Abstract
Objectives
Amelogenins are proposed to be responsible for enamel matrix derivative (EMD)‐induced periodontal regeneration; however, heterogeneity of amelogenins makes it challenging to purify the full‐length proteins. This study has been carried out to express and purify a recombinant full‐length human amelogenin protein (rHhAm175) in the eukaryotic yeast Pichia pastoris, and further compare biological responses of human periodontal ligament fibroblasts (PDLFs) to rHhAm175 and porcine EMD (pEMD).
Materials and methods
Human cDNA encoding a 175‐amino acid amelogenin was subcloned into the pPIC3.5K vector. The rHhAm175 expressed in P. pastoris GS115 (Mut+) was purified and characterized. We examined cell attachment, migration and proliferation responses of human PDLFs to rHhAm175 and pEMD respectively, and characterized associated changes of proliferation‐related intracellular signalling molecules, including extracellular signal response kinase (ERK) and Akt kinases/protein kinase B (Akt/PKB) kinases.
Results
The purified rHhAm175 was confirmed to be molecular mass 22 021.13 Da, phosphorylated human amelogenin, and alone significantly promoted proliferation and migration of human PDLFs to an extent comparable to that of pEMD. Cell attachment was increased over the first 60 min incubation with rHhAm175 or pEMD. Both rHhAm175 and pEMD induced PDLF mitogenesis via extracellular signal response kinase (ERK1/2), but not by Akt kinases/protein kinase B (Akt/PKB).
Conclusions
rHhAm175 modulated cell activities of human PDLFs, to a comparable extent as porcine EMD. These data suggest that rHhAm175 might be used to induce periodontal tissue regeneration.
Introduction
Periodontal diseases result in chronic and progressive destruction of periodontal tissues, and often lead to tooth loss. The ultimate goal of periodontal treatment is regeneration of all of the three periodontal tissues. As originally described by Hammarström et al., enamel matrix derivative (EMD) [comprised of acidic extracts of enamel matrix proteins (EMPs)] from porcine enamel, has been successfully employed to restore functional periodontal ligament, cementum and alveolar bone tissue both in vivo and in vitro 1, 2. Amelogenins, which compose 90% of developing EMPs, have been suggested to be responsible for EMD‐mediated periodontal regeneration 3. It has been known for several decades that amelogenins are a family of hydrophobic proteins that includes multiple alternatively spliced amelogenin isoforms and degradation products 4. This heterogeneity makes it extremely challenging to isolate and purify sufficiently large quantities of any one of the amelogenins present in developing tissues. Recombinant expression has been a valuable tool to overcome this problem 5, 6.
More recently, in vivo experimentation has shown that treatment with recombinant human amelogenin (rhAm) alone has led to substantial regeneration of peri‐odontal tissues 5. In a previous study, our team has used recombinant lentivirus to express the human full‐length amelogenin (hAm) gene in human bone marrow stromal cells (BMSCs) to explore its effects on cell differentiation, and our data revealed that it could regulate BMSC osteogenic/cementogenic differentiation 7. In addition, we observed formation of new cementum‐like tissue and adjacent fibrous tissue, along root slices in vivo; however, EMD‐treated samples had thicker, newly formed calcified tissue than did amelogenin‐transduced BMSCs (Jingchao Hu, Rong Shu, Zhongchen Song and Lan Cheng; unpublished data). Thus, localized concentration of amelogenin might be a key element in formation of new calcified tissue, and application of one of the amelogenins alone (for example, hAm), might help provide the appropriate growth stimulus in localized tissue. These findings suggested that amelogenin could serve as a bioactive analogue of EMD in periodontal regeneration.
We have previously reported that a recombinant porcine amelogenin (rpAm) in Escherichia coli had a selective regulation effect on various periodontal cells 8. However, the E. coli expression system largely lacked cell machinery to encourage correct folding and post‐translational modification of proteins, which is critical to their biological activities. Furthermore, the full‐length isoform of amelogenin and its single phosphate group (Ser‐16) have been reported to have profound association with newly formed enamel mineral and formation of calcium phosphates 9, 10. However, the mechanism of rHhAm175 in periodontal healing and regeneration was not explored. Here, we have devoted our investigation to find an appropriate system for expression of human amelogenin in its natural state, which has taken the past several years 11, 12. We report that 175‐amino acid human amelogenin (rHhAm175) was expressed in the eukaryotic yeast expression system, for the first time. We have characterized its phosphorylation state by linear ion trap‐Fourier transform ion cyclotron resonance mass spectrometry (LTQ‐FT MS), and further investigated its effects on cell signalling and further cell activities, in human PDLFs, in comparison to characterized native porcine EMD.
Materials and methods
Reagents
pPIC3.5K, P. pastoris strain GS115 (Mut+) (Invitrogen, Carlsbad, CA, USA); Ni‐NTA Superflow Cartridge (Qiagen, Dusseldorf, Germany); amelogenin C‐19 goat polyclonal antibody, amelogenin FL‐191 rabbit polyclonal antibody, his‐probe mouse monoclonal antibody (Santa Cruz Biotechnology, Santa Cruz, CA, USA); Dulbecco's modified Eagle's medium (DMEM), foetal bovine serum (FBS), antibiotics‐antimycotics (Gibco‐BRL, Gaithersburg, MD, USA); cell proliferation ELISA BrdU (colorimetric) (Roche, Basel, Switzerland); phospho‐p44/42 MAPK (ERK1/2) (Thr202/Tyr204) rabbit monoclonal antibody, p44/42 MAPK (ERK1/2) rabbit monoclonal antibody, phospho‐Akt (Ser473) (D9E) XP® rabbit monoclonal antibody, Akt (pan) (C67E7) rabbit monoclonal antibody, PD98059 (Cell Signaling Technology, Danvers, MA, USA) were used in this study. EMPs were extracted from porcine tooth buds according to the methods of Shu et al. 13.
