Abstract
Fibroblast growth factors (FGFs) are crucial signalling molecules involved in normal cell growth, differentiation and proliferation. Over the past few decades, a large body of research has illustrated effects of individual FGFs on tumour initiation and progression. Tumour development is commonly accompanied with generation of new blood and lymph vessels, which support enhanced cell proliferation. Moreover, acquisition of tumour cells of the epithelial–mesenchymal transition (EMT) phenotype, enhances tumour cell migration and invasion potentials, crucial steps in tumour metastasis. This review summarizes recent findings concerning roles of FGFs in angiogenesis, lymphangiogenesis and EMT.
1. Introduction
Fibroblast growth factors (FGFs) are a huge family of polypeptide cytokines displaying multiple functions. FGFs are involved in cell growth, angiogenesis, wound healing, tissue homoeostasis/regeneration and metabolism.1, 2 There are 23 FGF (FGF‐1–23) ligands in mammals, which are capable of binding to fibroblast growth factor receptors (FGFRs). Intriguingly, only four human FGFRs were found: FGFR‐1 to FGFR‐4. The function of FGFRs is triggered by a serial of processes that contribute to multiple isoforms through alternative initiation, alternative splicing and C‐terminal truncations.3 Recently, FGFR‐5 has also been found, but the function of this receptor still remains unclear.4 FGF family exists receptor specificities. On one hand, a part of FGFs only binds to their specific FGFRs. For example, FGF‐5 and FGF‐7 could only be combined with FGFR‐1 and FGFR‐2, respectively.5 FGF‐8 would bind to FGFR‐3 and FGFR‐4 but not bind to FGFR‐1 and FGFR‐2.6 By contrast, FGF‐10 exists high affinity with FGFR‐1/2 but not with FGFR‐3/4.7 On the other hand, there are also some FGFs that could bind with all types of FGFRs, such as FGF‐1, FGF‐2 and FGF‐4.8
The mammalian FGF family is classified into five paracrine‐ or autocrine‐acting subfamilies and one endocrine‐acting subfamily according to the sequence homology and phylogenetic and structural analysis.9 The paracrine–autocrine‐acting FGF subfamilies comprise FGF‐1 subfamily (FGF‐1 and FGF‐2), FGF‐4 subfamily (FGF‐4, FGF‐5 and FGF‐6), FGF‐8 subfamily (FGF‐8, FGF‐17 and FGF‐18), FGF‐9 subfamily (FGF‐9, FGF‐16 and FGF‐20) and FGF‐7 subfamily (FGF‐3, FGF‐7, FGF‐10 and FGF‐22). The endocrine‐acting FGFs include FGF‐15 (mouse)/FGF‐19 (human), FGF‐21 and FGF‐23 10 (Fig. 1). Different FGF subfamilies modulate a variety of biological functions and associate with various diseases, such as impaired wound healing, metabolic/chronic disease and cancer.10, 11 For instance, some paracrine–autocrine‐acting FGFs, such as FGF‐1 subfamily, FGF‐10 and FGF‐18, are involved in tissue repair, and the endocrine‐acting FGFs (FGF‐15/‐19, FGF‐21 and FGF‐23) regulate metabolism at postnatal stages.12 FGF‐15/‐19 and FGF‐21, respectively, govern bile acid metabolism in the liver and lipid metabolism in the white adipose tissue, and FGF‐23 modulates vitamin D and phosphate homoeostasis.13, 14 FGF‐2 is required for angiogenesis. Both FGF‐10 and FGF‐2 participate in epithelial–mesenchymal transition (EMT), which induces migration and invasion of tumour.15
Figure 1.
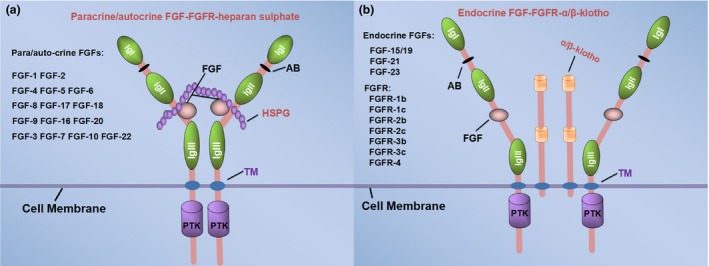
The different binding modes of paracrine and endocrine FGF ligands. The FGFs family are grouped into five paracrine–autocrine‐acting subfamilies (a) and one endocrine‐acting subfamily (b). FGF ligands is required for binding the cofactors to the receptors, and the cofactor of paracrine–autocrine FGF ligands is generally heparan sulphate proteoglycans (HSPGs), while Klotho is the co‐receptor of endocrine FGF ligands. The structure of FGFRs comprises three extracellular immunoglobulin‐like domains (Ig I to III), a transmembrane domain (TM) and an intracellular protein tyrosine kinase domain (PTK).118 The binding sites of FGFs and FGFRs located between IgII and Ig III, and the acid box (AB) composed of eight consecutive acidic residues located between IgI and Ig II. The extracellular domain of FGFRs generates the IIIb and IIIc isoforms through the alternative splicing.119, 120, 121 FGFR‐1, FGFR‐2 and FGFR‐3 all have different isoforms except FGFR‐4 (b). (These FGFRs in (b) are not endocrine FGF specific receptors, which binding with FGFs regardless of paracrine–autocrine or endocrine.) The binding of receptors with ligands leads to dimerization and activation of the tyrosine kinase domain
Cells that secrete FGF ligands also produce heparansulphate. The modified domains of heparan sulphate by sulfation and epimerization are the principal binding sites for FGFs. It is worth noting that cofactors are required for the interaction of FGFs with their receptors. Cofactor of paracrine FGF ligands is generally heparan sulphate proteoglycans (HSPGs), while Klotho is the co‐receptor of endocrine FGF ligands (Fig. 1). Binding of FGF/cofactor to FGFR results in receptor dimerization, receptor activation and finally phosphorylation of downstream molecules.
