Biological control using beneficial bacteria (PGPRs) represents an attractive and environment-friendly alternative to pesticides for controlling plant diseases. Different PGPR Bacillus species produce potent biofungicides and stimulate plant defense responses against phytopathogenic fungi. However, very little is known about how PGPRs recognize phytopathogens and process the antifungal response. Here, we report how B. subtilis triggers the induction of the stress-responsive sigma B transcription factor and the synthesis of the lipopeptide surfactin to fight the phytopathogen. Our findings show the participation of the stress-responsive regulon of PGPR Bacillus in the detection and biocontrol of a phytopathogenic fungus of agronomic impact.
KEYWORDS: Bacillus subtilis, bicontrol, fungi, sigma B, stress, surfactin
ABSTRACT
Different Bacillus species with PGPR (plant growth-promoting rhizobacterium) activity produce potent biofungicides and stimulate plant defense responses against phytopathogenic fungi. However, very little is known about how these PGPRs recognize phytopathogens and exhibit the antifungal response. Here, we report the antagonistic interaction between Bacillus subtilis and the phytopathogenic fungus Fusarium verticillioides. We demonstrate that this bacterial-fungal interaction triggers the induction of the SigB transcription factor, the master regulator of B. subtilis stress adaptation. Dual-growth experiments performed with live or dead mycelia or culture supernatants of F. verticillioides showed that SigB was activated and required for the biocontrol of fungal growth. Mutations in the different regulatory pathways of SigB activation in the isogenic background revealed that only the energy-related RsbP-dependent arm of SigB activation was responsible for specific fungal detection and triggering the antagonistic response. The activation of SigB increased the expression of the operon responsible for the production of the antimicrobial cyclic lipopeptide surfactin (the srfA operon). SigB-deficient B. subtilis cultures produced decreased amounts of surfactin, and B. subtilis cultures defective in surfactin production (ΔsrfA) were unable to control the growth of F. verticillioides. In vivo experiments of seed germination efficiency and early plant growth inhibition in the presence of F. verticillioides confirmed the physiological importance of SigB activity for plant bioprotection.
IMPORTANCE Biological control using beneficial bacteria (PGPRs) represents an attractive and environment-friendly alternative to pesticides for controlling plant diseases. Different PGPR Bacillus species produce potent biofungicides and stimulate plant defense responses against phytopathogenic fungi. However, very little is known about how PGPRs recognize phytopathogens and process the antifungal response. Here, we report how B. subtilis triggers the induction of the stress-responsive sigma B transcription factor and the synthesis of the lipopeptide surfactin to fight the phytopathogen. Our findings show the participation of the stress-responsive regulon of PGPR Bacillus in the detection and biocontrol of a phytopathogenic fungus of agronomic impact.
INTRODUCTION
In nature, microbes continuously interact with each other, while axenic or pure culture conditions occur only under laboratory conditions (1, 2). These microbial interactions can be synergistic, neutral, or antagonistic and might positively or negatively affect the colonization of a given niche or host by a specific microbe (2–4). In particular, an antagonistic interaction between bacteria and fungi would be beneficial to protect plants of agronomical importance against phytopathogenic fungi that reduces the yield and quality of crops (5–7). One strategy to achieve this goal is to control soilborne plant diseases via the utilization of plant growth-promoting rhizobacteria (PGPRs), which have the ability to maintain the population of plant-pathogenic microbes below the disease threshold level in soil and plant tissues (8–12). A large number of biocontrol agents have been identified, but to date, Bacillus species, in particular those that belong to the B. amyloliquefaciens/B. subtilis group, are considered the most efficient biocontrol bacteria, because these bacilli have the ability to produce long-lasting and robust biofilms that colonize and protect plant surfaces (i.e., the rhizosphere) and to produce spores that can survive in adverse environments (13). In addition to biofilm formation and sporulation proficiency, most PGPR Bacillus species produce cyclic lipopeptide molecules, mainly those of the iturin, surfactin, and fengycin families, which possess strong antifungal activity and the ability to induce systemic plant resistance against pathogens (8, 11, 12, 14).
B. subtilis is recognized as a PGPR for its ability to promote plant growth and provide protection against pests but also as an important model organism to investigate complex regulatory pathways and bacterial behaviors (15–20). We hypothesize that the antagonistic interaction with fungi represents a stressful situation for B. subtilis (because, for example, of the mutual fungus-bacterium competition for nutrient availability and sites to settle and due to the microbicidal metabolites produced by each microbe against the other) that might be genetically controlled. In B. subtilis and other bacilli, the genetic regulatory network that responds to danger (i.e., stress) is under the control of the alternative sigma factor of the RNA polymerase SigB (21). This transcription factor controls the general stress regulon, which involves more than 200 genes (∼5% of the genome) whose products confer resistance to multiple forms of stress in the bacterium (22, 23).
The activation of SigB is under the control of the partner-switching RsbV-RsbW-SigB module (22, 23). Under nonstress conditions, SigB is held inactive in a complex with the anti-sigma factor/kinase RsbW, and the third partner, the anti-anti-sigma factor RsbV, is inactive because of phosphorylation by RsbW (24–29). Under stress conditions, the release of SigB from the inactive SigB::RsbW complex is achieved by the dephosphorylated form of the anti-anti-sigma factor RsbV. In B. subtilis, activation (dephosphorylation) of RsbV, and therefore SigB activation, is achieved by alternative phosphatases that sense energy-related or environmental stress (the RsbP or RsbU phosphatase, respectively) (24–29). In addition, SigB is also activated by low temperature (30, 31) independently of RsbP, RsbU, and RsbV activities (30). However, to the best of our knowledge, the participation of SigB in the adaptive response of B. subtilis and its closest relatives (i.e., B. thuringiensis and B. amyloliquefaciens) to the presence of harmful organisms (for example, fungi) has not been previously explored (22, 23).
In particular, Fusarium verticillioides is a phytopathogenic fungus of economic importance because it is the most commonly reported fungal species that infects maize (32), in addition to causing stalk rot disease in sorghum (33) and Pokkah Boeng disease in sugarcane (34). The diseases caused by Fusarium spp. are difficult to control with existing fungicides, and many transgenic plants lack resistance to these diseases (35). In this work, we report that B. subtilis recognizes and responds to the presence of the phytopathogenic fungus F. verticillioides via the induction of SigB and increases production of the lipopeptide surfactin. We discuss the significance of these findings for the fitness of the bacterium and the enhancement of Bacillus-mediated plant protection.
RESULTS AND DISCUSSION
SigB is induced during the antagonistic interaction of B. subtilis with F. verticillioides.
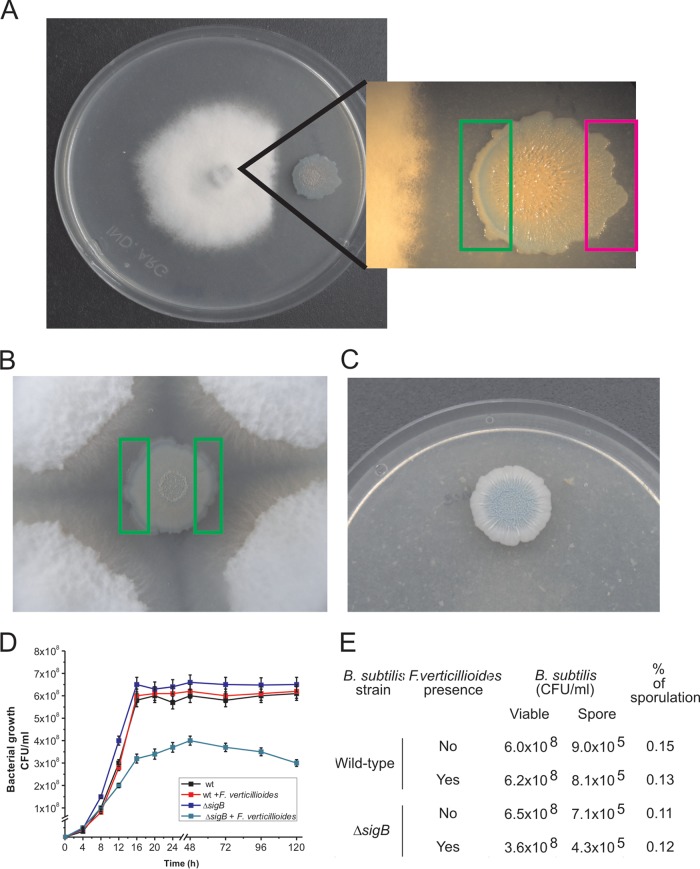
As shown in Fig. 1A, B. subtilis was able to repress the growth of F. verticillioides and SigB was activated, as suggested by the development of the blue color (because of the SigB-dependent ctc-lacZ activity) inside the B. subtilis colony (13, 16, 18) after 96 h of coincubation with the fungus. Interestingly, as observed in the figure, the activation of SigB was not uniform, increasing with the proximity of B. subtilis to the fungus. The B. subtilis cells situated in the part of the bacterial colony closest to the fungus and those furthest from the fungus (left and right rectangles in the figure, respectively) were scraped, and the SigB-dependent specific β-galactosidase activity was determined (see Materials and Methods for details). The specific β-galactosidase activities measured were 1,255 ± 25 and 234 ± 12 Miller units (MU) for the B. subtilis cells located closest and furthest from the fungus, respectively. Accordingly, when the B. subtilis colony was completely surrounded by the fungus, the induction of sigma B occurred uniformly throughout the boundary of the colony, and average values of 1,050 ± 18 MU were measured in the B. subtilis cells scraped after 96 h of coincubation from any of the two areas indicated in Fig. 1B. However, when B. subtilis developed in the absence of the fungus (Fig. 1C), sigma B induction occurred later (after 144 h of incubation) and only in the center of the bacterial colony because of the metabolic stress generated by the insufficient availability of nutrients in that inner part of the biofilm (36, 37).