Expression and purification of recombinant full‐length human amelogenin in the yeast P. pastoris
Human amelogenin X cDNA (Gen‐Bank Accession No. M86932), encoding mature amelogenin protein (175‐amino acid) with an additional N‐terminal containing a hexa‐histidine tag and a PreScission protease cleavage site, was subcloned into the yeast expression vector pPIC3.5K. The recombinant vector was sequenced to confirm the correct cDNA insertion and was named pPIC3.5K‐hAm. Linearized recombinant expression plasmid was transformed into the competent P. pastoris strain GS115 (Mut+) by electroporation (Bio‐Rad, Hercules, CA, USA). Stable subclones were subsequently screened.
His‐tagged amelogenin was induced by methanol, then purified using an Ni‐NTA Superflow Cartridge. Briefly, harvested cells were resuspended in 1:10 volume PBS with 25 U/ml lyticase, and lysed by ultra‐sonication. Precipitates were resuspended in 1:5 volume PBS with 1% Triton X‐100, and lysed twice using a high pressure homogenizer operated at 150 Mpa. Supernatants were then incubated using a pre‐equilibrated Ni‐NTA column. The column was washed in 20 column volumes (CV) TBS, 30 CV buffer 1 (500 mm NaCl; 25 mm Tris–HCl; 50 mm imidazole, pH 7.2), 20 CV buffer 2 (500 mm NaCl; 25 mm Tris–HCl; 100 mm imidazole, pH 7.2), and eluted using TBS containing 250 mm imidazole. The purified protein was analysed by sodium dodecyl sulphate‐polyacrylamide gel electrophoresis (SDS–PAGE) followed by Coomassie brilliant blue (CBB) staining, western blotting and mass spectrometry, as described below. The purified recombinant protein was named rHhAm175.
SDS–PAGE and western blot analysis
Fractions of proteins were denatured in SDS sample buffer and subjected to 15% SDS–PAGE and resultant gels were stained with CBB. Duplicate SDS–PAGE gels were transblotted on to PVDF membranes which were then blocked with 5% skimmed milk, and incubated successively with primary antibodies against human amelogenin or hexa‐histidine, then the fluorescently labelled secondary antibodies. Immunoreactive products were detected using the Odyssey Infrared Imaging System (LI‐COR Biotechnology, Lincoln, NE, USA).
Matrix‐assisted laser desorption/ionization‐time of flight mass spectrometry assay
After expression and purification, rHhAm175 samples were subjected to the Beijing Proteome Research Center for mass estimation. They were desalted using Amicon® Ultra‐0.5 centrifugal filters (3 kDa) (Millipore, Billerica, MA, USA) and eluted with 1% TFA. Mass spectra were conducted by matrix‐assisted laser desorption/ionization‐time of flight mass spectrometry (MALDI‐TOF MS) (ABI4800, Palo Alto, CA, USA).
Linear ion trap‐Fourier transform ion cyclotron resonance mass spectrometry assay
rHhAm175 samples were cleaved in solution using trypsin. Phosphopeptides enriched by TiO2 microcolumns were subjected to Linear ion trap‐Fourier transform ion cyclotron resonance mass spectrometry (LTQ‐FT MS) analysis (Thermo, Palo Alto, CA, USA) and peak lists were searched using the Mascot MS/MS Ion Search (Matrix Science, London, UK) against the IPI human database (V3.56).
Culture of human periodontal ligament fibroblasts
Human PDLFs were prepared from explants of periodontal ligaments of teeth extracted for orthodontic treatment (from healthy humans after obtaining informed consent), and under the protocol approved by our Institutional Review Board. Explants were cut into pieces and cultured in DMEM with 20% foetal bovine serum 14. When cells were approaching confluence, passages were performed; forth‐passage PDLFs were used for the following experiments.
Cell attachment analysis
The method modified from Jiang was performed to test for cell adhesion 15. 24‐well plates were first coated with rHhAm175 (10 μg/ml) or pEMD (200 μg/ml) overnight, and 0.02% BSA was used to block non‐specific binding sites. Before seeding, PDLFs were cultured in serum‐free medium for 24 h. Based on our previous investigations, this procedure was performed to keep cells quiescent and to reduce presence of pre‐existing serum factors 7. 2 × 104 cells per well were then seeded on coated 24‐well plates for 30, 60, 120, or 240 min. Attached cells were harvested and counted using a Z2™ Coulter Counter (Beckman, Fullerton, CA, USA).
