Abstract
Objectives
Oral cancer represents one of the most common malignancies in humans. Its prognosis is still poor, despite the most recent improvements in therapies. An increasing attention is placed on the role of programmed death ligand 1 (PD‐L1) in the tumour immunity and its potential function as a marker for tumour prognosis. Whether PD‐L1 expression is a prognostic factor for the poor outcomes in oral squamous cell carcinoma is still controversial. This study aimed to investigate, through a meta‐analysis, a potential correlation between PD‐L1 expression and the prognostic outcomes in patients with oral squamous cell carcinoma.
Materials and methods
The studies were identified by searching PubMed, SCOPUS, Web of Science and were assessed by two of the authors. After the selection process, 11 articles met eligibility criteria and were included in the meta‐analysis. Quality assessment of studies was performed according to the REMARK guidelines, and the risk of biases across studies was investigated through Q and I 2 tests. Meta‐analysis was performed to investigate the association between the PD‐L1 expression either overall survival (OS), disease‐free survival (DFS), disease‐specific survival (DSS), gender and lymph node metastasis.
Results
A total of 1060 patients were analysed in the 11 studies included in the meta‐analysis. Pooled analysis revealed that the expression of PD‐L1 did not correlate with poor OS (HR, 0.60; 95% CI: [0.33, 1.10]; P = 0.10), DFS (HR, 0.62; 95% CI: [0.21, 1.88]; P = 0.40), DSS (HR, 2.05; 95% CI: [0.53, 7.86]; P = 0.29 and lymph node metastasis (HR, 1.15; 95% CI: [0.74, 1.81]; P = 0.53). Furthermore, results of the meta‐analysis showed that high expression of PD‐L1 is two times more frequent in female patients (OR, 0.5; 95% CI: [0.36, 0.69]; P < 0.0001) compared to males. For all the three outcomes analysed, a high rate of heterogeneity was detected (I 2 > 50%).
Discussion
High PD‐L1 expression did not correlate with poor prognosis of patients suffering for oral squamous cell carcinoma. Studies published on the topic showed a significant variation in results, limiting the use of PD‐L1 expression by immunohistochemistry as prognostic biomarker in clinical practice.
Keywords: cancer, checkpoint inhibitors, immunity, meta‐analysis, mouth neoplasms, PD‐1, PD‐L1
1. INTRODUCTION
Oral squamous cells carcinoma (OSCC) represents one of the most common malignancies in humans.1 An annual incidence of about 200 000 new cases per year has been estimated worldwide.2 Both incidence and mortality rate are about 2.8 times higher in males than in females.3 The most known risk factors for the onset of OSCC are tobacco smoke, betel chew and alcohol consumption.4 The prognosis of OSCC is still poor, showing very little improvements in the last decades, despite advances in therapies.5 Recently, immunotherapy showed promising effects for the treatment of such patients.6 The results of several studies suggest an important role of immune evasion mechanisms in the pathogenesis of OSCC. For these reasons, a deeper understanding of molecules involved in the function of immune system is crucial for the development of future strategies of treatment.
As it is known, cancer cells can negatively regulate the immune response through the activation of inhibitory immune checkpoints. To date, different inhibitory immune checkpoints have been studied, including cytotoxic T‐lymphocyte protein 4 (CTLA4), programmed cell death protein 1 (PD‐1), lymphocyte activation gene‐3 (LAG3), T‐cell immunoglobulin‐3 (TIM3) and T‐cell immunoglobulin and ITIM domain (TIGIT).7 In this article, we focused on the PD‐1 immune checkpoint as the pharmacological inhibition of this immune checkpoints has recently demonstrated to improve the survival rate of patients with head and neck squamous cells carcinoma (HNSCC),8 while the power of evidence is still weak regarding the clinical efficacy of the pharmacological inhibition of the other immune checkpoints above mentioned. In particular, we reviewed studies focused on the analysis of the programmed cell death ligand‐1 (PD‐L1) as a prognostic factor of patients suffering for OSCC. PD‐L1 is a cell surface glycoprotein which induces both anergy and apoptosis of T cells through the activation of PD‐1 receptors located on their surface.9 The biological importance of the PD‐1 receptors influences significantly the immune responses because of a diffused ligand distribution in the body. In fact, such axis showed to play a crucial role in autoimmunity,10 tumour immunity,11 infectious immunity12 and allergy.13 PD‐L1 is commonly expressed in some healthy tissues since it is involved in the normal immunological homeostasis.14 However, in many types of cancer, the expression of PD‐L1 on tumour cells is remarkably higher. This overexpression seems to be present also in subsets of immune cells, including B and T cells, macrophages and dendritic cells.11 Several studies demonstrated a strong correlation between PD‐L1 expression on various tumour cells and a worse patients' prognosis.15, 16, 17, 18 Many studies have also been conducted to discover a possible role of the PD‐1/PD‐L1 axis in the biology of OSCC.19, 20 Its potential clinical and pathological implication has also been investigated providing, however, non‐homogeneous conclusions.
The aim of the present study was to systematically review the literature and perform a meta‐analysis on the available data in order to summarize the possible correlations between PD‐L1 expression and the prognosis of patients suffering for OSCC.
2. MATERIALS AND METHODS
2.1. Protocol and Registration
This systematic review has been carried out following the guidelines of the “Preferred Reporting Items for Systematic Reviews and Meta‐Analyses” (PRISMA) guidelines21 and the Cochrane Handbook.22 In addition, the protocol for the development of this review was prospectively registered on the online database PROSPERO (International prospective register of systematic reviews) with the registration number CRD42018090716.
2.2. Eligibility criteria
The inclusion criteria were the following: (a) both prospective and retrospective clinical cohort studies, written in English language, regarding the immunohistochemical evaluation of PD‐L1 expression in samples from OSCC patients; (b) at least 20 patients were included in each study; (c) studies which analysed the prognosis calculating the hazard ratio (HR) and its 95% confidence interval (95% CI) for at least one of the following: overall survival (OS), disease‐free survival (DFS), disease‐specific survival (DSS), gender and lymph node metastasis. Some studies reported the HR and 95% CI in the article. Others only reported the Kaplan‐Meier graph. In this case, the HR and 95% CI were extracted by Kaplan‐Meier graph using the method reported by Tierney et al.22 If the article did not report both HR and 95% CI, or the Kaplan‐Meier graph, author was contacted by email. By this last method, we got the HR and 95% CI for two studies.23, 24 Studies on non‐human model, case series with less than 20 patients and case reports were not considered for the inclusion in this review. No restrictions were applied about the year of publication.
