Abstract
Objectives
miR‐99a has been reported to function as a tumour suppressor in breast cancer. However, its role in the regulation of breast cancer stem cell (CSC) phenotype has up to now remained unknown.
Materials and methods
In this study, we isolated the side population (SP) cells by staining cultured MCF‐7 and MDA‐MB‐231 cells with fluorescent DNA‐binding dye Hoechst 33342, then by flow cytometric sorting. Next, we detected expression of miR‐99a in the SP cells compared to non‐SP cells using real‐time PCR, and explored effects of miR‐99a on the CSC phenotype of the breast cancer cells, including sphere formation, self‐renewal, tumourigenicity and cell migratory capability.
Results
We found that expression of miR‐99a was down‐regulated in the SP cells compared to non‐SP cells. Restoration of expression of miR‐99a inhibited cell migration and invasion, reduced sphere formation of breast SP cells in vitro, and suppressed tumour growth in vivo. Finally, bioinformatic prediction suggested the oncogene, mammalian target of rapamycin (mTOR) – a downstream effector of the PI3K/AKT signalling pathway, was a target gene of miR‐99a in SP cells. Further, quantitative RT‐PCR and western blot assays identified that overexpression of miR‐99a suppressed expression of mTOR and its downstream gene, HIF‐1α.
Conclusion
Collectively, these data suggest that miR‐99a reversed the breast cancer malignant CSC phenotype, probably by targeting the mTOR signalling pathway.
Introduction
Breast cancer is the leading cause of cancer‐related death in women 1. Recently, in the order of 200 000 new cases were diagnosed and approximately 40 000 deaths were recorded in the United States 2. Although great effort has been exerted to improve overall survival rate of this disease over the past number of decades, resistance to adjuvant chemotherapy remains a major obstacle in effective anti‐cancer treatment. Accumulating evidence has proposed that the existence of a subpopulation of cancer cells with stem cell‐like features, called cancer stem cells (CSCs), is the major reason of post‐treatment tumour recurrence 3, 4. In 2003, Al‐Hajj et al. first isolated breast CSCs from human breast tumour tissues and found that only these had the ability for tumourigenicity in NOD/SCID mice 5; this provided a new avenue for breast cancer research.
Cancer stem cells compose a small subpopulation of tumour cells with great capability for self‐renewal, differentiation and tumourigenicity, when transplanted into an appropriate animal host and emerging evidence has identified the existence of CSCs in a wide variety of human malignancies 5, 6. Over the past decade, several in vitro culture methods have been used to enrich CSCs from tumour tissues, one interesting method of which is isolation of side population (SP) cells 7, 8. Based on the ability of progenitor cells to efflux DNA‐binding dye Hoechst 33342, cells with stem cell properties have been characterized from established cancer cell lines 9. This so‐called SP, Hoechst low in the absence of inhibitors of ATP‐binding cassette (ABC) protein, can be identified and sorted by flow cytometry 8, 10. ABC protein, also known as breast cancer resistance protein, functions as an ATP‐dependent membrane transporter and can efficiently efflux chemotherapeutic agents 11, thus playing an important role in tumour recurrence and drug resistance.
Numerous studies in recent years have pointed to the involvement of microRNAs (miRNAs) in many biological and pathological processes 12, 13, as well as in regulation of CSC self‐renewal and differentiation 14. By profiling miRNAs in human breast CSCs and embryonal carcinoma cells, Shimono et al. found that the former had a concordant regulated subset of miRNAs to the latter 15. Moreover, miR‐93 can regulate proliferation and differentiation of normal and malignant breast stem cells, and its overexpression increases the CSC population in MCF‐7 cells 16. miR‐128 is down‐regulated in breast CSCs and directly targets Bmi‐1 and ABCC5, leading to cell chemotherapeutic resistance 17.
miR‐99a has been reported to be a tumour suppressor in various human cancers. In breast cancer cells, its overexpression not only induces apoptosis in vitro, but also inhibits tumourigenicity in vivo by targeting mammalian target of rapamycin (mTOR) 18. However, its definite function in breast CSCs has not previously been described. In this report, we describe that we isolated SP cells of human breast cancer cell lines, MCF‐7 and MDA‐MB‐231, and compared expression of miR‐99a between SP cells and their adherent non‐SP counterparts. We further investigated effects and relative mechanisms of miR‐99a on regulation of the malignant phenotype of the breast SP cells.
Materials and methods
SP isolation and fluorescence‐activated cell sorting assays
Side population isolation was performed according to the protocol described by Goodell et al. 19. Cells were suspended in DMEM at 1 × 105 cells/ml, containing 5% FBS, and 10 mm HEPES; 5 μg/ml Hoechst 33342 (Sigma‐Aldrich, St. Louis, MO, USA) was added, with or without ABCG transporter inhibitor, Fumitremorgin C (10 μm FTC; Sigma‐Aldrich, St. Louis, MO, USA), for 90 min at 37 °C. After incubation in a shaking water bath for 30 min, cells were placed on ice to terminate dye efflux, followed by washing in 2 ml cold HBSS containing 5% FBS (PAA, Pasching, Austria).
