Dietary protein intake is high in Western populations, which could result in potentially harmful metabolites in the gut from proteolysis. In an in vitro fermentation model, the addition of prebiotics reduced the negative consequences of high protein levels. Supplementation with a prebiotic resulted in a reduction of proteolytic metabolites in the model. A difference was seen in protein fermentation between omnivore and vegetarian gut microbiotas: bacteria from vegetarian donors grew more on soy and Quorn than on meat and casein, with reduced ammonia production. Bacteria from vegetarian donors produced less branched-chain fatty acids (BCFA).
KEYWORDS: diet, gut microbiota, prebiotics, protein fermentation, vegetarian
ABSTRACT
Metabolism of protein by gut bacteria is potentially detrimental due to the production of toxic metabolites, such as ammonia, amines, p-cresol, and indole. The consumption of prebiotic carbohydrates results in specific changes in the composition and/or activity of the microbiota that may confer benefits to host well-being and health. Here, we have studied the impact of prebiotics on proteolysis within the gut in vitro. Anaerobic stirred batch cultures were inoculated with feces from omnivores (n = 3) and vegetarians (n = 3) and four protein sources (casein, meat, mycoprotein, and soy protein) with and without supplementation by an oligofructose-enriched inulin. Bacterial counts and concentrations of short-chain fatty acids (SCFA), ammonia, phenol, indole, and p-cresol were monitored during fermentation. Addition of the fructan prebiotic Synergy1 increased levels of bifidobacteria (P = 0.000019 and 0.000013 for omnivores and vegetarians, respectively). Branched-chain fatty acids (BCFA) were significantly lower in fermenters with vegetarians’ feces (P = 0.004), reduced further by prebiotic treatment. Ammonia production was lower with Synergy1. Bacterial adaptation to different dietary protein sources was observed through different patterns of ammonia production between vegetarians and omnivores. In volunteer samples with high baseline levels of phenol, indole, p-cresol, and skatole, Synergy1 fermentation led to a reduction of these compounds.
IMPORTANCE Dietary protein intake is high in Western populations, which could result in potentially harmful metabolites in the gut from proteolysis. In an in vitro fermentation model, the addition of prebiotics reduced the negative consequences of high protein levels. Supplementation with a prebiotic resulted in a reduction of proteolytic metabolites in the model. A difference was seen in protein fermentation between omnivore and vegetarian gut microbiotas: bacteria from vegetarian donors grew more on soy and Quorn than on meat and casein, with reduced ammonia production. Bacteria from vegetarian donors produced less branched-chain fatty acids (BCFA).
INTRODUCTION
Dietary protein levels in western European populations can be as high as 105 g/day according to the Food and Agriculture Organization (1). However, the recommended dietary allowance (RDA) is 56 g/day for men and 46 g/day for women (2). This may result in high residual colonic nitrogen, with dietary protein having escaped digestion in the upper intestine entering the large gut where it can become a substrate for the colonic microbiota. Approximately 16 g of protein will be present in the colon following ingestion of 105 g protein/day, of which 8 g is endogenous and 8 g is exogenous (3, 4). Among the endogenous proteins, there are 69.2% bacterial proteins, 16.9% mucin, 7.65% enzymes, and 6.2% mucosal cells (5, 6).
Anaerobic metabolism of carbohydrate by gut bacteria produces short-chain fatty acids (SCFA) and gases from different pathways. Production of SCFA, mainly acetate, propionate, and butyrate, in the lumen is generally believed to mediate health benefits, such as maintaining colonic epithelial cell function, regulating energy intake and satiety, controlling inflammation, and defending against pathogen invasion (7). Microbial breakdown of protein generates not only SCFA and gases, however, but also ammonia, amines, indolic and phenolic compounds, and branched-chain fatty acids (BCFA) through the deamination and decarboxylation of amino acids (8). Though evidence in humans is scarce, in studies in rats and in ex vivo studies, ammonia at a physiologically relevant dose can harm colon barrier function and shorten the colonocyte life span and is cocarcinogenic in rats (9–11). Hydrogen sulfide can be produced from sulfur-containing amino acids and is toxic to colonocytes, damaging DNA and blocking the utilization of butyrate as an energy source (12–15). Metabolism of tyrosine, phenylalanine, and tryptophan produces phenol, indole, p-cresol, and skatole, which are potential carcinogens; phenol and p-cresol can reduce intestinal epithelial barrier function in vitro (10, 16, 17). BCFA are generated from branched-chain amino acids such as valine, leucine, and isoleucine, which make them biomarkers for bacterial proteolysis; however, there are no known human physiological roles for BCFA (18).
Thus, foods entering the colon can have a health impact on the host, possibly by changing gut microbiota composition and activity. The International Agency for Research on Cancer (19), an agency under the World Health Organization (WHO), published a press release in October 2015 where it classified red meat as “probably carcinogenic to humans” and processed meat as “carcinogenic to humans,” with concerns over colorectal cancer (19). Some epidemiological studies found reduced risk of colorectal cancer (CRC) with high consumption of dietary fiber, while red meat and processed meat had a positive correlation with CRC (20–23). Animal protein intake was associated with increased inflammatory bowel disease (IBD) risk in two Japanese and French studies (24, 25).
An increased consumption of prebiotics, which can reach the colon resulting in specific changes in the composition and/or activity in the gastrointestinal microflora, may counter the negative effects of gut microbial proteolysis in persons ingesting high-protein diets (26). Inulin-type fructans can resist hydrolytic enzymes in the human gastrointestinal (GI) tract and are resistant to small intestinal absorption; subsequently, they become a substrate source for the microbiota within the large intestine. The impact of inulin on the gut microbiome has been studied using in vitro and in vivo approaches (27–29). The aim of this study was to understand the metabolism of gut bacterial proteolysis in the distal colon and how prebiotics can affect the proteolysis to investigate the potential of consuming prebiotics to counteract the negative effect of a high-protein diet.
RESULTS
Bacterial enumeration.
Total bacteria and most microbial groups that were monitored in this study reached the highest number after 24 h of incubation. However, lactobacillus, Faecalibacterium prausnitzii, and Roseburia numbers only increased in the first 10 h, with lactobacillus numbers in particular declining after 10 h. Bacterial populations from omnivores and vegetarians responded differently to the proteins: fecal bacteria from omnivores had insignificantly higher counts on meat and casein than on soy protein and Quorn extract, while fecal bacteria from vegetarians had higher counts on soy protein and Quorn extract (8.75 ± 0.40 log10 CFU/ml) than on meat and casein (8.38 ± 0.47 log10 CFU/ml) (P = 0.03).
The vegetarian microbiota had higher bifidobacterium and lactobacillus counts at the beginning than omnivore microbiota (see Tables S1 and S2 in the supplemental material).