Cell migration assay
Cell migration was analysed using an in vitro wound healing model, as previously described 16. Human PDLFs (6 × 104 cells/well) were seeded in 12‐well plates and cultured under serum‐free conditions for 24 h. Confluent monolayers of cells were ‘wounded’ using a sterile 10 μl pipette tip, which produced a lane (<200 μm) devoid of cells. Wells were then exposed to medium containing rHhAm175 (10 μg/ml) or pEMD (200 μg/ml). During the course of wound healing, cell migration was photographed every 12 h for a total period of 48 h using an inverted phasecontrast microscope, equipped with a digital camera (Leica, Heerbrugg, Switzerland). Using the proprietary image analysis software, percentages of wound closure were qualitatively analysed.
Cell proliferation assay
Proliferation of human PDLFs was investigated using the 3‐(4, 5‐dimethylthiazol‐ 2‐yl)‐2, 5‐diphenyl tetrazolium bromide (MTT) assay 17. Cells were seeded at 5 × 104/ml in 96‐well plates and cultured in serum‐free medium. After 24 h, rHhAm175 (10 μg/ml) or pEMD (200 μg/ml) was added in triplicate assays. Wells lacking protein served as controls. Observation of cell proliferation lasted 8 days. At the end of each incubation period, 20 μl of MTT (0.5 mg/ml) was added and after being incubated for 4 h, absorbance was measured at 490 nm (Bio‐Tek, Winooski, VT, USA).
DNA synthesis
DNA synthesis was measured by overnight BrdU incorporation, in accordance with the manufacturer's guidelines. In brief, PDLFs were plated in 96‐well plates at 5 × 103 cells/well. After serum starvation for 24 h, media were replaced with various compounds (10 μg/ml rHhAm175, 200 μg/ml pEMD, PD98059 20 μm). Each experiment was performed in triplicate for each treatment group (DMEM, DMEM+PD98059, pEMD, pEMD+PD98059, rHhAm175 and rHhAm175 + PD98059). Cells were incubated for 24 h with BrdU and then BrdU incorporation was measured using BrdU ELISA.
Western blot analysis
Human PDLFs were stimulated with pEMD, rHhAm175 or PD98059, then solubilized in cell and tissue protein extraction reagent containing protease inhibitors, phosphatase inhibitors and phenylmethanesulphonyl fluoride. Protein content of lysates was determined using BCA reagent. Protein samples (~40 μg) were size‐fractionated by 10% SDS–PAGE and were then transferred to polyvinylidene fluoride membranes (0.22 μm). Target proteins were detected by probing with the following specific antibodies against p44/42 MAPK (ERK1/2), phospho‐p44/42 MAPK (ERK1/2) (Thr202/Tyr204), Akt or phospho‐Akt (Ser473).
Statistical analyses
Each whole experiment was repeated at least twice. To quantify data obtained, mean values and their standard errors were calculated. Multiple comparisons within and between groups were performed using analysis of variance (ANOVA) method and post‐hoc testing (Bonferroni's correction). Results were considered statistically significant at P < 0.01.
Results
Expression and purification of recombinant human full‐length amelogenin protein
hAm cDNA and the recombinant vector were confirmed by DNA sequencing. Transformed cells were determined by PCR from genomic DNA of monoclone (Fig. 1a). A 175‐amino acid recombinant human amelogenin was expressed in the eukaryotic yeast P. pastoris and was purified by metal affinity chromatography. Based on electrophoretic mobility of rHhAm175 relative to pre‐stained protein standards, SDS–PAGE analysis revealed a single 27 kDa band for the recombinant protein (Fig. 1b). All antibodies against human amelogenin and hexa‐histidine reacted strongly with rHhAm175, also resulting in detection of an approximately 27 kDa band (Fig. 1c–e). Presence of the C‐terminal portion of amelogenin was confirmed by antibodies against the 19 C‐terminal amino acids of human amelogenin (Fig. 1d). Occasionally, antibodies specific to hAm (FL191) also reacted with aggregation products in higher molecular weight regions of the gels (Fig. 1e).
Figure 1.
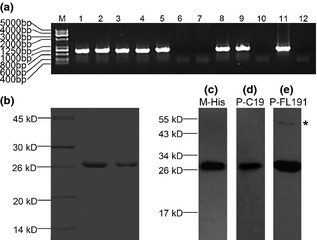
Expression and purification rHhAm175 in P . pastoris GS 115 (Mut+) cells. (a) PCR analysis of Pichia integrants. Amplification of rHhAm175 cDNA from isolated genomic DNA of 10 transformants (lane 1–10 represent 1–10# clone) and negative control clone of GS115 (Mut+) (lane 12). The recombinant vector was positive control (lane 11). (b) SDS–PAGE analysis of purified rHhAm175. Western blot analysis of rHhAm175 with (c) monoclonal antibody against hexa‐histidine (M‐His6), (d) a polyclonal antibody against 19 C‐terminal amino acids of human amelogenin (P‐C19) and (e) polyclonal antibody against human full‐length amelogenin (P‐FL191). Aggregates in the higher molecular weight region are marked by an asterisk (*).
