Abstract
Objectives
Degenerated disc disease is one of the most common medical conditions in patients suffering from low back pain. Recent studies have shown that microRNAs can regulate cell function in many pathological conditions. The aim of this study was to investigate expression and role of miR‐93 in disc degeneration.
Materials and methods
Quantitative RT‐PCR was employed to investigate level of miR‐93 in degenerative nucleus pulposus (NP) tissues. Then, functional analysis of miR‐93 in regulating collagen II expression was performed. Subsequently, western blotting and luciferase reporter assay were used to detect the target gene.
Results
We showed that miR‐93 was significantly down‐regulated in degenerative NP tissues and its levels were associated with grade of disc degeneration. Overexpression of miR‐93 stimulated type II collagen expression in NP cells. Moreover, MMP3 was identified as a putative target of miR‐93. MiR‐93 inhibited MMP3 expression by directly targeting its 3′UTR, and this was abolished by miR‐93 binding site mutations. Additionally, restoration of MMP3 in miR‐93‐overexpressed NP cells reversed effects of type II collagen expression. Expression of MMP3 inversely correlated with miR‐93 expression in degenerative NP tissues.
Conclusions
Taken together, we demonstrated that miR‐93 contributed to abnormal NP cell type II collagen expression by targeting MMP3, involved in intervertebral disc degeneration.
Introduction
Chronic low back pain affects up to 80% of adults at some point in their lives, with annual estimated direct and indirect costs upwards of $90 billion in the United States alone 1, 2. Causes of low back pain are multifactorial, and degenerated disc disease (DDD) is the most common medical condition in patients suffering from it 3. The causes of DDD are currently unknown, but candidate risk factors include genetic predisposition, lifestyle and ageing 4, 5. Current evidence implicates major pathological changes in a degenerating disc to begin with proteoglycan breakdown, cell loss and diminished water‐binding capacity of the nucleus pulposus (NP) 6, 7. This reduces ability of disc cells to synthesize extracellular matrix (ECM), leading to structural collapse and eventual loss of demarcation between the outer annulus fibrosus and inner NP tissues, and loading function of discs in advanced stages 7, 8. ECM metabolism is regulated by many factors, including degradative enzyme inhibitors, tissue inhibitors of metalloproteinases, degradative enzyme matrix metalloproteinases (MMPs) and aggrecanases 9, 10, 11. Increase in levels of MMP, especially MMP3, 7, 9 and 13, is a characteristic of DDD 12. MMPs mediate degradation of type II matrix collagen, but regulation of MMP3 expression in NP cells is unknown.
Recent evidence indicates that many cell processes, including proliferation, differentiation, apoptosis and cytokine release, are regulated by microRNAs (miRNAs), which are typically 21–23 nucleotides in length 13, 14. miRNAs are implicated in diverse pathological conditions, such as cancer, neurodegeneration and cardiovascular disease 15, 16, 17. They mediate their biological functions by binding to cis‐regulatory elements, mainly present in 3′ untranslated regions (3′UTR) of their target mRNAs, resulting in translational inhibition or degradation 18, 19. It has been estimated that miRNAs, which constitute only 1–3% of the human genome, can regulate up to approximately 30% of protein‐encoding genes in humans 20, 21. However, to date, only around four reports have attempted to explore the pathogenesis of DDD to in relation to miRNAs 22, 23, 24, 25.
Previous studies have shown that miR‐93 plays an important role in multiple cell processes, including proliferation, apoptosis, invasion and ECM degradation 26, 27, 28, 29. It has been reported that expression of miR‐93 is reduced in various tumour types such as hepatocellular carcinoma, colon cancer, non‐small lung cancer and gastric cancer 27, 28, 29, 30, 31. However, its role in the pathogenesis of DDD is still unknown. In this study, we demonstrated that miR‐93 was significantly down‐regulated in human degenerative NP tissues compared to those of patients with idiopathic scoliosis. Moreover, MMP3 was indicated to be its putative target. Overexpression of miR‐93 stimulated type II collagen expression in NP cells by directly binding to the 3′UTR of MMP3.
Materials and methods
Ethics statement
All experimental protocols were approved by the Clinical Research Ethics Committee of the Tianjin First Central Hospital. Human lumbar IVD samples were obtained from patients undergoing discectomy following approval from the Clinical Research Ethics Committee of the Tianjin First Central Hospital, with fully informed, written consent from the patients, or patients' parents on behalf of children.