2.3. Information sources and search strategy
Two authors (GT and KZ) performed an independent direct online search on the following databases: PUBMED, SCOPUS and Web of Science. The research process was carried out by two reviewers in an independent manner. MeSH terms and free text words were combined using Boolean operators (AND, OR). The following protocol was used: ((((PD‐L1 OR Programmed Death Ligand 1 OR checkpoint inhibitor OR immune system))) AND ((OSCC OR "oral cancer" OR Tongue OR gingiva))) AND ((survival OR prognosis OR biomarker)).
2.4. Study selection, data collection process and data items
The selection process was performed in two rounds. In the first round, authors screened the studies reading only title and abstract of publications, while in the second phase, a full‐text evaluation was performed. In case of disagreement between reviewers, a final decision for the inclusion was taken in a joint session with a third author (VCAC). This author also calculated a value of k‐statistic to show the level of reviewers' agreement. At the end of the selection process, papers fulfilling all inclusion criteria were included in the quantitative synthesis. Data extraction was performed using an ad hoc extraction sheet by two authors (VCAC and CA) in a joint session and controlled by a third author (GT). For each study, the following data were extracted: name of the first author, year of publication, name of the country where the study was performed, classification used for staging, number of patients included, cut‐off values, gender, staging, tumour size, rate of lymph node metastasis, HRs and 95% CI for the survival outcomes considered.
2.5. Risk of bias assessment
The risk of bias of the included studies was evaluated using a classification derived from the Reporting Recommendations for Tumour Marker Prognostic Studies (REMARK),25 as previously reported by Almangush et al.26 The scale consists of six parameters evaluating (a) samples, (b) clinical data of the cohort, (c) immunohistochemistry, (d) prognosis, (e) statistics and (f) classical prognostic factors. In addition, each parameter was considered as adequate, inadequate or not evaluable on the basis of the REMARKS guidelines. In addition, analysis of the risk of biases across studies was investigated through Q and I 2 tests. A P‐value of Q‐statistic <0.05 was considered significant for the presence of heterogeneity. The Higgins index was also assessed and classified as follows: low heterogeneity (<30%), medium heterogeneity (30%‐60%) and high heterogeneity (>60%).27
2.6. Summary measures and planned methods for analyses
For the pooled analysis of PD‐L1 expression as prognostic factor in OSCC patients, the natural logarithm of HR and its standard error (SE) were calculated and entered into the software: Review Manager version 5.2.8 (Cochrane Collaboration, Copenhagen, Denmark; 2014). The inverse of variance test was used to calculate the overall effect. Results of the meta‐analysis were summarized in forest plots, and a P‐value lower than 0.05 was considered as threshold of statistical significance for all the tests performed in this meta‐analysis. Sensitivity analyses were performed for the outcomes OS and DFS omitting articles on the basis of risk of bias, cut‐off and geography, hence repeating meta‐analysis through a random effect model.
3. RESULTS
3.1. Study selection
A total of 1137 records were screened by title and abstract. Of these, only 27 overcame the first selection process and were included in the full‐text evaluation. Among these, only 10 studies met the inclusion criteria and were included in the meta‐analysis.19, 23, 24, 28, 29, 30, 31, 32, 33, 34 The flow chart of the selection process is reported in Figure 1, while reasons for exclusion of the remaining 17 articles are provided in Table S2.33, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50 The value of k‐statistic was 0.8196 revealing an excellent level of agreement between reviewers (major details are available in Table S1).
Figure 1.
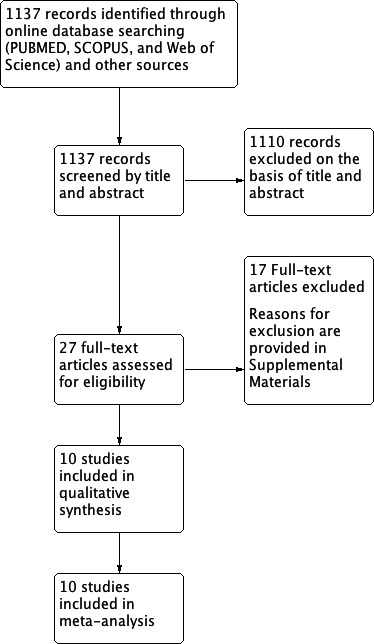
Flowchart for inclusion of studies in the meta‐analysis
3.2. Study features and risk of bias within studies
A total of 1060 patients were analysed in the 10 studies included in the meta‐analysis.19, 24, 28, 29, 30, 31, 32 Five studies were performed in Asia,19, 28, 29, 30, 33 two in Europe,32, 34 while the remaining three in other parts of the world (Brazil,24 Australia31 and USA23). The year of publication ranged from 2011 to 2018. Multivariate analysis was performed in two studies,24, 29 while the remaining eight19, 28, 30, 31, 32, 33, 34, 51 reported only results for univariate analysis. Three studies fully respected the REMARKS guidelines,19, 28, 29 while the remaining seven proved to be lacking in some of the parameters analysed.23, 24, 30, 31, 32, 33, 34, 51 Absence of risk of bias was detected only for the immunohistochemistry, while some deficiencies were present for the others parameters. Results of the risk of bias for each of the included study are reported in Table 1.
Table 1.