The Hoechst dye was excited using krypton ultraviolet laser at 337–356 nm, and its fluorescence was measured using two filters, 465/30 BP (Hoechst Blue) and 675 BP optical filter (Hoechst Red) with a FACScalibur flow cytometer (BD Biosciences, Franklin Lakes, NJ, USA). Hoechst low SP cells were sorted and subsequently incubated in serum‐free complete medium consisting of DMEM medium (PAA) supplemented with 20 ng/ml EGF (Cell Signaling Tech, Denver, MA, USA), 20 ng/ml bFGF (PeproTech, London, UK) and 5 μg/ml insulin.
Cell culture and transfection
The human breast cancer cell lines used, MCF‐7 and MDA‐MB‐231, were obtained from the American Type Culture Collection (ATCC). Both were maintained in DMEM medium (PAA) supplemented with 10% FBS (PAA), streptomycin (100 μg/ml) and penicillin (100 U/ml). SP cells were placed in serum‐free complete medium consisting of DMEM medium supplemented with EGF (20 ng/ml; Cell Signaling Tech), bFGF (20 ng/ml; PeproTech) and insulin (5 μg/ml). Cells were incubated in a humidified atmosphere of 5% CO2 at 37 °C. miR‐99a and scramble mimics were all purchased from Dharmacon (Austin, TX, USA). Oligonucleotides were transfected into cells to final concentration of 50 nm, by Dhamafect 1 (Dharmacon) according to manufacturer's instructions.
RNA extraction and quantitative real‐time PCR
Total RNA was extracted from SP and non‐SP cells using TRIzol reagent (Invitrogen, Life Technologies Inc., Darmstadt, Germany) according to the manufacturer's instructions. Expression level of GAPDH was used as internal control of mRNAs, and U6 level was regarded as internal control of miRNAs. First, total RNA was reverse transcribed using First‐Strand cDNA Synthesis kit (Invitrogen) with specific stem‐loop reverse transcription primers (shown in Table 1). Then, quantitative real‐time PCR was performed using Quanti‐TectSYBR Green PCR mixture on ABI PRISM 7900 Sequence Detection System (Applied Biosystems, Foster City, CA, USA). Relative expression levels were evaluated using the 2−ΔΔCt method and all experiments were run in triplicate. Primers used for reverse transcription and quantitative real‐time PCR assays are showed in Table 1.
Table 1.
Oligonucleotide primer sequences for PCR or reverse transcription
| Gene | Primer sequence |
|---|---|
| Primers for Real‐time PCR | |
| miR‐99a‐sense | 5′‐CGGAACCCGTAGATCCGAT‐3′ |
| miR‐99a‐antisense | 5′‐CAGTGCAGGGTCCGAGGT‐3′ |
| U6‐sense | 5′‐CTCGCTTCGGCAGCACATATACT‐3′ |
| U6‐anti‐sense | 5′‐ACGCTTCACGAATTTGCGTGTC‐3′ |
| mTOR‐sense | 5′‐CGGACTATGACCACTTGACTC‐3′ |
| mTOR‐antisense | 5′‐CCAAACCGTCTCCAATGAAAGA‐3′ |
| GAPDH‐sense | 5′‐TCAACGACCACTTTGTCAAGCTCA‐3′ |
| GAPDH‐antisense | 5′‐GCTGGTGGTCCAGGGGTCTTACT‐3′ |
| Primers for reverse transcription | |
| miR‐99a | 5′‐GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGCACAAG‐3′ |
| U6 | 5′‐AAAATATGGAACGCTTCACGAATTTG‐3′ |
| mTOR | 5′‐TTTTTTTTTTTTTTTTTT‐3′(Oligo(dT)) |
| GAPDH | 5′‐TTTTTTTTTTTTTTTTTT‐3′(Oligo(dT)) |
| Primers for mTOR luciferase reporter | |
| mTOR‐F | 5′‐AGGCTTGATTTGGTTCCCA‐3′ |
| mTOR‐R | 5′‐CAGACCTTCCCTGTGTTCA‐3′ |
Tumourigenicity assay
Five‐week‐old BALB/c female NOD/SCID nude mice were purchased from Beijing Huafu Biotechnology Inc. (Beijing, China). All animals were housed and maintained in pathogen‐free conditions at our laboratory and experiments were performed according to our institute guidelines for animal welfare.
For tumourigenicity assay, 2 × 104 MCF‐7‐SP cells were subcutaneously injected into fat pads of nude mice. When tumours reached 100 mm3, 10 mice with approximately equal tumour volumes were selected for further experimentation and were separated into two groups, each of five mice. miR‐99a or scramble mimic (5 μg) was suspended in 100 μl Dharmafect 1 solution and directly injected into each tumour. Injections were performed for a total of seven times, every 3 days. Tumour diameters were first measured at the time of the first injection, then subsequently every 3 days. Twenty‐eight days after first injection, all mice were killed and tumours were excised and weighed. Approximate tumour volume was calculated as follows: length × width2 ×1/2.