FIG 1.
Antagonistic response of B. subtilis confronted with F. verticillioides. (A and B) The coculture of a B. subtilis NCIB3610 isogenic strain harboring a ctc-lacZ fusion as a reporter of SigB activity (strain DG555) (Table 1) with F. verticillioides allowed observation of the antagonistic fungus-bacterium interaction. The pattern of induction of SigB is evidenced by the development of blue color (derived from expression of the ctc-lacZ fusion) inside the bacterial colony. The areas represented by the rectangles correspond to the colony areas that were used to quantify the level of SigB-directed β-galactosidase activity (see the text for details). (C) Pattern of SigB expression when the DG555 strain was developed in the absence of F. verticillioides. For panels A to C, cells were grown on PDA plates supplemented with X-Gal (60 µg/ml) for 96 h at 28°C. (D and E) Planktonic growth and sporulation proficiency of the wild-type (wt) strain NCIB3610 and its isogenic SigB-deficient derivative (ΔsigB; strain DG559) (Table 1) in the absence and presence of live F. verticillioides (see Materials and Methods for details). Typical results from five independent experiments performed in duplicate are shown for panels A to E.
The results presented in Fig. 1A to C suggested that the antagonistic interaction of B. subtilis with F. verticillioides would represent a stressful (or threatening) condition that could be sensed by the bacterium, which induced SigB activity as a response. In order to test this hypothesis, we separately cultured the wild-type strain NCIB3610 and its isogenic ΔsigB derivative, deficient in SigB activity (strain DG599) (Table 1), in Luria-Bertani (LB) broth (see Materials and Methods) with shaking at 28°C until the mid-logarithmic phase of growth. At that time, we divided each bacterial culture into two flasks and added to one of them live mycelia from a F. verticillioides culture grown for 24 h (see Materials and Methods for details). As shown in Fig. 1D, there were no significant differences between the kinetics of growth and the final cellular yields (CFU ml−1) of the SigB-proficient and SigB-deficient B. subtilis cultures grown in the absence of the fungus. However, confirming the hypothesized antagonistic interaction between the bacterium and the fungus, there was an appreciable decrease of the rate of growth and final cellular yield of the SigB-deficient B. subtilis strain cocultured with the fungus compared to the bacterial yield reached in coculture of the SigB-proficient B. subtilis strain with the fungus (Fig. 1D). The overall results (Fig. 1A to D) suggest that B. subtilis senses the presence of F. verticillioides as a hostile situation and induces SigB as an adaptive response. The other adaptive pathway that B. subtilis might employ to protect itself from the fungal presence is the onset of sporulation (38). However, sporulation is largely prevented in nutrient-enriched media because it is subject to nitrogen and carbon catabolite control (38). Under our experimental growth conditions in rich media (i.e., LB), the sporulation frequency of the wild-type and ΔsigB B. subtilis strains after 96 h of growth was approximately 0.15%; neither of these strains' sporulation values was significantly affected by the presence of F. verticillioides (Fig. 1E). Therefore, the sporulation pathway does not significantly influence the B. subtilis fungal adaptation under our experimental conditions.
TABLE 1.
Strains used in this work
| Strain | Relevant phenotype and/or genotype | Comment(s) and/or sourceb (reference) |
|---|---|---|
| F. verticillioides | Wild-type isolate | CEREMICa |
| B. subtilis | ||
| NCIB3610 | Wild-type Marburg isolate, surfactin (Srf) proficient and biofilm formation (Bio) proficient | Laboratory collection (18) |
| MR101 | JH642 amyE::Pctc-lacZ::cat | Laboratory collection (31) |
| DG555 | amyE::Pctc-lacZ::cat, similar to NCIB3610 but also has a reporter of SigB activity | This work, MR101→NCIB3610 |
| DG5572 | JH642 ΔrsbU::kan | Laboratory collection (30) |
| DG5573 | JH642 ΔrsbP::spc | Laboratory collection (30) |
| DG5574 | JH642 ΔrsbUP::kan-spc | Laboratory collection (30) |
| DG556 | ΔrsbU::kan, similar to NCIB3610 but deficient in the environmental pathway of SigB activation | This work, DG5572→NCIB3610 |
| DG557 | ΔrsbP::spc, similar to NCIB3610 but deficient in the energy-related pathway of SigB activation | This work, DG5573→NCIB3610 |
| DG558 | ΔrsbUP::kan-spc, similar to NCIB3610 but deficient in the energy-related and environmental pathways of SigB activation | This work, DG556→DG558 |
| MR644 | JH642 ΔsigB::neo | Laboratory collection (31) |
| DG559 | ΔsigB::neo, similar to NCIB3610 but deficient in SigB activity (strain sensitive to stress) | This work, MR644→NCIB3610 |
| DG560 | NCIB3610 ΔsrfAA::ery, idem to NCIB3610 but deficient in surfactin synthesis (also deficient in biofilm formation) | Laboratory collection (18) |
| MR760 | JH642 amyE::Psrf-lacZ::cat | Laboratory collection (18) |
| DG561 | amyE::Psrf-lacZ::cat, similar to NCIB3610 but also has a reporter of surfactin production | This work, MR760→NCIB3610 |
| DG562 | ΔsigB::neo amyE::Psrf-lacZ::cat | This work, DG559→DG561 |
| DG563 | ΔrsbU::kan amyE::Psrf-lacZ::cat | This work, DG556→DG561 |
| DG564 | ΔrsbP::spc amyE::Psrf-lacZ::cat | This work, DG557→DG561 |
Centro de Referencia en Micología, Universidad Nacional de Rosario.
Donor DNA→receptor strain.
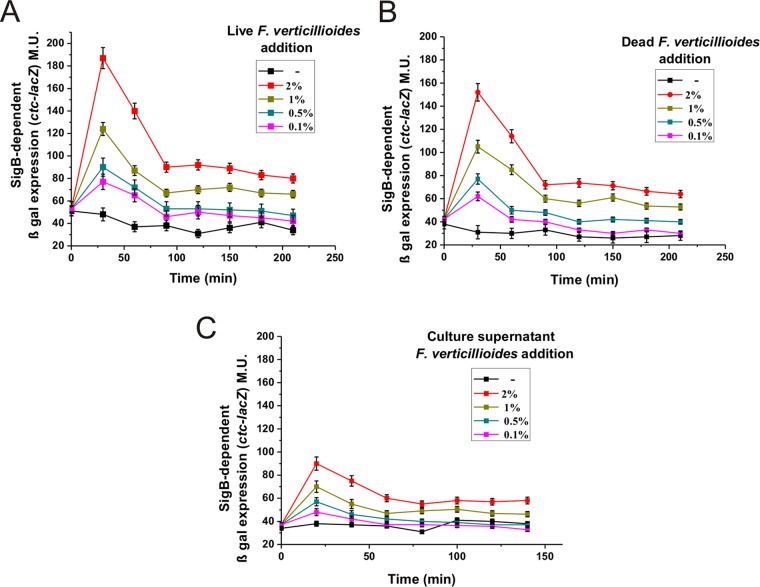
To obtain new insights into the kinetics of the induction of SigB in response to the presence of the fungus, we exposed subcultures of the DG555 strain to different amounts of live mycelia from a F. verticillioides culture grown for 24 h (see Materials and Methods for details). As shown in Fig. 2A, 30 min after the fungal addition, there was a rapid and dose-dependent induction of SigB in response to the presence of different amounts of fungal mycelia. One hour after the peak of fungus-induced β-galactosidase activity, the SigB activity decreased and became stable but was significantly higher than the SigB-directed β-galactosidase activity of the untreated culture even after prolonged incubation (2 h and beyond in Fig. 2A).