Characterization of recombinant full‐length human amelogenin by mass spectrometry assay
SDS–PAGE and western blots validated successful expression of rHhAm175. However, exact mass and any post‐translational or chemical modifications of rHhAm175 expressed in P. pastoris yeast needed to be further identified. Thus, we performed MADLI‐TOF MS and LTQ‐FT MS assay to characterize its molecular mass and phosphorylation state. Molecular mass of purified recombinant full‐length human amelogenin with his‐tag arm, and protease cleavage site was confirmed as 22 021.13 Da, by MADLI‐TOF MS (Fig. 2a–c), which was in the region of 80 Da higher than the calculated mass (21 944.37 Da). The observed mass shift suggested that post‐translational modification might occur in this rHhAm175 expressed in P. pastoris yeast. Phosphorylated proteolytic peptides were analysed using LTQ‐FT MS and MS/MS fragmentation data showed that this recombinant amelogenin expressed in P. pastoris yeast was phosphorylated on Ser 16, as illustrated for the peptide pSYEVLTPLK in Fig. 2d; this supported previous reports that amelogenin contained a single phosphorylation on serine 16.
Figure 2.
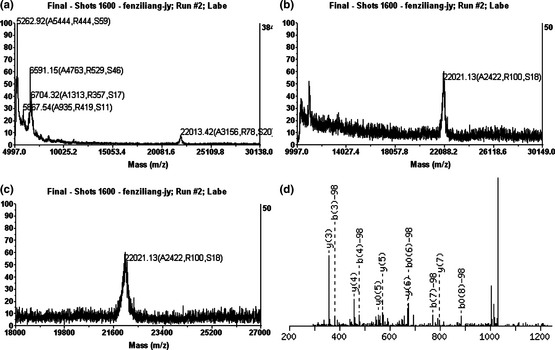
Characterization of full‐length rHhAm175 by mass spectrometry assay. (a–c) MALDI‐TOF MS analysis of recombinant amelogenin detected a protein with molecular mass 22 021.13 Da. (d) MS/MS Fragmentation of phosphopeptide pSYEVLTPLK. Matches were found in IPI00013959, Tax_Id = 9606 Gene_Symbol = AMELX Isoform 1 of Amelogenin, X isoforms. S1: Phospho (ST), with neutral losses 97.9769.
Cell attachment and migration
This experiment has shown that the attachment level of human PDLFs was approximately 80%, 2 h after seeding (Fig. 3a). We also found that both 10 μg/ml rHhAm175 and 200 μg/ml pEMD enhanced attachment of PDLFs over the first 60 min treatment, but that results on coated surfaces did not differ significantly from controls at later time points.
Figure 3.
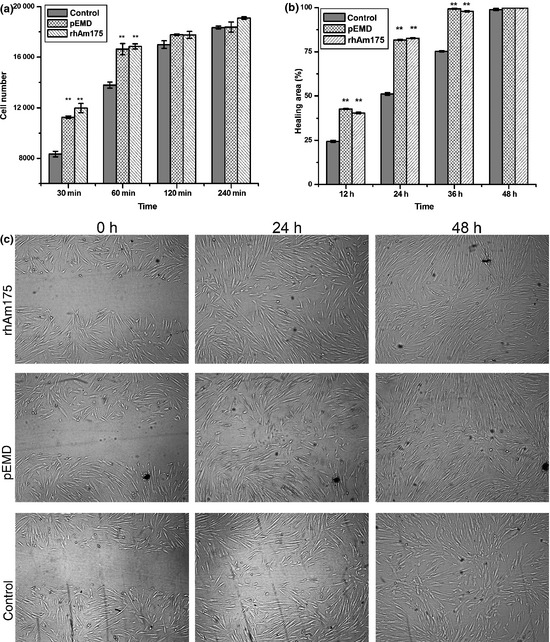
Effect of 10 μg/ml rHhAm175 on cell attachment and migration of human PDLF s in comparison to 200 μg/ml p EMD . **P < 0.01. Bars represent standard deviation (SD). (a) Effect of rHhAm175 on attachment of human PDLFs was similar to that of pEMD, at all time points (30, 60, 120 and 240 min). (b) Treatment with rHhAm175 or pEMD promoted migration of PDLFs. (c) Migration of PDLFs in the presence and absence of rHhAm175 or pEMD. Cells were visualized by microscopy, and data were digitally recorded (×200).
Next, effects of rHhAm175 on cell migration were assessed and compared to those of pEMD. Percentage of wound closure was 40% in the presence of rHhAm175 or pEMD 12 h after scratching, and reached over 80% by 24 h later (Fig. 3b,c). For negative control, closure was less than 25% at the end of 12 h and took a further 24 h to reach approximately 75%. These results showed that, similar to pEMD, rHhAm175 could increase level of migration of human PDLFs (P < 0.01).
Cell proliferation
Incubation of human PDLFs with rHhAm175 or pEMD visibly promoted replication of the cells in a dose‐dependent manner (data not shown). Figure 4a shows that 10 μg/ml rHhAm175 or 200 μg/ml pEMD caused higher levels of cell proliferation compared to the control group at day 3 (P < 0.01). This increase was persistent up to the 8th day and reached a plateau around day 6.
Figure 4.