Western blot analysis
For western blot assays, cells were harvested in ice‐cold PBS 48 h after transfection and lysed on ice cold modified radioimmunoprecipitation buffer, supplemented with protease inhibitors. Protein concentration was determined by BCA Protein Assay Kit (Bio‐Rad, Hercules, CA, USA) and equal amounts of protein were separated on SDS‐PAGE (8, 10, or 12%) and gels were electroblotted on to PVDF membranes (Millipore, Darmstadt, Germany). Non‐specific binding sites were blocked by incubation with TBST containing 5% non‐fat dried milk, in Tris‐buffered saline containing 0.1% Tween‐20, for 2 h, then incubated at 4 °C overnight with primary antibody. Detection was performed with peroxidase‐conjugated secondary antibodies using enhanced chemiluminescence system (Millipore). Primary antibodies employed were agianst mTOR, p‐mTOR, HIF‐1α, Oct4, C‐myc and GAPDH (Cell Signaling, Danvers, MA, USA). Bands were assessed by semi‐quantification using Image J software.
Cell proliferation analysis
For analysis of cell proliferation, cells were seeded into 24‐well plates at 5 × 103 cells per well. They were then incubated in 10% Cell Counting Kit‐8 (CCK‐8, Dojindo, Mashikimachi, Japan) and diluted in normal culture medium at 37 °C, until visual colour conversion occurred. Proliferation level was determined at 0, 24, 48 and 72 h after transfection. Absorbance in each well was measured using a microplate reader set at 450 and 630 nm. All experiments were performed in quadruplicate.
Mammosphere formation assay
For analysis of capability of mammosphere formation, single‐cell suspensions of 5 × 103 cells per millilitre were seeded into six‐well non‐adherent plates, in serum‐free DMEM, supplemented with 2% B27 (Gibco, Grand Island, NY, USA), 20 ng/ml EGF (Cell Signaling Tech), 20 ng/ml bFGF (PeproTech), 0.4% BSA (Sigma‐Aldrich, St. Louis, MO, USA), and 5 μg/ml insulin. After culturing for 8 days, number of mammospheres was counted for all cases.
Cell migration and invasion assays
For the migration assay, modified Boyden chambers (BD Biosciences, San Jose, CA, USA) with 8 μm pore filters were inserted into 24‐well plates. 1 × 104 SP cells were added to upper chambers, and medium containing 20% FBS was added to lower chambers as chemoattractant. For the invasion assay, transwell migration chambers were coated with Matrigel (BD Biosciences) and incubated at 37 °C for 1 h allowing it solidify. After 24 h post transfection, 7 × 103 SP cells suspended in serum‐free DMEM were added to upper chambers. After 24 h transfection, non‐filtered cells were gently removed with cotton swabs. Filtered cells located on lower sides of chambers were stained with crystal violet, air dried, and photographed.
Vector construction and luciferase assays
The whole 3′‐UTR of mTOR gene was amplified from genomic DNA and cloned into the pGL‐3‐vector (Promega, San Luis, CA, USA) immediately downstream of the Renilla luciferase gene. According to the QuickChange Site‐Directed Mutagenesis kit (Stratagene, Santa Clara, CA, USA), mutation in the 3′‐UTR of mTOR gene was generated. The two constructs were identified by sequencing. Twenty four hours before transfection, in the order of 1 × 105 cells/well were seeded into 24‐well plates. Cells were co‐transfected with pGL‐3 firefly luciferase reporter (50 ng), pRL‐TK Renilla luciferase reporter (10 ng) and miR‐125b mimic or scramble mimic (50 nm), using Lipofectamine 2000 (Invitrogen). Moreover, a luciferase reporter construct containing miR‐99a consensus target sequence, served as positive control and pRL‐TK vector served as internal control, respectively. Forty eight hours after transfection, cell lysates were prepared with Passive Lysis Buffer (Promega). Finally, respective firefly and Renilla luciferase activities were measured using Dual‐Luciferase Reporter Assay (Promega), and results were normalized to Renilla luciferase. Final results were expressed as relative luciferase activity (Firefly LUC/Renilla LUC).
Statistical analysis
Data were expressed as mean ± SD of at least three independent experiments. Statistical analysis was carried out using SPSS 15.0 software (SPSS Inc., Chicago, IL, USA). Student's t‐test (two‐tailed) was performed to analyse the data. P‐values <0.05 were considered significant.