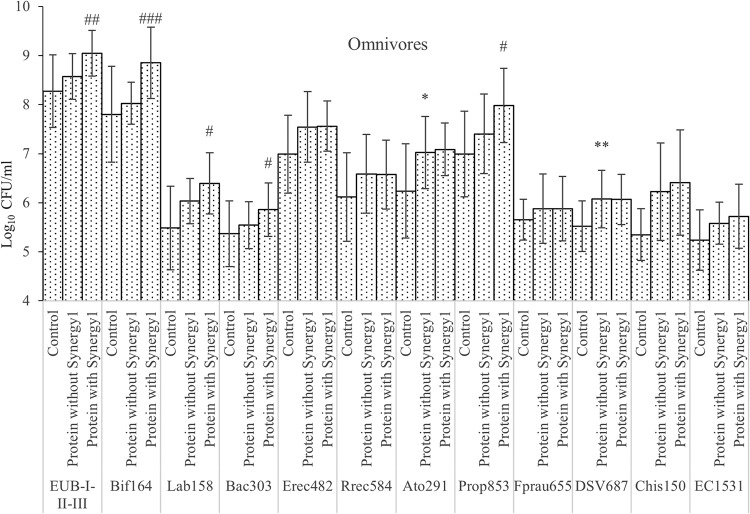
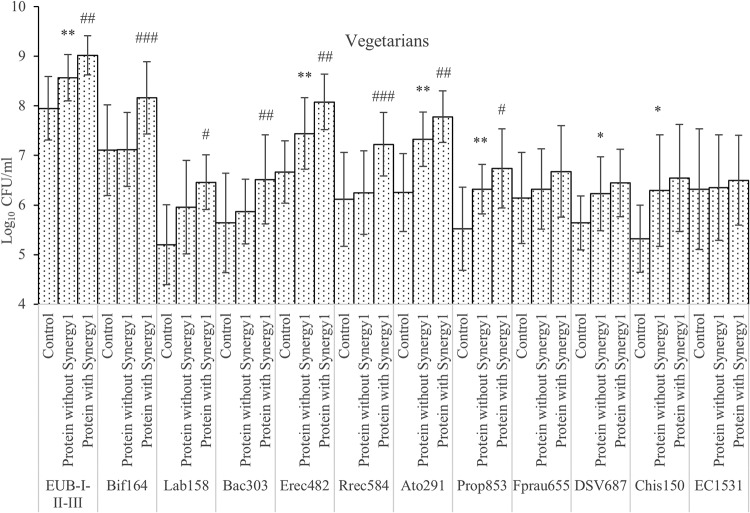
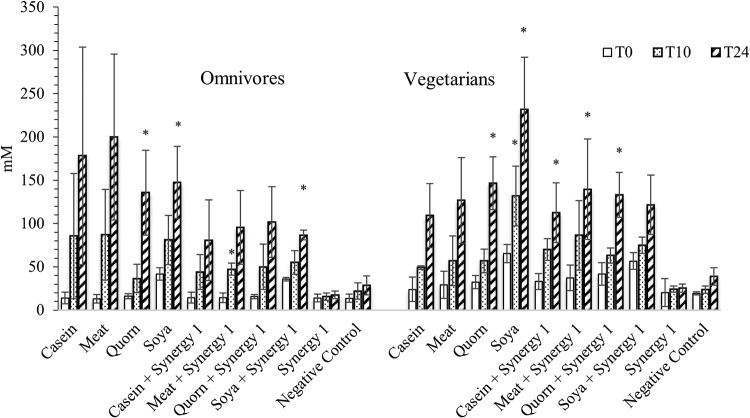
To investigate proteolytic bacteria, independent t tests were performed to compare samples with protein addition (casein, meat, mycoprotein, and soy protein) and controls at 24 and 48 h (Fig. 1 and 2). Though there are studies confirming that many Bacteroides spp. are proteolytic (30), we found no significant changes in Bacteroides spp. on protein substrates. Clostridium coccoides, Eubacterium rectale, and Clostridium clusters XIVa and XIVb grew on protein substrates: bacteria from omnivore donors had higher counts than the control group (P = 0.055), while those from vegetarian donors were significantly higher (P < 0.01). Roseburia numbers did not change with protein added. The Atopobium cluster from both omnivore and vegetarian donors grew on protein substrates, with statistical significance. Clostridial cluster IX populations in cultures inoculated with samples from vegetarian donors increased significantly on the protein substrates, while cultures with omnivore samples were not statistically different. Lower counts of clostridial cluster IX in vegetarian donors’ controls might explain the difference. Desulfovibrio counts were significantly higher with protein from both omnivore and vegetarian donors. Clostridium clusters I and II also grew more on proteins; however, growth only reached statistical significance with inocula from vegetarians.
FIG 1.
Bacterial counts in the single-stage batch culture as analyzed by FISH. Values are mean values at 24 and 48 h of fermentation from 3 omnivores’ microbiota ± standard deviations. *, mean values were significantly different between control and protein without Synergy1 (P < 0.05); **, mean values were significantly different between control and protein without Synergy1 (P < 0.01); #, mean values were significantly different between protein with and without Synergy1 (P < 0.05); ##, mean values were significantly different between protein with and without Synergy1 (P < 0.01); ###, mean values were significantly different between protein with and without Synergy1 (P < 0.001).
FIG 2.
Bacterial counts in the single-stage batch culture as analyzed by FISH. Values are mean values at 24 and 48 h of fermentation from 3 vegetarians’ microbiota ± standard deviations. *, mean values were significantly different between control and protein without Synergy1 (P < 0.05); **, mean values were significantly different between control and protein without Synergy1 (P < 0.01); #, mean values were significantly different between protein with and without Synergy1 (P < 0.05); ##, mean values were significantly different between protein with and without Synergy1 (P < 0.01); ###, mean values were significantly different between protein with and without Synergy1 (P < 0.001).
To investigate how prebiotics may modify the microbiota, independent t tests were used to compare cultures with prebiotics and without after 24 and 48 h of fermentation (Fig. 1 and 2). Synergy1 addition significantly boosted the growth of total bacteria, bacteroides, clostridial cluster IX, bifidobacteria, and lactobacilli with both omnivore and vegetarian inocula, with bifidobacteria displaying the highest growth on Synergy1. In cultures with vegetarian donor samples, Clostridium coccoides, Eubacterium rectale, Clostridium clusters XIVa and XIVb, Roseburia, Faecalibacterium prausnitzii, and Atopobium also had significant higher counts with prebiotics than without. There were no inhibitory effects of prebiotics found on any of bacterial groups monitored in this study.
Organic acids.
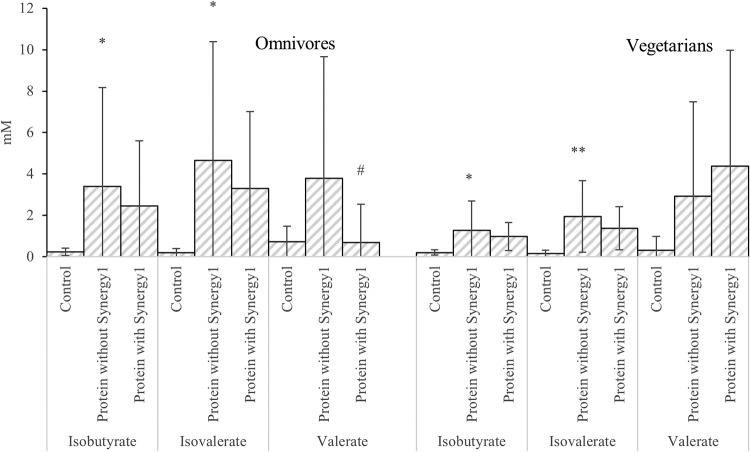
Most organic acids accumulated during fermentation and reached their highest concentrations at 24 or 48 h of fermentation, with the exception of lactate, which transiently increased during the first 10 h and then gradually decreased to below 1 mM at 48 h. Branched amino acids such as leucine and isoleucine can be metabolized by fecal bacteria to produce BCFA, indicating proteolytic fermentation. Omnivores had higher BCFA production (4.03 ± 5.25 mM), while vegetarians had little production (1.61 ± 1.60 mM) (P = 0.004). For instance, while growing on casein, bacteria from omnivores produced 10.19 ± 8.62 and 13.13 ± 10.93 mM isobutyrate and isovalerate, respectively, while bacteria from vegetarians produced 2.03 ± 2.16 and 3.52 ± 3.29 mM isobutyrate and isovalerate, respectively (Tables S1 and S2).
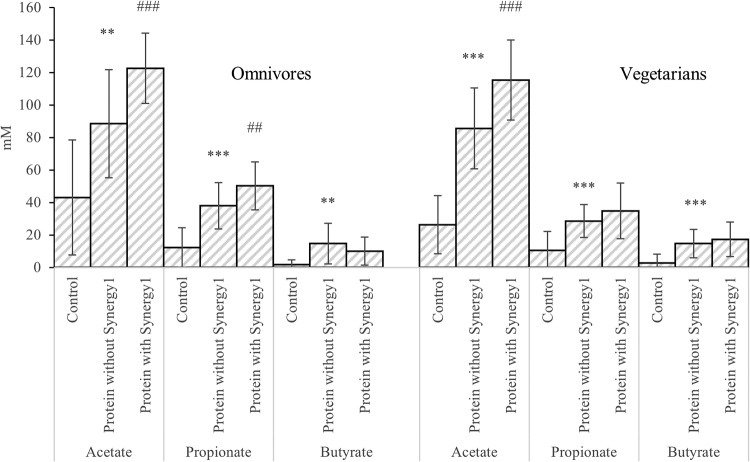
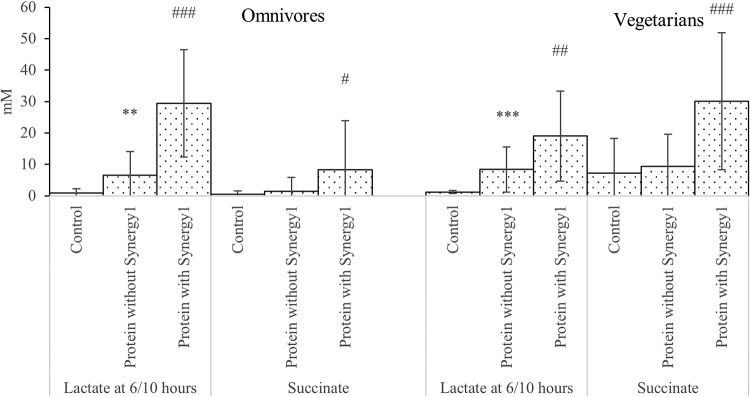
Comparing samples with protein and without at 24 and 48 h, cultures inoculated with both omnivore and vegetarian donors had significantly higher concentrations of acetate, propionate, isobutyrate, butyrate, and isovalerate on protein (Fig. 3 and 4). However, fermentation samples with prebiotics had significantly elevated concentrations of acetate and succinate at 24 and 48 h, and significantly more lactate at 6 and 10 h (Fig. 3 and 5).
