Abstract
Objectives
Nedd4 (neural precursor cell expressed developmentally down‐regulated protein 4) is a member of the HECT E3 ubiquitin ligases, and is elevated in prostate, bladder and colorectal cancers, and promotes colonic cell population expansion. Up to now, molecular mechanisms of how Nedd4 functions, have not been well understood.
Materials and methods
In this study, shRNA was used to reduce expression of Nedd4 in the human prostate carcinoma cell line DU145. To analyse effects of Nedd4 on cell proliferation, MTT and colony formation assays were performed. DAPI staining and FACS analysis were used to investigate outcomes of Nedd4 activity, on apoptosis. Results of Nedd4 expression on lysosomal membrane permeabilization and autophagy were further investigated using acridine orange (AO) staining, immunofluorescence and western blot analysis.
Results
We found that in HeLa cells, expression of Nedd4 promoted cell proliferation, whereas its knockdown inhibited colony formation and induced apoptosis in DU145 cells. Furthermore, down‐regulation of Nedd4 in DU145 cells promoted lysosomal membrane permeabilization. We also found that down‐regulation of Nedd4 inhibited autophagy in both DU145 and A549 cells. Investigation into mechanisms involved revealed that knockdown of endogenous Nedd4 expression notably increased activated mTOR (p‐mTOR) levels, which suggests that mTOR signalling was involved in the Nedd4‐mediated autophagy.
Conclusions
Our results indicate that expression of Nedd4 promoted cell proliferation and colony formation but prevented apoptosis. Moreover, Nedd4 promoted autophagy and was associated with the mTOR signalling pathway.
Introduction
Ubiquitination is a common post‐translational modification to proteins, which is catalysed by a series of ubiquitination enzymes: ubiquitin activation enzyme (E1), ubiquitin conjugation enzymes (E2s), and ubiquitin ligases (E3s) 1, 2. Ubiquitination and deubiquitination of proteins plays an important role in tumourigenesis 3. Ubiquitination occurs through a multiple enzyme cascade, where recognition is specified by the ubiquitin ligases (E3s); they specifically recognize a substrate for modification temporally and spatially, and have important roles in cell regulation. There are more than 500 E3s common to mammalian cells, which contain either a RING (really interesting new gene) finger domain or a HECT (homologous to E6‐AP COOH terminus) domain.
The Nedd4 family belongs to HECT type E3 ubiquitin ligases. Protein structure of Nedd4 contains three functional domains, N‐terminal C2 domain for Ca2+/membrane binding; a central region containing WW domains for protein–protein interaction (which contains two conservative tryptophan (W)s separated by 21 amino acids) and a C‐terminal HECT domain for transfer ubiquitin from its cysteine residues to lysine residues of the substrate protein. It is elevated in prostate, bladder and colorectal cancers and has been shown to promote colonic cell growth 4, 5. Predominant phenotype of Nedd4 knockout mice is of growth retardation, associated with perinatal lethality. Mouse embryonic fibroblasts (MEFs) isolated from Nedd4 knockout embryos have slower turnover 6. One previous study showed that polyubiquitination by Nedd4 was thought to target PTEN for proteasomal degradation 7, while covalent attachment of a single ubiquitin molecule favoured its nuclear translocation 8. An alternative inactivation pathway of PTEN during tumourigenesis may occur as a result of Nedd4 overexpression in human bladder and prostate carcinomas, in which aberrant degradation of PTEN promotes AKT signalling and ultimately provides an advantage to cell survival. Other reports however have shown that Nedd4 knocking‐out did not influence degradation nor subcellular localization of PTEN 9. Thus, it remained a challenge for future research to further validate a role of Nedd4 in tumourigenesis.
A series of studies has suggested that occurrence of a tumour is closely related to changes in various lysosomal characteristics 10, 11. Alteration of lysosomal membrane permeabilization and release of hydrolytic enzymes are important for death of tumour cells. The lysosome plays a key role in apoptosis, and lysosomal permeabilization appears to be an early event in the apoptotic cascade, preceding other hallmarks of apoptosis such as mitochondrial destabilization and caspase activation 12, 13, 14, 15. In this process, proteases released from the lysosomal compartment trigger initiating events of apoptosis, generally termed the ‘lysosomal pathway of apoptosis’.
Recently, it has been proposed that Nedd4 has extended roles in cancer development 16, but the molecular mechanisms have not been well understood. Here, we demonstrate that expression of Nedd4 promoted cell proliferation and autophagy in DU145 cells, and further studies indicated that mTOR signal activation was involved in Nedd4‐mediated promotion of autophagy.
Materials and methods
Materials and antibodies
All materials were from Sigma (St. Louis, MO, USA)unless otherwise stated. Commercial antibodies were purchased from the following sources: rabbit anti‐Nedd4 (Abcam, Cambridge, UK), rabbit anti‐LC3B (Sigma), rabbit anti‐caspase‐3, rabbit anti‐caspase‐8 and rabbit anti‐caspase‐9 (Santa Cruz Biotechnology, Santa Cruz, CA, USA). Fluorophore and HRP‐conjugated secondary antibodies were obtained from Invitrogen (Carlsbad, CA, USA), while pEGFP–Nedd4 plasmid was prepared in our laboratory.