Results
miR‐99a was down‐regulated in breast SP cells
As the SP cells had been identified as a population of cancer stem‐like cells, we sought to use this subclone to explore effects of miR‐99a on CSC phenotype of our breast cancer cells. First, we had isolated SP cells by staining cultured MCF‐7 and MDA‐MB‐231 cells with fluorescent DNA‐binding dye Hoechst 33342, then FACS sorting them. Their flow cytometric analysis for blue and red fluorescence demonstrated a small population with low fluorescence, implying active extrusion of the dye (Fig. 1a). SP characteristics of such cells was confirmed by ‘normal fluorescence’ (Fig. 1b) following treatment with the inhibitor of ABC transporters, fumitremorgin C. Using fumitremorgin C for characterization, 1.5 ± 0.2% total cells in the MDA‐MB‐231 culture, and 1.8 ± 0.2% total cells in the MCF‐7 culture, respectively, demonstrated SP characteristics. We next detected expression of miR‐99a in isolated SP populations of each cell line using Taqman real‐time PCR assay. Subsequent analyses indicated that expression of miR‐99a was significantly lower in the SP cells compared to non‐SP cells in each cell line (Fig. 1c).
Figure 1.
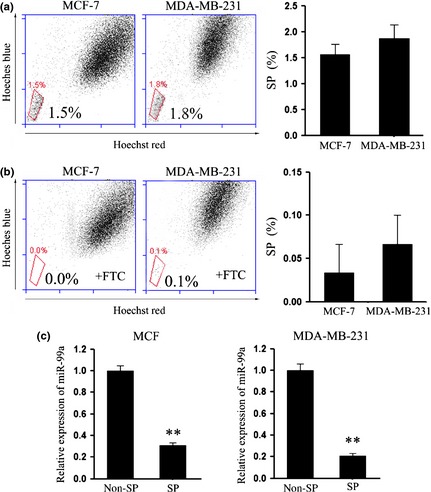
Cells with side population characteristics in breast cancer cell lines. Human breast cancer cells, MCF‐7 and MDA‐MB‐231, were stained with Hoechst 33342 for 30 min, in the absence (a) or presence of fumitremorgin C (b). A small population of cells with side population characteristics (low staining) is shown in (a), largely lost in the presence of inhibitors of ATP binding cassette (ABC) transporter fumitremorgin C (b). Figures are representative of three independent experiments. (c) Expression of miR‐99a in MCF‐7 and MDA‐MB‐231 SP and non‐SP cells was detected by quantitative RT‐PCR. Data are shown as log10 relative ratio change in miR‐99a relative to U6; Data representative of three independent experiments. Error bars represent SEM **P < 0.01.
miR‐99a suppressed self‐renewal of breast SP cells
As maintenance of undifferentiated status and self‐renewal are the most important features of CSCs, we further explored effects of miR‐99a on the malignant phenotype of our breast SP cells after sorting by flow cytometry. First, to explore effects of miR‐99a on self‐renewal of the SP cells, we restored expression of miR‐99a in MCF‐7 and MDA‐MB‐231 cells, and detected percentages of SP cells on transfection. As shown in Fig. 2a and 2b, induction of miR‐99a was positively associated with significant reduction in percentage of SP cells in each cell line, as assessed by FACS assay. Next, we further detected effects of miR‐99a on sphere formation capacity of the breast SP cells. Interestingly, under stem cell growth conditions, the MCF‐7 SP cell culture formed spheres that reached 65 μm in diameter after 8 days, while administration of miR‐99a suppressed sphere formation in both number and size (Fig. 2c). The generality of this observation was further confirmed in MDA‐MB‐231 cells. These results suggest that miR‐99a regulated CSC characteristics of these cell types, including capacities of self‐renewal and sphere formation.
Figure 2.
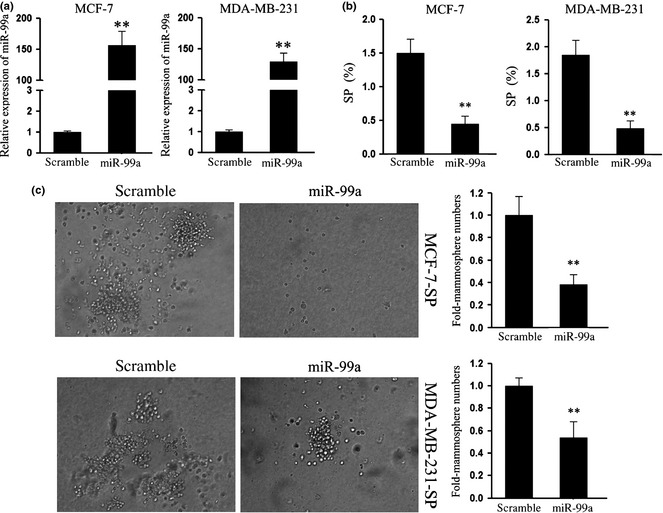
miR‐99a suppressed self‐renewal of SP cells in the breast cancer cell lines. (a) RT‐PCR was performed to detect expression of miR‐99a in MCF‐7 and MDA‐MB‐231 breast cancer cell lines upon transfection with miR‐99a mimic; (b) Using FACS assays, SP population cells upon transfection with miR‐99a mimic were detected. Overexpression of miR‐99a reduced the percentage of SP cells in both cell lines; (c) Sphere formation assays were performed to explored effects of miR‐99a on self‐renewal of SP cells. Administration of miR‐99a reduced SP cell sphere formation of both cell lines. Data are representative of three independent experiments. Error bars represent SEM **P < 0.01.