FIG 3.
SCFA differences between samples with and without protein in the single-stage batch culture. Values are mean values at 24 and 48 h of fermentation from 3 omnivores’ microbiota and 3 vegetarians’ microbiota ± standard deviations. **, mean values were significantly different between control and protein without Synergy1 (P < 0.01); ***, mean values were significantly different between control and protein without Synergy1 (P < 0.001); #, mean values were significantly different between protein with and without Synergy1 (P < 0.05); ##, mean values were significantly different between protein with and without Synergy1 (P < 0.01); ###, mean values were significantly different between protein with and without Synergy1 (P < 0.001).
FIG 4.
BCFA and valerate differences between samples with and without protein in the single-stage batch culture. Values are mean values at 24 and 48 h of fermentation from 3 omnivores’ microbiota and 3 vegetarians’ microbiota ± standard deviations. *, mean values were significantly different between control and protein without Synergy1 (P < 0.05); **, mean values were significantly different between control and protein without Synergy1 (P < 0.01); #, mean values were significantly different between protein with and without Synergy1 (P < 0.05).
FIG 5.
Lactate and succinate differences between samples with and without protein in the single-stage batch culture. Values are mean values at 24 and 48 h of fermentation unless specified from 3 omnivores’ microbiota and 3 vegetarians’ microbiota ± standard deviations. **, mean values were significantly different between control and protein without Synergy1 (P < 0.01); ***, mean values were significantly different between control and protein without Synergy1 (P < 0.001); #, mean values were significantly different between protein with and without Synergy1 (P < 0.05); ##, mean values were significantly different between protein with and without Synergy1 (P < 0.01); ###, mean values were significantly different between protein with and without Synergy1 (P < 0.001).
Butyric acid production was low in this study, and no changes were found in cultures with omnivore samples; this correlates with the lack of differences in populations of butyrate-producing bacteria (Roseburia and Faecalibacterium prausnitzii). In samples with vegetarian donors’ inocula, butyrate producers (Clostridium coccoides, Eubacterium rectale, Clostridium clusters XIVa and XIVb, Roseburia, and Faecalibacterium prausnitzii) had significantly higher counts; however, butyrate production was not significantly increased.
Concentrations of BCFA were lower on prebiotics, although without statistical significance. Variation in BCFA production between donors was seen in this study; therefore, a two-way analysis of variance (ANOVA) for isovalerate and isobutyrate was used to examine the effect of both treatment and donor on production. A significant influence of donor on isobutyrate and isovalerate was found with six donors (P < 0.01). Donor variation may indicate that a larger sample size is needed to observe the inhibitory effect of prebiotics on BCFA production (see Table S3).
Volatile organic compounds.
This study quantified four potentially detrimental volatile organic compounds (VOCs), which were indole, phenol, p-cresol, and skatole. The production of these compounds varied with individual donors, and the effect of prebiotics on VOC production also varied according to donor diet. The production of VOCs, from highest to lowest, was indole, phenol, p-cresol, and skatole in most cases. However, with soy protein, phenol production was higher than indole production. With all donors, comparing negative and positive controls, the production of volatile compounds was reduced by Synergy1. However, comparing cultures on protein plus Synergy1 with cultures on the corresponding protein, the production of indole, phenol, p-cresol, and skatole was inhibited by Synergy1 after 48 h of fermentation with inocula from omnivore donor 1, omnivore donor 2, and vegetarian donor 1. Inocula from these three donors resulted in the production of relatively high levels of phenol and indole on protein (292.20 ± 521.76 μg/ml) compared with those from others (28.92 ± 23.61 μg/ml) (P = 0.02). In fermentation models inoculated with these high VOC producers, Synergy1 plus protein models produced significantly less phenol and indole (113.21 ± 227.94 μg/ml) (P = 0.046).
The protein source affected the production of VOCs. According to this study, casein resulted in the highest concentration of VOCs in five donors; this was probably because casein is high in aromatic amino acids, which are the main substrates for bacteria to produce phenolic and indolic compounds. Omnivore donor 3 had low phenolic production from casein, correlating with this donor’s low total bacterial count (see Table S4).
Ammonia.
Ammonia is a major metabolite of protein fermentation by fecal bacteria. Ammonia concentrations increased gradually during fermentation on all substrates as well as with the negative control. Ammonia concentrations on Synergy1, however, remained at low levels (17.55 ± 4.53 mM at 48 h for omnivores and 25.47 ± 4.55 mM for vegetarians) compared to those with all protein treatments in this study. The volunteer diet also influenced the selective fermentation of fecal substrates. With fecal samples from omnivores, fermentation resulted in higher ammonia levels on casein and meat extract; however, with fecal samples from vegetarians, soy protein and Quorn extract resulted in more ammonia production (Fig. 6).
FIG 6.
Changes in ammonia concentrations in batch culture samples over time. Values are mean values at three time points from 3 omnivore and 3 vegetarian fecal donors ± standard deviations. *, mean values were significantly different from 0-h fermentation samples (P < 0.05).
Fermentation on protein for 24 h resulted in significantly higher concentrations of ammonia than fermentation without protein using both omnivore and vegetarian samples (P < 0.001). Fermentation on prebiotics resulted in significantly lower concentrations of ammonia in cultures with omnivore donors’ fecal bacteria (Table 1).
TABLE 1.
Ammonia concentration in samples in the single stage batch culture
| Group | Ammonia concn (mM)a
|
||
|---|---|---|---|
| Control (n = 6) | Protein without Synergy1 (n = 12) | Protein with Synergy1 (n = 12) | |
| Omnivores | 23.07 ± 9.58 | 165.24 ± 77.44b | 91.16 ± 33.24c |
| Vegetarians | 32.02 ± 8.97 | 153.53 ± 62.69b | 126.64 ± 35.76 |
Values are mean values at 24 h of fermentation from 3 omnivores’ microbiota and 3 vegetarians’ microbiota ± standard deviations.
Mean values were significantly different between control and protein without Synergy1 (P < 0.001).
Mean values were significantly different between protein with and without Synergy1 (P < 0.01).
DISCUSSION
Lactate production peaked at 10 h of fermentation, while other organic acid concentrations kept increasing. This coincided with counts of lactobacilli and was to be expected, as lactate can be utilized by several bacteria to produce other SCFA. Changes in propionic acid-producing Bacteroides and Clostridium cluster IX populations were seen and propionic acid increased in vessels containing Synergy1, with the difference reaching significance with omnivore donors’ samples (P = 0.006). Succinate is an intermediate product for propionate production, the succinate pathway being widely present in bacteroides (31). The significantly higher levels of succinate in samples with Synergy1 might be associated with propionate production by bacteroides.
Fecal bacteria responded differently on various substrates in pH-controlled stirred batch cultures. Total bacterial numbers from vegetarians were significantly higher on soy protein and Quorn than on meat and casein. Host dietary habits may explain a preference for different protein sources. The growth of proteolytic bacteria from the human gut supported this: Clostridium coccoides and Eubacterium rectale from omnivore microbiota and vegetarian microbiota grew on meat/casein and soy/Quorn, respectively (see Tables S1 and S2 in the supplemental material). Ammonia concentrations also indicated that an omnivore microbiota and a vegetarian microbiota favor different protein sources based on their host diet. A possible reason is differences in amino acid compositions among various proteins: bacteria that have adapted to the host diet can break down peptides, metabolize amino acids, or utilize coupled Stickland amino acid fermentation.
By observing the fermentation characteristics of the negative controls, we noted that saccharolytic bifidobacterial growth at 6 h with omnivore feces occurred, indicating that there was a small amount of undigested saccharides within the omnivore fecal sample. However, this was not seen from the vegetarian donors.
Even when total bacteria tend to be more saccharolytic, there were some proteolytic bacteria present in the gut microbiota. The genus Clostridium contains more than 100 species, and these bacteria can be saccharolytic, proteolytic, or both. Within clostridial clusters I and II, there are saccharolytic species such as C. butyricum and C. beijerinckii; C. sporogenes and C. acetobutylicum are both saccharolytic and proteolytic, and there are proteolytic species such as C. limosum and C. histolyticum (32). This might explain why Clostridium spp. grew on prebiotics with a vegetarian microbiota: saccharolytic types from this genus were likely to be stimulated by prebiotics. This would also imply that these fecal bacteria from vegetarians are more saccharolytic than clostridia from omnivore donors.