Evaluation criteria used to assess the quality of studies included in the meta‐analysis according to the REMARK guidelines are reported in the Almangush et al25 article.—Included Studies were evaluated as A: Adequate; I: Inadequate; N/A: no description
| Author (year) | Country | Samples | Clinical data | Immunohistochemistry | Prognostication | Statistics | Classical Prognostic Factors |
|---|---|---|---|---|---|---|---|
| Ahn (2016) | Korea | A | A | A | A | A | A |
| Cho (2011) | Korea | A | A | A | A | A | A |
| Kogashiwa (2017) | Japan | A | A | A | A | A | A |
| Lin (2015) | Taiwan | I | A | A | A | A | A |
| Oliveira‐Costa (2015) | Brazil | I | A | A | I | I | A |
| Satgunaseelan (2016) | Australia | A | A | A | I | I | I |
| Straub (2016) | Germany | A | A | A | I | I | I |
| Hirai (2016) | Japan | I | A | A | I | A | A |
| Troeltzsch (2016) | Germany | A | A | A | I | A | A |
| Mattox (2017) | USA | I | I | A | I | I | I |
3.3. Synthesis of results and risk of bias across studies
Meta‐analysis of seven studies revealed no significant correlation between high/low expression of PD‐L1 and OS (HR, 0.60; 95% CI: [0.33, 1.10]; P = 0.10). A high rate of heterogeneity was detected (I 2 = 89%), and for such reason, a random effects model was used. Meta‐analysis of studies for DFS revealed no statistical significant differences between the expression of PD‐L1 in the tumour cells and DFS (HR, 0.62; 95% CI: [0.21, 1.88]; P = 0.40). Also for DFS, results obtained on the analysis of three studies showed a high rate of heterogeneity (I2 = 81%). No significant differences were also detected for the rate of lymph node metastasis (HR, 1.15; 95% CI: [0.74, 1.81]; P = 0.53). On the basis of the extracted data, meta‐analysis was also performed for the secondary outcomes: gender and tumour size. Results for DSS (Figure S1) revealed the absence of a statistical difference between the high and low expression of PD‐L1 (HR, 2.05; 95% CI: [0.53, 7.86]; P = 0.29).The cumulative Odds Ratio (OR) for gender status showed that high expression of PD‐L1 is two times more frequent in female patients (OR, 0.5; 95% CI: [0.36, 0.69]; P < 0.0001). The rate of heterogeneity was I 2 = 0%, and for such reason, a fixed effects model was used. Summary effect size for OS did not substantially change in sensitivity analyses performed including only studies at low risk of bias (HR = 0.55 [0.24, 1.28] P = 0.17), with an equal cut‐off (intensity > 2) (HR = 0.73 [0.27, 1.98] P = 0.54) and performed only in Asia (HR = 0.55 [0.24, 1.28] P = 0.17) (Figure S2). Sensitivity analysis was not performed for DFS and DSS because of the little number of studies included. Characteristics of included studies and their relative results are summarized in Tables 2, 3 and 4.
Table 2.
Main characteristics of included studies
| Study | Year | Country | No of patients | Staging edition | Detection method | Cut‐off |
|---|---|---|---|---|---|---|
| Ahn H. | 2017 | South Korea | 68 | 7th AJCC | IHC | Intensity >2 |
| Cho Y‐A. | 2011 | South Korea | 45 | 7th AJCC | IHC | Score >2 |
| Kogashiwa Y. | 2017 | Japan | 84 | N/A | IHC | >5% of tumour cells |
| Lin Y‐M. | 2015 | Taiwan | 305 | 7th AJCC | IHC | Score >2 |
| Oliveira‐Costa J. P. | 2015 | Brazil | 96 | N/A | IHC | >5% of tumour cells |
| Satgunaseelan L. | 2016 | Australia | 217 | 7th AJCC | IHC | >5% of tumour cells |
| Straub M. | 2016 | Germany | 80 | 7th AJCC | IHC | >5% of tumour cells |
| Mattox A. K. | 2017 | USA | 53 | N/A | IHC | >1% of membranous PD‐L1 expression by tumour and/or immune cells |
| Hirai M. | 2016 | Japan | 24 | N/A | IHC | >10% of tumour cells |
| Troeltzsch M. | 2016 | Germany | 88 | 7th AJCC | IHC | Score >2 |
N/A: not reported.
Table 3.
Synthesis of data extracted from the included studies related to outcomes pooled in the meta‐analysis
| Study | Follow‐up | Overall survival | Disease‐free survival | HR estimation | ||
|---|---|---|---|---|---|---|
| HR | 95% CI | HR | 95%CI | |||
| Ahn H. | 44.3 mean (2.1 to 122 months) | 0.32 | 0.11‐0.94 | 0.25 | 0.06‐1.12 | Reported |
| Cho Y‐A. | over 125 months/not reported | 1.10 | N/A | N/A | N/A | Calculated |
| Kogashiwa Y. | 40.6 mean (3.8 to 89.6 months) | 0.256 | 0.101‐0.646 | N/A | N/A | Reported |
| Lin Y‐M. | 45,6 mean (1,2 to 133,2 months) | 1.209 | 0.890‐1.643 | N/A | N/A | Reported |
| Oliveira‐Costa J. P. | 20 mean (4 to 108 months) | 0.426 | 0.186‐0.977 | N/A | N/A | Reported |
| Satgunaseelan L. | 22 median (1 to 144 months) | N/A | N/A | 1.46 | N/A | Calculated |
| Straub M. | 31 mean (2 to 63 months) | N/A | N/A | 2.11 | 1.00‐4.43 | Calculated |
| Mattox A. K. | N/A | 1.622 | 0.5‐4.464 | N/A | N/A | Reported |
| Hirai M. | N/A | N/A | N/A | N/A | N/A | N/A |
| Troeltzsch M. | N/A | N/A | N/A | N/A | N/A | N/A |
N/A: not reported.
Table 4.
PD‐L1 expression in Lymph node metastasis (LNM) and Gender Status patients
| PD‐L1 Expression | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Study | Country | High with LNM | Low with LNM | High in male | Low in male | High in female | Low in female | High expression | Low expression |
| Ahn H. | South Korea | N/A | N/A | N/A | N/A | N/A | N/A | 45 | 23 |
| Cho Y‐A. | South Korea | 8 | 8 | 18 | 14 | 8 | 5 | 26 | 19 |
| Kogashiwa Y. | Japan | 31 | 29 | 24 | 33 | 20 | 7 | 44 | 40 |
| Lin Y‐M. | Taiwan | 52 | 64 | 93 | 143 | 40 | 29 | 133 | 172 |
| Oliveira‐Costa J. P. | Brazil | Unclear | Unclear | Unclear | Unclear | Unclear | Unclear | Unclear | Unclear |
| Satgunaseelan L. | Australia | 18 | 76 | 17 | 113 | 23 | 64 | 40 | 177 |
| Straub M. | Germany | 26 | 19 | 23 | 31 | 13 | 13 | 36 | 44 |
| Hirai M. | Japan | 1 | 4 | N/A | N/A | N/A | N/A | 13 | 11 |
| Troeltzsch M. | Germany | 18 | 27 | 13 | 35 | 13 | 27 | 26 | 62 |
| Mattox A. K. | USA | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A |
N/A: not reported; unclear: data were reported but they were not clear.