FIG 2.
F. verticillioides induces the stress-responsive SigB regulon. β-Galactosidase activity of LB cultures of the wild-type strain DG555 in response to different amounts of live (A), dead (B), or cell-free supernatant (C) of F. verticillioides. β-Galactosidase values are expressed in MU ± standard errors of the means (SEM), and time zero corresponds to the moment that the bacterial cultures reached the mid-logarithmic phase of growth (OD600 of 0.5) and fungal addition (see Materials and Methods for details). A typical output from three independent experiments performed in parallel is shown.
To obtain more information regarding the novel fungus-directed SigB induction, we performed two modified versions of the experiment described in Fig. 2A. In one case, the mycelia of the F. verticillioides culture grown for 24 h were heat killed (i.e., autoclaved) before being added to the SigB reporter strain DG555 (Fig. 2B). In the other type of experiment, we used the culture supernatant of a 24-h culture of the fungus (Fig. 2C). In both cases (Fig. 2B and C), SigB was activated and the induction of SigB was once again rapid and dose dependent. The values for β-galactosidase activity obtained in these experiments (Fig. 2B and C) were slightly lower than those obtained after the addition of live mycelia (Fig. 2A). This observation suggested that the fungal metabolite(s) responsible for the activation of SigB was not completely heat resistant and/or was intrinsically unstable; therefore, the metabolite must be continuously replenished by the fungus to obtain maximal SigB induction. Regardless, the overall results (Fig. 2) were consistent with the results shown in Fig. 1 and indicated that physical contact of the fungus with the bacterium was not necessary to induce SigB.
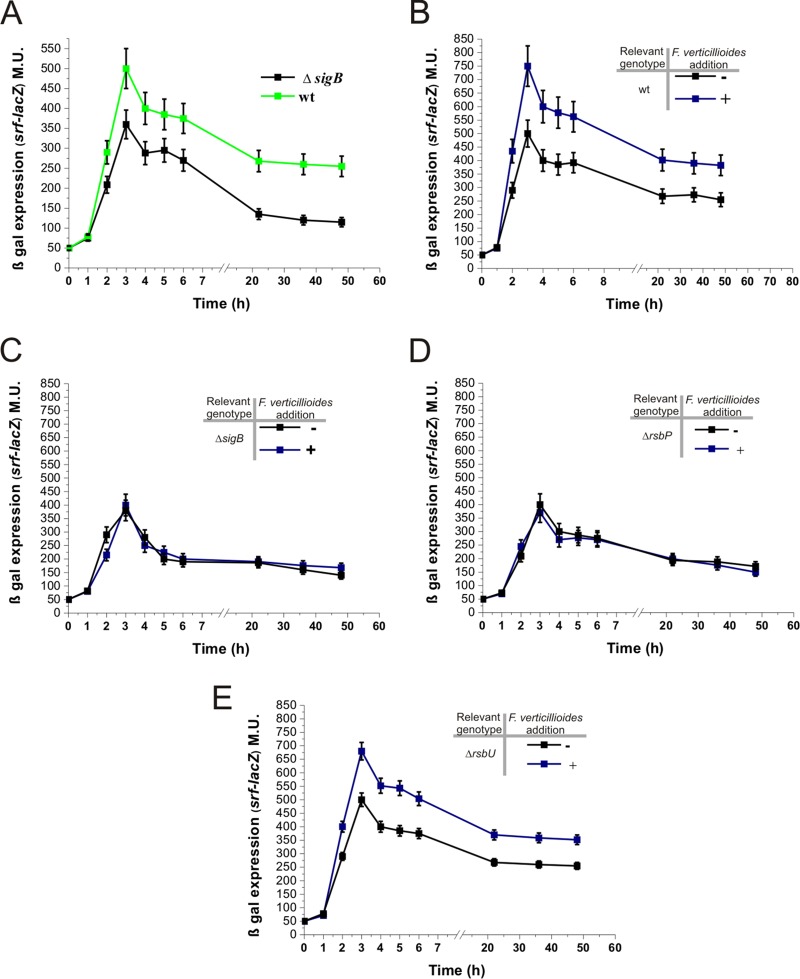
F. verticillioides activates the RsbP-dependent pathway of the SigB-controlling cascade.
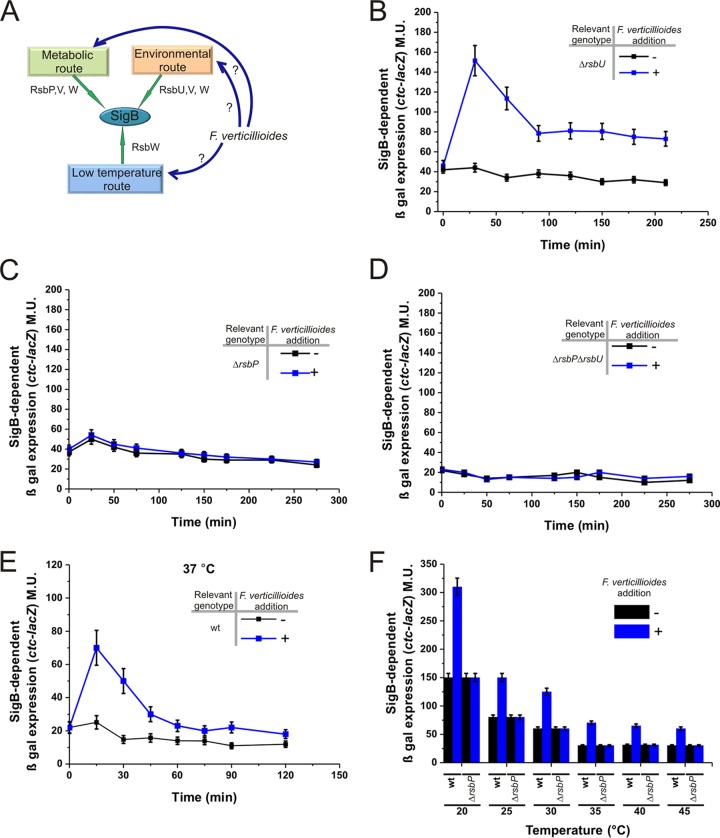
What is the SigB activation pathway (Fig. 3A) that controls the activation of the alternative sigma factor when the bacterium is confronted with the fungus? To answer this question, we monitored the expression of the SigB-dependent ctc-lacZ reporter fusion in isogenic B. subtilis strains whose SigB activation pathways had been altered (i.e., the DG556-ΔrsbU, DG557-ΔrsbP, and DG558-ΔrsbUP strains; details can be found in Table 1 and the introduction). For the ΔrsbU strain, the SigB reporter fusion (ctc-lacZ) was induced in the presence of mycelia from F. verticillioides (Fig. 3B), and the level and kinetics of β-galactosidase expression were very similar to those exhibited by wild-type DG555 cells exposed to the fungus (Fig. 2A). Interestingly, the B. subtilis ΔrsbP (DG557) and ΔrsbUP (DG558) mutant strains were unable to induce SigB in response to the presence of the fungus (Fig. 3C and D). These results strongly suggested that during the antagonistic interaction between B. subtilis and F. verticillioides, stress sensed by the RsbP pathway represented a unique or primary input responsible for the activation of SigB when B. subtilis is confronted with the fungus. The involvement of the RsbP-dependent pathway (Fig. 3A), and the previously described results (Fig. 2), suggested that the fungus produces one or more metabolites that interfere with the ability of B. subtilis to produce and/or utilize energy. Notably, many Fusarium species, including F. verticillioides, produce the mycotoxin fusaric acid. In addition to its ability to modulate the production of antifungal bacterial metabolites (39–41), it has also been reported that, in treated plants, this mycotoxin blocked mitochondrial respiration, decreased the ATP concentration, and destroyed membrane integrity (42). However, our preliminary results indicated that the addition of fusaric acid to B. subtilis does not induce SigB (data not shown).
FIG 3.
B. subtilis recognizes F. verticillioides via the energy-dependent pathway of the SigB regulatory cascade. (A) A cartoon summarizing the three known pathways of SigB activation, one of which is likely responsible for sensing the presence of the fungus (see the text for details). (B to F) β-Galactosidase activity of NCIB3610 isogenic strains harboring the ctc-lacZ fusion in wild-type background (strain DG555) (E and F) or affected in the different pathways of SigB activation: ΔrsbU (strain DG556) (B), ΔrsbP (strain DG557) (C), and ΔrsbUP (strain DG558) (D and F). Each bacterial culture was grown in LB broth with shaking at 28°C (B to D), 37°C (E), or the indicated temperatures (F) until the mid-logarithmic phase, at which time (time zero) the culture was divided into two subcultures and F. verticillioides was added to one of them (final fungal concentration, 1%). The incubation was continued as shown in the figure, and aliquots for the determination of β-galactosidase activity were taken at the indicated times and processed. For the experiment shown in panel F, β-galactosidase activity was determined 40 min after time zero. For panels B to F, a typical output from three independent experiments performed in parallel is shown.