miR‐99a suppressed tumourigenicity of the breast SP cells in vivo
MCF‐7‐SP cells had then been examined for their tumour initiating ability, by injecting them into mammary fat pads of nude mice. Our results confirmed that isolated SP cells had strong ability to generate tumours, as those formed reached 100 mm3 by 7 days after being injected. Moreover, we found that miR‐99a treatment suppressed MCF‐7‐SP xenograft tumour growth, resulting in significant reduction in average tumour weight and volume (Fig. 3a,b). As ability to form tumours is a critical and standard criterion for identification of CSC, this suggests that miR‐99a suppressed tumourigenicity of these breast CSCs in vivo.
Figure 3.
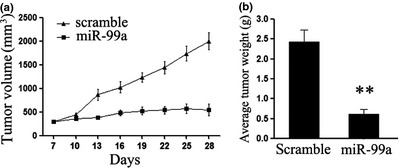
(a) Graph representing approximate tumour volumes at the indicated days; six mice per group; (b) Graph representing average tumour weight for each mouse group at the end of the experiment. Data are representative of three independent experiments. Error bars represent SEM **P < 0.01.
miR‐99a blocked migration and invasion of SP cells
Migration and invasion promote tumour metastasis, the major cause of cancer death. Thus, we further explored effects of miR‐99a on migratory and invasive capacity of the SP cells of each cell line. As shown in Fig. 4a, we found that miR‐99a significantly suppressed migration of MCF‐7 SP cells, and the generality of this observation was further confirmed in MDA‐MB‐231 cells. Furthermore, invasive capacity of SP cells transfected with miR‐99a or scramble mimic was evaluated by Matrigel invasion chamber assay. Restored expression of miR‐99a in the SP cells clearly suppressed cells passing through chambers coated with Matrigel (Fig. 4b).
Figure 4.
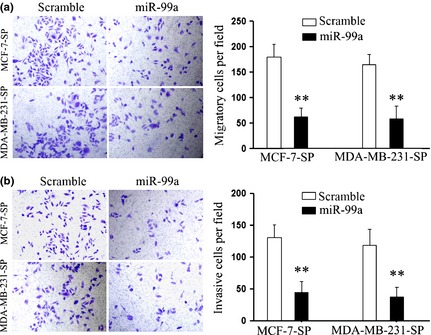
miR‐99a suppressed migration and invasion of SP cells in vitro . (a, b) Transwell chamber assays were performed to detect effects of miR‐99a on migration and invasion of SP cells. Panel (a) shows results of migration; Panel (b) shows results of invasion. Chambers coated with Matrigel, which functions as extracellular cell matrix. Ectopic expression of miR‐99a inhibited cells passing through the membrane in both assays. Data are representative of three independent experiments. Error bars represent SEM **P < 0.01.
miR‐99a suppressed the mTOR signalling pathway in the breast SP cells
As miRNAs can function as tumour activators or indeed as suppressors, through targeting the 3′UTR of relative suppressor genes or oncogene mRNAs, we searched putative targets of miR‐99a using prediction programs, TargetScan, PicTar and miRanda. Among predicted targets of miR‐99a, mTOR was selected out for its important role in the PI3K/AKT/mTOR signalling pathway, suppression of which has previously been reported to inhibit human breast CSC proliferation in vitro and in vivo 20. As shown in Fig. 5a, there exists a putative binding site of miR‐99a in the 3′UTR of mTOR gene. To determine whether this putative binding site would perform any function in breast CSCs, we performed luciferase assays. Restored expression of miR‐99a suppressed luciferase activity in SP cells, while suppression was not significant in CSCs transfected with mutant construct (Fig. 5b). Next, we explored the mRNA and protein level of mTOR in the SP cells, on transfection. Administration of miR‐99a mimic suppressed expression of both mRNA and protein (Fig. 5c,d). Additionally, we also detected protein level of HIF‐1α, which has been reported to be the downstream gene of mTOR signalling and to simultaneously mediate important signalling in CSC progression. As expected, expression of HIF‐1α, was consistent with its respective downstream transcription factors, and Oct‐4 and c‐Myc, were also suppressed 21, 22 (Fig. 5d). These results indicate that miR‐99a regulated CSC characteristics of breast cancer, at least partially, by targeting the mTOR signalling pathway.
Figure 5.