4. DISCUSSION
PD‐L1, also known as B7‐H1 or CD274, is a cell surface glycoprotein, which leads to T‐cell inactivity or apoptosis by binding PD‐1, a receptor expressed by the T lymphocytes.19 The interaction between PD‐1/PD‐L1 leads to immune system impairment through a range of mechanisms, which often differs between tumour types. Once PD‐1 binds to PD‐L1, an inhibitory signal is induced. This happens through the phosphorylation of the tyrosine residue in the immunoreceptor tyrosine‐based switch motif, leading to the recruitment of SH2‐domain containing tyrosine phosphatase 2 (SHP‐2) to the cytoplasmic domain of PD‐1, which then down‐regulates CD28‐mediated PI3K activity. These events, ultimately, lead to reduction of Akt activation, which is involved in the proliferation and cytokine production from the immunity cells.52, 53 PD‐1 activation is also linked to inhibition of the anti‐apoptotic protein Bcl‐xL.54 In OSCC, many studies showed different links between PD‐1/PD‐L1 pathway and other molecules. Chen et al reported in an in vitro study that IFN‐γ causes an increase of PD‐L1 expression on the surface of the OSCC cell line, through PKD2 signalling pathway.36 However, this seems to contradict the description of the inhibitory effect of INF‐γ on cancer proliferation, showing an opposite role as cancer immune resistance.55 Ahn et al performed an immunohistochemical study on OSCC samples demonstrating that miR‐197 expression is inversely correlated with PD‐L1 expression. This relation had been already shown in non‐small cell lung cancer (NSCLC), where miR‐197 blocks the cyclin‐dependent kinase CKS1B, which is linked to PD‐L1 expression through STAT3 signal.28 Jingjing et al56 reported that protein level of PD‐L1 in OSCC cell line is higher than normal oral mucosa cell line, while no differences were highlighted in the PD‐L1 mRNA. They justified these statements by showing that ubiquitination could be the main mechanism involved in the PD‐L1 expression in OSCC cell lines, targeting USP9X as the main molecule acting as deubiquitinase, and this mechanism leads to the PD‐L1 protein accumulation.
The literature is still lacking studies regarding action and role of PD‐1/PD‐L1 pathway in OSCC cells. Recently, there has been growing interest about the PD‐L1 expression in tumour‐associated macrophages (TAM) and fibroblasts. Next studies should integrate findings coming from both tumour and peritumoral microenvironment PD‐L1 expression to improve the understanding of its role in OSCC prognosis.
Different studies showed that tumour cells could express on their surface PD‐L1, suggesting a potential role of this protein in reducing the anti‐cancer immune response.19 These findings improved the research in anti‐cancer drug development, which could interact with the PD‐1/PD‐L1 pathway.
On November 2016, the FDA approved a new pharmacological principle, nivolumab for the treatment of recurrent or metastatic head and neck squamous cell carcinoma. Nivolumab stands for a human IgG4 PD‐1 immune checkpoint inhibitor antibody, which selectively counters the link between PD‐1 and its ligand (PD‐L1), promoting the action of T‐cell function.57 Although the promising role of these new drugs, there are still problems about their controversial activity, above all the different mechanisms, in which PD‐1/PD‐L1 could also be involved in different cancer types. For example, for the NSCLC, not all tumours expressing PD‐L1 respond to PD‐1/PD‐L1 inhibitors. Conversely, some PD‐L1‐negative tumours can respond to these agents.58 However, the predictive role of PD‐L1 expression in tumour samples is still controversial.59
In this study, we focused on the analysis of PD‐L1 expression in OSCC tissue as a prognostic (and not predictive) biomarker. In fact, such marker has demonstrated to be an independent prognostic factor in different cancer types, including NSCLC,60 renal cell carcinoma61 and breast cancer.62 However, there are conflicting evidences in relation to the prognostic value of PD‐L1 in different types of cancer.63, 64, 65 Results of this study failed to reveal a correlation between the expression of PD‐L1 in tissues and a poor prognostic of OSCC patients. For both OS and DFS, the rate of heterogeneity among studies resulted to be very high, demonstrating that the results of the included studies are strongly conflicting among each other (Figures 1 and 2). No differences were also detected for the rate of lymph node metastasis in patients with higher PD‐L1 expression (Figure 3). Our findings are in discordance with the results of a previous meta‐analysis on head and neck cancers in which authors revealed a significant association between PD‐L1 expression and poor prognosis in a subgroup analysis.66 Such discrepancy is in part due to the inclusion of the meta‐analysis of two recently published studies in which PD‐L1 expression correlated with a better prognosis.28, 29
Figure 2.

(A/B) Forest plot for the association of higher PD‐L1 expression with overall survival (A) and disease‐free survival (B)
Figure 3.