Up to this point, all the experiments have been performed at 28°C, a growth temperature that not only favors fungal growth but also activates the RsbUP-independent low-temperature pathway of SigB activation (Fig. 3A) (30, 31). Therefore, the incubation temperature at which the former experiments were performed (28°C) indicated the possibility that the low-temperature pathway of SigB activation also was involved in the specific fungal detection (30, 31). However, as shown in Fig. 3E, experiments similar to those described for Fig. 3B but performed at 37°C (a growth temperature that inactivated the low-temperature-dependent pathway of SigB activation), and other incubation temperatures higher than 37°C (Fig. 3F), in wild-type and rsbP-negative backgrounds confirmed that SigB was specifically induced by the presence of the fungus independently of the incubation temperature.
SigB is required for the proficiency of B. subtilis to control F. verticillioides growth.
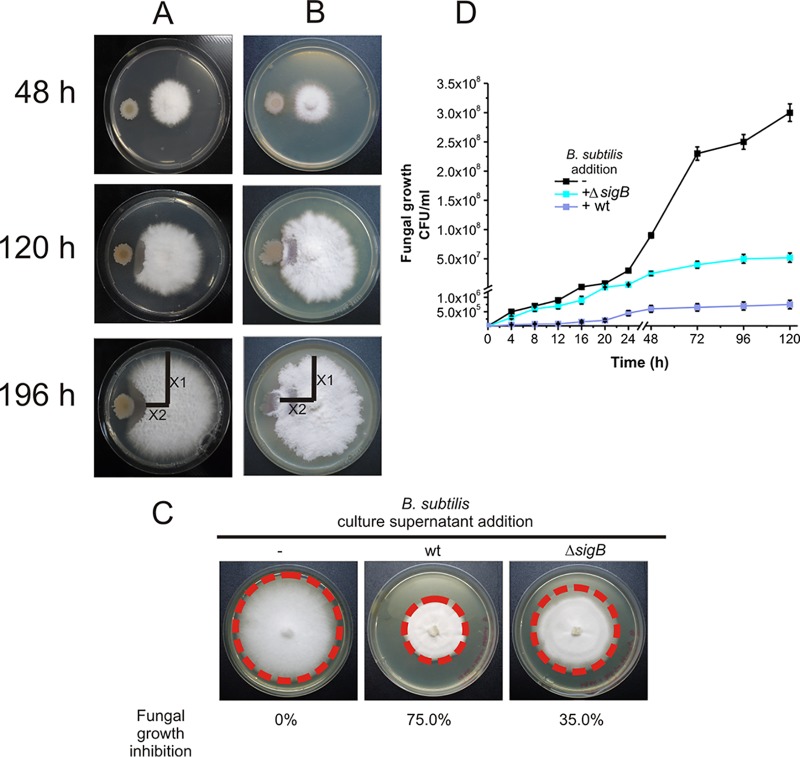
B. subtilis is known for its ability to inhibit fungal growth (8, 14), and SigB is induced when the fungus F. verticillioides or its metabolites are present (Fig. 1 and 3). Therefore, we wondered whether SigB plays a role in fungal biocontrol. To answer this question, we monitored the growth of F. verticillioides in dual-growth experiments where the fungus was grown in the presence of the wild-type strain NCIB3610 or in the presence of the isogenic strain DG559, which lacked SigB activity (ΔsigB strain) (Table 1). As shown in Fig. 4A, the wild-type B. subtilis strain NCIB3610 was able to control the growth of the fungus even after more than 1 week of coincubation, and it showed a fungal growth inhibition index of 62.5% (see Materials and Methods) (40). In the case of the DG559 strain, which lacked SigB activity (ΔsigB), the inhibition of the mycelial growth of F. verticillioides was weaker (the fungal growth inhibition index was 17.5%) than that observed with NCIB3610 (Fig. 4B). Furthermore, the culture supernatants of B. subtilis cultures deficient in SigB activity were less efficient at controlling the growth of the fungus than the culture supernatants of B. subtilis cultures proficient in SigB activity (35% and 75% of fungal growth inhibition, respectively) (Fig. 4C). These results showed that SigB plays a previously unidentified and important, although not essential, role in the biocontrol of F. verticillioides growth. To confirm this conclusion, we quantified the fungal growth (CFU ml−1) in dual cultures of F. verticillioides developed in LB broth in the presence of the wild-type strain NCIB3610 or the isogenic ΔsigB derivative, with shaking at 28°C for 5 days. As shown in Fig. 4D, the fungal yield was significantly decreased in the coculture of F. verticillioides with the wild-type (SigB-proficient) B. subtilis strain (4 × 105 CFU ml−1) compared to the final cellular yield reached by the fungus developed in the absence of bacteria (3 × 108 CFU ml−1). As expected from the results shown in Fig. 4B and C, the fungal yield obtained in coculture with the SigB-deficient B. subtilis strain was intermediate (5 × 107 CFU ml−1) between the cellular yield reached by the fungus grown in the absence or presence of the wild-type B. subtilis strain (Fig. 4D).
FIG 4.
Role of SigB in the in vitro antifungal activity of B. subtilis. (A and B) F. verticillioides (A) and the wild-type B. subtilis strain NCIB3610 and its isogenic ΔsigB strain (DG559) (B) were grown on PDA plates at 28°C as indicated in Materials and Methods. (C) Four-day growth of F. verticillioides inoculated in the middle of a PDA plate without supplementation (left plate) or supplemented with 10% culture supernatant from NCIB3610 (wild type) or DG599 (ΔsigB) B. subtilis cultures. (D) Growth of F. verticillioides under axenic conditions or cocultured with the wild-type strain NCIB3610 or the isogenic SigB-deficient derivative (ΔsigB). Cultures were developed in LB broth with shaking at 28°C, and fungal quantification (CFU ml−1 ± SEM) at different times of growth was carried out as described in Materials and Methods. The results from five independent experiments performed in duplicate are shown in panel D, and panels A through C show representative results.
F. verticillioides induces surfactin production via activation of SigB.
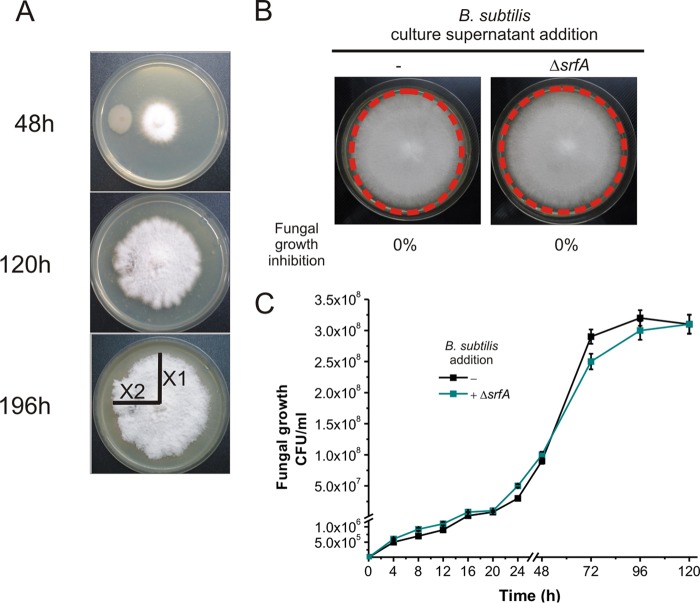
B. subtilis and its closest relatives (i.e., B. amyloliquefaciens) are recognized to constitute powerful biological weapons against phytopathogenic fungi because of their proficiency in synthesizing natural compounds with strong antifungal activity (i.e., the cyclic lipopeptides iturin, fengycin, and surfactin) (8, 43–45). The NCIB3610 strain does not produce iturins but harbors the operons for surfactin and plipastatin (a member of the fengycin family) production, i.e., the srfA and ppsB operons, respectively (46). However, it has been reported that the promoter of the plipastatin operon in the NCIB3610 isolate and its isogenic derivatives (i.e., strain 168) is weak; thus, surfactin would be the only lipopeptide produced in significant amounts by this wild isolate under laboratory conditions (47, 48). Therefore, we investigated whether the ability of B. subtilis NCIB3610 to control the growth of F. verticillioides depended on the proficiency of this strain in producing surfactin. To this end, we constructed an NCIB3610 isogenic strain deficient in surfactin production (ΔsrfA, DG560 strain) (Table 1). Interestingly, the surfactin-deficient strain DG560 completely lost the ability to control the mycelial growth of F. verticillioides (Fig. 5A and B, 0% fungal growth inhibition index). Accordingly, the fungal yield in coculture with the surfactin-deficient (ΔsrfA) B. subtilis strain was unaffected and indistinguishable from the final cellular yield reached by the fungus developed in the absence of bacteria (Fig. 5C).