Cell culture
DU145 cells were grown at 37 °C under 5% CO2 in RPMI 1640 medium, supplemented with 10% foetal bovine serum (FBS) and HEK293T and HeLa cells were grown at 37 °C under 5% CO2 in DMEM containing 10% FBS. A549 cells were grown at 37 °C under 5% CO2 in F12K containing 10% FBS. Cell transfections were performed using TurboFect (Thermo Scientific, Waltham, MA, USA) and Lipofectamine 2000 (Invitrogen) transfection reagents according to the manufacturer's instructions.
RNA interference
Lentiviral vectors expressing five Nedd4‐shRNAs (TRCN0000272424 CCGGCGGTTGGAGAATGTAGCAATACTCGAGTATTGCTACATTCTCCAACCGTTTTTC; TRCN0000272425 CCGGAGTGCTACTCGCAGCTATTTACTCGA GTAAATAGCTGCGAGTAGCACTTTTTTC; TRCN0000272476 CCGGGCTGA ACTATACGGTTCAAATCTCGAGATTTGAACCGTATAGTTCAGCTTTTTC; TRCN0000272477 CCGGTACGTGAGAGTGACGTTATATCTCGAGATATAACGT CACTCTCACGTATTTTTC; TRCN0000284755 CCGGCCGGAGAATTATGGG TGTCAACTCGAGTTGACACCCATAATTCTCCGGTTTTTC) and a non‐target shRNA control vector (SHC002) were obtained from Sigma. Lentiviruses were produced according to the manufacturer's manual and cells were infected once, 3 days prior to start of the experiments. DU145 cells were transduced overnight then selected in 0.5 μg/ml puromycin to obtain stable Nedd4 knockdown cell lines.
Stable cell lines
DU145 cell lines with stably knocked down Nedd4 (Nedd4‐shRNA2‐1#, Nedd4‐shRNA2‐2#) were obtained by infection of Nedd4‐shRNAs followed by selection in 0.5 μg/ml puromycin for 2 weeks.
Western blotting
Cultured cells were washed twice in PBS before addition of ice cold lysis buffer (50 mm Tris–HCl pH 7.5, 150 mm NaCl, 1 mm NaF, 1 mm phenylmethylsulphonyl fluoride, 4 mg/ml leupeptin and 1 mg/ml aprotinin, and 1% Nonidet P‐40). Equal amounts of protein were loaded into each well and separated by 7%, 10% or 12% SDS‐PAGE gel, followed by transfer to nitrocellulose membranes. These were blocked in PBST buffer containing 5% non‐fat dry milk for 1 hour at room temperature. Blots were then incubated overnight at 4 °C with primary antibodies. Secondary antibody incubation was for 1 hour at room temperature. Finally, Odyssey infrared laser imaging system (LI‐COR Biosciences, Lincoln, NE, USA) was used to image the experimental results.
MTT assay
Cell proliferation was assessed using the MTT (3‐[4,5‐dimethylthiazol‐2‐yl]‐2,5‐diphenyltetrazolium bromide; thiazolyl blue) assay. Cells were plated (5000 cells/well) in triplicate with 200 μl growth medium, in 96‐well plates, and cultured for the indicated period. Freshly prepared MTT was added at final concentration of 0.5 mg/ml, in PBS pH 7.4, and plates were incubated in the dark for 4 h at 37 °C. Culture medium was then removed and remaining blue precipitate was solubilized in dimethylsulphoxide (DMSO). Absorbance was read at 570 nm using a microplate reader.
Colony formation assay
Cells were plated (800 cells/well) in triplicate with 2 ml growth medium, in six‐well plates. After 20 days, colonies were either left unstained, or stained with 5 mg/ml MTT (Sigma) and photographed.
Apoptosis assay
Treated cells were collected and washed in PBS, and apoptosis was detected using Annexin V‐FITC Apoptosis Detection Kit (Cat. No KGA106 KeyGEN BioTECH, Nanjing, China) following the manufacturer's instructions. Cells were detected by flow cytometry and data were analysed using CFlow 1.0.264.15 software.
Acridine orange (AO) staining
Cells were seeded on glass coverslips in six‐well plates. After treatment, they were washed twice in PBS then incubated with 1 μm AO for 15 min at 37 °C. After staining, cells were washed once in PBS. Coverslips were counted over glass slides with PBS and immediately observed and photographed using a fluorescence microscope (Olympus BX53F).
Immunofluorescence microscopy
Cells were grown on coverslips and fixed in 100% methanol at 20 °C for 5 min. Specimens were incubated for 60 min at RT with primary antibodies diluted in PBS containing 0.5 mg/ml BSA, then washed three times in PBS and incubated for a further 30 min at RT, with fluorophore‐conjugated secondary antibodies diluted in PBS/BSA. Specimen coverslips were mounted in Mowiol, allowed to dry and analysed using an Olympus FV1000 confocal microscope with DT72 camera and FV10‐ASW 3.0 Viewer software.
Statistical analysis
Statistical analysis was performed using the χ2 test or Fisher's exact test and Spearman's rank correlation coefficient analysis. P < 0.05 indicated significant difference, and P < 0.01 indicated that the difference was highly significant.