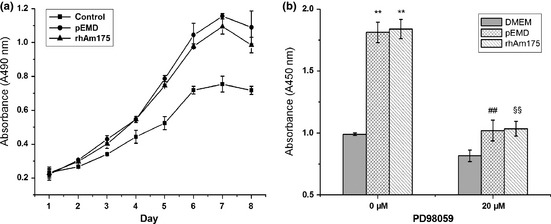
Mitogenic responses of human PDLF s to 10 μg/ml rHhAm175 or 200 μg/ml p EMD . (a) rHhAm175‐treated group had higher levels of proliferation compared to controls from day 3 (P < 0.01), and this upward trend was maintained until day 8. There were no significant differences between rHhAm175‐ and pEMD‐treated cells at each time point. (b) Increased DNA synthesis of human PDLFs in response to pEMD or rHhAm175 and their inhibition by 20 μm PD98059. **P < 0.01, pEMD versus DMEM; rHhAm175 versus DMEM. ##P < 0.01, pEMD versus pEMD+PD98059. §§P < 0.01, rHhAm175 versus rHhAm175 + PD98059. Bars represent standard deviation (SD).
We then analysed association between mitogenesis of PDLFs and stimulation of rHhAm175, as assessed by BrdU incorporation. As shown in Fig. 4b, rHhAm175 or pEMD significantly enhanced DNA synthesis of the cells, but 20 μm PD98059 (a specific inhibitor of ERK kinase) abrogated mitogenic responses induced by either rHhAm175 or pEMD.
Activation of intracellular signalling molecules
We examined activation of ERK1/2 and Akt/PKB in PDLFs stimulated by 10 μg/ml rHhAm175 or 200 μg/ml pEMD. As shown in Fig. 5a, both proteins stimulated dual‐phosphorylation of ERK1/2. Maximal effects were observed by 15 min, and effects gradually weakened over the subsequent 45 min. Activation of ERK1/2 by rHhAm175 or pEMD could be abrogated by 20 μm PD98059 (Fig. 5b). In contrast, phosphorylated Akt/PKB was barely detectable and showed no visible increase following exposure to the stimulus (rHhAm175 or pEMD) (Fig. 5c).
Figure 5.
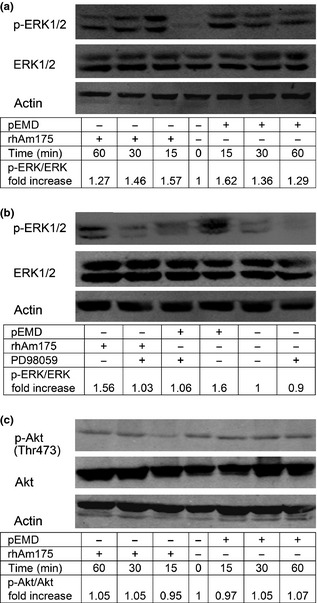
Western blot analysis of total and phosphorylated ERK 1/2 and Akt/ PKB in human PDLF s after stimulation with rHhAm175 (10 μg/ml) or p EMD (200 μg/ml). Ratio between treatment group and negative control, obtained by densitometric analysis, is displayed under each lane. Experiments were performed in duplicate or triplicate and representative results are shown. (a) Quiescent cells were stimulated with either pEMD or rHhAm175 for 0–60 min. Lower bands showed abundance of total ERK as the loading control. (b) Quiescent cells (60‐min pre‐treatment of 20‐μm PD98059) were analysed 15 min after addition of rHhAm175 or pEMD to cells. Induced ERK phosphorylation could be abrogated by PD98059 and basal p‐ERK levels were not altered by the inhibitor. (c) Levels of total and phosphorylated Akt/PKB in human PDLFs at 0, 15, 30 or 60 min after stimulation with rHhAm175 or pEMD.
Discussion
EMD, composed of natural extracts of EMPs, has been reported to be efficacious in periodontal regeneration therapy. Recently, amelogenin, the major protein present in EMPs and EMD, has been proposed to be responsible for its therapeutic effects during wound healing and regeneration 18. Haze et al. found that rhAm alone caused significant regeneration of all three tooth‐supporting tissues: alveolar bone, periodontal ligament and cementum 5. Our data show that rpAm might play an important role in periodontal regeneration by activation of PDLFs and inhibition of the epithelial cell component 8.
Amelogenin isoforms are derived from a single gene and amount to 90% of developing EMPs. During enamel development, original full‐length amelogenin is sequentially and discretely degraded by specific proteases, which gives rise to a range of heterogeneous proteins and peptides 19. Post‐translational modifications and post‐secretory processing, as well as time and tissue‐specific expression of EMPs, limit tissue sources of EMD or amelogenin, and add to the difficulties found in their isolation and purification. We have reported on hAm gene transduction by recombinant lentivirus mediated BMSC osteogenic/cementogenic differentiation 7, however, it was unable to induce cementum‐like calcified tissue to the same degree as EMD (unpublished data). We have conjectured that yield of amelogenin in gene‐transduced BMSCs might limit any inductive effect and additionally, presence of other cytokines in EMD has been documented 20, 21. Thus, achieving relatively high amounts of pure amelogenin may help overcome these difficulties, ensuring presence of the appropriate proteins to provide growth stimulus in localized tissue.
Expression of rhAm has been reported in different expression systems 22, 23, whereas, there has been no previous report so far, on phosphorylation state of these recombinant proteins. Taylor et al. expressed rhAm in the eukaryotic baculovirus expression system; however, no unambiguous indication of phosphorylation was detected 23. Here, we have expressed rHhAm175 in P. pastoris yeast and characterized phosphorylation modification of purified rHhAm175 for the first time. The yeast expression system has several notable benefits, including high levels of expression, secretory expression of foreign proteins and presence of cell mechanics to encourage correct folding, and post‐translational modification of the protein; this is more conducive to production of biologically active proteins. One potential disadvantage of the yeast system is presence of active proteases; thus, proteolysis could be a problem for secreted rhAm, already susceptible to proteolysis. Due to potential problems of rhAm aggregation and proteolysis, we preferred to use a non‐secreting, yeast system.