Patients and samples
Human lumbar NP specimens were collected from patients with idiopathic scoliosis (n = 4; average age 19 ± 2.53, range 16–20 years) and from patients with DDD (n = 54; average age 48.18 ± 8.81, range 29–64 years). Routine MRI scans of the lumbar spine were taken of these patients before surgery and degree of disc degeneration was graded from T2‐weighted images, using modified Pfirrmann classification.
Isolation and primary culture of human NP cells
Nucleus pulposus cells were isolated as previously described 32, 33. Tissue specimens were collected from patients with idiopathic scoliosis (n = 4; average age 19 ± 2.53, range 16–20 years) and were first washed twice in PBS and NP was separated from NF (viewed using a stereotaxic microscope) and cut into pieces (2–3 mm3). NP cells were released from the NP tissues by incubation with 0.25 mg/ml type II collagenase (Invitrogen, Carlsbad, CA, USA) for 8 h at 37 °C in Dulbecco's modified Eagle's medium (DMEM; Gibco, Grand Island, NY, USA). After isolation, NP cells were resuspended in DMEM containing 10% FBS (Gibco), 100 μg/ml streptomycin, 100 U/ml penicillin and 1% l‐glutamine and then incubated at 37 °C in a humidified atmosphere in the presence of 5% CO2. Confluent cells were detached by trypsinization, seeded into 35 mm tissue culture dishes in complete culture medium (DMEM supplemented with 10% FBS, 100 μg/ml streptomycin and 100 U/ml penicillin), and incubated in a 37 °C, 5% CO2 environment. Medium was changed every 3 days and second passage cells were used for subsequent experiments.
Dual luciferase assays
Cells were co‐transfected with 0.4 μg reporter construct, 0.2 μg pGL‐3 control vector, and either miR‐93 or a negative control. Cells were harvested 24 h post‐transfection and then assayed using Dual Luciferase Assay (Promega, WI, USA), according to the manufacturer's instructions. Firefly luciferase values were normalized to the Renilla signal and ratio of Firefly/Renilla values was reported. All transfection assays were carried out in triplicate.
Oligonucleotides, constructs and transfections
MiR‐93 mimics and negative controls were synthesized by GenePharma (Shanghai, China) and were transfected into the cells with final oligonucleotide concentration of 20 nmol/l. All cell transfections were performed using DharmaFECT1 reagent (Dharmacon, Austin, TX, USA), according to the manufacturer's instructions. For each cell transfection, two or three replicate experiments were performed.
RNA isolation, reverse transcription and quantitative RT‐PCR
Total RNA was extracted from harvested cells and tissues using Trizol reagent (Invitrogen) according to the manufacturer's protocol. MicroRNAs were quantitated with real‐time PCR using TaqMan microRNA Assays (Invitrogen). First‐strand complementary DNA synthesis was carried out with 1 μg total RNA in 12 μl final volume, that contained 2 m the stem‐loop primer and 10 mm dNTP mix (Invitrogen). This was incubated at 65 °C for 5 min then combined with 5× RT buffer, 0.1 m DTT, 200 U/μl MultiScribe reverse transcriptase and 40 U/μl RNase inhibitor (Invitrogen). The mix was incubated at 37 °C for 55 min, 70 °C for 15 min, then held at −20 °C. Real‐time PCR was performed by following a standard TaqMan PCR protocol. 20 μl PCRs included 1 μl RT product, 1× Universal TaqMan Master Mix and 1× TaqMan probe/primer mix (Invitrogen, Table S2). All RT reactions, including reactions containing no‐template controls, were run in triplicate. All miRNA quantification data were normalized to U6 expression and mRNA quantification data were normalized to GAPDH. Relative amounts of transcript were calculated using the comparative Ct method.
Western blotting
Western blot analysis was conducted using standard methods. Proteins were separated on 10% SDS‐PAGE gel then transferred to PVDF membranes (Amersham, Buckinghamshire, UK), which were blocked using 5% non‐fat dried milk for 2 h, and incubated for 12 h with anti‐type II collagen antibody (1:1000; Abcam, Cambridge, UK), anti‐MMP3 antibody (1:1000; Bioworlde, Minneapolis, MN, USA) or anti‐GAPDH antibody (1:50 000; Proteintech, Chicago, IL, USA). After washing in TBST (10 mm Tris, pH 8.0, 150 mm NaCl and 0.1% Tween 20), membranes were incubated for 2 h in goat anti‐rabbit antibody (zsgb‐bio, Beijing, China; 1:5000 or 1:50 000).