Results
Nedd4 was overexpressed in prostate cancer cell line DU145
To profile expression of Nedd4 protein across different cancer cell lines, western blot analysis was performed on a selected panel of cell lysates. As shown in Fig. 1, it was most highly expressed in DU145 prostate cancer cells, with medium levels in MDA‐MB‐231, MGC‐803 and SW620 cells, and lower expression levels detected in HeLa cells. Endogenous Nedd4 in DU145 cells was then depleted by RNA interference, to study roles and molecular mechanisms of Nedd4 in development of this type of cancer.
Figure 1.
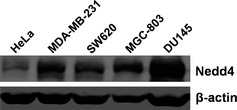
Expression of Nedd4 protein in a panel of selected cancer cell lines. Cervical cancer cell line HeLa, breast cancer cell line MDA‐MB‐231, colon cancer cell line SW620, gastric cancer cell line MGC‐803 and prostate cancer cell line DU145 were cultured in DMEM or RPMI 1640 medium. Cells were collected and cell lysates were analysed by western blotting, with antibodies to Nedd4, and β‐actin as loading control.
Knockdown of endogenous Nedd4 in DU145 cells by RNA interference
Previous studies have revealed that overexpression of Nedd4 is abnormally reactivated in many prostate and bladder cancers. To investigate the biological function of Nedd4, shRNA was used to reduce its expression in DU145 cells. Five pLKO.1/puro lentiviral vectors expressing shRNAs (shRNA1#, shRNA2#, shRNA3#, shRNA4# and shRNA5#) were designed to target various regions in Nedd4 transcripts. The Nedd4‐shRNAs were individually co‐transfected with lentiviral packaging plasmids into HEK293T cells. Virus‐containing supernatant was harvested and used to infect the DU145 cells. Knockdown efficiency of Nedd4 protein was analysed by western blot analysis (Fig. 2a) and results showed that its expression was significantly reduced after infection with Nedd4‐shRNA2# virus‐containing supernatant (Fig. 2b). Thus, lentiviral vector containing Nedd4‐shRNA#2 was selected to deplete Nedd4 and to further investigate its role in tumourigenesis.
Figure 2.
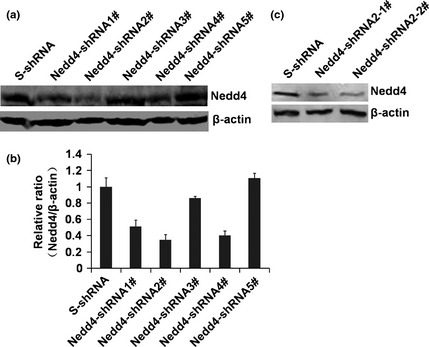
Knockdown of Nedd4 expression in DU 145 cells by sh RNA . (a) DU145 cells were infected with 200 μl lentiviruses expressing Nedd4‐shRNAs, and expression of Nedd4 was detected by western blotting. (b) Nedd4 protein levels were detected by western blotting, and quantitated using ImageJ software. (c) expression of Nedd4 in Nedd4‐shRNA2‐1# and Nedd4‐shRNA2‐2# stable knockdown cell lines, were detected by western blotting.
Nedd4‐shRNA#2 was used to generate cell lines stably interfering with expression of Nedd4, and stable cell lines generated were named Nedd4‐shRNA#2‐1 and shRNA#2‐2. DU145 cells infected with scrambled shRNA virus, S‐shRNA, were used as negative control. Western blot analysis confirmed depletion of Nedd4 in the Nedd4 knockdown stable cell lines (Fig. 2c). These results showed that expression of Nedd4 protein was reduced by RNA interference using Nedd4‐shRNA2#, and that Nedd4 stably knocked‐down cell lines would be useful tools for subsequent functional analysis.
Expression of Nedd4 promoted cell proliferation
To analyse effects of Nedd4 on cell proliferation, MTT and colony formation assays were performed. MTT assays showed that Nedd4‐shRNA2# and shRNA4# transiently infected cell populations grew much more slowly compared to negative control S‐shRNA cells (Fig. 3a, b), and a similar effect was also observed using stably knocked‐down Nedd4 in DU145 cells (Fig. 3c, d). Furthermore, MTT cell assays indicated that overexpression of Nedd4 in HeLa cells promoted proliferation (Fig. 3e, f). Colony formation assays also showed that cells stably expressing Nedd4‐shRNA formed far fewer colonies compared to control (Fig. 3g, h). These results indicate that Nedd4 plays an important role in the process of cell proliferation and colony formation.
Figure 3.