In this study, rHhAm175 was representative of unprocessed, full‐length amelogenin and the principal amelogenin complex existing in human secretory stage enamel 24. The histidine tag at the N‐terminus enabled effective, one‐step purification of rHhAm175 on Ni‐NTA resin (Fig. 1b). Purified recombinant amelogenin was further confirmed to be full‐length human amelogenin, molecular mass 22 021.13 Da, phosphorylated at Ser‐16. Our data show that the apparent mass (~27 kDa) of rHhAm175 observed by SDS–PAGE analysis was higher than the actual mass (~22 kDa) as measured by MS. This anomalous migration is a unique characteristic of amelogenin and has been well documented 25, 26. Occasionally, we could detect aggregation products, which most likely resulted from amelogenin self‐assembly (Fig. 1e) 27, 28.
Next, we examined biological functions of rHhAm175 in human PDLFs in comparison to the well‐studied pEMD. We have previously demonstrated that 200 μg/ml pEMD is a biologically active concentration (13, 15 and 17), consistent with previous studies, in which 100–200 μg/ml EMD significantly affected behaviour of periodontal cells. In addition, according to results of one of our preliminary studies (data not shown), optimum concentration of pEMD used in the following experiments was 200 μg/ml. As porcine full‐length amelogenin is composed of approximately 7.4% EMD 29, we chose to investigate effects of 10 μg/ml rHhAm175 (human homologue of the porcine molecule) on attachment, proliferation and migration of human PDLFs. Sequential events of periodontium‐related cells in response to 10 μg/ml rpAm were probably dependent upon the cell type 8. Here, we found that rHhAm175 significantly promoted cell proliferation and migration of human PDLFs in a manner comparable to that of pEMD. Rate of human PDLF adhesion was increased during the first 60 min of treatment.
Generally, cell adhesion and migration play a pivotal role in tissue formation and regeneration 30. These activities of human PDLFs are also critical in initiation of periodontal wound healing. As reported previously, human PDLFs attached and spread much better and faster than gingival fibroblasts 31, and they had the ability of migrating directionally 32. These unique characteristics of human PDLFs contribute to periodontal regeneration. Our data show that cell attachment was accelerated only during the first 60 min of treatment and wound healing in vitro was accelerated in the presence of rHhAm175 or pEMD. These observations are in accordance with our previous findings that cell adhesion and migration of human PDLFs have differences only at earlier times after stimulation 8, and are further supported the unique cell responses of PDLFs to the stimuli. Thus, effects of rHhAm175 on cell adhesion and migration rate of PDLFs might contribute to their preferential occupation of root surfaces by PDLFs in initiation of tissue repair, a crucial step in periodontal regeneration.
Cell proliferative effects may be important in early stages of healing, to enhance cell reserves in a wound space, and are a prerequisite for regeneration at later stages. Our results show that rHhAm175 had the same stimulative effect on proliferation as pEMD in a dose‐dependent manner (data not shown). These data provide verification of previous reports regarding effects of EMD or amelogenin, on PDLF proliferation 33, 34. Various studies have indicated that EMD might act as a matrix for cells at a periodontal regenerative site. Similarly, our findings revealed that rHhAm175 created a positive environment for cell proliferation and migration, as required for regeneration of the periodontium.
To further investigate and compare mechanisms of rHhAm175 and pEMD in PDLF responses, we examined effects of these treatments on relevant intracellular signalling molecules, including ERK1/2 and Akt/PKB kinases. We found that activation of ERK1/2 was a crucial signalling event for the mitogenic effect of rHhAm175, as its inhibition could abolish proliferation of human PDLFs. As Fig. 5 indicates, the wave of ERK1/2 dual‐phosphorylation stimulated by rHhAm175 or pEMD peaked at 15 min, and this was abrogated by a specific MAPK inhibitor. This observation was in accord with findings that EMD induced PDLF mitogenesis via the ERK1/2 pathway, but not by activation of other MAPKs 35, 36. We further found that the Akt/PKB signalling pathway was not involved in cellular responses of human PDLFs stimulated by rHhAm175 or pEMD, such as proliferation and/or migration. These results reveal that rHhAm175 alone activated the same intracellular signalling pathway as pEMD.
Our previous studies have characterized biological activities of pEMD on periodontal regeneration in vitro and in vivo (13, 15 and 17). Here, we have verified that rHhAm175 has significant positive effects on responses of human PDLFs, to an extent comparable to pEMD. Although rHhAm175‐induced osteogenic/cementogenic differentiation and cementogenesis need to be further explored, its stimulative effect on proliferation and migration of PDLFs is helpful in the regenerating periodontal apparatus.
In conclusion, we expressed and purified a human recombinant full‐length amelogenin (rHhAm175) in P. pastoris yeast, for the first time. Its molecular mass and phosphorylation status were characterized by mass spectrometry assay. We found that rHhAm175 and pEMD had the same impact in vitro on cell activities and intracellular signalling molecules, in human PDLFs, suggesting that rHhAm175 is a potential candidate to be a bioactive substitute for EMD. Nevertheless, rHhAm175 needs to be further investigated in regulation of regeneration and mineralization processes in in vivo animal models, before clinical application for periodontal wound healing and tissue regeneration can take place.