Immunofluorescence staining
Coverslips were placed into 24‐well plates. Then, medium was removed and cells were washed twice in PBS and fixed in 3.5% formaldehyde for 30 min at 37 °C. They were then rinsed three times in PBS, permeabilized with 0.1% (v/v) Triton X‐100 in PBS for 20 min, and blocked with 3% (w/v) BSA and 0.05% (v/v) Tween 20, in PBS for 30 min at room temperature. After blocking, cells were incubated overnight at 4 °C with primary antibody (PBS used as control), rabbit monoclonal anti‐type II collagen (dilution ratio 1:500; Bioworlde). Cells were then treated with Alexa Fluor 488 goat anti‐rabbit IgG (1:500; Bioworlde) for 2 h at room temperature. Fluorescence images were acquired using a Leica TCS SP2 confocal microscope (Leica, Mannheim, Germany) using Leica Confocal Software.
Statistical analysis
Statistical analyses were performed using the spss 17.0 statistical software program (IBM, Chicago, IL, USA). For these human studies, Kruskal–Wallis test was used to assess difference in expression of miR‐93 in disc specimens from different herniation types, and independent t‐testing was used to assess differences between specimens of different genders. Correlation between expression of miR‐93 and age, BMI and expression of MMP3, of the patients was determined by Pearson's test. Correlation between expression of miR‐93 and duration of symptoms was determined by Spearman's test. Data were expressed as mean ± SD. Western blot results were normalized using GAPDH. Independent experiments were performed twice. Statistical analysis was performed using Student's t‐test. P values <0.05 were considered statistically significant.
Results
MiR‐93 expression was down‐regulated in degenerative NP tissues and correlated with grade of degeneration
Our results demonstrated that miR‐93 was significantly down‐regulated in degenerative NP tissues in comparison with controls (Fig. 1a). To further study the relationship of miR‐93 to degenerative NP development, expression of miR‐93 was detected in a further 50 clinical patients using real‐time PCR. As shown in Table S1 and Fig. 1, no significant difference was observed between samples from different herniation types, genders, duration of symptoms or age of the patients. However, miR‐93 level was positively correlated with disc degeneration grade (r = 0.87, P < 0.001).
Figure 1.
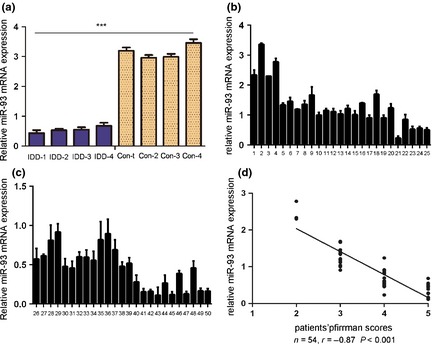
Expression of miR‐93 was down‐regulated in human degenerative nucleus pulposus tissues. (a) Expression of miR‐93 in four degenerative nucleus pulposus tissues and four idiopathic scoliosis nucleus pulposus tissues. These degenerative NP tissues exhibited significantly reduced expression of miR‐93; total RNA was extracted from them. (b) and (c) TaqMan RT‐PCR analysis of miR‐93 expression in the human nucleus pulposus tissue of 50 patients; total RNA was extracted from them. (d) Correlation between expression of miR‐93 and disc degeneration grade of patients. Error bars represent SD. ***Indicates P < 0.001.
MiR‐93 induced expression of type II collagen in NP cells
Nucleus pulposus cells were transfected with scrambled control oligo or with miR‐93 mimic, both of which had high transfection efficiency (Fig. 2a). Real‐time PCR assay demonstrated that type II collagen was increased in cells that were transfected with miR‐93 mimics compared to scrambled oligo‐transfected cells or untreated cells (Fig. 2b). This effect was further confirmed by immunohistochemistry. As shown in Fig. 2c, there was a significant increase in expression levels of type II collagen in the miR‐93 mimic‐transfected group, compared to control or untreated groups.
Figure 2.