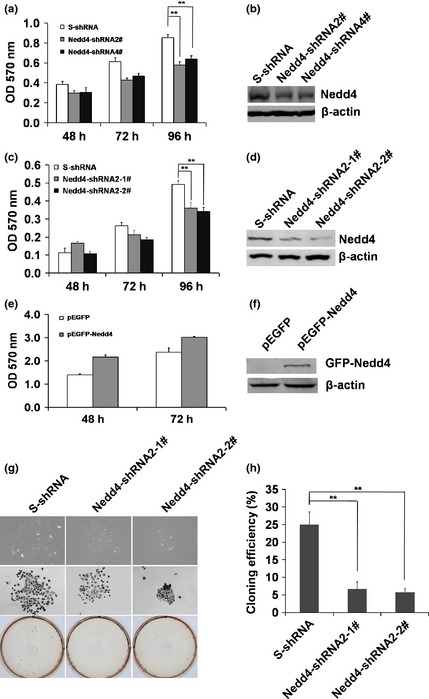
Expression of Nedd4 promoted cell proliferation. (a), (b) DU145 cells were infected with 200 μl lentiviruses expressing Nedd4‐shRNA2#, shRNA4#, control S‐shRNA, and the cell proliferation was analysed by MTT assay at 48 h, 72 h and 96 h post‐infection. Expression of Nedd4 was detected by western blotting. Values represent mean ± S.E.M. of at least three independent experiments, **P < 0.01. (c), (d) Cell proliferation of DU145 cells stably expressing Nedd4‐shRNA2‐1#, shRNA2‐2# and the control S‐shRNA was measured by MTT assay 48 h, 72 h and 96 h after culture initiation. Expression of Nedd4 was detected by western blotting. Values represent mean ± S.E.M. of at least three independent experiments, **P < 0.01. (e), (f) Effects of Nedd4 expression on cell proliferation of HeLa cells were determined by MTT assay. Results are representative of three independent experiments. (g) DU145 cells stably expressing Nedd4‐shRNA2‐1#, shRNA2‐2# and control S‐shRNA were used for colony formation assays after 20 days culture, then plates were photographed. (h) Quantification of efficiency of colony formation by statistical analysis. Values represent mean ± S.E.M. of at least three independent experiments, **P < 0.01.
Knockdown of endogenous Nedd4 induced apoptosis in DU145 cells
To investigate whether shRNA‐mediated knockdown of Nedd4 in DU145 cells induced apoptosis, DU145 cells stably expressing Nedd4‐shRNAs were treated with staurosporine, a widely used chemotherapeutic agent, to induce apoptosis in various types of cancer cells. As shown in Fig. 4a and 4b, knockdown of Nedd4 increased frequency of staurosporine‐induced apoptosis in DU145 cells as revealed by DAPI staining. Similar results were also found using annexin V/PI staining with 0.6% apoptosis detected in S‐shRNA‐expressing cells and 13.2% and 16.1% apoptosis in Nedd4‐shRNA#2‐1‐ and shRNA#2‐2‐expressing cells, respectively (Fig. 4c). These results indicate that depletion of Nedd4 in DU145 cells promoted apoptosis.
Figure 4.
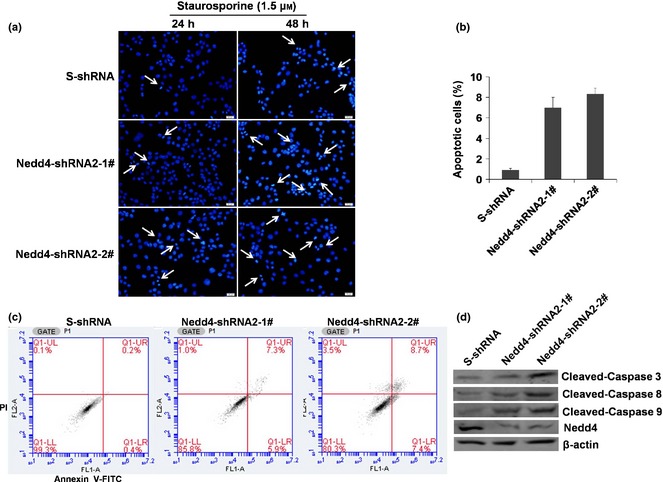
Knockdown of endogenous Nedd4 expression induced apoptosis in DU 145 cells. (a) DU145 cells stably expressing Nedd4‐shRNA2‐1#, shRNA2‐2# and control S‐shRNA, were treated with 1.5 μm staurosporine for 24 h and 48 h, then stained with DAPI. Scale bar, 20 μm. (b) Apoptotic cells were quantified by counting a minimum of four fields at 100 cells per field. (c) DU145 cells stably expressing Nedd4‐shRNA2‐1#, shRNA2‐2# and control S‐shRNA, were treated with 1.5 μm staurosporine for 48 h, then double‐labelled with annexin V/PI staining. Percentage of apoptotic cells is sum of lower and upper right panels. (d) DU145 cells stably expressing Nedd4‐shRNA2‐1#, shRNA2‐2# and control S‐shRNA were treated with 1.5 μm staurosporine for 6 h, and analysed by western blotting using antibodies against caspases ‐3, ‐8, ‐9, Nedd4 and β‐actin.
Apoptosis is triggered by two types of apoptotic caspases, namely initiator caspases ‐2, ‐8, ‐9 and ‐10, and effector caspases ‐3, ‐6 and ‐7. Initiator caspases cleave and activate inactive forms of effector caspases, and these activated effector caspases subsequently cleave other protein substrates to trigger the apoptotic process. To further assess Nedd4 knockdown‐induced apoptosis in DU145 cells, activity levels of these effector caspases were investigated. We found that level of cleaved caspase‐3 increased in Nedd4‐shRNA#2‐1‐ and shRNA#2‐2‐expressing cells, and that knockdown of Nedd4 significantly increased levels of cleaved caspases ‐8 and ‐9 (Fig. 4d). These data suggest that Nedd4 knockdown‐induced apoptosis in DU145 cells acted through both death receptor and mitochondria‐dependent pathways.