Vegetarian donor 1 had the highest production of phenolic and indolic compounds together with the highest Escherichia coli population, which correlates with the ability of E. coli to produce phenolic compounds (33). Indole and p-cresol are conjugated as indoxyl sulfate and p-cresol sulfate in the human body; before they are excreted via urine, they are toxic to human endothelial cells, can reflect the progression of chronic kidney diseases, and increase cardiovascular disease risk for such patients (34–37). Therefore, reduced production of indole and p-cresol can benefit human health in many ways.
Studies in which rats were fed with different protein sources did not find higher colonic toxicity of casein than soybean, which is contrary to the phenol and p-cresol results in this study (38, 39). Feeding the rats red meat resulted in higher DNA damage than feeding them casein (40). Similar effects were found in human epidemiological research: the consumption of dairy products was inversely correlated with colorectal cancer in Finnish men and New York University women; it was speculated that this protective effect may result from other nutrients in the dairy products but not from macronutrients such as protein (41, 42). Mycoprotein is a relatively new protein source from the filamentous fungus Fusarium venenatum source under the trademark of Quorn (43). Quorn products contain all the essential amino acids, are low in fat, and are high in dietary fiber. However, in terms of protein fermentation by gut microbiota, Quorn was no different than other proteins.
The use of pH-controlled stirred batch culture systems allowed rapid analyses of different protein fermentations by gut microbiota and the impact of prebiotics. This fermentation system is limited, however; SCFA would be absorbed from the human colon, and the digesta supply would be continuous.
Some animal studies and human studies have revealed an inhibitory effect of proteolysis by prebiotics such as resistant starch, fructooligosaccharide (FOS), and xylooligosaccharide (XOS) (44–49). These were investigated by analyzing indolic/phenolic compounds or nitrogen secretion in the urine and feces. One of these studies also compared DNA damage with and without resistant starch in rat colonic cells and found that the starch protected cells from DNA damage (46). One possible mechanism of decreased proteolytic fermentation in the presence of prebiotics is through the enhanced growth of saccharolytic bacteria requiring more amino acids for growth, reducing the amino acid availability for proteolytic bacteria.
Differences between the gut microbiotas from vegetarian and omnivore donors are not clear with three donors; however, fermentation patterns on different substrates were seen in this study, such as the differences in BCFA, ammonia, and total bacteria. In terms of protein fermentation by fecal bacteria, based on the different ammonia production and bacteria growth responses to different protein sources, microbiota from vegetarian donors have adapted to vegetarian protein sources and can utilize these proteins more efficiently. In addition, in this study, lower BCFA production was found with vegetarians’ gut bacteria; this might suggest that these donors had lower branched-chain amino acids in their diet. Prebiotic supplementation lowered proteolytic metabolites more in cultures with omnivores’ samples than in cultures with vegetarians’ bacteria: vegetarian donors are more likely to be on a high-fiber diet and may need a higher dose of Synergy1 to see a prebiotic effect (50).
The addition of Synergy1 at the beginning of 48-h batch culture fermentation changed the microbiota to a more saccharolytic nature by the stimulation of bifidobacteria and lactobacilli without a significant change of Clostridium and E. coli. Supplementation with Synergy1 also reduced the concentrations of protein metabolites (ammonia with significance and BCFA without reaching significance); in those donors with high production of VOCs, inhibition was also found with Synergy1. An inulin-rich diet could be beneficial in individuals with high protein diet; however, this effective dose of inulin is relatively difficult to achieve, especially in people consuming a Western diet (51, 52). Therefore, adding fructan prebiotics might potentially reduce the negative consequences of ingesting high-protein diets, although this would need to be demonstrated in vivo. The EFSA has approved the use of chicory inulin at a dosage of 12 g per day to maintain normal bowel function; however, the effective dose of prebiotics to regulate bacterial proteolysis is unknown (53). In this study, 5 g of inulin-type fructans were effective in vitro, but the production of metabolites such as phenol and indole was inhibited in only some of the donors. This needs to be validated in vivo, and a higher dose might have a better inhibitory effect and cover more of the population. This study also revealed the importance of host habitual diet on the metabolic function of human gut microbiome. This implies that host diet shapes the gut bacteria in a profound way. The individual difference is significant, which again might be due to individual diet difference.
MATERIALS AND METHODS
Proteins.
The protein substrates used were casein hydrolysate (Sigma-Aldrich, Poole, UK), meat extract for microbiology (Sigma-Aldrich, Poole, UK), soy protein acid hydrolysate powder (Sigma-Aldrich, Poole, UK), and mycoprotein which was extracted from a commercial product (Quorn) purchased from a local supermarket.
Prebiotic.
Inulin-type fructan was a mixture of oligofructose and inulin: 50% ± 10% degree of polymerization (DP) of 3 to 9 and 50% ± 10% DP of ≥10 (Orafti Synergy1; BENEO-Orafti, Tienen, Belgium).
Protein extraction.
Mycoproteins were extracted from Quorn based on the method described by Williams et al. (54). Quorn mince (500 g) was mixed with 1,200 ml water and then homogenized in a blender. Sixty milliliters of formic acid was added after homogenization, and the pH was lowered to 1.6. Afterwards, 5 g pepsin was added, and the solution was incubated at 37°C for 48 h. Samples were centrifuged at 3,000 × g for 15 min, and the supernatants were freeze-dried for later use. After extraction, the nitrogen content of mycoproteins was quantified using the Kjeldahl method (Campden BRI, UK) and was found to be 10.3%. The remaining mycoprotein was stored at −20°C.
Protein dose determination.
Based on previous validation work from in vitro batch culture experiments and in human trials, the dose of 1% of substrate (wt/vol) equates to 5 g inulin reaching the colon (27, 55). Synergy1 (1% wt/vol) was used in this study to investigate the prebiotic effect. The approach used in 150-ml batch culture experiments to simulate high protein ingestion is shown in Table 2. The amounts of casein, meat extract, mycoprotein, and soy protein were adjusted based on their true protein contents, which are shown in Table 3.
TABLE 2.
Endogenous and exogenous protein dosage to simulate the in vivo effect of 105-g dietary protein per day consumption for the 150-ml batch culture experiment
| Category | In vitro fermentation dosage (g) |
|---|---|
| Dietary protein | 2.4 |
| Mucin | 0.57 |
| Digestive enzymesa | 0.18 |
Digestive enzyme is a mixture of 0.107 g pepsin, 0.022 g pancreatin, and 0.00079 g α-amylase based on an in vitro upper gut digestion simulation paper (57).
TABLE 3.
Protein dose that is equivalent to 2.4 g dietary protein responding with protein content
| Protein | Protein content (%) | Protein dose (g) |
|---|---|---|
| Casein | 68.75 | 3.5 |
| Soy protein | 75 | 3.2 |
| Meat extract | 76 | 3.2 |
| Mycoprotein | 64.2 | 3.7 |
In vitro batch culture fermentation.
(i) Fecal sample preparation. Ethical approval of collecting fecal samples from healthy volunteers was obtained from University of Reading University Research Ethics Committee in 2014. Fecal samples were obtained from three healthy meat-eating individuals and three healthy vegetarian volunteers between the ages of 18 and 60 (vegetarians, 34.44 ± 6.03 years old; omnivores, 29.33 ± 3.06 years) who had not taken antibiotics for at least 6 months prior to the experiment and had no history of gastrointestinal disorders. None were taking prebiotic supplements. All volunteers had been following their diet for at least 5 years.
Fecal samples were diluted 1 in 10 (wt/vol) using 1 M (pH 7.4) anaerobically prepared phosphate-buffered saline (PBS; Oxoid, Hampshire, UK). This solution was homogenized in a stomacher (stomacher 80, Biomaster; Seward) for 120 s at normal speed. Fifteen milliliters of this was then immediately used to inoculate batch culture vessels.
(ii) Batch culture basal nutrient medium. Basal nutrient medium was prepared with chemicals obtained from Sigma-Aldrich (Poole, UK) unless otherwise stated. One liter contained 2 g peptone water, 2 g yeast extract (Oxoid, Hampshire, UK), 0.1 g NaCl, 0.04 g K2HPO4 (BDH, Poole, UK), 0.04 g KH2PO4 (BDH), 0.01 g MgSO3·7H2O (Fischer Scientific, Loughborough, UK), 0.01 g CaCl2·6H2O, 2 g NaHCO3 (Fischer), 0.5 g l-cystine HCl, 2 ml Tween 80, 10 μl vitamin K1, 0.05 g hemin, 0.05 g bile salts (Oxoid), and 4 ml resazurin (pH 7).