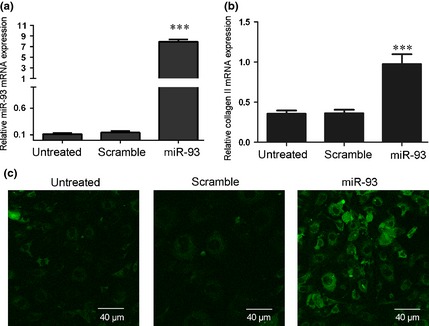
Overexpression of miR‐93 promoted type II collagen expression in NP . (a) Expression levels were examined using real‐time PCR for non‐transfected cells or after transfection of 20 nmol/l of miR‐93 mimic or scramble control, after 24 h. (b) Overexpression of miR‐93 promoted type II collagen mRNA expression using real‐time PCR. NP cells were transfected with 20 nmol/l of miR‐93 mimic or scramble control or remained non‐transfected after 24 h. Type II collagen was detected using real‐time PCR. (c) Overexpression of miR‐93 promoted type II collagen protein expression as demonstrated by immunohistochemistry. NP cells were transfected with 20 nmol/l miR‐93 mimic or scramble control or remained non‐transfected after 24 h. Type II collagen was detected by immunohistochemical analysis. Images were acquired using laser scanning confocal microscopy under a 40× objective lens. Values are presented as mean ± SD. ***Indicates P < 0.001.
MiR‐93 repressed MMP3 expression
As predicted by PicTar, there was complementarity between miR‐93 and MMP3 3′UTR (Fig. 3a). Overexpression of miR‐93 reduced both MMP3 protein and mRNA levels in NP cells (Fig. 3c,d). Next, the effect of miR‐93 on translation of MMP3 mRNA into protein was assessed, using luciferase reporter assay (Fig. 3b). Overexpression of miR‐93 significantly reduced luciferase activity of the reporter gene in wild type, but not mutant, MMP3 3′UTR, indicating that miR‐93 directly targeted the MMP3 3′UTR.
Figure 3.
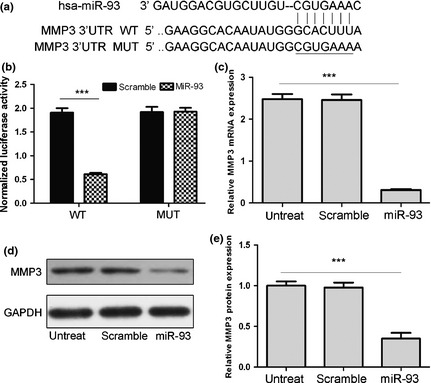
MMP3 is a direct target of miR‐93. (a) Schematic representation of MMP3 3′UTR showing putative miRNA target site. (b) Luciferase activities of wild‐type (WT‐UTR) and mutant (MUT‐UTR) constructs. (c) Real‐time PCR analysis showed that miR‐93 reduced mRNA level of MMP3. (d) MMP3 protein expression in NP cells transfected with 20 nmol/l of indicated miRNA. Values presented as mean ± SD. GAPDH was loading control. Compared to control, ***P < 0.001.
MMP3 was required for miR‐93 induced type II collagen expression in NP cells
We performed rescue experiments to further validate that targeting MMP3 was involved in the function of miR‐93 in NP cells. MMP3 expression vector was used to restore MMP3 expression. Up‐regulation of type II collagen mRNA and protein expression, upon overexpression of miR‐93, was significantly attenuated by re‐introduction of MMP3 (Fig. 4).
Figure 4.
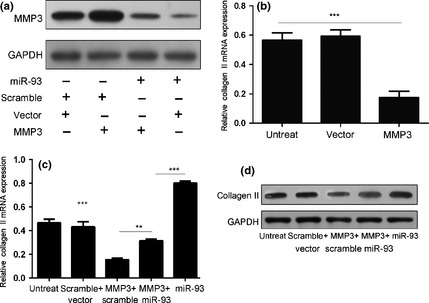
Overexpression of MMP3 partially rescued miR‐93‐induced type II collagen expression. (a) Western blot analysis of MMP3 in NP cells co‐transfected with either miR‐93 mimic (20 nm) or scrambled miRNA (20 nm) and either MMP3 vector (2 μg) or empty vector (2 μg). (b) Overexpression of MMP3 reduced type II collagen mRNA expression. NP cells were transfected with MMP3 vector or empty control. Type II collagen expression was detected using real‐time PCR; relative ratio of cells per field is shown. (c) Real‐time PCR assays of type II collagen mRNA expression with NP cells treated as described in (a). (d) Western blotting analyses of type II collagen protein expression of NP cells treated as described in (a). Bars represent mean ± SD. GAPDH was loading control. **Indicates P < 0.01, ***Indicates P < 0.001.
MMP3 was inversely expressed compared to miR‐93 in degenerative NP tissues
Our results demonstrated that expression level of MMP3 was significantly up‐regulated in degenerative NP tissues in comparison to controls (Fig 5c). To study the relationship of MMP3 in degenerative NP development, expression of MMP3 was detected in a further 50 clinical patients. As shown in Fig. 5, expression level of MMP3 positively correlated with disc degeneration grade (r = 0.81, P < 0.001). MiR‐93 expression negatively correlated with level of MMP3 mRNA in degenerative NP samples.