FIG 5.
Surfactin has an essential role in the in vitro antifungal activity of B. subtilis. (A and B) Absence of antifungal activity of an NCIB3610 isogenic strain deficient in surfactin production (ΔsrfA, strain DG560) (Table 1). (C) Growth of F. verticillioides in the absence or presence of the surfactin-deficient derivative (ΔsrfA) DG560 in LB broth with shaking at 28°C as described in Materials and Methods. The results of five representative experiments are shown.
Because the biocontrol ability of SigB-deficient B. subtilis DG559 was partial (Fig. 4) and surfactin synthesis was essential for the control of fungal growth (Fig. 5), we analyzed whether this defective biocontrol phenotype of the SigB-deficient strain was due to decreased surfactin production. As shown in Fig. 6A, the expression of a lacZ reporter fused to the promoter of the surfactin operon (srf-lacZ) was lower in the strain deficient in SigB activity than in the wild-type strain proficient in SigB activity (strains DG562 and DG561, respectively). Therefore, SigB plays a previously unknown, direct or indirect, positive, although nonessential, role in surfactin production. Interestingly, the β-galactosidase activity driven by srf-lacZ in wild-type and ΔsigB mutant cultures confronted with F. verticillioides (Fig. 6B and C, respectively) indicated that SigB was essential for the augmentation of surfactin production in the presence of the fungus. Accordingly, the RsbP-dependent pathway of SigB activation, but not the RsbU-dependent pathway, was required for the increase in surfactin production triggered by the presence of the fungus (Fig. 6D and E, respectively). The overall results indicate that F. verticillioides induces the energy-related RsbP-dependent activation pathway of SigB (see “Conclusions” below), which in turn increases the expression of the operon that controls surfactin synthesis.
FIG 6.
SigB-dependent surfactin production. β-Galactosidase activity of NCIB3610 isogenic strains, proficient and deficient in SigB activity, harboring srf-lacZ::amyE as a reporter of surfactin production (strains DG561 [wt], DG562 [ΔsigB], DG563 [ΔrsbP], and DG564 [ΔrsbU]). Each bacterial culture (with or without 1% fungal addition) was grown in LB broth with shaking at 28°C and processed as indicated in Materials and Methods.
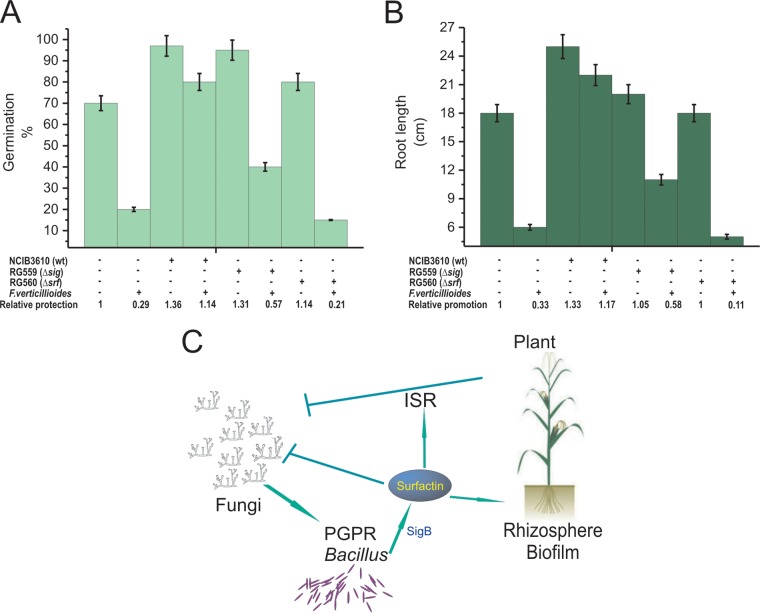
To evaluate the in vivo importance of SigB to protect plants against infections by F. verticillioides, we used Zea mays (i.e., maize) as a model plant for the infective assays. Untreated and bacillus-treated maize seeds were sowed in vermiculite infected with F. verticillioides, and the germination efficiency and early plant development was monitored (see Materials and Methods for details). The average inhibition of germination rate produced by F. verticillioides was 70% and 15% for untreated and treated seeds with the wild-type strain NCIB3610 (Fig. 7A). The proficiency of B. subtilis to protect the seeds from the fungal attack significantly decreased in seeds treated with the SigB-deficient (ΔsigB) B. subtilis strain (58% of germination inhibition compared to germination of uninfected ΔsigB strain-treated seeds) and was null in the case of seeds treated with the surfactin-defective (ΔsrfA) strain (90% of germination inhibition compared with the germination of uninfected ΔsrfA strain-treated seeds) (Fig. 7A). Consequently, the in vivo PGPR activity of wild-type, ΔsigB, and ΔsrfA B. subtilis cells, evaluated by the average root length of emerged plants infected by F. verticillioides, confirmed the important and essential roles of SigB and surfactin, respectively, for the biocontrol proficiency of the bacterium against phytopathogens of agronomical importance (Fig. 7B).
FIG 7.
Biocontrol proficiency of PGPR B. subtilis: in vivo roles of SigB and surfactin. (A and B) Germination efficiency (A) and plant growth (root length) (B) of Zea mays infected with F. verticillioides in the absence or presence of B. subtilis (see Materials and Methods for details). A typical output of three independent experiments is shown. (C) A cartoon summarizing the beneficial interactions between surfactin-producing PGPR B. subtilis cells and plants to resist phytopathogenic fungi. For simplicity, the stimulatory effect of plant polysaccharides on biofilm formation and surfactin synthesis is not indicated (see the text for details).
Conclusions.
From the present results, we demonstrated that the interaction of B. subtilis with fungi induced SigB and its stress-responsive regulon (Fig. 1 and 2). The activation of SigB was required for biocontrol when B. subtilis is cocultured with F. verticillioides (Fig. 4). The activation of SigB depended on the functionality of the RsbP route (Fig. 3) that senses energy depletion in planktonic pure cultures of B. subtilis (22, 23). Therefore, it could be expected that F. verticillioides is able to interfere with energy production in B. subtilis. Another possibility for the nature of the fungus-mediated signal working on SigB activation that cannot be excluded is that RsbP senses a novel and unknown signal, not related to energy depletion, that is exclusively present when B. subtilis is cocultivated with other organisms (i.e., fungi) and absent (or weaker) in axenic B. subtilis cultures. Unfortunately, all studies on regulation of SigB activity have so far been uniquely confined to the planktonic and axenic growth style of B. subtilis but not related to its life style with other organisms (22, 23, 49).
The biocontrol of the growth of F. verticillioides by B. subtilis depended on the proficiency of surfactin production (Fig. 5), and SigB upregulated this synthesis (Fig. 6). Accordingly, with the in vitro results, wild-type (surfactin- and SigB-proficient) B. subtilis completely protected a model plant (maize) from the fungal attack (Fig. 7A and B). How SigB regulates srfA expression is an unsolved question. The simplest scenario for the influence of SigB on srfA expression would be direct binding of SigB to the srfA promoter. The expression of the srfA operon is known to be regulated by the ComPA two-component regulatory system and the RapC phosphatase (50), but to the best of our knowledge, SigB does not regulate ComPA or RapC (22, 23). Moreover, even the most comprehensive characterization of the transcriptional landscape of B. subtilis performed to date (49) did not reveal a SigB-dependent promoter directly upstream of srfA. Therefore, it is likely that SigB activates srfA expression indirectly via an unidentified pathway.
Pesticides have been extensively used to combat plant diseases (51). However, the increased use of pesticides has had negative impacts on human health and the environment (52, 53). Therefore, many efforts have been directed at improving biological control, and biocontrol using PGPRs represents an attractive and environment-friendly alternative approach for controlling plant diseases (54). The main strategies that PGPRs use to control phytopathogenic fungi are the production of natural antifungal compounds (45) and/or the induction of plant systemic resistance (55). Bacillus species are able to use both strategies (8, 14, 45, 56). Bacillus species can produce three types of antifungal lipopeptides: iturins, fengycins, and surfactins (8). In addition, surfactins are very important molecules for the proficiency of bacilli at establishing robust and persistent beneficial biofilms in the plant rhizosphere (Fig. 7C) and work synergistically with fengycins and bacillomycins (a member of the iturin family) against fungi (57–59). Additionally, surfactins, but not iturins or fengycins, are able to induce plant systemic resistance (ISR) against pathogens (Fig. 7C) (55). As a specific mode of positive feedback from the plant to the bacterium, it has been reported that plant polysaccharides stimulate B. subtilis biofilm formation (60) and induce the synthesis of surfactins by Bacillus spp. (61). Therefore, the selection and use of a Bacillus strain or a cocktail of bacilli that overexpress SigB and produce important amounts of surfactins would be of interest for environmental applications (Fig. 7C).