Acknowledgements
This research was funded by the National Natural Science Foundation of China (Project Number: 81070838), Shanghai Jiao Tong University (YG2011MS31) and 9th People's Hospital, Shanghai Jiao Tong University School of Medicine. The authors declare no financial relationships between any author and a commercial firm that may pose a conflict of interest.
References
- 1. Hammerström L, Heijl L, Gestrelius S (1997) Periodontal regeneration in a buccal dehiscence model in monkeys after application of enamel matrix proteins. J. Clin. Periodontol. 24, 669–677. [DOI] [PubMed] [Google Scholar]
- 2. Gestrelius S, Anderson C, Lidström D, Hammerström L, Somerman M (1997) In vitro studies on periodontal ligament cells and enamel matrix derivative. J. Clin. Periodontol. 24, 685–692. [DOI] [PubMed] [Google Scholar]
- 3. Grandin HM, Gemperli AC, Dard M (2012) Enamel matrix derivative: a review of cellular effects in vitro and a model of molecular arrangement and functioning. Tissue Eng. Part B Rev. 18, 181–202. [DOI] [PubMed] [Google Scholar]
- 4. Brookes SJ, Robinson C, Kirkham J, Bonass WA (1995) Biochemistry and molecular biology of amelogenin proteins of developing dental enamel. Arch. Oral Biol. 40, 1–14. [DOI] [PubMed] [Google Scholar]
- 5. Haze A, Taylor AL, Haegewald S, Leiser Y, Shay B, Rosenfeld E, et al (2009) Regeneration of bone and periodontal ligament induced by rHAM after periodontitis. J. Cell Mol. Med. 13, 1110–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Huang YC, Tanimoto K, Tanne Y, Kamiya T, Kunimatsu R, Michida M, et al (2010) Effects of human full‐length amelogenin on the proliferation of human mesenchymal stem cells derived from bone marrow. Cell Tissue Res. 342, 205–212. [DOI] [PubMed] [Google Scholar]
- 7. Hu JC, Shu R, Song ZC, Cheng L (2011) Human amelogenin up‐regulates osteogenic gene expression in human bone marrow stroma cells. Biochem. Biophys. Res. Commun. 408, 437–441. [DOI] [PubMed] [Google Scholar]
- 8. Li X, Shu R, Liu D, Jiang S (2010) Different effects of 25‐kDa amelogenin on the proliferation, attachment and migration of various periodontal cells. Biochem. Biophys. Res. Commun. 394, 581–586. [DOI] [PubMed] [Google Scholar]
- 9. Uchida T, Tanabe T, Fukae M, Shimizu M, Yamada M, Miake K et al (1991) Immunochemical and immunohistochemical studies, using antisera against porcine 25 kDa amelogenin, 89 kDa enamelin and the 13‐17 kDa nonamelogenins, on immature enamel of the pig and rat. Histochemistry 96, 129–138. [DOI] [PubMed] [Google Scholar]
- 10. Kwak SY, Wiedemann‐Bidlack FB, Beniash E, Yamakoshi Y, Simmer JP, Litman A et al (2009) Role of 20‐kDa amelogenin P148 phosphorylation in calcium phosphate formation in vitro. J. Biol. Chem. 284, 18972–18979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cheng L, Shu R, Xue JL, Chen JZ, Tian L, Fang YX (2008) Expression and purification of recombinant human amelogenin in E. coli . J. Shanghai Jiaotong University (Med. Sci.) 28, 811–814. [Google Scholar]
- 12. Cheng L, Shu R, Zhang XL, Tian L (2011) Construction of recombinant human amelogenin eukaryon expression vector and its stable expression in NIH3T3 cells. J. Shanghai Jiaotong University (Med. Sci.) 31, 1008–1112. [Google Scholar]
- 13. Song AM, Shu R, Xie YF, Song ZC, Li HY, Liu XF, et al (2007) A study of enamel matrix proteins on differentiation of porcine bone marrow stromal cells into cementoblasts. Cell Prolif. 40, 381–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mailhot JM, Schuster GS, Garnick JJ, Hanes PJ, Lapp CA, Lewis JB (1995) Human periodontal ligament and gingival fibroblast response to TGF‐beta 1 stimulation. J. Clin. Periodontol. 22, 679–685. [DOI] [PubMed] [Google Scholar]
- 15. Jiang SY, Shu R, Xie YF (2011) The effects of enamel matrix proteins on proliferation, differentiation and attachment of human alveolar osteoblasts. Cell Prolif. 44, 372–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lemire JM, Merrilees MJ, Braun KR, Wight TN (2002) Overexpression of the V3 variant of versican alters arterial smooth muscle cell adhesion, migration, and proliferation in vitro . J. Cell. Physiol. 190, 38–45. [DOI] [PubMed] [Google Scholar]