Figure 5.
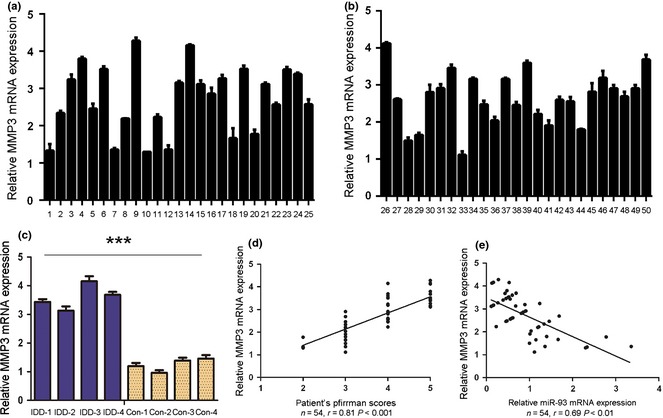
MMP3 was inversely expressed compared to miR‐93 expression in degenerative NP tissue. (a) and (b) Real‐time PCR analysis of MMP3 mRNA expression in human nucleus pulposus tissue of 50 patients. (c) Expression of MMP3 in four degenerative nucleus pulposus tissues and four idiopathic scoliosis nucleus pulposus tissues. Degenerative NP tissues exhibited higher expression of MMP3 compared to controls. (d) Correlation between expression of MMP3 and disc degeneration grade of the patients. (e) Analysis of correlation of miR‐93 and MMP3 expression in degenerative NP tissues. (Two‐tailed Pearson's correlation analysis, r = −0.69; P < 0.01). Error bars represent SD. ***Indicates P < 0.001.
Discussion
Increasing evidence has suggested miRNAs to be salient regulators of diverse biological and pathological processes, including cell growth, differentiation, apoptosis and carcinogenesis 34, 35, 36. However, roles of miRNAs in disc degeneration remain largely uncharacterized. In the present study, miR‐93 was found to be significantly down‐regulated in human degenerative NP tissues. Moreover, overexpression of miR‐93 increased expression of type II collagen in NP cells. We also identified MMP3 to be a novel and direct target of miR‐93. MMP3 was up‐regulated in human degenerative NP tissues compared to normal NP tissues and expression of MMP3 inversely correlated with miR‐93 expression. Furthermore, increased expression of type II collagen after MMP3 overexpression in was almost completely blocked by miR‐93.
MiR‐93 has frequently been reported to be implicated in multiple cell processes, including proliferation, apoptosis, invasion and ECM degradation 26, 27, 28, 29. miR‐93 is low in various tumour types, including hepatocellular carcinoma, colon cancer, non‐small lung cancer and gastric cancer 27, 28, 29, 30, 31. However, expression of miR‐93 in degenerative NP tissues and its role in pathogenesis of DDD are still unknown. In the present study, miR‐93 levels were significantly reduced in degenerative NP and significantly associated with disc degeneration grade (Fig. 1). Reduced levels of miR‐93 may associate with post‐trauma reactions or local inflammation in the intervertebral disc. Previous studies have shown that type II collagen degradation contributes to development of DDD. The ECM provides both mechanical and biochemical signals for NP cells to regulate their survival, morphology and differentiation 37, 38. Current evidence implicates that loss of type II collagen is considered to be an early indicator of DDD 39. To further investigate the function of miR‐93 in development of DDD, we performed functional analysis of miR‐93 to investigate its relationship with type II collagen. Overexpression of miR‐93 significantly increased expression of type II collagen in NP cells (Fig. 2). These findings suggest that reduced type II collagen expression induced by down‐regulation of miR‐93 might participate in DDD development.