MATERIALS AND METHODS
Bacterial strains, media, and general conditions.
The different NCIB3610 isogenic B. subtilis strains used in this work (Table 1) were streaked on Luria-Bertani (LB) agar plates and cultured at 37°C for 14 h. A single colony was transferred to 30 ml of SM broth (Difco, USA) and grown on a reciprocating shaker (150 rpm) at 37°C for 36 h to obtain a high titer of mature spores. This 36-h-old culture was heat treated at 80°C for 15 min to remove nonsporulated cells, diluted to a concentration of 1 × 107 CFU ml−1, and stored at −20°C until use. The fungus F. verticillioides (strain HFV1904ccc143-2005) was obtained from CEREMIC (Centro de Referencia en Micología, Universidad Nacional de Rosario) and maintained on potato dextrose agar (PDA) plates. Bacterial and fungal quantification (CFU ml−1) was performed on LB or PDA plates incubated at 37°C or 28°C, respectively, for 48 h. For sporulation efficiency, cells were grown in LB or SM broth with shaking (150 rpm) for 96 h at 28°C or 36 h at 37°C and then diluted and plated on LB agar plates before and after heat treatment at 80°C for 15 min to quantify the number of viable cells and spores, expressed as CFU ml−1 (18, 31).
When appropriate, antibiotics were included at the following final concentrations: 1 μg/ml erythromycin (Ery), 5 μg/ml kanamycin (Kan), 5 μg/ml chloramphenicol (Cm), and 50 μg/ml spectinomycin (Spc). Transformation of B. subtilis to obtain isogenic derivatives of the parental strains was carried out as previously described (18, 31). The specific β-galactosidase activity is expressed in Miller units (MU) and was calculated as previously reported (18, 31). The cultures used to measure β-galactosidase activity were grown in LB or PDA medium at the indicated temperatures.
In vitro antifungal activity.
The B. subtilis strain NCIB3610 and its isogenic derivatives were subjected to an in vitro antifungal activity assay against mycelia of F. verticillioides. PDA medium was used as the basal medium, and PDA plates supplemented with 60 µg/ml 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) (as an indicator of β-galactosidase) were used as indicated. A plug (0.3 cm in diameter) containing mycelia of the phytopathogen F. verticillioides was taken from an 8-day-old fungus developed on PDA and placed at the center of a fresh 100-mm PDA petri dish. A single 3-µl inoculum containing 3 × 107 spores of NCIB3610 (or its isogenic strains) was placed 1 cm away from the edge of the mycelium-inoculated PDA or PDA–X-gal plate. The PDA plates were incubated at 28°C, and the evolution of fungal growth was monitored daily for a 2-week period. The fungal growth inhibition index was calculated from measurements of fungal radial growth toward (X1) versus perpendicular to (X2) the bacterial colony according to the formula [1 − (X1/X2)] × 100 (40). The coinoculated PDA–X-gal plate was incubated at 28°C for 96 h. To obtain culture supernatants, the bacteria were grown at 28°C for 18 h before filtering with 0.22-µm sterile filters.
Coculture of bacteria and fungi.
The coculture of B. subtilis and F. verticillioides was initiated by mixing 1 × 105 CFU of the corresponding B. subtilis strain and 5 × 105 CFU of F. verticillioides per ml of LB broth. The cocultivation was prolonged during the indicated times with shaking (150 rpm) at 28°C. Aliquots of the coculture were taken at the indicated times, diluted, plated on LB or PDA, and incubated for 36 h at 37°C or 28°C for bacterial and fungal cellular yield determination (CFU ml−1), respectively. For the determination of the β-galactosidase activity in cocultures of bacteria and fungi, 500 ml of the indicated B. subtilis strain was cultured in LB broth in 2-liter Erlenmeyer flasks at 28°C with shaking (125 rpm) until the mid-logarithmic phase of growth (optical density at 600 nm [OD600] of 0.5). At this time, the bacterial culture was divided in five 0.5-liter Erlenmeyer flasks. Four of them, containing 98.0 ml, 99.0 ml, 99.5 ml, and 99.9 ml of the B. subtilis culture, were supplemented with 2 ml, 1 ml, 0.5 ml, or 0.1 ml, respectively, of a live or dead (autoclaved) or cell-free supernatant culture of F. verticillioides grown for 24 h at 28°C in LB broth. The final fungal concentration in each 0.5-liter Erlenmeyer flask was of 2%, 1%, 0.5%, and 0.1%, respectively. The fifth 0.5-liter Erlenmeyer flask only contained 100.0 ml of the B. subtilis culture (positive-control culture). The fungus-inoculated and noninoculated bacterial cultures were incubated at 28°C with shaking, as shown in the corresponding experiments, and aliquots for the determination of β-galactosidase activity were taken at the indicated times and processed.
In vivo antifungal activity.
Maize (Zea mays) seeds were surface disinfected by dipping in 70% (vol/vol) ethanol for 2 min, followed by 10 mM sodium hypochlorite for 5 min, rinsed three times with sterile water, and soaked for 10 min in sterile water containing a B. subtilis cell suspension at a concentration of 5 × 107 CFU ml−1 or with sterile water alone. Finally, the treated seeds were dried under a filter-sterilized airflow at room temperature. To quantify the average number of B. subtilis cells adhered per seed, fifteen B. subtilis-treated seeds were dipped in a Falcon tube of 50-ml capacity containing 15 ml of sterile water and shaken during 60 min at 75 rpm at room temperature. At this time, without shaking, and after the seeds completely decanted to the bottom of the Falcon tube, 1 ml of the suspension, containing the eluted bacteria, was used to make serial dilutions before being plated on LB agar plates and incubated at 37°C for 36 h. The average number of B. subtilis cells per seed was 1 × 106 CFU. The bacterium-treated seeds were sown in plastic pots (30-cm diameter and 50-cm depth) containing sterilized vermiculite previously infected with F. verticillioides by adding a suspension of mycelial fragments to obtain a final fungal concentration of 5 × 104 CFU g−1 of vermiculite. For each treatment with the different B. subtilis strains, ten seeds were placed in each pot and seven pods were used. The pots were incubated in a room set at 28°C, 95% relative humidity, with a photoperiod of 16 h. Seedling emergence and root lengths were recorded after 14 days of plant development (or plant appearance).
The in vitro and in vivo antifungal experiments of each treatment were repeated five times, and the statistical analysis to evaluate the effect of the organisms in vitro and on plants was carried out using one-way analysis of variance (ANOVA) (P < 0.01).
ACKNOWLEDGMENTS
This work was funded by national grants from FONCyT (PID2011-0051 and PICT2014 start-up 3777) and CONICET to R.G.
We thank the Fulbright (Washington, DC) and Pew (Philadelphia) foundations for their support and David Zeigler from BGSC (Bacillus Genetic Stock Center, Ohio) for providing strains and technical support. M.B., S.C., D.V., C.B., and W.R. carried out the experiments. All authors contributed to the experimental design and concepts, and all authors contributed to the text. R.G. designed the experiments and wrote the main text, with contributions from all other authors. We have no conflicts of interest to declare.