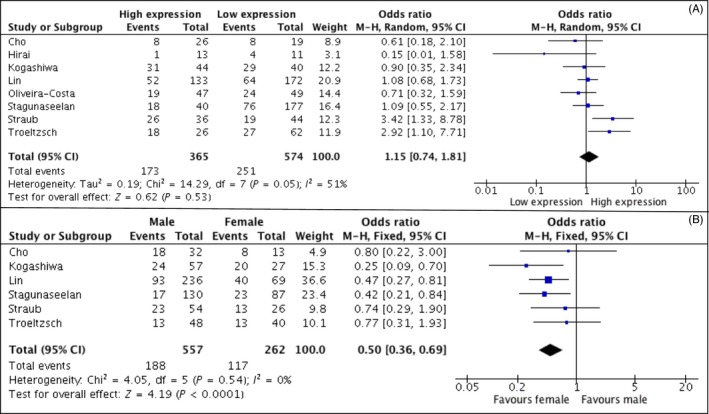
(A/B) Forest plot showing the association of higher PD‐L1 expression with lymph node metastasis (A) and gender status (B)
The lack of correlation between PD‐L1 expression and OS appears to contrast with the prognostic value that is attributed to this marker, based on its immunosuppressive function. Several studies regarding other tumour types found the same results, suggesting a more complex function of PD‐L1 in immunosurveillance signalling.67 A possible explanation is that PD‐L1 expression by cancer cells can be considered as a marker of an active host anti‐tumour immune response.68 Another way to address the issue is to consider the heterogeneity of tumour microenvironment in different tumour types. In fact, a classification of tumours into 4 types based on the presence of PD‐L1 positivity and/or tumour‐infiltrating lymphocytes has been proposed.68, 69 In some tumours like NSCLC, oncogenes may be more important drivers of tumour PD‐L1 expression compared to other tumours, like melanoma, in which it seems more influenced by infiltrating immune cells.69 Furthermore, as reported by Lyford‐Pike et al, in head and neck squamous cell carcinoma, the expression of PD‐L1 may be driven by both oncogenic and adaptive immune resistance mechanisms in the same lesion.70
Therefore, the evaluation of PD‐L1 expression alone as prognostic marker can be misleading, suggesting the need for the integration of other immune markers to obtain a better patient stratification. This action should consider the different phases, which are linked to patients' management. In this view, according to Bigras et al,71 the use of small biopsies misclassified up to the 35% of PD‐L1 assessments in advanced NSCLC. The biopsy sample undergoes different processes for the evaluation of PD‐L1 expression. De Meulenaere et al72 reported that pathologists can find hurdles in the choice of assay, antibody and cut‐off/score selection of PD‐L1 expression. In this study, authors compared the results of PD‐L1 expression coming from biopsy samples versus resection specimens and a poor agreement emerged. Another study,73 on the other hand, showed that the VENTANA PD‐L1 (SP263) assay was characterized by high reproducibility, meanwhile tumour‐infiltrating PD‐L1 immune cells were more variable within and between blocks and across cut‐offs. These data are important for the concept of precise medicine, according also to the evidence that microenvironment has an important role in PD‐L1 expression and tumour behaviour, as showed in other kinds of cancer74 and in OSCC.33, 37, 40 According to these statements, future research should focus on the validation and standardization of all steps, from biopsy, IHC assay and tumour microenvironment evaluation for the selection of patients, who can undergo anti‐PD‐L1 therapy.
In order to investigate the influence of specific parameters on the results of this study, we also performed sensitivity analysis for risk of bias, cut‐off values and geography. Summary effect size did not substantially change in sensitivity analyses performed including only studies at low risk of bias, performed in Asia and reporting the same cut‐off value. Results of this study revealed a significant association between PD‐L1 expression and female gender. In fact, in women, higher expression of PD‐L1 seems to be more common as already reported for NSCLC.75, 76 In these studies, the female subset of patient also corresponds to patients who are more likely to harbour EGFR mutations, suggesting a relationship between PD‐L1 expression and altered EGFR signalling pathway.77 A recent meta‐analysis revealed that the magnitude of benefit of patients treated with immunotherapy is sex‐dependent, and in particular, women have lower rates of positive response to the treatment.78 However, it is not clear whether such different outcomes are due to the more frequent expression of PD‐L1 in females or to other sex‐related mechanism. Such findings underline the importance of performing future studies aiming to compare sex‐related expression as independent prognostic factor, in order to clarify whether PD‐L1 could be considered a prognostic factor in men but not in women.
Furthermore, as previously mentioned, there is a complex relationship between PD‐L1 expression and the presence and pattern of inflammatory infiltrate. This must be considered in the evaluation of prognostic significance of PD‐L1, because peritumoral inflammatory process seems to be more intense in female patients with OSCC, mainly due to postmenopausal inflammatory state.19
Analysis of risk of bias in the included studies revealed deficiencies in some parameters of the REMARKS guidelines. In particular, the authors recorded ambiguity in some of the included studies in the distinction between OS and disease‐specific survival. As it is known, in the calculation of OS, death for any reason is taken into consideration while in disease‐specific survival only deaths for cancer are considered. It is to underline that direct contact of authors helped to clarify such discrepancy for two of the included studies.23, 24 To note, such meta‐analysis presents some limits, first of all, it relied on published results rather than on individual patients' data. In addition, it presented, for the survival outcomes considered in the meta‐analysis, a very high rate of heterogeneity was detected, that strongly limits the quality of evidences despite the inclusion of an adequate number of studies performed in a good quality manner. Such heterogeneity could reflect the wide variation of PD‐L1 expression in the population that limits its use as prognostic biomarker in clinical practice. It should be stressed that such results are not related to the analysis of PD‐L1 expression as predictor of response to checkpoint inhibitors, such topic should be evaluated in further studies with different design.
5. CONCLUSION
High PD‐L1 expression did not correlate with poor prognosis of patients suffering for OSCC. The studies published on the topic showed a significant variation in results. Hence, results from the current available literature limit the use of PD‐L1 expression by immunohistochemistry as prognostic biomarker in clinical practice. Higher levels of PD‐L1 expression are more frequent in females than in males, and such factor should encourage future studies on the sex‐related role of this biomarker.
CONFLICT OF INTEREST
The authors declare that they have no conflict of interests.
Supporting information
Troiano G, Caponio VCA, Zhurakivska K, et al. High PD‐L1 expression in the tumour cells did not correlate with poor prognosis of patients suffering for oral squamous cells carcinoma: A meta‐analysis of the literature. Cell Prolif. 2019;52:e12537 10.1111/cpr.12537
Vito C. A. Caponio and Khrystyna Zhurakivska contributed equally.