(iii) pH-controlled stirred batch culture fermentation. Vessels with an operating volume of 300 ml were set up. One hundred thirty-five milliliters of basal nutrient medium was autoclaved (121°C for 15 min) and aseptically poured into sterile vessels. This system was left overnight with oxygen-free nitrogen sparging into the medium at a rate of 15 ml/min. After 4 h of fermentation, the nitrogen flow was stopped and gas outlets were clamped to trap gas. pH meters (Electrolab pH controller; Electrolab, Tewksbury, UK) were connected to each vessel to regulate pH 6.7 to 6.9 with the aid of 0.5 M HCl or NaOH.
Each vessel was also temperature controlled at 37°C and stirred using a magnetic stirrer. Prebiotic and relative protein treatments were added to the vessels prior to inoculation with 15 ml of fecal inoculum. For each donor, 10 vessels were prepared for 10 treatments: casein, meat extract, Quorn, soy protein, casein plus Synergy1, meat extract plus Synergy1, Quorn plus Synergy1, soy protein plus Synergy1, Synergy1, and a negative control.
Samples were removed from the fermentors after 0, 6, 10, 24, and 48 h of incubation.
Enumeration of fecal microbial populations by flow cytometry fluorescence in situ hybridization.
A 750-μl sample of batch culture fluid was centrifuged at 11,337 × g for 5 min, and the supernatant was discarded. The pellet was then suspended in 375 μl filtered 0.1 M PBS solution. Filtered cold (4°C) 4% paraformaldehyde (PFA) (1,125 μl) was added, and the samples were stored at 4°C for 4 h. These were then washed thoroughly with PBS to remove PFA and resuspended in a mixture containing 300 μl PBS and 300 μl 99% ethanol. Samples were then stored at −20°C prior to fluorescence in situ hybridization (FISH) analysis by flow cytometry. Filtered cold (4°C) 0.1 M PBS (500 μl) was mixed with fixed samples (75 μl) before centrifuging at 11,337 × g for 3 min. The pellets were then resuspended in 100 μl of TE-FISH (1 M Tris-HCl [pH 8], 0.5 M EDTA [pH 8], and filtered distilled water with the ratio of 1:1:8) containing lysozyme solution (1 mg/ml of 50,000 U/mg protein). Samples were then incubated in the dark at room temperature for 10 min and then centrifuged at 11,337 × g for 3 min. Pellets were washed with 500 μl filtered cold PBS and then washed with 150 μl hybridization buffer (5 M NaCl, 1 M Tris-HCl [pH 8], formamide, double-distilled water [ddH2O], and 10% SDS with the ratio of 180:20:300:499:1) and centrifuged at 11,337 × g for 3 min. Pellets were then resuspended in 1 ml of hybridization buffer. Aliquots (50 μl) with 4 μl of different probes (50 ng · μl−1) were incubated at 35°C for at least 10 h. The probes used in this study are listed in Table 4. Fluorescent Alexa Fluor 488 was attached to the 5′ ends of Non Eub and Eub338I, -II, and -III, and Alexa Fluor 647 was attached to other specific probes. Fluorescent Alexa Fluor 647 was attached to the 5′ ends of a set of Non Eub and Eub338I, -II, and -III as the controls. For samples to detect specific groups, 4 μl of Eub338I, -II, and -III was added together with 4 μl specific probes. Hybridization buffer (150 μl) was added to each aliquot after incubation, followed by 3-min centrifugation at 11,337 × g. Supernatants (150 μl) were carefully removed before the samples were centrifuged at 11,337 × g for 3 min. The remaining supernatant was then removed, and pellets were resuspended in 200 μl washing buffer. Washing buffer was prepared as 12.8 μl of 5 M NaCl, 20 μl of 1 M Tris-HCl (pH 8), 10 μl of 0.5 M EDTA (pH 8), and 1 μl of 10% SDS in 956.2 μl of filtered cold distilled water. Samples were then incubated at 37°C for 20 min and centrifuged at 11,337 × g for 3 min. After supernatant removal, the pellets were resuspended in different volumes of filtered cold PBS based on flow cytometry load. Bacterial counts were then calculated with the consideration of flow cytometry reading and PBS dilution.
TABLE 4.
Name, sequence, and target group of oligonucleotide probes used in this study for FISH of bacterial enumeration
| Probe name | Sequence (5′→3′) | Target group(s) | Reference |
|---|---|---|---|
| Non Eub | ACTCCTACGGGAGGCAGC | Control probe complementary to Eub338 | 58 |
| Eub338I | GCTGCCTCCCGTAGGAGT | Most Bacteria | 59 |
| Eub338II | GCAGCCACCCGTAGGTGT | Planctomycetales | 60 |
| Eub338III | GCTGCCACCCGTAGGTGT | Verrucomicrobiales | 60 |
| Bif164 | CATCCGGCATTACCACCC | Bifidobacterium spp. | 61 |
| Lab158 | GGTATTAGCAYCTGTTTCCA | Lactobacillus and Enterococcus | 62 |
| Bac303 | CCAATGTGGGGGACCTT | Most Bacteroidaceae and Prevotellaceae, some Porphyromonadaceae | 63 |
| Erec482 | GCTTCTTAGTCARGTACCG | Most of the Clostridium coccoides-Eubacterium rectale group (Clostridium clusters XIVa and XIVb) | 64 |
| Rrec584 | TCAGACTTGCCGYACCGC | Roseburia genus | 65 |
| Ato291 | GGTCGGTCTCTCAACCC | Atopobium cluster | 66 |
| Prop853 | ATTGCGTTAACTCCGGCAC | Clostridial cluster IX | 65 |
| Fprau655 | CGCCTACCTCTGCACTAC | Faecalibacterium prausnitzii and relatives | 67 |
| DSV687 | TACGGATTTCACTCCT | Desulfovibrio genus | 68 |
| Chis150 | TTATGCGGTATTAATCTYCCTTT | Most of the Clostridium histolyticum group (Clostridium cluster I and II) | 64 |
| EC 1531 | CACCGTAGTGCCTCGTCATCA | Escherichia coli BJ4 | 69 |
Short-chain fatty acid analysis by gas chromatography.
Samples were centrifuged at 11,337 × g for 10 min to remove all particulate matter. Supernatants were then filtered through a 0.2-μm polycarbonate syringe filter (VWR, Farlington, UK). Extraction was done with some modifications of a method from Richardson et al. (56). The filtered sample (500 μl) was transferred into a labeled 100-mm by 16-mm glass tube (International Scientific Supplies Ltd., Bradford, England) with 25 μl of 2-ethylbutyric acid (0.1 M, internal standard; Sigma-Aldrich, Poole, UK). Concentrated HCl (250 μl) and 1 ml diethyl ether were added to each glass tube, and the samples were vortexed for 1 min. Samples were then centrifuged at 2,000 × g for 10 min. The diethyl ether (upper) layer of each sample was transferred to a labeled clean glass tube. A second extraction was conducted by adding another 0.5 ml diethyl ether, followed by vortexing and centrifugation. The diethyl ether layers were pooled. Pooled ether extract (400 μl) and 50 μl N-(tert-butyldimethylsilyl)-N-methyltrifluoroacetamide (MTBSTFA; Sigma-Aldrich, Poole, UK) were added into a gas chromatography (GC) screw-cap vial. Samples were left at room temperature for 72 h to allow lactic acid in the samples to completely derivatize.
An Agilent/HP 6890 gas chromatograph (Hewlett Packard, UK) using an HP-5MS 30-m by 0.25-mm column with a 0.25-μm coating crosslinked (5%-phenyl)-methylpolysiloxane; (Hewlett Packard, UK) was used for analysis of SCFA. The temperature of the injector and detector was 275°C, with the column programmed from 63°C for 0 min to 190°C at 15°C · min−1 and held at 190°C for 3 min. Helium was the carrier gas (flow rate, 1.7 ml · min−1; head pressure, 133 kPa). A split ratio of 100:1 was used. Quantification of the samples was obtained through calibration curves of lactic acid and acetic, propionic, butyric, valeric, and branched SCFA (isobutyric and isovaleric) in concentrations between 12.5 and 100 mM.
Volatile organic compound analysis by GC-MS.