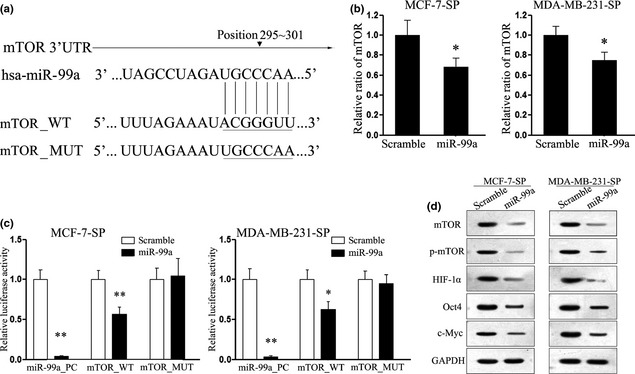
miR‐99a targeting mTOR gene in SP cells. (a) Schematic representation of mTOR 3′UTR showing putative miR‐99a target site; (b) Quantitative RT‐PCR assays performed to detect expression of mTOR upon transfection with miR‐99a mimic or scramble mimic; (c) Relative luciferase activity of indicated mTOR reporter construct in SP cells, co‐transfected with miR‐99a mimic or scramble mimic; (d) Western blot analysis showing expression levels of mTOR, p‐mTOR, HIF1‐α, Oct4 and c‐Myc proteins in SP cells treated with miR‐99a mimic. Data are representative of three independent experiments. Error bars represent SEM *P < 0.05, **P < 0.01.
Discussion
Although miR‐99a has been reported to be a tumour suppressor in breast cancer, its definite function in breast CSCs had not previously been described. In this study, we showed that expression of miR‐99a was reduced in MCF‐7 and MDA‐MB‐231 SP cells, and overexpression of miR‐99a significantly reversed their CSC phenotype and suppressed the mTOR/HIF1‐α signalling pathway.
Previously, CSC‐like SP cells have been isolated from the MCF‐7 breast cancer cell line, and have been demonstrated to have greater tumourigenic activity than the majority of the other cells of this cell line 23. Here, we isolated SP cells from both MCF‐7 and MDA‐MB‐231 cell lines and recapitulated previous findings. Thus, we believe that the isolated SP cells are stem‐like cells.
Mature miR‐99a is individually encoded by genes harboured on chromosome 21, trisomy of which is one of the most common types of chromosomal aneuploidy of live born infants 24. Kuhn et al. have also demonstrated overexpression of miR‐99a in human foetal hippocampus and heart samples in individuals with Down syndrome 25. Moreover, miR‐99a has been found to be specific to human mesenchymal stem cells, but not to differentiated osteoblasts 26, which suggests that miR‐99a might play a vital role in cell differentiation. On the other hand, miR‐99a has been reported to be involved in TGF‐β‐induced epithelial to mesenchymal transition (EMT) of normal murine mammary gland cells 27. As EMT is considered to be a crucial event in the metastatic process [that involves acquisition of a migratory mesenchymal phenotype 28], and also to be an important process during embryonic development in many mammalian species 29, we hypothesized that miR‐99a might play an important role in acquisition of CSC characteristics of breast cancer. To address this question, we compared expression of miR‐99a between SP cells and non‐SP cells. As expected, expression of miR‐99a was significantly suppressed in SP cells relative to their counterpart non‐SP cells.
Next, we further detected effects of miR‐99a on the CSC phenotype of the breast cancer cells. Spheroid formation is an important index of self‐renewal capacity, which is a major property of stem cells 30. As would be expected from the functional studies, our results showed significant reduction in cell number and sphere formation of both types of SP cell upon transfection with miR‐99a. Furthermore, our results also confirmed that the isolated SP cells had strong ability to generate tumours, and treatment with miR‐99a suppressed tumourigenicity of the SP cells in vivo. These results suggest that miR‐99a might play a key role in regulating CSC phenotype of breast cancer.
Cancer stem cells from many tumours have been shown to be endowed with great capabilities of migration and invasion, leading to cancer metastasis. For example, CD44+/CD24− cells in breast cancer are enriched with CSCs, which express higher levels of pro‐invasive genes and exhibit highly invasive properties 31. In this article, we also explored effects of miR‐99a on migration and invasion capacity of our breast SP cells. We also demonstrated that administration of miR‐99a significantly inhibited high migratory and invasive capacity of the breast SP cells.