MMP3 was identified as the direct target of miR‐93 in NP cells. A sequence complementary to miR‐93 was identified at the 3′UTR of MMP3 mRNA and overexpression of miR‐93 led to significant reduction in MMP3 expression. Overexpression of miR‐93 also suppressed MMP3 3′UTR luciferase reporter activity, and this was abolished by mutation of the miR‐93 binding site. These results indicate that miR‐93 could play an important role in development of DDD, in part by repressing MMP3 expression. Induction of MMP production plays a central role in pathophysiology of IVD degeneration 40. Previous studies have shown that IVD cells in lumbar herniated disc tissues express MMPs, especially MMP3 and MMP‐7, which can degrade collagen and the core protein of aggrecan 41. Hallmarks of DDD are progressive loss of ECM macromolecules aggrecan and collagen II. High levels of MMP‐1 and MMP3 have been found in degenerating IVDs 42. MMP3 can degrade ECM components such as type II collagen and aggrecan 43. It can also amplify the degradative process by activation of secreted MMPs, suggesting that its regulation may be important in maintaining NP homoeostasis 44. We also found that MMP3 was up‐regulated in human degenerative NP tissues compared to normal NP, and expression level of MMP3 inversely correlated with miR‐93 expression. It is considered that reduced miR‐93 expression increased MMP3, degrading ECM components such as type II collagen and aggrecan, might be the potential mechanism in human degenerative processes. One of the drawbacks of this study, however, has been lack of age‐matched non‐degenerate discs as controls. NP cells derived from idiopathic scoliosis patients may not necessarily reflect the in vivo scenario. Comparison of reaction of idiopathic scoliosis and normal NP cells to miR‐93 stimulation could provide further information for potential involvement of miR‐93 in DDD development.
In conclusion, our data suggest that miR‐93 was lower in human degenerative NP tissues and that its level was associated with disc degeneration grade. In addition, overexpression of miR‐93 increased expression of type II collagen expression by targeting MMP3. These results have shed new light on the role of miR‐93 in the pathogenesis of DDD.
Conflicts of interest
The authors declare no conflict of interest. No benefits in any form have been or will be received from any commercial party related directly or indirectly to the subject of this manuscript.
Supporting information
Table S1. Clinical finding in 54 patients with degenerated disc
Table S2. Primer/mimics/probe sequence
Acknowledgements
We would like to thank all volunteers who took part in this study. This work was supported by grants from the Science & Technology Fund of Tianjin Municipal Commission of Health and Family Planning (2014KZ025).
References
- 1. Frymoyer JW, Cats‐Baril WL (1991) An overview of the incidences and costs of low back pain. Orthop. Clin. North Am. 22, 263–271. [PubMed] [Google Scholar]
- 2. Nakamura M, Nishiwaki Y, Ushida T, Toyama Y (2011) Prevalence and characteristics of chronic musculoskeletal pain in Japan. J. Orthop. Sci. 16, 424–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hangai M, Kaneoka K, Kuno S, Hinotsu S, Sakane M, Mamizuka N et al (2008) Factors associated with lumbar intervertebral disc degeneration in the elderly. Spine J. 8, 732–740. [DOI] [PubMed] [Google Scholar]
- 4. Inoue N, Espinoza Orias AA (2011) Biomechanics of intervertebral disk degeneration. Orthop. Clin. North Am. 42, 487–499, vii. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Samartzis D, Karppinen J, Mok F, Fong DY, Luk KD, Cheung KM (2011) A population‐based study of juvenile disc degeneration and its association with overweight and obesity, low back pain, and diminished functional status. J. Bone Joint Surg. Am. 93, 662–670. [DOI] [PubMed] [Google Scholar]
- 6. Song YQ, Karasugi T, Cheung KM, Chiba K, Ho DW, Miyake A et al (2013) Lumbar disc degeneration is linked to a carbohydrate sulfotransferase 3 variant. J. Clin. Invest. 123, 4909–4917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ogawa T, Matsuzaki H, Uei H, Nakajima S, Tokuhashi Y, Esumi M. (2005) Alteration of gene expression in intervertebral disc degeneration of passive cigarette‐ smoking rats: separate quantitation in separated nucleus pulposus and annulus fibrosus. Pathobiology 72, 146–151. [DOI] [PubMed] [Google Scholar]