(i) Entrapment of volatile compounds. All fermentation samples were adjusted to a pH of 7.0 ± 0.3 using hydrochloric acid or sodium chloride prior to volatile entrapment. Each sample (1 g) was placed in a 250-ml conical flask fitted with a Dreschel head. The flask was placed in a water bath maintained at a temperature of 30°C for 1 h. The flask was attached to oxygen-free nitrogen (40 ml/min) which swept volatile compounds from the headspace above the sample onto a glass trap (4-mm inside diameter [i.d.], 6.35-mm outside diameter [o.d.] by 90 mm long), containing 85 mg of Tenax TA poly (a porous polymer absorbent based on 2,6-diphenylene-oxide; Supelco, Poole, UK). Following volatile extraction, 1 μl of 1,2-dichlorobenzene in methanol (653 ng/μl) was added to each trap as an internal standard, and the trap was then flushed with oxygen-free nitrogen to remove moisture (100 ml/min) for 10 min.
(ii) Gas chromatography and mass spectrometry. Volatile compounds collected on the Tenax adsorbent were analyzed using a PerkinElmer Clarus 500 gas chromatograph-mass spectrometer (GC-MS) attached to an automated thermal desorber (Turbomatrix ATD; PerkinElmer, Beaconsfield, UK). Tenax traps were desorbed at 300°C for 10 min, and the volatiles were cryofocused on the internal cold trap held at −30°C. After desorption, the cold trap was heated to 300°C at 40°C/s to release volatile material onto the GC column. GC separation was carried out on a DB-5 nonpolar column (60-m by 0.32-mm i.d., 1-μm film thickness; J&W Scientific from Agilent). Helium at 145 kPa was used as the carrier gas. The GC oven temperature program was 2 min at 40°C followed by an increase at 4°C/min up to 260°C, where it was held for 10 min. All data were collected and stored using Turbomatrix software (version 3.5, PerkinElmer). Compounds were identified from their mass spectra, and identities were confirmed by comparing the retention times (linear retention index [LRI]) and mass spectra with those of authentic compounds analyzed from an online library database. Analyses were carried out using an Agilent 6890/5975 GC-MS system (Agilent Technologies, Palo Alto, CA, USA) fitted with a Turbomatrix ATD.
Indole, p-cresol, and phenol (Sigma-Aldrich, Poole, UK) were diluted using the same internal standard, which was 1,2-dichlorobenzene in methanol (653 ng/μl). Quantification of the samples was obtained through calibration curves of phenol, p-cresol, indole, and skatole in concentrations between 25 and 100 μg/ml.
Ammonia analysis.
Samples at 0, 10, and 24 h were diluted 1 in 50 (vol/vol) prior to analysis. The ammonia concentrations in diluted fermentation samples were analyzed using a FluoroSelect ammonia kit (Sigma-Aldrich, Poole, UK). Reagent was prepared by combining 100 μl assay buffer, 4 μl reagent A, and 4 μl reagent B in the kit. Ten microliters H2O (blank) and 10 μl sample were added to each glass vial. Afterwards, 100 μl reagent was added to each tube. Samples were kept in the dark for 15 min at room temperature before they were read in the fluorometer. Ammonia standards were prepared by diluting 20 mmol/liter NH4Cl in distilled water, and the concentration range was 0.25 to 1 mmol/liter.
Statistical analysis.
All statistical tests were performed with the use of IBM SPSS Statistics version 24 (IBM Corp., USA). Results are presented as means ± standard deviations (SDs). Changes in specific bacterial groups, organic acids, and ammonia were assessed among different treatments and time points using two-way ANOVAs. Significant differences were assessed by post hoc Tukey’s honestly significant difference (HSD) test. In addition, to monitor the influence of protein and prebiotics, independent t tests were used for all variables. For branched-chain fatty acid and ammonia, a two-way ANOVA was used to assess treatment effect and donor difference.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge the partial financial support from BENEO.
We thank the technicians from Food and Nutritional Sciences department at University of Reading for their support for the study. We also thank Angelika Kristek for flow cytometer training.
S. Theis and M. Sailer are/were employees of BENEO/Suedzucker Group. We declare no other conflicts of interest.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.02749-18.
REFERENCES
- 1.Food and Agriculture Organization of the United Nations. 2017. Food balance/food balance sheets. http://faostat3.fao.org/browse/FB/FBS/E. Accessed 15 December 2017.
- 2.Institute of Medicine. 2005. Dietary reference intakes for energy, carbohydrate, fiber, fat, fatty acids, cholesterol, protein, and amino acids. National Academies Press, Washington, DC. [DOI] [PubMed] [Google Scholar]
- 3.Gibson JA, Sladen GE, Dawson AM. 1976. Protein absorption and ammonia production: the effects of dietary protein and removal of the colon. Br J Nutr 35:61–65. doi: 10.1079/BJN19760009. [DOI] [PubMed] [Google Scholar]
- 4.Moughan PJ, Butts CA, Rowan AM, Deglaire A. 2005. Dietary peptides increase endogenous amino acid losses from the gut in adults. Am J Clin Nutr 81:1359–1365. doi: 10.1093/ajcn/81.6.1359. [DOI] [PubMed] [Google Scholar]
- 5.Miner-Williams W, Deglaire A, Benamouzig R, Fuller MF, Tomé D, Moughan PJ. 2014. Endogenous proteins in the ileal digesta of adult humans given casein-, enzyme-hydrolyzed casein- or crystalline amino-acid-based diets in an acute feeding study. Eur J Clin Nutr 68:363–369. doi: 10.1038/ejcn.2013.270. [DOI] [PubMed] [Google Scholar]
- 6.Miner-Williams W, Deglaire A, Benamouzig R, Fuller MF, Tomé D, Moughan PJ. 2012. Endogenous proteins in terminal ileal digesta of adult subjects fed a casein-based diet. Am J Clin Nutr 96:508–515. doi: 10.3945/ajcn.111.033472. [DOI] [PubMed] [Google Scholar]
- 7.Russell WR, Hoyles L, Flint HJ, Dumas M-E. 2013. Colonic bacterial metabolites and human health. Curr Opin Microbiol 16:246–254. doi: 10.1016/j.mib.2013.07.002. [DOI] [PubMed] [Google Scholar]
- 8.Smith EA, Macfarlane GT. 1997. Dissimilatory amino acid metabolism in human colonic bacteria. Anaerobe 3:327–337. doi: 10.1006/anae.1997.0121. [DOI] [PubMed] [Google Scholar]
- 9.Clinton SK, Bostwick DG, Olson LM, Mangian HJ, Visek WJ. 1988. Effects of ammonium acetate and sodium cholate on N-methyl-N'-nitro-N-nitrosoguanidine-induced colon carcinogenesis of rats. Cancer Res 48:3035–3039. [PubMed] [Google Scholar]
- 10.Hughes R, Kurth MJ, McGilligan V, McGlynn H, Rowland I. 2008. Effect of colonic bacterial metabolites on Caco-2 cell paracellular permeability in vitro. Nutr Cancer 60:259–266. doi: 10.1080/01635580701649644. [DOI] [PubMed] [Google Scholar]
- 11.Lin HC, Visek WJ. 1991. Colon mucosal cell-damage by ammonia in rats. J Nutr 121:887–893. doi: 10.1093/jn/121.6.887. [DOI] [PubMed] [Google Scholar]
- 12.Attene-Ramos MS, Nava GM, Muellner MG, Wagner ED, Plewa MJ, Gaskins HR. 2010. DNA damage and toxicogenomic analyses of hydrogen sulfide in human intestinal epithelial FHs 74 Int cells. Environ Mol Mutagen 51:304–314. doi: 10.1002/em.20546. [DOI] [PubMed] [Google Scholar]
- 13.Attene-Ramos MS, Wagner ED, Gaskins HR, Plewa MJ. 2007. Hydrogen sulfide induces direct radical-associated DNA damage. Mol Cancer Res 5:455–459. doi: 10.1158/1541-7786.MCR-06-0439. [DOI] [PubMed] [Google Scholar]
- 14.Attene-Ramos MS, Wagner ED, Plewa MJ, Gaskins HR. 2006. Evidence that hydrogen sulfide is a genotoxic agent. Mol Cancer Res 4:9–14. doi: 10.1158/1541-7786.MCR-05-0126. [DOI] [PubMed] [Google Scholar]