To investigate whether the lysosomal pathway was involved in Nedd4‐mediated apoptosis, lysosome membrane permeabilization (LMP) was measured by lysosomotropic agent acridine orange (AO) staining and by detection of cathepsin D in the cell cytosolic fraction. AO is a weak base that moves freely across membranes when uncharged, and accumulates in acid compartments such as lysosomes, in its protonated form, where it forms bright red fluorescent aggregates. LMP is associated with proton release, which renders lysosomes more alkaline and results in decreased red fluorescence. Fluorescence microscopy analysis showed that higher percentages of DU145 cells expressing control S‐shRNA, had intact acidic lysosomes (high levels of red fluorescence corresponding to accumulation of AO). In contrast, knockdown of Nedd4 led to reduced percentage of cells with red fluorescence (Fig. 5a).
Figure 5.
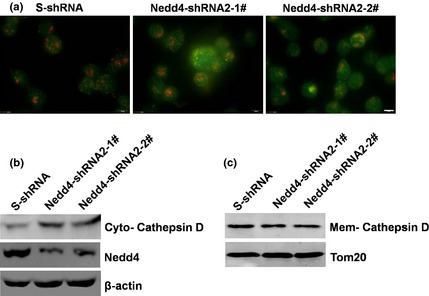
Knockdown of endogenous Nedd4 expression induced lysosome membrane permeabilization. (a) DU145 cells stably expressing Nedd4‐shRNA2‐1#, shRNA2‐2# and control S‐shRNA were treated with 1.5 μm staurosporine for 24 h. LMP was detected by AO staining with visualization by fluorescence microscopy. Scale bar, 10 μm. (b, c) DU145 cells stably expressing Nedd4shRNA2‐1#, shRNA2‐2# and control S‐shRNA were treated with 1.5 μm staurosporine for 24 h. Release of cathepsin D into the cytosol and membranes were detected by western blotting.
Lysosomal proteases, such as cathepsin D, are released from lysosomes into the cytosol under conditions of lysosome membrane permeabilization. Western blot analysis indicated that knockdown of Nedd4 led to significant release of cathepsin D into the cytosol (Fig. 5b, c). This further suggests that the lysosomal pathway was involved in down‐regulation of Nedd4‐induced apoptosis.
Expression of Nedd4 promoted autophagy in DU145 cells
To further investigate effects of down‐regulation of Nedd4 on autophagy in DU145 cells, immunofluorescence and western blotting were employed. Fluorescence microscopy analysis showed that formation of autophagic punctate structures with GFP‐LC3 proteins was reduced in Nedd4‐shRNA#2‐1‐ and shRNA#2‐2‐expressing cells after treatment with rapamycin (Fig. 6a), and a similar effect was also observed using endogenous LC3 by immunofluorescence in A549 cells (Fig 3b). Western blot data indicated that depletion of Nedd4 reduced conversion of LC3B‐I to LC3B‐II (Fig. 6c). Furthermore, overexpression of Nedd4 in HeLa cells promoted conversion of LC3B‐I to LC3B‐II after cell treatment with rapamycin (Fig. 6d). These results suggest that expression of Nedd4 promoted rapamycin‐induced autophagy.
Figure 6.
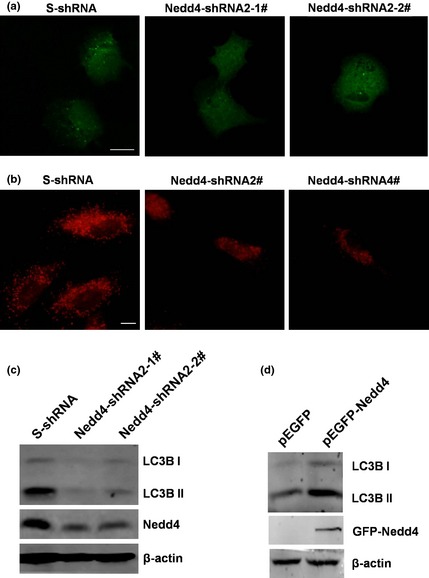
Expression of Nedd4 promoted autophagy. (a) DU145 cells stably expressing Nedd4‐shRNA2‐1#, shRNA2‐2# and control S‐shRNA were transfected with pEGFP‐LC3 plasmid, then treated with 3 μm rapamycin for 3 h before detection of GFP‐LC3 expression with immunofluorescence microscopy. Scale bar, 10 μm. (b) A549 cells transiently infected with Nedd4‐shRNA2#, shRNA4# and control S‐shRNA, then treated with 3 μm rapamycin for 3 h before detection of endogenous LC3 by immunofluorescence microscopy. Scale bar, 10 μm. (c) DU145 cells stably expressing Nedd4‐shRNA2‐1#, shRNA2‐2# and control S‐shRNA were treated with 3 μm rapamycin for 3 h, and conversion of LC3B‐I into LC3B‐II was detected by western blotting. (d) HeLa cells expressing Nedd4 and the control were treated with 3 μm rapamycin for 3 h and conversion of LC3B‐I into LC3B‐II was detected by western blotting.