- 17. Song ZC, Shu R, Zhang XL (2010) Cellular responses and expression profiling of human bone marrow stromal cells stimulated with enamel matrix proteins in vitro. Cell Prolif. 43, 84–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bosshardt DD (2008) Biological mediators and periodontal regeneration: a review of enamel matrix proteins at the cellular and molecular levels. J. Clin. Periodontol. 35(Suppl. 8), 87–105. [DOI] [PubMed] [Google Scholar]
- 19. Simmer JP, Hu JC (2002) Expression, structure, and function of enamel proteinases. Connect. Tissue Res. 43, 441–449. [DOI] [PubMed] [Google Scholar]
- 20. Saito K, Konishi I, Nishiguchi M, Hoshino T, Fujiwara T (2008) Amelogenin binds to both heparan sulfate and bone morphogenetic protein 2 and pharmacologically suppresses the effect of noggin. Bone 43, 371–376. [DOI] [PubMed] [Google Scholar]
- 21. Suzuki S, Nagano T, Yamakoshi Y, Gomi K, Arai T, Fukae M, Katagiri T, Oida S (2005) Enamel matrix derivative gel stimulates signal transduction of BMP and TGFbeta. J. Dent. Res. 84, 510–514. [DOI] [PubMed] [Google Scholar]
- 22. Deutsch D, Chityat E, Hekmati M, Palmon A, Farkash Y, Dafni L (1996) High expression of human amelogenin in E. coli . Adv. Dent. Res. 10, 187–194. [DOI] [PubMed] [Google Scholar]
- 23. Taylor AL, Haze‐Filderman A, Blumenfeld A, Shay B, Dafni L, Rosenfeld E, et al (2006) High yield of biologically active recombinant human amelogenin using the baculovirus expression system. Protein Expr. Purif. 45, 43–53. [DOI] [PubMed] [Google Scholar]
- 24. Fincham AG, Belcourt AB, Termine JD (1983) Molecular composition of the protein matrix of developing human dental enamel. J. Dent. Res. 62, 11–15. [DOI] [PubMed] [Google Scholar]
- 25. Chen WY, BelL AW, Simmer JP, Smith CE (2000) Mass spectrometry of native rat amelogenins: primary transcripts, secretory isoforms and C‐terminal degradation. J. Dent. Res. 79, 840–849. [DOI] [PubMed] [Google Scholar]
- 26. Sun Z, Ahsan MM, Wang HJ, Du C, Abbott C, Moradian‐Oldak J (2006) Assembly and processing of an engineered amelogenin proteolytic product (rP148). Eur. J. Oral Sci. 114(Suppl. 1), 59–63. [DOI] [PubMed] [Google Scholar]
- 27. Moradian‐Oldak J, Simmer JP, Lau EC, Diekwisch T, Slavkin HC, Fincham AG (1995) A review of the aggregation properties of a recombinant amelogenin. Connect. Tissue Res. 32, 125–130. [DOI] [PubMed] [Google Scholar]
- 28. Du C, Falini G, Fermani S, Abbott C, Moradian‐Oldak J (2005) Supramolecular assembly of amelogenin nanospheres into birefringent microribbons. Science 307, 1450–1454. Erratum in: Science, 309, 2166. [DOI] [PubMed] [Google Scholar]
- 29. Wen HB, Moradian‐Oldak J, Leung W, Bringas P Jr, Fincham AG (1999) Microstructures of an amelogenin gel matrix. J. Struct. Biol. 126, 42–51. [DOI] [PubMed] [Google Scholar]
- 30. Dubin‐Thaler BJ, Giannone G, Dobereiner HG, Sheetz MP (2004) Nanometer analysis of cell spreading on matrix‐coated surfaces reveals two distinct cell states and STEPs. Biophys. J. 86, 1794–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Palioto DB, Coletta RD, Graner E, Joly JC, de Lima AF (2004) The influence of enamel matrix derivative associated with insulin‐like growth factor‐1 on periodontal ligament fibroblasts. J. Periodontol. 75, 498–504. [DOI] [PubMed] [Google Scholar]
- 32. Kwon SM, Kim SA, Fujii S, Maeda H, Ahn SG, Yoon JH (2011) Transforming growth factor β1 promotes migration of human periodontal ligament cells through heat shock protein 27 phosphorylation. Biol. Pharm. Bull. 34, 486–489. [DOI] [PubMed] [Google Scholar]
- 33. Rodrigues TL, Marchesan JT, Coletta RD, Novaes AB Jr, Grisi MF, Souza SL, et al (2007) Effects of enamel matrix derivative and transforming growth factor‐b1 on human periodontal ligament fibroblasts. J. Clin. Periodontol. 34, 514–522. [DOI] [PubMed] [Google Scholar]
- 34. Kunimatsu R, Tanimoto K, Tanne Y, Kamiya T, Ohkuma S, Huang YC, et al (2011) Amelogenin enhances the proliferation of cementoblast lineage cells. J. Periodontol. 82, 1632–1638. [DOI] [PubMed] [Google Scholar]
- 35. Zeldich E, Koren R, Nemcovsky C, Weinreb M (2007) Enamel matrix derivate stimulates human gingival fibroblast proliferation via ERK. J. Dent. Res. 86, 41–46. [DOI] [PubMed] [Google Scholar]
- 36. Zeldich E, Koren R, Dard M, Nemcovsky C, Weinreb M (2008) EGFR in enamel matrix derivative‐induced gingival fibroblast mitogenesis. J. Dent. Res. 87, 850–855. [DOI] [PubMed] [Google Scholar]