MicroRNAs (miRNAs) compose a large family of non‐coding single‐stranded RNAs 32, which regulate stability or translational efficiency of targeted mRNAs by complementary or partial interaction with 3′‐untranslated regions (3′UTR) of target genes. Previous work has reported that miR‐99a blocked invasiveness and tumourigenic potential of breast cancer cells by targeting mTOR. mTOR is a downstream effector of the phosphatidyl inositol‐3 kinase/protein kinase B (PI3K/AKT) signalling pathway 18, whose vital role has been widely explored in CSC regulation, including of pancreatic and breast CSCs 20, 33. Zhu et al. found that suppression of the mTOR pathway by everolimus has effective inhibitory effects on HER2‐overexpressing stem cell proliferation in vitro and in vivo 20. Here, we have identified mTOR as a critical downstream target of miR‐99a in the tested breast SP cells. In support of this notion, we explored mRNA and protein levels of mTOR and its downstream gene, hypoxia‐inducible factor 1α (HIF‐1α) 34. Ectopic expression of miR‐99a significantly suppressed expression of mTOR and HIF‐1α simultaneously. As the HIF‐1α signalling pathway has been reported to play important roles in maintaining the biology of CSCs and regulating EMT progression 35, by targeting a variety of embryonic stem cell‐like transcriptional genes, such as OCT4 and c‐Myc 21, 22, we also explored expression of Oct4 and c‐Myc in SP cells after treatment of miR‐99a mimic. As expected, expression of Oct4 and C‐myc were consistently reduced upon transfection, suggesting that the mTOR signalling pathway might be involved in miR‐99a‐mediated suppression. However, further studies are required to identify whether mTOR and its downstream genes are directly involved in regulation of the CSC phenotype of breast cancer.
Collectively, our study has demonstrated that miR‐99a plays an important role in regulating the CSC phenotype of breast cancer. This function isprobably mediated by targeting the mTOR/HIF‐1α signalling pathway. This finding may not only help us understand the molecular mechanism of breast carcinogenesis, but also provide solid foundation for utilization of miR‐99a as an important biomarker for breast cancer prognosis and anti‐cancer therapy (specially when there is recurrence), in the future.
Conflicts of interest
The authors declare no conflict of interest.
Acknowledgements
This research was supported through Fundamental Research Funds for the National Natural Science Foundation of China (30972932, 81173601).
References
- 1. Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D et al (2011) Global cancer statistics. CA Cancer J. Clin. 61, 69–90. [DOI] [PubMed] [Google Scholar]
- 2. Siegel R, Naishadham D, Jemal A (2013) Cancer statistics. CA Cancer J. Clin. 63, 11–30. [DOI] [PubMed] [Google Scholar]
- 3. Guzman ML, Swiderski CF, Howard DS, Grimes BA, Rossi RM, Szilvassy SJ et al (2002) Referential induction of apoptosis for primary human leukemic stem cells. Proc. Natl. Acad. Sci. USA 99, 16220–16225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dalerba P, Cho RW, Clarke MF (2007) Cancer stem cells: models and concepts. Annu. Rev. Med. 58, 267–284. [DOI] [PubMed] [Google Scholar]
- 5. Al‐Hajj M, Wicha MS, Benito‐Hernandez A, Morrison SJ, Clarke MF (2003) Prospective identification of tumorigenic breast cancer cells. Proc. Natl. Acad. Sci. USA 100, 3983–3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ponti D, Costa A, Zaffaroni N, Pratesi G, Petrangolini G, Coradini D et al (2005) Isolation and in vitro propagation of tumorigenic breast cancer cells with stem/progenitor cell properties. Cancer Res. 65, 5506–5511. [DOI] [PubMed] [Google Scholar]
- 7. Chiba T, Kita K, Zheng YW, Yokosuka O, Saisho H, Iwama A et al (2006) Side population purified from hepatocellular carcinoma cells harbors cancer stem cell‐like properties. Hepatology 44, 240–251. [DOI] [PubMed] [Google Scholar]
- 8. Salcido CD, Larochelle A, Taylor BJ, Dunbar CE, Varticovski L (2010) Molecular characterisation of side population cells with cancer stem cell‐like characteristics in small‐cell lung cancer. Br. J. Cancer 102, 1636–1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kim M, Morshead CM (2003) Distinct populations of forebrain neural stem and progenitor cells can be isolated using side‐population analysis. J. Neurosci. 23, 10703–10709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Montanaro F, Liadaki K, Volinski J, Flint A, Kunkel LM (2003) Skeletal muscle engraftment potential of adult mouse skin side population cells. Proc. Natl. Acad. Sci. USA 100, 9336–9341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Singh A, Wu H, Zhang P, Happel C, Ma J, Biswal S (2010) Expression of ABCG2 (BCRP) is regulated by Nrf2 in cancer cells that confers side population and chemoresistance phenotype. Mol. Cancer Ther. 9, 2365–2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Palanichamy JK, Rao DS (2014) miRNA dysregulation in cancer: towards a mechanistic understanding. Front Genet. 