mTOR signalling was involved in Nedd4‐mediated autophagy
Previous studies have revealed that mTOR signalling is involved in the process of autophagy 17, 18. To study the mechanisms underlying Nedd4‐mediated promotion of autophagy induced by rapamycin, the mTOR signal pathway was examined using Nedd4‐depleted DU145 cells. Although overall mTOR protein levels were similar in Nedd4‐shRNA#2‐1‐, shRNA#2‐2‐ and S‐shRNA‐expressing cells, specific levels of phosphorylated mTOR (p‐mTOR) protein species were notably higher in Nedd4‐depleted cells compared to control (Fig. 7). This suggests that expression of Nedd4 in DU145 cells promoted autophagy by down‐regulation of mTOR signalling.
Figure 7.
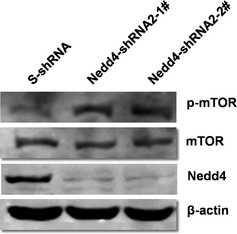
mTOR signalling was involved in Nedd4‐mediated autophagy. DU145 cells stably expressing Nedd4‐shRNA2‐1#, shRNA2‐2# and control S‐shRNA were treated with 3 μm rapamycin for 3 h, and levels of mTOR and p‐mTOR were detected by western blotting.
Discussion
Nedd4 is the product of neural precursor cell expressed developmentally down‐regulated gene 4 (Nedd4), originally implicated in regulation of fluid and electrolyte homoeostasis, by controlling surface abundance of epithelial cell sodium channel subunits. Recent studies have revealed its role in cancer development; however, its precise mechanisms of action are not well characterized. To address functional significance of Nedd4 in cancer cells, we designed shRNA lentiviral vectors targeting Nedd4, and tested the effect of Nedd4 down‐regulation on cell proliferation, colony formation and apoptosis, in DU145 cells. We found that expression of Nedd4 was inhibited by more than 65%, as assessed by western blot analysis, after infection with lentiviruses expressing Nedd4‐shRNA#2 (Fig. 2). Furthermore, knockdown of Nedd4 inhibited cell proliferation and colony formation and induced apoptosis in DU145 cells (Figs. 3 and 4).
Lysosomal extracts, as well as selected purified cathepsins, have previously been shown to cleave, then activate pro‐apoptotic Bcl‐2‐like protein, Bid. Cathepsin D triggers Bax activation and translocation to mitochondria, where it induces opening of pores on outer mitochondrial membranes, and then directs induction of mitochondrial damage as well as direct activation of procaspases by lysosomal enzymes 19, 20, 21. We have shown here that down‐regulation of Nedd4 in DU145 cells promoted lysosome membrane permeabilization and led to release of cathepsin D into the cytosol (Fig. 5). Considering that down‐regulation of Nedd4 induces apoptosis, we propose that a lysosomal pathway for apoptosis involved in the Nedd4 down‐regulation, induced inhibition of cell population growth.
Autophagy plays a complex role in tumourigenesis and drug treatment responsiveness. It may act in tumour‐suppression during early stages of tumourigenesis, however, in established tumours, the cancer cells may be promoting autophagy to survive under metabolic and therapeutic stresses associated with malignant transformation 22, 23, 24. In this study, we found that formation of autophagic punctate structures with GFP‐LC3 proteins was reduced and conversion of LC3B‐I to LC3B‐II was lowered in Nedd4‐depleted cells (Fig. 6a, b). This indicates that knockdown of endogenous Nedd4 expression inhibited autophagy in DU145 cells. These results suggest that autophagy may be tumour‐promoting in established prostate cancer cells, and that expression of Nedd4 in DU145 and A549 cells promotes autophagy and cell number expansion.
mTOR‐mediated signalling is recognized to be the most important pathway in autophagy, a process required for recycling damaged organelles and cell adaptation to nutrient starvation 25. Constitutive activation of PI3K‐mTOR signalling observed in cancer cells strongly inhibits autophagy. In mammals, mTORC1 (mammalian target of rapamycin complex 1) directly phosphorylates and suppresses ULK1/Atg13/FIP200 (Unc‐51‐like kinase 1/mammalian autophagy‐related gene 13/focal adhesion kinase family‐interacting protein of 200 kDa), a kinase complex required to initiate autophagy 26, 27, 28. Consistently, we found that knockdown of endogenous Nedd4 expression significantly increased levels of activated mTOR (p‐mTOR) (Fig. 7), which suggests that mTOR signalling is involved in Nedd4‐mediated autophagy.
Other studies have shown that involvement of mTOR in autophagy occurs in a rapamycin independent manner 29, 30, 31, and it would be particularly interesting from a mechanistic viewpoint, to determine rapamycin‐sensitivity of Nedd4 promotion of autophagy in prostate cancer cells and other cell types.
Here, to explore mechanisms that underlie Nedd4‐mediated promotion of autophagy, the mTOR signal pathway was examined in Nedd4‐depleted DU145 cells. Data show that down‐regulation of Nedd4 inhibited rapamycin‐induced autophagy partly by up‐regulation of p‐mTOR (Fig. 7). But reasons why down‐regulation of Nedd4 lead to regulation of mTOR requires more detailed studies, specifically to identify substrates of Nedd4 E3 ubiquitin ligase, involved in mTOR activation.