REFERENCES
- 1.Palková Z. 2004. Multicellular microorganisms: laboratory versus nature. EMBO Rep 5:470–476. doi: 10.1038/sj.embor.7400145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Deng Y-J, Wang SY. 2016. Synergistic growth in bacteria depends on substrate complexity. J Bacteriol 54:23–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balbontín R, Vlamakis H, Kolter R. 2014. Mutualistic interaction between Salmonella enterica and Aspergillus niger and its effects on Zea mays colonization: Salmonella-Aspergillus interaction on maize. Microb Biotechnol 7:589–600. doi: 10.1111/1751-7915.12182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou L, Slamti L, Nielsen-LeRoux C, Lereclus D, Raymond B. 2014. The social biology of quorum sensing in a naturalistic host pathogen system. Curr Biol 24:2417–2422. doi: 10.1016/j.cub.2014.08.049. [DOI] [PubMed] [Google Scholar]
- 5.Jayaswal RK, Fernandez MA, Schoerder R. 1990. Isolation and characterization of a Pseudomonas strain that restricts growth of various phytopathogenic fungi. Appl Environ Microbiol 56:1053–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Palm ME. 2001. Systematics and the impact of invasive fungi on agriculture in the United States. Bioscience 51:141–147. doi: 10.1641/0006-3568(2001)051[0141:SATIOI]2.0.CO;2. [DOI] [Google Scholar]
- 7.Ricroch A, Harwood W, Svobodová Z, Sági L, Hundleby P, Badea EM, Rosca I, Cruz G, Salema Fevereiro MP, Marfà Riera V, Jansson S, Morandini P, Bojinov B, Cetiner S, Custers R, Schrader U, Jacobsen H-J, Martin-Laffon J, Boisron A, Kuntz M. 2016. Challenges facing European agriculture and possible biotechnological solutions. Crit Rev Biotechnol 36:875–883. doi: 10.3109/07388551.2015.1055707. [DOI] [PubMed] [Google Scholar]
- 8.Cawoy H, Debois D, Franzil L, De Pauw E, Thonart P, Ongena M. 2015. Lipopeptides as main ingredients for inhibition of fungal phytopathogens by Bacillus subtilis/amyloliquefaciens: lipopeptides as inhibitors of phytopathogens. Mol Microbiol 8:281–295. doi: 10.1111/1751-7915.12238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Compant S, Clément C, Sessitsch A. 2010. Plant growth-promoting bacteria in the rhizo- and endosphere of plants: their role, colonization, mechanisms involved and prospects for utilization. Soil Biol Biochem 42:669–678. doi: 10.1016/j.soilbio.2009.11.024. [DOI] [Google Scholar]
- 10.Lim SM, Yoon M-Y, Choi GJ, Choi YH, Jang KS, Shin TS, Park HW, Yu NH, Kim YH, Kim J-C. 2017. Diffusible and volatile antifungal compounds produced by an antagonistic Bacillus velezensis G341 against various phytopathogenic fungi. Plant Pathol J 33:488–498. doi: 10.5423/PPJ.OA.04.2017.0073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gu Q, Yang Y, Yuan Q, Shi G, Wu L, Lou Z, Huo R, Wu H, Borriss R, Gao X. 2017. Bacillomycin D produced by Bacillus amyloliquefaciens is involved in the antagonistic interaction with the plant-pathogenic fungus Fusarium graminearum. Appl Environ Microbiol 83:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu L, Wu H, Chen L, Xie S, Zang H, Borriss R, Gao X. 2014. Bacilysin from Bacillus amyloliquefaciens BFZB42 has specific bactericidal activity against harmful algal bloom species. Appl Environ Microbiol 80:7512–7520. doi: 10.1128/AEM.02605-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vlamakis H, Chai Y, Beauregard P, Losick R, Kolter R. 2013. Sticking together: building a biofilm the Bacillus subtilis way. Nat Rev Microbiol 11:157–168. doi: 10.1038/nrmicro2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ongena M, Jacques P. 2008. Bacillus lipopeptides: versatile weapons for plant disease biocontrol. Trends Microbiol 16:115–125. doi: 10.1016/j.tim.2007.12.009. [DOI] [PubMed] [Google Scholar]
- 15.Piggot PJ, Coote JG. 1976. Genetic aspects of bacterial endospore formation. Bacteriol Rev 40:908–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abee T, Kovács ÁT, Kuipers OP, van der Veen S. 2011. Biofilm formation and dispersal in Gram-positive bacteria. Curr Opin Biotechnol 22:172–179. doi: 10.1016/j.copbio.2010.10.016. [DOI] [PubMed] [Google Scholar]
- 17.Losick R. 2015. A love affair with Bacillus subtilis. J Biol Chem 290:2529–2538. doi: 10.1074/jbc.X114.634808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grau RR, de Oña P, Kunert M, Leñini C, Gallegos-Monterrosa R, Mhatre E, Vileta D, Donato V, Hölscher T, Boland W, Kuipers OP, Kovács ÁT. 2015. A duo of potassium-responsive histidine kinases govern the multicellular destiny of Bacillus subtilis. mBio 6:e00581-15. doi: 10.1128/mBio.00581-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ayala F, Bauman C, Cogliati S, Lenini C, Bartolini M, Grau R. 2017. Microbial flora, probiotics, Bacillus subtilis and the search for a long and healthy human longevity. Microb Cell 4:133–136. doi: 10.15698/mic2017.04.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cairns LS, Hobley L, Stanley-Wall NR. 2014. Biofilm formation by Bacillus subtilis: new insights into regulatory strategies and assembly mechanisms: regulation and assembly of Bacillus subtilis biofilms. Mol Microbiol 93:587–598. doi: 10.1111/mmi.12697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haldenwang WG, Losick R. 1980. Novel RNA polymerase sigma factor from Bacillus subtilis. Proc Natl Acad Sci U S A 77:7000–7004. doi: 10.1073/pnas.77.12.7000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Price CW. 2002. Bacillus subtilis and its closest relatives. From genes to cells, p 369–384. In Sonenshein AL, Hoch JA, Losick R (ed), General stress response. ASM Press, Washington, DC. [Google Scholar]
- 23.Hecker M, Pané-Farré J, Uwe V. 2007. SigB-dependent general stress response in Bacillus subtilis and related Gram-positive bacteria. Annu Rev Microbiol 61:215–236. doi: 10.1146/annurev.micro.61.080706.093445. [DOI] [PubMed] [Google Scholar]
- 24.Benson AK, Haldenwang WG. 1993. Bacillus subtilis σB is regulated by a binding protein (RsbW) that blocks its association with core RNA polymerase. Proc Natl Acad Sci U S A 90:2330–2334. doi: 10.1073/pnas.90.6.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dufour A, Haldenwang WG. 1994. Interactions between a Bacillus subtilis anti-sigma factor (RsbW) and its antagonist (RsbV). J Bacteriol 176:1813–1820. doi: 10.1128/jb.176.7.1813-1820.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wise AA, Price CW. 1995. Four additional genes in the sigB operon of Bacillus subtilis that control activity of the general stress factor sigB in response to environmental signals. J Bacteriol 177:123–133. doi: 10.1128/jb.177.1.123-133.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Voelker U, Voelker A, Maul B, Hecker M, Dufour A, Haldenwang WG. 1995. Separate mechanisms activate σB of Bacillus subtilis in response to environmental and metabolic stresses. J Bacteriol 177:3771–3780. doi: 10.1128/jb.177.13.3771-3780.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang X, Kang CM, Brody MS, Price CW. 1996. Opposing pairs of serine protein kinases and phosphatases transmit signals of environmental stress to activate a bacterial transcription factor. Genes Dev 10:2265–2275. doi: 10.1101/gad.10.18.2265. [DOI] [PubMed] [Google Scholar]
- 29.Vijay K, Brody MS, Fredlund E, Price CW. 2000. A PP2C phosphatase containing a PAS domain is required to convey signals of energy stress to the sigma B transcription factor of Bacillus subtilis. Mol Microbiol 35:180–188. doi: 10.1046/j.1365-2958.2000.01697.x. [DOI] [PubMed] [Google Scholar]
- 30.Brigulla M, Hoffmann T, Krisp A, Volker A, Bremer E, Volker U. 2003. Chill induction of the SigB-dependent general stress response in Bacillus subtilis and its contribution to low-temperature adaptation. J Bacteriol 185:4305–4314. doi: 10.1128/JB.185.15.4305-4314.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Méndez MB, Orsaria LM, Philippe V, Pedrido ME, Grau RR. 2004. Novel roles of the master transcription factors Spo0A and sigma B for survival and sporulation of Bacillus subtilis at low growth temperature. J Bacteriol 186:989–1000. doi: 10.1128/JB.186.4.989-1000.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oren L, Ezrati S, Cohen D, Sharon A. 2003. Early events in the Fusarium verticillioides-maize interaction characterized by using a green fluorescent protein-expressing transgenic isolate. Appl Environ Microbiol 69:1695–1701. doi: 10.1128/AEM.69.3.1695-1701.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tesso TT, Ochanda N, Little CR, Claflin L, Tuinstra MR. 2010. Analysis of host plant resistance to multiple Fusarium species associated with stalk rot disease in sorghum [Sorghum bicolor (L.) Moench]. Field Crops Res 118:177–182. doi: 10.1016/j.fcr.2010.05.010. [DOI] [Google Scholar]