REFERENCES
- 1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7‐30. [DOI] [PubMed] [Google Scholar]
- 2. Troiano G, Mastrangelo F, Caponio V, Laino L, Cirillo N, Lo ML. Predictive prognostic value of tissue‐based microRNA expression in oral squamous cell carcinoma: a systematic review and meta‐analysis. J Dent Res. 2018;97(7):759‐766. [DOI] [PubMed] [Google Scholar]
- 3. Shield KD, Ferlay J, Jemal A, et al. The global incidence of lip, oral cavity, and pharyngeal cancers by subsite in 2012. CA Cancer J Clin. 2017;67(1):51‐64. [DOI] [PubMed] [Google Scholar]
- 4. Rivera C. Essentials of oral cancer. Int J Clin Exp Pathol. 2015;8(9):11884‐11894. [PMC free article] [PubMed] [Google Scholar]
- 5. Khammissa RA, Meer S, Lemmer J, Feller L. Oral squamous cell carcinoma in a South African sample: race/ethnicity, age, gender, and degree of histopathological differentiation. J Cancer Res Ther. 2014;10(4):908‐914. [DOI] [PubMed] [Google Scholar]
- 6. Ferris RL, Blumenschein G Jr, Fayette J, et al. Nivolumab for recurrent squamous‐cell carcinoma of the head and neck. N Engl J Med. 2016;375(19):1856‐1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Anderson AC, Joller N, Kuchroo VK. Lag‐3, Tim‐3, and TIGIT: co‐inhibitory receptors with specialized functions in immune regulation. Immunity. 2016;44(5):989‐1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ferris R, Gillison ML. Nivolumab for squamous‐cell cancer of head and neck. N Engl J Med. 2017;376(6):596. [DOI] [PubMed] [Google Scholar]
- 9. Okazaki T, Honjo T. PD‐1 and PD‐1 ligands: from discovery to clinical application. Int Immunol. 2007;19(7):813‐824. [DOI] [PubMed] [Google Scholar]
- 10. Kobayashi M, Kawano S, Hatachi S, et al. Enhanced expression of programmed death‐1 (PD‐1)/PD‐L1 in salivary glands of patients with Sjogren's syndrome. J Rheumatol. 2005;32(11):2156‐2163. [PubMed] [Google Scholar]
- 11. Iwai Y, Ishida M, Tanaka Y, Okazaki T, Honjo T, Minato N. Involvement of PD‐L1 on tumor cells in the escape from host immune system and tumor immunotherapy by PD‐L1 blockade. Proc Natl Acad Sci U S A. 2002;99(19):12293‐12297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Urbani S, Amadei B, Tola D, et al. PD‐1 expression in acute hepatitis C virus (HCV) infection is associated with HCV‐specific CD8 exhaustion. J Virol. 2006;80(22):11398‐11403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. James ES, Harney S, Wordsworth BP, Cookson WO, Davis SJ, Moffatt MF. PDCD1: a tissue‐specific susceptibility locus for inherited inflammatory disorders. Genes Immun. 2005;6(5):430‐437. [DOI] [PubMed] [Google Scholar]
- 14. Dong H, Zhu G, Tamada K, Chen L. B7–H1, a third member of the B7 family, co‐stimulates T‐cell proliferation and interleukin‐10 secretion. Nat Med. 1999;5(12):1365‐1369. [DOI] [PubMed] [Google Scholar]
- 15. Ohigashi Y, Sho M, Yamada Y, et al. Clinical significance of programmed death‐1 ligand‐1 and programmed death‐1 ligand‐2 expression in human esophageal cancer. Clin Cancer Res. 2005;11(8):2947‐2953. [DOI] [PubMed] [Google Scholar]
- 16. Wu C, Zhu Y, Jiang J, Zhao J, Zhang XG, Xu N. Immunohistochemical localization of programmed death‐1 ligand‐1 (PD‐L1) in gastric carcinoma and its clinical significance. Acta Histochem. 2006;108(1):19‐24. [DOI] [PubMed] [Google Scholar]
- 17. Ghebeh H, Mohammed S, Al‐Omair A, et al. The B7–H1 (PD‐L1) T lymphocyte‐inhibitory molecule is expressed in breast cancer patients with infiltrating ductal carcinoma: correlation with important high‐risk prognostic factors. Neoplasia. 2006;8(3):190‐198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hamanishi J, Mandai M, Iwasaki M, et al. Programmed cell death 1 ligand 1 and tumor‐infiltrating CD8+ T lymphocytes are prognostic factors of human ovarian cancer. Proc Natl Acad Sci U S A. 2007;104(9):3360‐3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cho YA, Yoon HJ, Lee JI, Hong SP, Hong SD. Relationship between the expressions of PD‐L1 and tumor‐infiltrating lymphocytes in oral squamous cell carcinoma. Oral Oncol. 2011;47(12):1148‐1153. [DOI] [PubMed] [Google Scholar]
- 20. Tsushima F, Tanaka K, Otsuki N, et al. Predominant expression of B7–H1 and its immunoregulatory roles in oral squamous cell carcinoma. Oral Oncol. 2006;42(3):268‐274. [DOI] [PubMed] [Google Scholar]
- 21. Liberati A, Altman DG, Tetzlaff J, et al. The PRISMA statement for reporting systematic reviews and meta‐analyses of studies that evaluate health care interventions: explanation and elaboration. J Clin Epidemiol. 2009;62(10):e1–e34. [DOI] [PubMed] [Google Scholar]
- 22. Higgins J, Green S, Cochrane Collaboration . Cochrane handbook for systematic reviews of interventions. Chichester, UK; Hoboken, NJ: Wiley‐Blackwell; 2008. [Google Scholar]
- 23. Mattox AK, Lee J, Westra WH, et al. PD‐1 expression in head and neck squamous cell carcinomas derives primarily from functionally anergic CD4(+) TILs in the presence of PD‐L1(+) TAMs. Cancer Res. 2017;77(22):6365‐6374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Oliveira‐Costa JP, de Carvalho AF, da Silveira da GG, et al. Gene expression patterns through oral squamous cell carcinoma development: PD‐L1 expression in primary tumor and circulating tumor cells. Oncotarget. 2015;6(25):20902‐20920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Altman DG, McShane LM, Sauerbrei W, Taube SE. Reporting Recommendations for Tumor Marker Prognostic Studies (REMARK): explanation and elaboration. PLoS Med. 2012;9(5):e1001216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Almangush A, Heikkinen I, Makitie AA, et al. Prognostic biomarkers for oral tongue squamous cell carcinoma: a systematic review and meta‐analysis. Br J Cancer. 2017;117(6):856‐866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Higgins JP, Thompson SG. Quantifying heterogeneity in a meta‐analysis. Stat Med. 2002;21(11):1539‐1558. [DOI] [PubMed] [Google Scholar]