Consistent with our results, a recent study has reported that Nedd4 functions as an E3 ligase of HER3 and regulates steady‐state HER3 levels. Knockdown of Nedd4 in MCF‐7 and DU145 cells resulted in elevated HER3 levels and enhanced NRG‐1‐dependent HER3 downstream AKT and ERK signalling 32. However, other reports indicate that accumulation of Nedd4 inhibited PTEN leads to activation of the downstream oncogenic mTOR/Akt pathway 33. Potential reasons for these apparent discrepancies are likely to be that Nedd4 has numerous physiological targets 7, 32, 33, 34, 35, 36, 37, some of which may have different roles in the mTOR/Akt pathway under given microenvironments.
Taken together, we have demonstrated in this study that expression of Nedd4 promoted cell proliferation and colony formation and inhibited apoptosis in DU145 cells. Moreover, Nedd4 promoted autophagy and associated with the mTOR signalling pathway. These results will help researchers understand modes of action of Nedd4 in tumourigenesis and to provide important information concerning feasibility of developing reagents for therapeutic down‐regulation of this protein in human cancers.
Conflict of interest
None declared.
Acknowledgements
We are grateful to Dr. Edward McKenzie (University of Manchester, UK) for critical reading of the manuscript. This research is supported by the National Natural Science Foundation of China (81402952), program for Changjiang University Scholars and Innovative Research Team (IRT1166) and a Tianjin Natural Science Foundation grant (10JCZDJC16800).
References
- 1. Pickart CM, Fushman D (2004) Polyubiquitin chains: polymeric protein signals. Curr. Opin. Chem. Biol. 8, 610–616. [DOI] [PubMed] [Google Scholar]
- 2. Schwartz AL, Ciechanover A (1999) The ubiquitin‐proteasome pathway and pathogenesis of human diseases. Annu. Rev. Med. 50, 57–74. [DOI] [PubMed] [Google Scholar]
- 3. Guardavaccaro D, Pagano M (2004) Oncogenic aberrations of cullin‐dependent ubiquitin ligases. Oncogene 23, 2037–2049. [DOI] [PubMed] [Google Scholar]
- 4. Chen C, Matesic LE (2007) The Nedd4‐like family of E3 ubiquitin ligases and cancer. Cancer Metastasis Rev. 26, 587–604. [DOI] [PubMed] [Google Scholar]
- 5. Eide PW, Cekaite L, Danielsen SA, Eilertsen IA, Kjenseth A, Fykerud TA et al (2013) NEDD4 is overexpressed in colorectal cancer and promotes colonic cell growth independently of the PI3K/PTEN/AKT pathway. Cell. Signal. 25, 12–18. [DOI] [PubMed] [Google Scholar]
- 6. Cao XR, Lill NL, Boase N, Shi PP, Croucher DR, Shan H et al (2008) Nedd4 controls animal growth by regulating IGF‐1 signaling. Sci. Signal. 1, ra5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wang X, Trotman LC, Koppie T, Alimonti A, Chen Z, Gao Z et al (2007) NEDD4‐1 is a proto‐oncogenic ubiquitin ligase for PTEN. Cell 128, 129–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Trotman LC, Wang X, Alimonti A, Chen Z, Teruya‐Feldstein J, Yang H et al (2007) Ubiquitination regulates PTEN nuclear import and tumor suppression. Cell 128, 141–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fouladkou F, Landry T, Kawabe H, Neeb A, Lu C, Brose N et al (2008) The ubiquitin ligase Nedd4‐1 is dispensable for the regulation of PTEN stability and localization. Proc. Natl Acad. Sci. USA 105, 8585–8590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fehrenbacher N, Jaattela M (2005) Lysosomes as targets for cancer therapy. Cancer Res. 65, 2993–2995. [DOI] [PubMed] [Google Scholar]
- 11. Guicciardi ME, Leist M, Gores GJ (2004) Lysosomes in cell death. Oncogene 23, 2881–2890. [DOI] [PubMed] [Google Scholar]
- 12. Liu N, Raja SM, Zazzeroni F, Metkar SS, Shah R, Zhang M et al (2003) NF‐kappaB protects from the lysosomal pathway of cell death. EMBO J. 22, 5313–5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Boya P, Gonzalez‐Polo RA, Poncet D, Andreau K, Vieira HL, Roumier T et al (2003) Mitochondrial membrane permeabilization is a critical step of lysosome‐initiated apoptosis induced by hydroxychloroquine. Oncogene 22, 3927–3936. [DOI] [PubMed] [Google Scholar]
- 14. Boya P, Andreau K, Poncet D, Zamzami N, Perfettini JL, Metivier D et al (2003) Lysosomal membrane permeabilization induces cell death in a mitochondrion‐dependent fashion. J. Exp. Med. 197, 1323–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bidere N, Lorenzo HK, Carmona S, Laforge M, Harper F, Dumont C et al (2003) Cathepsin D triggers Bax activation, resulting in selective apoptosis‐inducing factor (AIF) relocation in T lymphocytes entering the early commitment phase to apoptosis. J. Biol. Chem. 278, 31 401–31 411. [DOI] [PubMed] [Google Scholar]