- 34.Lin Z, Wang J, Bao Y, Guo Q, Powell CA, Xu S, Chen B, Zhang M. 2016. Deciphering the transcriptomic response of Fusarium verticillioides in relation to nitrogen availability and the development of sugarcane pokkah boeng disease. Sci Rep 6:29692. doi: 10.1038/srep29692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Borah SN, Goswami D, Sarma HK, Cameotra SS, Deka S. 2016. Rhamnolipid biosurfactant against Fusarium verticillioides to control stalk and ear rot disease of maize. Front Microbiol 7:1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bartolini M, Cogliati S, Vileta D, Bauman C, Rateni L, Leñini C, Argañaraz F, Francisco M, Villalba JM, Steil L, Völker U, Grau R. 2018. Regulation of biofilm aging and dispersal in Bacillus subtilis by the alternative sigma factor SigB. J Bacteriol 201:e00473-18. doi: 10.1128/JB.00473-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu J, Prindle A, Humphries J, Gabalda-Sagarra M, Asally M, Lee DY, Ly S, Garcia-Ojalvo J, Süel GM. 2015. Metabolic co-dependence gives rise to collective oscillations within biofilms. Nature 523:550–554. doi: 10.1038/nature14660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang L, Grau R, Perego M, Hoch JA. 1997. A novel histidine kinase inhibitor regulating development in Bacillus subtilis. Genes Dev 11:2569–2579. doi: 10.1101/gad.11.19.2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Notz R, Maurhofer M, Dubach H, Haas D, Defago G. 2002. Fusaric acid-producing strains of Fusarium oxysporum alter 2,4-diacetylphloroglucinol biosynthetic gene expression in Pseudomonas fluorescens CHA0 in vitro and in the rhizosphere of wheat. Appl Environ Microbiol 68:2229–2235. doi: 10.1128/AEM.68.5.2229-2235.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Quecine M, Kidarsa TA, Goebel NC, Shaffer BT, Henkels MD, Zabriskie TM, Loper JE. 2016. An interspecies signaling system mediated by fusaric acid has parallel effects on antifungal metabolite production by Pseudomonas protegens strain Pf-5 and antibiosis of Fusarium spp. Appl Environ Microbiol 82:1372–1382. doi: 10.1128/AEM.02574-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bacon CW, Hinton DM, Hinton A. 2006. Growth-inhibiting effects of concentrations of fusaric acid on the growth of Bacillus mojavensis and other biocontrol Bacillus species. J Appl Microbiol 100:185–194. doi: 10.1111/j.1365-2672.2005.02770.x. [DOI] [PubMed] [Google Scholar]
- 42.Telles-Pupulin AR, Diniz SPSS, Bracht A, Ishii-Iwamoto EL. 1996. Effects of fusaric acid on respiration in maize root mitochondria. Biol Plant 38:421–429. doi: 10.1007/BF02896673. [DOI] [Google Scholar]
- 43.Bottone EJ, Peluso RW. 2003. Production by Bacillus pumilus (MSH) of an antifungal compound that is active against Mucoraceae and Aspergillus species: preliminary report. J Med Microbiol 52:69–74. doi: 10.1099/jmm.0.04935-0. [DOI] [PubMed] [Google Scholar]
- 44.Yu G, Sinclair J, Hartman G, Bertagnolli B. 2002. Production of iturin A by Bacillus amyloliquefaciens suppressing Rhizoctonia solani. Soil Biol Biochem 34:955–963. doi: 10.1016/S0038-0717(02)00027-5. [DOI] [Google Scholar]
- 45.Ongena M, Jacques P, Touré Y, Destain J, Jabrane A, Thonart P. 2005. Involvement of fengycin-type lipopeptides in the multifaceted biocontrol potential of Bacillus subtilis. Appl Microbiol Biotechnol 69:29–38. doi: 10.1007/s00253-005-1940-3. [DOI] [PubMed] [Google Scholar]
- 46.Nye TM, Schroeder JW, Kearns DB, Simmons LA. 2017. Complete genome sequence of undomesticated Bacillus subtilis strain NCIB 3610. Genome Announc 5:e00364-17. doi: 10.1128/genomeA.00364-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gao L, Han J, Liu H, Qu X, Lu Z, Bie X. 2017. Plipastatin and surfactin coproduction by Bacillus subtilis pB2-L and their effects on microorganisms. Antonie Van Leeuwenhoek 110:1007–1018. doi: 10.1007/s10482-017-0874-y. [DOI] [PubMed] [Google Scholar]
- 48.McLoon AL, Guttenplan SB, Kearns DB, Kolter R, Losick R. 2011. Tracing the domestication of a biofilm-forming bacterium. J Bacteriol 193:2027–2034. doi: 10.1128/JB.01542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nicolas P, Mäder U, Dervyn E, Rochat T, Leduc A, Pigeonneau N, Bid-Nenko E, Marchadier E, Hoebeke M, Aymerich S, Becher D, Bisicchia P, Botella E, Delumeau O, Doherty G, Denham EL, Fogg MJ, Fromion V, Goelzer A, Hansen A, Härtig E, Harwood CR, Homuth G, Jarmer H, Jules M, Klipp E, Le Chat L, Lecointe F, Lewis P, Liebermeister W, March A, Mars RA, Nannapaneni P, Noone D, Pohl S, Rinn B, Rügheimer F, Sappa PK, Samson F, Schaffer M, Schwikowski B, Steil L, Stülke J, Wiegert T, Devine KM, Wilkinson AJ, van Dijl JM, Hecker M, Völker U, Bessières P, Noirot P. 2012. Condition-dependent transcriptome reveals high-level regulatory architecture in Bacillus subtilis. Science 335:1103–1106. doi: 10.1126/science.1206848. [DOI] [PubMed] [Google Scholar]
- 50.Core L, Perego M. 2003. TPR-mediated interaction of RapC with ComA inhibits response regulator-DNA binding for competence development in Bacillus subtilis. Mol Microbiol 49:1509–1522. doi: 10.1046/j.1365-2958.2003.03659.x. [DOI] [PubMed] [Google Scholar]
- 51.Pingali PL. 2012. Green revolution: impacts, limits, and the path ahead. Proc Natl Acad Sci U S A 109:12302–12308. doi: 10.1073/pnas.0912953109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tago D, Andersson H, Treich N. 2014. Pesticides and health: a review of evidence on health effects, valuation of risks, and benefit-cost analysis, p 203–295. In Blomquist GC, Bolin K (ed), Advances in health economics and health services research. Emerald Group Publishing Ltd, Bingley, United Kingdom. [PubMed] [Google Scholar]
- 53.Kim K-H, Kabir E, Jahan SA. 2017. Exposure to pesticides and the associated human health effects. Sci Total Environ 575:525–535. doi: 10.1016/j.scitotenv.2016.09.009. [DOI] [PubMed] [Google Scholar]
- 54.Raja N. 2013. Biopesticides and biofertilizers: ecofriendly sources for sustainable agriculture. J Biofertil Biopestici 4:1–2. [Google Scholar]
- 55.Cawoy H, Mariutto M, Henry G, Fisher C, Vasilyeva N, Thonart P, Dommes J, Ongena M. 2014. Plant defense stimulation by natural isolates of Bacillus depends on efficient surfactin production. Mol Plant Microbe Interact 27:87–100. doi: 10.1094/MPMI-09-13-0262-R. [DOI] [PubMed] [Google Scholar]
- 56.Raaijmakers JM, De Bruijn I, Nybroe O, Ongena M. 2010. Natural functions of lipopeptides from Bacillus and Pseudomonas: more than surfactants and antibiotics. FEMS Microbiol Rev 34:1037–1062. doi: 10.1111/j.1574-6976.2010.00221.x. [DOI] [PubMed] [Google Scholar]
- 57.Aleti G, Lehner S, Bacher M, Compant S, Nikolic B, Plesko M, Schuhmacher R, Sessitsch A, Brader G. 2016. Surfactin variants mediate species-specific biofilm formation and root colonization in Bacillus: surfactins mediate species-specific signaling. Environ Microbiol 18:2634–2645. doi: 10.1111/1462-2920.13405. [DOI] [PubMed] [Google Scholar]
- 58.Zeriouh H, de Vicente A, Pérez-García A, Romero D. 2014. Surfactin triggers biofilm formation of Bacillus subtilis in melon phylloplane and contributes to the biocontrol activity. Environ Microbiol 16:2196–2211. doi: 10.1111/1462-2920.12271. [DOI] [PubMed] [Google Scholar]
- 59.Luo C, Zhou H, Zou J, Wang X, Zhang R, Xiang Y, Chen Z. 2015. Bacillomycin L and surfactin contribute synergistically to the phenotypic features of Bacillus subtilis 916 and the biocontrol of rice sheath blight induced by Rhizoctonia solani. Appl Microbiol Biotechnol 99:1897–1910. doi: 10.1007/s00253-014-6195-4. [DOI] [PubMed] [Google Scholar]
- 60.Beauregard PB, Chai Y, Vlamakis H, Losick R, Kolter R. 2013. Bacillus subtilis biofilm induction by plant polysaccharides. Proc Natl Acad Sci U S A 110:1621–1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Debois D, Fernandez O, Franzil L, Jourdan E, de Brogniez A, Willems L, Clément C, Dorey S, De Pauw E, Ongena M. 2015. Plant polysaccharides initiate underground crosstalk with bacilli by inducing synthesis of the immunogenic lipopeptide surfactin: Bacillus lipopeptides induced by plant polymers. Environ Microbiol Rep 7:570–582. doi: 10.1111/1758-2229.12286. [DOI] [PubMed] [Google Scholar]