikumar S, Kroger C, Hebrard M, Colgan A, Owen SV, Sivasankaran SK, Cameron AD, Hokamp K, Hinton JC. 2015. RNA-seq brings new insights to the intra-macrophage transcriptome of Salmonella Typhimurium. PLoS Pathog 11:e1005262. doi: 10.1371/journal.ppat.1005262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Troxell B, Sikes ML, Fink RC, Vazquez-Torres A, Jones-Carson J, Hassan HM. 2011. Fur negatively regulates hns and is required for the expression of HilA and virulence in Salmonella enterica serovar Typhimurium. J Bacteriol 193:497–505. doi: 10.1128/JB.00942-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stojiljkovic I, Baumler AJ, Hantke K. 1994. Fur regulon in gram-negative bacteria. Identification and characterization of new iron-regulated Escherichia coli genes by a Fur titration assay. J Mol Biol 236:531–545. doi: 10.1006/jmbi.1994.1163. [DOI] [PubMed] [Google Scholar]
- 25.Hamed MY, Neilands JB, Huynh V. 1993. Binding of the ferric uptake regulation repressor protein (Fur) to Mn(II), Fe(II), Co(II), and Cu(II) ions as co-repressors: electronic absorption, equilibrium, and 57Fe Mossbauer studies. J Inorg Biochem 50:193–210. doi: 10.1016/0162-0134(93)80025-5. [DOI] [PubMed] [Google Scholar]
- 26.Gravina F, Sanchuki HS, Rodrigues TE, Gerhardt ECM, Pedrosa FO, Souza EM, Valdameri G, de Souza GA, Huergo LF. 2018. Proteome analysis of an Escherichia coli ptsN-null strain under different nitrogen regimes. J Proteomics 174:28–35. doi: 10.1016/j.jprot.2017.12.006. [DOI] [PubMed] [Google Scholar]
- 27.Troxell B, Fink RC, Porwollik S, McClelland M, Hassan HM. 2011. The Fur regulon in anaerobically grown Salmonella enterica sv. Typhimurium: identification of new Fur targets. BMC Microbiol 11:236. doi: 10.1186/1471-2180-11-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karimova G, Pidoux J, Ullmann A, Ladant D. 1998. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc Natl Acad Sci U S A 95:5752–5756. doi: 10.1073/pnas.95.10.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sharma R, Shimada T, Mishra VK, Upreti S, Sardesai AA. 2016. Growth inhibition by external potassium of Escherichia coli lacking PtsN (EIIANtr) is caused by potassium limitation mediated by YcgO. J Bacteriol 198:1868–1882. doi: 10.1128/JB.01029-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kortman GA, Boleij A, Swinkels DW, Tjalsma H. 2012. Iron availability increases the pathogenic potential of Salmonella Typhimurium and other enteric pathogens at the intestinal epithelial interface. PLoS One 7:e29968. doi: 10.1371/journal.pone.0029968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nagy TA, Moreland SM, Andrews-Polymenis H, Detweiler CS. 2013. The ferric enterobactin transporter Fep is required for persistent Salmonella enterica serovar Typhimurium infection. Infect Immun 81:4063–4070. doi: 10.1128/IAI.00412-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tuttobene MR, Cribb P, Mussi MA. 2018. BlsA integrates light and temperature signals into iron metabolism through Fur in the human pathogen Acinetobacter baumannii. Sci Rep 8:7728. doi: 10.1038/s41598-018-26127-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mussi MA, Gaddy JA, Cabruja M, Arivett BA, Viale AM, Rasia R, Actis LA. 2010. The opportunistic human pathogen Acinetobacter baumannii senses and responds to light. J Bacteriol 192:6336–6345. doi: 10.1128/JB.00917-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wosten MM, Kox LF, Chamnongpol S, Soncini FC, Groisman EA. 2000. A signal transduction system that responds to extracellular iron. Cell 103:113–125. doi: 10.1016/S0092-8674(00)00092-1. [DOI] [PubMed] [Google Scholar]
- 35.Perez JC, Groisman EA. 2007. Acid pH activation of the PmrA/PmrB two-component regulatory system of Salmonella enterica. Mol Microbiol 63:283–293. doi: 10.1111/j.1365-2958.2006.05512.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zaharik ML, Vallance BA, Puente JL, Gros P, Finlay BB. 2002. Host-pathogen interactions: host resistance factor Nramp1 up-regulates the expression of Salmonella pathogenicity island-2 virulence genes. Proc Natl Acad Sci U S A 99:15705–15710. doi: 10.1073/pnas.252415599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hood MI, Skaar EP. 2012. Nutritional immunity: transition metals at the pathogen-host interface. Nat Rev Microbiol 10:525–537. doi: 10.1038/nrmicro2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paradkar PN, De Domenico I, Durchfort N, Zohn I, Kaplan J, Ward DM. 2008. Iron depletion limits intracellular bacterial growth in macrophages. Blood 112:866–874. doi: 10.1182/blood-2007-12-126854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Plant J, Glynn AA. 1974. Natural resistance to Salmonella infection, delayed hypersensitivity and Ir genes in different strains of mice. Nature 248:345–347. doi: 10.1038/248345a0. [DOI] [PubMed] [Google Scholar]
- 40.Watanabe T, Ogata Y, Chan RK, Botstein D. 1972. Specialized transduction of tetracycline resistance by phage P22 in Salmonella typhimurium: I. Transduction of R factor 222 by phage P22. Virology 50:874–882. doi: 10.1016/0042-6822(72)90441-2. [DOI] [PubMed] [Google Scholar]
- 41.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ellermeier CD, Janakiraman A, Slauch JM. 2002. Construction of targeted single copy lac fusions using lambda Red and FLP-mediated site-specific recombination in bacteria. Gene 290:153–161. doi: 10.1016/S0378-1119(02)00551-6. [DOI] [PubMed] [Google Scholar]
- 43.Merighi M, Ellermeier CD, Slauch JM, Gunn JS. 2005. Resolvase-in vivo expression technology analysis of the Salmonella enterica serovar Typhimurium PhoP and PmrA regulons in BALB/c mice. J Bacteriol 187:7407–7416. doi: 10.1128/JB.187.21.7407-7416.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Karimova G, Ullmann A, Ladant D. 2001. Protein-protein interaction between Bacillus stearothermophilus tyrosyl-tRNA synthetase subdomains revealed by a bacterial two-hybrid system. J Mol Microbiol Biotechnol 3:73–82. [PubMed] [Google Scholar]
- 45.Miller JH. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Plainview, NY. [Google Scholar]
- 46.Lucas RL, Lee CA. 2000. Unravelling the mysteries of virulence gene regulation in Salmonella typhimurium. Mol Microbiol 36:1024–1033. doi: 10.1046/j.1365-2958.2000.01961.x. [DOI] [PubMed] [Google Scholar]
- 47.Hanahan D. 1983. Studies on transformation of Escherichia coli with plasmids. J Mol Biol 166:557–580. doi: 10.1016/S0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 48.Soncini FC, Vescovi EG, Groisman EA. 1995. Transcriptional autoregulation of the Salmonella typhimurium phoPQ operon. J Bacteriol 177:4364–4371. doi: 10.1128/jb.177.15.4364-4371.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.