- 16. Bernassola F, Karin M, Ciechanover A, Melino G (2008) The HECT family of E3 ubiquitin ligases: multiple players in cancer development. Cancer Cell 14, 10–21. [DOI] [PubMed] [Google Scholar]
- 17. Jung CH, Ro SH, Cao J, Otto NM, Kim DH (2010) mTOR regulation of autophagy. FEBS Lett. 584, 1287–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nazio F, Strappazzon F, Antonioli M, Bielli P, Cianfanelli V, Bordi M et al (2013) mTOR inhibits autophagy by controlling ULK1 ubiquitylation, self‐association and function through AMBRA1 and TRAF6. Nat. Cell Biol. 15, 406–416. [DOI] [PubMed] [Google Scholar]
- 19. Zhao M, Antunes F, Eaton JW, Brunk UT (2003) Lysosomal enzymes promote mitochondrial oxidant production, cytochrome c release and apoptosis. Eur. J. Biochem. 270, 3778–3786. [DOI] [PubMed] [Google Scholar]
- 20. Foghsgaard L, Wissing D, Mauch D, Lademann U, Bastholm L, Boes M et al (2001) Cathepsin B acts as a dominant execution protease in tumor cell apoptosis induced by tumor necrosis factor. J. Cell Biol. 153, 999–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Roberg K, Johansson U, Ollinger K (1999) Lysosomal release of cathepsin D precedes relocation of cytochrome c and loss of mitochondrial transmembrane potential during apoptosis induced by oxidative stress. Free Radic. Biol. Med. 27, 1228–1237. [DOI] [PubMed] [Google Scholar]
- 22. Onodera J, Ohsumi Y (2005) Autophagy is required for maintenance of amino acid levels and protein synthesis under nitrogen starvation. J. Biol. Chem. 280, 31 582–31 586. [DOI] [PubMed] [Google Scholar]
- 23. Rabinowitz JD, White E (2010) Autophagy and metabolism. Science 330, 1344–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. White E, DiPaola RS (2009) The double‐edged sword of autophagy modulation in cancer. Clin. Cancer Res. 15, 5308–5316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Laplante M, Sabatini DM (2012) mTOR signaling in growth control and disease. Cell 149, 274–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ganley IG, du Lam H, Wang J, Ding X, Chen S, Jiang X (2009) ULK1.ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy. J. Biol. Chem. 284, 12 297–12 305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hosokawa N, Hara T, Kaizuka T, Kishi C, Takamura A, Miura Y et al (2009) Nutrient‐dependent mTORC1 association with the ULK1‐Atg13‐FIP200 complex required for autophagy. Mol. Biol. Cell 20, 1981–1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jung CH, Jun CB, Ro SH, Kim YM, Otto NM, Cao J et al (2009) ULK‐Atg13‐FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol. Biol. Cell 20, 1992–2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Thoreen CC, Kang SA, Chang JW, Liu Q, Zhang J, Gao Y et al (2009) An ATP‐competitive mammalian target of rapamycin inhibitor reveals rapamycin‐resistant functions of mTORC1. J. Biol. Chem. 284, 8023–8032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Laplante M, Sabatini DM (2013) Regulation of mTORC1 and its impact on gene expression at a glance. J. Cell Sci. 126, 1713–1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Laplante M, Sabatini DM (2009) mTOR signaling at a glance. J. Cell Sci. 122, 3589–3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Huang Z, Choi BK, Mujoo K, Fan X, Fa M, Mukherjee S et al (2014) The E3 ubiquitin ligase NEDD4 negatively regulates HER3/ErbB3 level and signaling. Oncogene 9, 1105–1115. [DOI] [PubMed] [Google Scholar]
- 33. Liu J, Wan L, Liu P, Inuzuka H, Wang Z, Wei W (2014) SCF(beta‐TRCP)‐mediated degradation of NEDD4 inhibits tumorigenesis through modulating the PTEN/Akt signaling pathway. Oncotarget 5, 1026–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Persaud A, Alberts P, Hayes M, Guettler S, Clarke I, Sicheri F et al (2011) Nedd4‐1 binds and ubiquitylates activated FGFR1 to control its endocytosis and function. EMBO J. 30, 3259–3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pham N, Rotin D (2001) Nedd4 regulates ubiquitination and stability of the guanine‐nucleotide exchange factor CNrasGEF. J. Biol. Chem. 276, 46 995–47 003. [DOI] [PubMed] [Google Scholar]
- 36. Blot V, Perugi F, Gay B, Prevost MC, Briant L, Tangy F et al (2004) Nedd4.1‐mediated ubiquitination and subsequent recruitment of Tsg101 ensure HTLV‐1 Gag trafficking towards the multivesicular body pathway prior to virus budding. J. Cell Sci. 117, 2357–2367. [DOI] [PubMed] [Google Scholar]
- 37. Zeng F, Xu J, Harris RC (2009) Nedd4 mediates ErbB4 JM‐a/CYT‐1 ICD ubiquitination and degradation in MDCK II cells. FASEB J. 23, 1935–1945. [DOI] [PMC free article] [PubMed] [Google Scholar]