- 8. Crean JK, Roberts S, Jaffray DC, Eisenstein SM, Duance VC (1997) Matrix metalloproteinases in the human intervertebral disc: role in disc degeneration and scoliosis. Spine (Phila Pa 1976) 22: 2877–2884. [DOI] [PubMed] [Google Scholar]
- 9. Loreto C, Musumeci G, Castorina A, Martinez G (2011) Degenerative disc disease of herniated intervertebral discs is associated with extracellular matrix remodeling, vimentin‐positive cells and cell death. Ann. Anat. 193, 156–162. [DOI] [PubMed] [Google Scholar]
- 10. Schram K, De Girolamo S, Madani S, Munoz D, Thong F, Sweeney G (2010) Leptin regulates MMP‐2, TIMP‐1 and collagen synthesis via p38 MAPK in HL‐1 murine cardiomyocytes. Cell. Mol. Biol. Lett. 15, 551–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rutges JP, Nikkels PG, Oner FC, Ottink KD, Verbout AJ, Castelein RJ et al (2010) The presence of extracellular matrix degrading metalloproteinases during fetal development of the intervertebral disc. Eur. Spine J. 19, 1340–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rutges JP, Kummer JA, Oner FC, Verbout AJ, Castelein RJ, Roestenburg HJ et al (2008) Increased MMP‐2 activity during intervertebral disc degeneration is correlated to MMP‐14 levels. J. Pathol. 214, 523–530. [DOI] [PubMed] [Google Scholar]
- 13. Kim J, Kang Y, Kojima Y, Lighthouse JK, Hu X, Aldred MA et al (2013) An endothelial apelin‐FGF link mediated by miR‐424 and miR‐503 is disrupted in pulmonary arterial hypertension. Nat. Med. 19, 74–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Banaudha KK, Verma M (2012) The role of microRNAs in the management of liver cancer. Methods Mol. Biol. 863, 241–251. [DOI] [PubMed] [Google Scholar]
- 15. Vaira V, Faversani A, Dohi T, Montorsi M, Augello C, Gatti S et al (2012) miR‐296 regulation of a cell polarity‐cell plasticity module controls tumor progression. Oncogene 31, 27–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ponomarev ED, Veremeyko T, Barteneva NS (2011) Visualization and quantitation of the expression of microRNAs and their target genes in neuroblastoma single cells using imaging cytometry. BMC Res. Notes 4, 517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Aghabozorg Afjeh SS, Ghaderian SM (2013) The role of micro‐RNAs in cardiovascular disease. Int. J. Mol. Cell. Med. 2, 50–57. [PMC free article] [PubMed] [Google Scholar]
- 18. Catuogno S, Cerchia L, Romano G, Pognonec P, Condorelli G, de Franciscis V (2013) miR‐34c may protect lung cancer cells from paclitaxel‐induced apoptosis. Oncogene 32, 341–351. [DOI] [PubMed] [Google Scholar]
- 19. Chuang JC, Jones PA (2007) Epigenetics and microRNAs. Pediatr. Res. 61, 24R–29R. [DOI] [PubMed] [Google Scholar]
- 20. Jansson MD, Lund AH (2012) MicroRNA and cancer. Mol. Oncol. 6, 590–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mo YY (2012) MicroRNA regulatory networks and human disease. Cell. Mol. Life Sci. 69, 3529–3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yu X, Li Z, Shen J, Wu WK, Liang J, Weng X et al (2013) MicroRNA‐10b promotes nucleus pulposus cell proliferation through RhoC‐Akt pathway by targeting HOXD10 in intervetebral disc degeneration. PLoS One 8, e83080. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 23. Wang HQ, Yu XD, Liu ZH, Cheng X, Samartzis D, Jia LT et al (2011) Deregulated miR‐155 promotes Fas‐mediated apoptosis in human intervertebral disc degeneration by targeting FADD and caspase‐3. J. Pathol. 225, 232–242. [DOI] [PubMed] [Google Scholar]
- 24. Zhao B, Yu Q, Li H, Guo X, He X (2014) Characterization of microRNA expression profiles in patients with intervertebral disc degeneration. Int. J. Mol. Med. 33, 43–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liu H, Huang X, Liu X, Xiao S, Zhang Y, Xiang T et al (2014) miR‐21 promotes human nucleus pulposus cell proliferation through PTEN/AKT signaling. Int. J. Mol. Sci. 15, 4007–4018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fang L, Deng Z, Shatseva T, Yang J, Peng C, Du WW et al (2011) MicroRNA miR‐93 promotes tumor growth and angiogenesis by targeting integrin‐beta8. Oncogene 30, 806–821. [DOI] [PubMed] [Google Scholar]