5, 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yang Y, Gu X, Zhou M, Xiang J, Chen Z (2013) Serum microRNAs: a new diagnostic method for colorectal cancer. Biomed. Rep. 1, 495–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yu Z, Pestell TG, Lisant MP, Pestell RG (2012) Cancer stem cells. Int. J. Biochem. Cell Biol. 44, 2144–2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Shimono Y, Zabala M, Cho RW, Lobo N, Dalerba P, Qian D et al (2009) Downregulation of miRNA‐200c links breast cancer stem cells with normal stem cells. Cell 138, 592–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Liu S, Patel SH, Ginestier C, Ibarra I, Martin‐Trevino R, Bai S et al (2012) MicroRNA93 regulates proliferation and differentiation of normal and malignant breast stem cells. PLoS Genet. 8, e1002751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhu Y, Yu F, Jiao Y, Feng J, Tang W, Yao H et al (2011) Reduced miR‐128 in breast tumor‐initiating cells induces chemotherapeutic resistance via Bmi‐1 and ABCC5. Clin. Cancer Res. 17, 7105–7115. [DOI] [PubMed] [Google Scholar]
- 18. Hu Y, Zhu Q, Tang L (2014) MiR‐99a antitumor activity in human breast cancer cells through targeting of mTOR expression. PLoS One 9, e92099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Goodell MA, Brose K, Paradis G, Conner AS, Mulligan RC (1996) Isolation and functional properties of murine hematopoietic stem cells that are replicating in vivo. J. Exp. Med. 183, 1797–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhu Y, Zhang X, Liu Y, Zhang S, Liu J, Ma Y et al (2012) Antitumor effect of the mTOR inhibitor everolimus in combination with trastuzumab on human breast cancer stem cells in vitro and in vivo. Tumour Biol. 33, 1349–1362. [DOI] [PubMed] [Google Scholar]
- 21. Mathieu J, Zhang Z, Zhou W, Wang AJ, Heddleston JM, Pinna CM et al (2011) HIF induces human embryonic stem cell markers in cancer cells. Cancer Res. 71, 4640–4652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gordan JD, Lal P, Dondeti VR, Letrero R, Parekh KN, Oquendo CE et al (2008) HIF‐alpha effects on c‐Myc distinguish two subtypes of sporadic VHL‐deficient clear cell renal carcinoma. Cancer Cell 14, 435–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Engelmann K, Shen H, Finn OJ (2008) MCF7 side population cells with characteristics of cancer stem/progenitor cells express the tumor antigen MUC1. Cancer Res. 68, 2419–2126. [DOI] [PubMed] [Google Scholar]
- 24. Centers for Disease Control and Prevention (CDC) (2006) Improved national prevalence estimates for 18 selected major birth defects – United States, 1999–2001. MMWR Morb. Mortal. Wkly Rep. 54, 1301–1305. [PubMed] [Google Scholar]
- 25. Kuhn DE, Nuovo GJ, Martin MM, Malana GE, Pleister AP, Jiang J et al (2008) Human chromosome 21‐derived miRNAs are overexpressed in down syndrome brains and hearts. Biochem. Biophys. Res. Commun. 370, 473–477. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 26. Vimalraj S, Selvamurugan N (2014) MicroRNAs expression and their regulatory networks during mesenchymal stem cells differentiation toward osteoblasts. Int. J. Biol. Macromol. 66, 194–202. [DOI] [PubMed] [Google Scholar]
- 27. Turcatel G, Rubin N, El‐Hashash A, Warburton D (2012) MIR‐99a and MIR‐99b modulate TGF‐beta induced epithelial to mesenchymal plasticity in normal murine mammary gland cells. PLoS One 7, e31032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yang J, Weinberg RA (2008) Epithelial‐mesenchymal transition: at the crossroads of development and tumor metastasis. Dev. Cell 14, 818–829. [DOI] [PubMed] [Google Scholar]
- 29. Kiesslich T, Berr F, Alinger B, Kemmerling R, Pichler M, Ocker M et al (2012) Current status of therapeutic targeting of developmental signalling pathways in oncology. Curr. Pharm. Biotechnol. 13, 2184–2220. [DOI] [PubMed] [Google Scholar]
- 30. Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY et al (2008) The epithelial‐mesenchymal transition generates cells with properties of stem cells. Cell 133, 704–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Korpal M, Lee ES, Hu G, Kang Y (2008) The miR‐200 family inhibits epithelial‐mesenchymal transition and cancer cell migration by direct targeting of E‐cadherin transcriptional repressors ZEB1 and ZEB2. J. Biol. Chem. 283, 14910–14914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Vislovukh A, Vargas TR, Polesskaya A, Groisman I (2014) Role of 3′‐untranslated region translational control in cancer development, diagnostics and treatment. World J. Biol. Chem. 5, 40–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Matsubara S, Ding Q, Miyazaki Y, Kuwahata T, Tsukasa K, Takao S (2013) mTOR plays critical roles in pancreatic cancer stem cells through specific and stemness‐related functions. Sci. Rep. 3, 3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Briest F, Grabowski P (2014) PI3K‐AKT‐mTOR‐signaling and beyond: the complex network in gastroenteropancreatic neuroendocrine neoplasms. Theranostics 4, 336–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bao B, Azmi AS, Ali S, Ahmad A, Li Y, Banerjee S et al (2012) The biological kinship of hypoxia with CSC and EMT and their relationship with deregulated expression of miRNAs and tumor aggressiveness. Biochim. Biophys. Acta 1826, 272–296. [DOI] [PMC free article] [PubMed] [Google Scholar]