- 15.Roediger WEW, Duncan A, Kapaniris O, Millard S. 1993. Sulfide impairment of substrate oxidation in rat colonocytes: a biochemical basis for ulcerative colitis? Clin Sci (Lond) 85:623–627. doi: 10.1042/cs0850623. [DOI] [PubMed] [Google Scholar]
- 16.McCall IC, Betanzos A, Weber DA, Nava P, Miller GW, Parkos CA. 2009. Effects of phenol on barrier function of a human intestinal epithelial cell line correlate with altered tight junction protein localization. Toxicol Appl Pharmacol 241:61–70. doi: 10.1016/j.taap.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cerini C, Dou L, Anfosso F, Sabatier F, Moal S, Glorieux G, De Smet R, Vanholder R, Dignat-George F, Sampol J, Berland Y, Brunet P. 2004. p-Cresol, a uremic retention solute, alters the endothelial barrier function in vitro. Thromb Haemost 92:140–150. doi: 10.1160/TH03-07-0491. [DOI] [PubMed] [Google Scholar]
- 18.Verbeke KA, Boobis AR, Chiodini A, Edwards CA, Franck A, Kleerebezem M, Nauta A, Raes J, van Tol EAF, Tuohy KM. 2015. Towards microbial fermentation metabolites as markers for health benefits of prebiotics. Nutr Res Rev 28:42–66. doi: 10.1017/S0954422415000037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.IARC. 2015. Monographs evaluate consumption of red meat and processed meat. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 20.Young TB, Wolf DA. 1988. Case-control study of proximal and distal colon cancer and diet in Wisconsin. Int J Cancer 42:167–175. doi: 10.1002/ijc.2910420205. [DOI] [PubMed] [Google Scholar]
- 21.Cottet V, Bonithon-Kopp C, Kronborg O, Santos L, Andreatta R, Boutron-Ruault MC, Faivre J, European Cancer Prevention Organisation Study Group . 2005. Dietary patterns and the risk of colorectal adenoma recurrence in a European intervention trial. Eur J Cancer Prev 14:21–29. doi: 10.1097/00008469-200502000-00004. [DOI] [PubMed] [Google Scholar]
- 22.Willett WC, Stampfer MJ, Colditz GA, Rosner BA, Speizer FE. 1990. Relation of meat, fat, and fiber intake to the risk of colon cancer in a prospective-study among women. N Engl J Med 323:1664–1672. doi: 10.1056/NEJM199012133232404. [DOI] [PubMed] [Google Scholar]
- 23.Gonzalez CA, Riboli E. 2010. Diet and cancer prevention: contributions from the European Prospective Investigation into Cancer and Nutrition (EPIC) study. Eur J Cancer 46:2555–2562. doi: 10.1016/j.ejca.2010.07.025. [DOI] [PubMed] [Google Scholar]
- 24.Shoda R, Matsueda K, Yamato S, Umeda N. 1996. Epidemiologic analysis of Crohn disease in Japan: increased dietary intake of n-6 polyunsaturated fatty acids and animal protein relates to the increased incidence of Crohn disease in Japan. Am J Clin Nutr 63:741–745. doi: 10.1093/ajcn/63.5.741. [DOI] [PubMed] [Google Scholar]
- 25.Jantchou P, Morois S, Clavel-Chapelon F, Boutron-Ruault M-C, Carbonnel F. 2010. Animal protein intake and risk of inflammatory bowel disease: the E3N prospective study. Am J Gastroenterol 105:2195–2201. doi: 10.1038/ajg.2010.192. [DOI] [PubMed] [Google Scholar]
- 26.Gibson GR, Hutkins R, Sanders ME, Prescott SL, Reimer RA, Salminen SJ, Scott K, Stanton C, Swanson KS, Cani PD, Verbeke K, Reid G. 2017. Expert consensus document: the International Scientific Association for Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of prebiotics. Nat Rev Gastroenterol Hepatol 14:194–2201. doi: 10.1038/nrgastro.2017.75. [DOI] [PubMed] [Google Scholar]
- 27.Wang X, Gibson GR. 1993. Effects of the in-vitro fermentation of oligofructose and inulin by bacteria growing in the human large-intestine. J Appl Bacteriol 75:373–380. doi: 10.1111/j.1365-2672.1993.tb02790.x. [DOI] [PubMed] [Google Scholar]
- 28.Kolida S, Tuohy K, Gibson GR. 2002. Prebiotic effects of inulin and oligofructose. Br J Nutr 87:S193–S197. doi: 10.1079/BJN/2002537. [DOI] [PubMed] [Google Scholar]
- 29.Vandeputte D, Falony G, Vieira-Silva S, Wang J, Sailer M, Theis S, Verbeke K, Raes J. 2017. Prebiotic inulin-type fructans induce specific changes in the human gut microbiota. Gut 66:1968–1974. doi: 10.1136/gutjnl-2016-313271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Macfarlane GT, Cummings JH, Allison C. 1986. Protein-degradation by human intestinal bacteria. J Gen Microbiol 132:1647–1656. doi: 10.1099/00221287-132-6-1647. [DOI] [PubMed] [Google Scholar]
- 31.Reichardt N, Duncan SH, Young P, Belenguer A, Leitch CM, Scott KP, Flint HJ, Louis P. 2014. Phylogenetic distribution of three pathways for propionate production within the human gut microbiota. ISME J 8:1323–1335. doi: 10.1038/ismej.2014.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vos P, Garrity GM, Jones D, Krieg NR, Ludwig W, Rainey FA, Schleifer K-H, Whitman WB. (ed) 2009. Bergey’s manual of systematic bacteriology. Volume three: the Firmicutes, 2nd ed Springer New York, New York, NY. [Google Scholar]
- 33.Nicholson JK, Holmes E, Kinross J, Burcelin R, Gibson G, Jia W, Pettersson S. 2012. Host-gut microbiota metabolic interactions. Science 336:1262–1267. doi: 10.1126/science.1223813. [DOI] [PubMed] [Google Scholar]
- 34.Dou L, Bertrand E, Cerini C, Faure V, Sampol J, Vanholder R, Berland Y, Brunet P. 2004. The uremic solutes p-cresol and indoxyl sulfate inhibit endothelial proliferation and wound repair. Kidney Int 65:442–451. doi: 10.1111/j.1523-1755.2004.00399.x. [DOI] [PubMed] [Google Scholar]
- 35.Dou L, Jourde-Chiche N, Faure V, Cerini C, Berland Y, Dignat-George F, Brunet P. 2007. The uremic solute indoxyl sulfate induces oxidative stress in endothelial cells. J Thromb Haemost 5:1302–1308. doi: 10.1111/j.1538-7836.2007.02540.x. [DOI] [PubMed] [Google Scholar]
- 36.Aronov PA, Luo FJG, Plummer NS, Quan Z, Holmes S, Hostetter TH, Meyer TW. 2011. Colonic contribution to uremic solutes. J Am Soc Nephrol 22:1769–1776. doi: 10.1681/ASN.2010121220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vanholder R, Schepers E, Pletinck A, Nagler EV, Glorieux G. 2014. The uremic toxicity of indoxyl sulfate and p-cresyl sulfate: a systematic review. J Am Soc Nephrol 25:1897–1907. doi: 10.1681/ASN.2013101062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Govers MJAP, Lapré JA, de Vries HT, van der Meer R. 1993. Dietary soybean protein compared with casein damages colonic epithelium and stimulates colonic epithelial proliferation in rats. J Nutr 123:1709–1713. doi: 10.1093/jn/123.10.1709. [DOI] [PubMed] [Google Scholar]
- 39.Toden S, Bird AR, Topping DL, Conlon MA. 2007. Differential effects of dietary whey, casein and soya on colonic DNA damage and large bowel SCFA in rats fed diets low and high in resistant starch. Br J Nutr 97:535–543. doi: 10.1017/S0007114507336817. [DOI] [PubMed] [Google Scholar]
- 40.Toden S, Bird AR, Topping DL, Conlon MA. 2006. Resistant starch prevents colonic DNA damage induced by high dietary cooked red meat or casein in rats. Cancer Biol Ther 5:267–272. doi: 10.4161/cbt.5.3.2382. [DOI] [PubMed] [Google Scholar]
- 41.Pietinen P, Malila N, Virtanen M, Hartman TJ, Tangrea JA, Albanes D, Virtamo J. 1999. Diet and risk of colorectal cancer in a cohort of Finnish men. Cancer Causes Control 10:387–396. doi: 10.1023/A:1008962219408. [DOI] [PubMed] [Google Scholar]
- 42.Kato I, Akhmedkhanov A, Koenig K, Toniolo PG, Shore RE, Riboli E. 1997. Prospective study of diet and female colorectal cancer: the New York University women's health study. Nutr Cancer 28:276–281. doi: 10.1080/01635589709514588. [DOI] [PubMed] [Google Scholar]
- 43.Wiebe MG. 2004. Quorn myco-protein—overview of a successful fungal product. Mycologist 18:17–20. doi: 10.1017/S0269915X04001089. [DOI] [Google Scholar]