- 27. Yu XF, Zou J, Bao ZJ, Dong J (2011) miR‐93 suppresses proliferation and colony formation of human colon cancer stem cells. World J. Gastroenterol. 17, 4711–4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Du L, Zhao Z, Ma X, Hsiao TH, Chen Y, Young E et al (2013) miR‐93‐directed downregulation of DAB2 defines a novel oncogenic pathway in lung cancer. Oncogene 33, 4307–4315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Xu D, He XX, Chang Y, Sun SZ, Xu CR, Lin JS (2012) Downregulation of MiR‐93 expression reduces cell proliferation and clonogenicity of HepG2 cells. Hepatogastroenterology 59, 2367–2373. [DOI] [PubMed] [Google Scholar]
- 30. Chen L, Jiang M, Yuan W, Tang H (2012) Prognostic value of miR‐93 overexpression in resectable gastric adenocarcinomas. Acta Gastroenterol. Belg. 75, 22–27. [PubMed] [Google Scholar]
- 31. Zhu W, He J, Chen D, Zhang B, Xu L, Ma H et al (2014) Expression of miR‐29c, miR‐93, and miR‐429 as potential biomarkers for detection of early stage non‐small lung cancer. PLoS One 9, e87780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Li Z, Shen J, Wu WK, Yu X, Liang J, Qiu G et al (2012) Leptin induces cyclin D1 expression and proliferation of human nucleus pulposus cells via JAK/STAT, PI3K/Akt and MEK/ERK pathways. PLoS One 7, e53176. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 33. Li Z, Shen J, Wu WK, Yu X, Liang J, Qiu G et al (2013) The role of leptin on the organization and expression of cytoskeleton elements in nucleus pulposus cells. J. Orthop. Res. 31, 847–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ma L, Teruya‐Feldstein J, Weinberg RA (2007) Tumour invasion and metastasis initiated by microRNA‐10b in breast cancer. Nature 449, 682–688. [DOI] [PubMed] [Google Scholar]
- 35. Chen L, Zhang J, Feng Y, Li R, Sun X, Du W et al (2012) MiR‐410 regulates MET to influence the proliferation and invasion of glioma. Int. J. Biochem. Cell Biol. 44, 1711–1717. [DOI] [PubMed] [Google Scholar]
- 36. Zhao WH, Wu SQ, Zhang YD (2013) Downregulation of miR‐124 promotes the growth and invasiveness of glioblastoma cells involving upregulation of PPP1R13L. Int. J. Mol. Med. 32, 101–107. [DOI] [PubMed] [Google Scholar]
- 37. Hayes AJ, Benjamin M, Ralphs JR (2001) Extracellular matrix in development of the intervertebral disc. Matrix Biol. 20, 107–121. [DOI] [PubMed] [Google Scholar]
- 38. He F, Pei M (2012) Rejuvenation of nucleus pulposus cells using extracellular matrix deposited by synovium‐derived stem cells. Spine (Phila Pa 1976) 37, 459–469. [DOI] [PubMed] [Google Scholar]
- 39. Li S, Jia X, Duance VC, Blain EJ (2011) The effects of cyclic tensile strain on the organisation and expression of cytoskeletal elements in bovine intervertebral disc cells: an in vitro study. Eur. Cell Mater. 21, 508–522. [DOI] [PubMed] [Google Scholar]
- 40. Yurube T, Takada T, Suzuki T, Kakutani K, Maeno K, Doita M et al (2012) Rat tail static compression model mimics extracellular matrix metabolic imbalances of matrix metalloproteinases, aggrecanases, and tissue inhibitors of metalloproteinases in intervertebral disc degeneration. Arthritis. Res. Ther. 14, R51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Huang AC, Hsu SC, Kuo CL, Liao CL, Lai KC, Lin TP et al (2009) Involvement of matrix metalloproteinases in the inhibition of cell invasion and migration through the inhibition of NF‐(kappa)B by the new synthesized ethyl 2‐(N‐p‐chlorobenzyl‐(2'‐methyl))anilino‐4‐oxo‐4,5‐dihydrofuran‐3‐carboxylate (JOTO1007) in human cervical cancer Ca ski cells. In Vivo 23, 613–619. [PubMed] [Google Scholar]
- 42. Zigouris A, Batistatou A, Alexiou GA, Pachatouridis D, Mihos E, Drosos D et al (2011) Correlation of matrix metalloproteinases‐1 and ‐3 with patient age and grade of lumbar disc herniation. J. Neurosurg. Spine 14, 268–272. [DOI] [PubMed] [Google Scholar]
- 43. Wang X, Li F, Fan C, Wang C, Ruan H (2011) Effects and relationship of ERK1 and ERK2 in interleukin‐1beta‐induced alterations in MMP3, MMP13, type II collagen and aggrecan expression in human chondrocytes. Int. J. Mol. Med. 27, 583–589. [DOI] [PubMed] [Google Scholar]
- 44. Lisboa RA, Andrade MV, Cunha‐Melo JR (2013) Toll‐like receptor activation and mechanical force stimulation promote the secretion of matrix metalloproteinases 1, 3 and 10 of human periodontal fibroblasts via p38, JNK and NF‐kB. Arch. Oral Biol. 58, 731–739. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Clinical finding in 54 patients with degenerated disc
Table S2. Primer/mimics/probe sequence