- 28. Ahn H, Yang JM, Kim H, et al. Clinicopathologic implications of the miR‐197/PD‐L1 axis in oral squamous cell carcinoma. Oncotarget. 2017;8(39):66178‐66194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kogashiwa Y, Yasuda M, Sakurai H, et al. PD‐L1 expression confers better prognosis in locally advanced oral squamous cell carcinoma. Anticancer Res. 2017;37(3):1417‐1424. [DOI] [PubMed] [Google Scholar]
- 30. Lin YM, Sung WW, Hsieh MJ, et al. High PD‐L1 expression correlates with metastasis and poor prognosis in oral squamous cell carcinoma. PLoS One. 2015;10(11):e0142656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Satgunaseelan L, Gupta R, Madore J, et al. Programmed cell death‐ligand 1 expression in oral squamous cell carcinoma is associated with an inflammatory phenotype. Pathology. 2016;48(6):574‐580. [DOI] [PubMed] [Google Scholar]
- 32. Straub M, Drecoll E, Pfarr N, et al. CD274/PD‐L1 gene amplification and PD‐L1 protein expression are common events in squamous cell carcinoma of the oral cavity. Oncotarget. 2016;7(11):12024‐12034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hirai M, Kitahara H, Kobayashi Y, et al. Regulation of PD‐L1 expression in a high‐grade invasive human oral squamous cell carcinoma microenvironment. Int J Oncol. 2017;50(1):41‐48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Troeltzsch M, Woodlock T, Pianka A, et al. Is there evidence for the presence and relevance of the PD‐1/PD‐L1 pathway in oral squamous cell carcinoma? Hints from an immunohistochemical study. J Oral Maxillofac Surg. 2017;75(5):969‐977. [DOI] [PubMed] [Google Scholar]
- 35. Chen TC, Wu CT, Wang CP, et al. Associations among pretreatment tumor necrosis and the expression of HIF‐1alpha and PD‐L1 in advanced oral squamous cell carcinoma and the prognostic impact thereof. Oral Oncol. 2015;51(11):1004‐1010. [DOI] [PubMed] [Google Scholar]
- 36. Chen J, Feng Y, Lu L, et al. Interferon‐gamma‐induced PD‐L1 surface expression on human oral squamous carcinoma via PKD2 signal pathway. Immunobiology. 2012;217(4):385‐393. [DOI] [PubMed] [Google Scholar]
- 37. Foy JP, Bertolus C, Michallet MC, et al. The immune microenvironment of HPV‐negative oral squamous cell carcinoma from never‐smokers and never‐drinkers patients suggests higher clinical benefit of IDO1 and PD1/PD‐L1 blockade. Ann Oncol. 2017;28(8):1934‐1941. [DOI] [PubMed] [Google Scholar]
- 38. Fuse H, Tomihara K, Heshiki W, et al. Enhanced expression of PD‐L1 in oral squamous cell carcinoma‐derived CD11b(+)Gr‐1(+) cells and its contribution to immunosuppressive activity. Oral Oncol. 2016;59:20‐29. [DOI] [PubMed] [Google Scholar]
- 39. Hanna GJ, Woo SB, Li YY, Barletta JA, Hammerman PS, Lorch JH. Tumor PD‐L1 expression is associated with improved survival and lower recurrence risk in young women with oral cavity squamous cell carcinoma. Int J Oral Maxillofac Surg. 2018;47(5):568‐577. [DOI] [PubMed] [Google Scholar]
- 40. Jiang C, Yuan F, Wang J, Wu L. Oral squamous cell carcinoma suppressed antitumor immunity through induction of PD‐L1 expression on tumor‐associated macrophages. Immunobiology. 2017;222(4):651‐657. [DOI] [PubMed] [Google Scholar]
- 41. Katou F, Ohtani H, Watanabe Y, Nakayama T, Yoshie O, Hashimoto K. Differing phenotypes between intraepithelial and stromal lymphocytes in early‐stage tongue cancer. Cancer Res. 2007;67(23):11195‐11201. [DOI] [PubMed] [Google Scholar]
- 42. Kubota K, Moriyama M, Furukawa S, et al. CD163(+)CD204(+) tumor‐associated macrophages contribute to T cell regulation via interleukin‐10 and PD‐L1 production in oral squamous cell carcinoma. Sci Rep. 2017;7(1):1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lanzel EA, Paula Gomez Hernandez M, Bates AM, et al. Predicting PD‐L1 expression on human cancer cells using next‐generation sequencing information in computational simulation models. Cancer Immunol Immunother. 2016;65(12):1511‐1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Malaspina TS, Gasparoto TH, Costa MR, et al. Enhanced programmed death 1 (PD‐1) and PD‐1 ligand (PD‐L1) expression in patients with actinic cheilitis and oral squamous cell carcinoma. Cancer Immunol Immunother. 2011;60(7):965‐974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Poropatich K, Fontanarosa J, Swaminathan S, et al. Comprehensive T‐cell immunophenotyping and next‐generation sequencing of human papillomavirus (HPV)‐positive and HPV‐negative head and neck squamous cell carcinomas. J Pathol. 2017;243(3):354‐365. [DOI] [PubMed] [Google Scholar]
- 46. Ritprajak P, Azuma M. Intrinsic and extrinsic control of expression of the immunoregulatory molecule PD‐L1 in epithelial cells and squamous cell carcinoma. Oral Oncol. 2015;51(3):221‐228. [DOI] [PubMed] [Google Scholar]
- 47. Stasikowska‐Kanicka O, Wagrowska‐Danilewicz M, Danilewicz M. Immunohistochemical analysis of Foxp3(+), CD4(+), CD8(+) cell infiltrates and PD‐L1 in oral squamous cell carcinoma. Pathol Oncol Res. 2017;24(3):497‐505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Takahashi H, Sakakura K, Kudo T, et al. Cancer‐associated fibroblasts promote an immunosuppressive microenvironment through the induction and accumulation of protumoral macrophages. Oncotarget. 2017;8(5):8633‐8647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Weber M, Wehrhan F, Baran C, et al. PD‐L1 expression in tumor tissue and peripheral blood of patients with oral squamous cell carcinoma. Oncotarget. 2017;8(68):112584‐112597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wu L, Deng WW, Huang CF, et al. Expression of VISTA correlated with immunosuppression and synergized with CD8 to predict survival in human oral squamous cell carcinoma. Cancer Immunol Immunother. 2017;66(5):627‐636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Maruse Y, Kawano S, Jinno T, et al. Significant association of increased PD‐L1 and PD‐1 expression with nodal metastasis and a poor prognosis in oral squamous cell carcinoma. Int J Oral Maxillofac Surg. 2018;47(7):836–845. [DOI] [PubMed] [Google Scholar]