- 44.Younes H, Demigne C, Behr S, Remesy C. 1995. Resistant starch exerts a lowering effect on plasma urea by enhancing urea N transfer into the large intestine. Nutr Res 15:1199–1210. doi: 10.1016/0271-5317(95)00079-X. [DOI] [Google Scholar]
- 45.Heijnen MLA, Beynen AC. 1997. Consumption of retrograded (RS3) but not uncooked (RS2) resistant starch shifts nitrogen excretion from urine to feces in cannulated piglets. J Nutr 127:1828–1832. doi: 10.1093/jn/127.9.1828. [DOI] [PubMed] [Google Scholar]
- 46.Toden S, Bird AR, Topping DL, Conlon MA. 2005. Resistant starch attenuates colonic DNA damage induced by higher dietary protein in rats. Nutr Cancer 51:45–51. doi: 10.1207/s15327914nc5101_7. [DOI] [PubMed] [Google Scholar]
- 47.Wutzke KD, Lotz M, Zipprich C. 2010. The effect of pre- and probiotics on the colonic ammonia metabolism in humans as measured by lactose-[15N2] ureide. Eur J Clin Nutr 64:1215–1221. doi: 10.1038/ejcn.2010.120. [DOI] [PubMed] [Google Scholar]
- 48.Lecerf JM, Depeint F, Clerc E, Dugenet Y, Niamba CN, Rhazi L, Cayzeele A, Abdelnour G, Jaruga A, Younes H, Jacobs H, Lambrey G, Abdelnour AM, Pouillart PR. 2012. Xylo-oligosaccharide (XOS) in combination with inulin modulates both the intestinal environment and immune status in healthy subjects, while XOS alone only shows prebiotic properties. Br J Nutr 108:1847–1858. doi: 10.1017/S0007114511007252. [DOI] [PubMed] [Google Scholar]
- 49.De Preter V, Vanhoutte T, Huys G, Swings J, De Vuyst L, Rutgeerts P, Verbeke K. 2007. Effects of Lactobacillus casei Shirota, Bifidobacterium breve, and oligofructose-enriched inulin on colonic nitrogen-protein metabolism in healthy humans. Am J Physiol Gastrointest Liver Physiol 292:G358–G368. doi: 10.1152/ajpgi.00052.2006. [DOI] [PubMed] [Google Scholar]
- 50.Clarys P, Deliens T, Huybrechts I, Deriemaeker P, Vanaelst B, De Keyzer W, Hebbelinck M, Mullie P. 2014. Comparison of nutritional quality of the vegan, vegetarian, semi-vegetarian, pesco-vegetarian and omnivorous diet. Nutrients 6:1318–1332. doi: 10.3390/nu6031318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van Loo J, Coussement P, de Leenheer L, Hoebregs H, Smits G. 1995. On the presence of inulin and oligofructose as natural ingredients in the western diet. Crit Rev Food Sci Nutr 35:525–552. doi: 10.1080/10408399509527714. [DOI] [PubMed] [Google Scholar]
- 52.Moshfegh AJ, Friday JE, Goldman JP, Ahuja JKC. 1999. Presence of inulin and oligofructose in the diets of Americans. J Nutr 129:1407S–1411S. doi: 10.1093/jn/129.7.1407S. [DOI] [PubMed] [Google Scholar]
- 53.EFSA Panel on Dietetic Products, Nutrition and Allergies. 2015. Scientific opinion on the substantiation of a health claim related to “native chicory inulin” and maintenance of normal defecation by increasing stool frequency pursuant to article 13.5 of regulation (EC) no. 1924/2006. EFSA J 13:3951. doi: 10.2903/j.efsa.2015.3951. [DOI] [Google Scholar]
- 54.Williams RJH, Brownsell VL, Andrews AT. 2001. Application of the plastein reaction to mycoprotein: I. Plastein synthesis. Food Chem 72:329–335. doi: 10.1016/S0308-8146(00)00233-8. [DOI] [Google Scholar]
- 55.Buddington RK, Williams CH, Chen SC, Witherly SA. 1996. Dietary supplement of neosugar alters the fecal flora and decreases activities of some reductive enzymes in human subjects. Am J Clin Nutr 63:709–716. doi: 10.1093/ajcn/63.5.709. [DOI] [PubMed] [Google Scholar]
- 56.Richardson AJ, Calder AG, Stewart CS, Smith A. 1989. Simultaneous determination of volatile and non-volatile acidic fermentation products of anaerobes by capillary gas chromatography. Lett Appl Microbiol 9:5–8. doi: 10.1111/j.1472-765X.1989.tb00278.x. [DOI] [Google Scholar]
- 57.Mills DJ, Tuohy KM, Booth J, Buck M, Crabbe MJ, Gibson GR, Ames JM. 2008. Dietary glycated protein modulates the colonic microbiota towards a more detrimental composition in ulcerative colitis patients and non-ulcerative colitis subjects. J Appl Microbiol 105:706–714. doi: 10.1111/j.1365-2672.2008.03783.x. [DOI] [PubMed] [Google Scholar]
- 58.Wallner G, Amann R, Beisker W. 1993. Optimizing fluorescent in situ hybridization with rRNA-targeted oligonucleotide probes for flow cytometric identification of microorganisms. Cytometry 14:136–143. doi: 10.1002/cyto.990140205. [DOI] [PubMed] [Google Scholar]
- 59.Amann RI, Binder BJ, Olson RJ, Chisholm SW, Devereux R, Stahl DA. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Environ Microbiol 56:1919–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Daims H, Bruhl A, Amann R, Schleifer KH, Wagner M. 1999. The domain-specific probe EUB338 is insufficient for the detection of all Bacteria: development and evaluation of a more comprehensive probe set. Syst Appl Microbiol 22:434–444. doi: 10.1016/S0723-2020(99)80053-8. [DOI] [PubMed] [Google Scholar]
- 61.Langendijk PS, Schut F, Jansen GJ, Raangs GC, Kamphuis GR, Wilkinson MHF, Welling GW. 1995. Quantitative fluorescence in situ hybridization of Bifidobacterium spp with genus-specific 16S rRNA-targeted probes and its application in fecal samples. Appl Environ Microbiol 61:3069–3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Harmsen HJM, Elfferich P, Schut F, Welling GW. 1999. A 16S rRNA-targeted probe for detection of lactobacilli and enterococci in faecal samples by fluorescent in situ hybridization. Microb Ecol Health Dis 11:3–12. [Google Scholar]
- 63.Manz W, Amann R, Ludwig W, Vancanneyt M, Schleifer KH. 1996. Application of a suite of 16S rRNA-specific oligonucleotide probes designed to investigate bacteria of the phylum cytophaga-flavobacter-bacteroides in the natural environment. Microbiology 142:1097–1106. doi: 10.1099/13500872-142-5-1097. [DOI] [PubMed] [Google Scholar]
- 64.Franks AH, Harmsen HJ, Raangs GC, Jansen GJ, Schut F, Welling GW. 1998. Variations of bacterial populations in human feces measured by fluorescent in situ hybridization with group-specific 16S rRNA-targeted oligonucleotide probes. Appl Environ Microbiol 64:3336–3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Walker AW, Duncan SH, Leitch ECM, Child MW, Flint HJ. 2005. pH and peptide supply can radically alter bacterial populations and short-chain fatty acid ratios within microbial communities from the human colon. Appl Environ Microbiol 71:3692–3700. doi: 10.1128/AEM.71.7.3692-3700.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Harmsen HJM, Wildeboer-Veloo ACM, Grijpstra J, Knol J, Degener JE, Welling GW. 2000. Development of 16S rRNA-based probes for the Coriobacterium group and the Atopobium cluster and their application for enumeration of Coriobacteriaceae in human feces from volunteers of different age groups. Appl Environ Microbiol 66:4523–4527. doi: 10.1128/AEM.66.10.4523-4527.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Suau A, Rochet V, Sghir A, Gramet G, Brewaeys S, Sutren M, Rigottier-Gois L, Doré J. 2001. Fusobacterium prausnitzii and related species represent a dominant group within the human fecal flora. Syst Appl Microbiol 24:139–145. doi: 10.1078/0723-2020-00015. [DOI] [PubMed] [Google Scholar]
- 68.Ramsing NB, Fossing H, Ferdelman TG, Andersen F, Thamdrup B. 1996. Distribution of bacterial populations in a stratified fjord (Mariager Fjord, Denmark) quantified by in situ hybridization and related to chemical gradients in the water column. Appl Environ Microbiol 62:1391–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Poulsen LK, Lan FS, Kristensen CS, Hobolth P, Molin S, Krogfelt KA. 1994. Spatial distribution of Escherichia coli in the mouse large intestine inferred from rRNA in situ hybridization. Infect Immun 62:5191–5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.