- 52. Riella LV, Paterson AM, Sharpe AH, Chandraker A. Role of the PD‐1 pathway in the immune response. Am J Transplant. 2012;12(10):2575‐2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Parry RV, Chemnitz JM, Frauwirth KA, et al. CTLA‐4 and PD‐1 receptors inhibit T‐cell activation by distinct mechanisms. Mol Cell Biol. 2005;25(21):9543‐9553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Chemnitz JM, Parry RV, Nichols KE, June CH, Riley JL. SHP‐1 and SHP‐2 associate with immunoreceptor tyrosine‐based switch motif of programmed death 1 upon primary human T cell stimulation, but only receptor ligation prevents T cell activation. J Immunol. 2004;173(2):945‐954. [DOI] [PubMed] [Google Scholar]
- 55. Nicolini A, Carpi A, Rossi G. Cytokines in breast cancer. Cytokine Growth Factor Rev. 2006;17(5):325‐337. [DOI] [PubMed] [Google Scholar]
- 56. Jingjing W, Wenzheng G, Donghua W, Guangyu H, Aiping Z, Wenjuan W. Deubiquitination and stabilization of programmed cell death ligand 1 by ubiquitin‐specific peptidase 9, X‐linked in oral squamous cell carcinoma. Cancer Med. 2018;7(8):4004‐4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Casaluce F, Sgambato A, Sacco PC, et al. Emerging drugs targeting PD‐1 and PD‐L1: reality or hope? Expert Opin Emerg Drugs. 2014;19(4):557‐569. [DOI] [PubMed] [Google Scholar]
- 58. Aguiar PN Jr, Santoro IL, Tadokoro H, et al. A pooled analysis of nivolumab for the treatment of advanced non‐small‐cell lung cancer and the role of PD‐L1 as a predictive biomarker. Immunotherapy. 2016;8(9):1011‐1019. [DOI] [PubMed] [Google Scholar]
- 59. Teng F, Meng X, Kong L, Yu J. Progress and challenges of predictive biomarkers of anti PD‐1/PD‐L1 immunotherapy: a systematic review. Cancer Lett. 2018;414:166‐173. [DOI] [PubMed] [Google Scholar]
- 60. Ma G, Deng Y, Jiang H, Li W, Wu Q, Zhou Q. The prognostic role of programmed cell death‐ligand 1 expression in non‐small cell lung cancer patients: an updated meta‐analysis. Clin Chim Acta. 2018;482:101‐107. [DOI] [PubMed] [Google Scholar]
- 61. Wang Z, Peng S, Xie H, et al. Prognostic and clinicopathological significance of PD‐L1 in patients with renal cell carcinoma: a meta‐analysis based on 1863 individuals. Clin Exp Med. 2018;18(2):165‐175. [DOI] [PubMed] [Google Scholar]
- 62. Zhang M, Sun H, Zhao S, et al. Expression of PD‐L1 and prognosis in breast cancer: a meta‐analysis. Oncotarget. 2017;8(19):31347‐31354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Tsao MS, Le Teuff G, Shepherd FA, et al. PD‐L1 protein expression assessed by immunohistochemistry is neither prognostic nor predictive of benefit from adjuvant chemotherapy in resected non‐small cell lung cancer. Ann Oncol. 2017;28(4):882‐889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Qu HX, Zhao LP, Zhan SH, et al. Clinicopathological and prognostic significance of programmed cell death ligand 1 (PD‐L1) expression in patients with esophageal squamous cell carcinoma: a meta‐analysis. J Thorac Dis. 2016;8(11):3197‐3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Zhong A, Xing Y, Pan X, Shi M, Xu H. Prognostic value of programmed cell death‐ligand 1 expression in patients with non‐small‐cell lung cancer: evidence from an updated meta‐analysis. Onco Targets Ther. 2015;8:3595‐3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Li J, Wang P, Xu Y. Prognostic value of programmed cell death ligand 1 expression in patients with head and neck cancer: a systematic review and meta‐analysis. PLoS One. 2017;12(6):e0179536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Topalian SL, Taube JM, Anders RA, Pardoll DM. Mechanism‐driven biomarkers to guide immune checkpoint blockade in cancer therapy. Nat Rev Cancer. 2016;16(5):275‐287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Taube JM, Anders RA, Young GD et al. Colocalization of inflammatory response with B7–h1 expression in human melanocytic lesions supports an adaptive resistance mechanism of immune escape. Sci Transl Med. 2012;4(127):127ra37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Teng MW, Ngiow SF, Ribas A, Smyth MJ. Classifying cancers based on T‐cell infiltration and PD‐L1. Cancer Res. 2015;75(11):2139‐2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Lyford‐Pike S, Peng S, Young GD, et al. Evidence for a role of the PD‐1:PD‐L1 pathway in immune resistance of HPV‐associated head and neck squamous cell carcinoma. Cancer Res. 2013;73(6):1733‐1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Bigras G, Mairs S, Swanson PE, Morel D, Lai R, Izevbaye I. Small Biopsies misclassify up to 35% of PD‐L1 assessments in advanced lung non‐small cell lung carcinomas. Appl Immunohistochem Mol Morphol. 2018. [Epub ahead of print]. 10.1097/PAI0000000000000698 [DOI] [PubMed] [Google Scholar]
- 72. De Meulenaere A, Vermassen T, Creytens D, et al. Importance of choice of materials and methods in PD‐L1 and TIL assessment in oropharyngeal squamous cell carcinoma. Histopathology. 2018;73(3):500‐509. [DOI] [PubMed] [Google Scholar]
- 73. Scorer P, Scott M, Lawson N, et al. Consistency of tumor and immune cell programmed cell death ligand‐1 expression within and between tumor blocks using the VENTANA SP263 assay. Diagn Pathol. 2018;13(1):47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Silva MA, Ryall KA, Wilm C, Caldara J, Grote HJ, Patterson‐Kane JC. PD‐L1 immunostaining scoring for non‐small cell lung cancer based on immunosurveillance parameters. PLoS One. 2018;13(6):e0196464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Azuma K, Ota K, Kawahara A, et al. Association of PD‐L1 overexpression with activating EGFR mutations in surgically resected nonsmall‐cell lung cancer. Ann Oncol. 2014;25(10):1935‐1940. [DOI] [PubMed] [Google Scholar]
- 76. D'Incecco A, Andreozzi M, Ludovini V, et al. PD‐1 and PD‐L1 expression in molecularly selected non‐small‐cell lung cancer patients. Br J Cancer. 2015;112(1):95‐102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Akbay EA, Koyama S, Carretero J, et al. Activation of the PD‐1 pathway contributes to immune escape in EGFR‐driven lung tumors. Cancer Discov. 2013;3(12):1355‐1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Conforti F, Pala L, Bagnardi V, et al. Cancer immunotherapy efficacy and patients' sex: a systematic review and meta‐analysis. Lancet Oncol. 2018;19(6):737‐746. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