2. FGF‐mediated intracellular signalling pathways
Activation of FGFRs by tyrosine residue autophosphorylation transmits extracellular signals into multiple cytoplasmic signal transduction pathways, such as phospholipase Cγ (PLCγ),16 phosphatidylinositol‐3 kinase (PI3K)/AKT, RAS/mitogen‐activated protein kinase (MAPK), as well as signal transducer and activator of transcription (STAT) and NFκB pathways (Fig. 2), which are involved in tumour cell proliferation and migration.17
Figure 2.
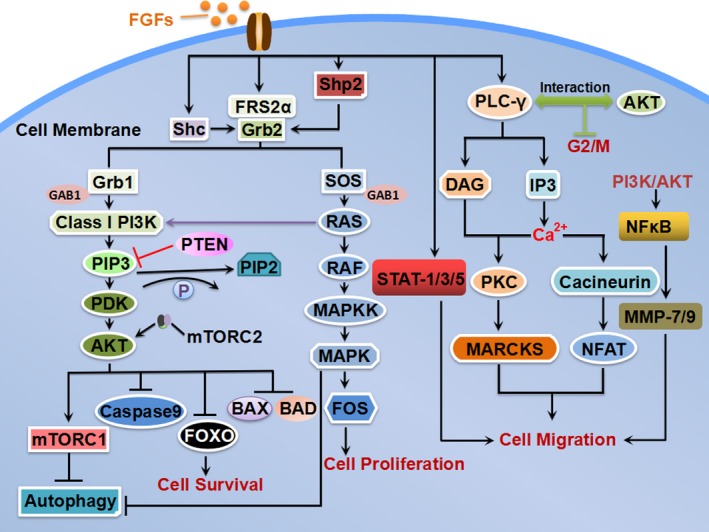
Intracellular signal transduction pathways of FGFs. Activation of FGFRs by tyrosine phosphorylation leads to signal transduction through multiple pathways, including phospholipase Cγ (PLCγ), PI3K–AKT pathway, RAS–MAPK pathway and STAT and NFκB pathway. Tumour suppressor Lipid phosphatase (PTEN) negatively regulates the PI3K signalling pathway,122 which can prevent the signal transduction by promoting dephosphorylation PIP3 into PIP2. Class I PI3K can also be stimulated by RAS, which directly binds to the p110 catalytic subunit of PI3K. Activation of the mTORC2–AKT and ERK signalling pathways promotes cell survival and invasion. AKT promotes cells survival by inhibiting BCL‐2 antagonist of cell death (BAD and BAX),forkhead box class O (FOXO) transcription factors and Caspase 9. The PI3K–Akt‐regulated mTOR is crucial for the FGFs signalling axis to suppress autophagy. Recruited SOS activates RAS GTPase, which stimulates activation of the mitogen‐activated protein kinase (MAPK) cascade and downstream FOS to induce cell proliferation. FGFR substrate 2α (FRS2α) is major substrates of FGFR kinases, which is constitutively associated with the receptor kinase and phospholipase Cγ1 (PLCγ1). Activated PLCγ1 catalyses the hydrolysis of the membrane phospholipid phosphatidylinositol‐4,5‐bisphosphate (PtdIns 4, 5(P2) into diacylglycerol (DAG) and inositol‐1,4,5,‐trisphosphate (IP3). Ca2+ can activate Cacineurin to regulate transcription factor NFAT and promote cell migration. Whereas Ca2+ together with DAG stimulates cytosolic protein kinase C (PKC), which further activates its substrate myristoylatedalanine‐rich C‐kinase substrate (MARCKS) by phosphorylation. δ‐PKC‐mediated MARCKS phosphorylation is important for cancer cell migration and adhesion. In addition, PLCγ–AKT interaction regulates the M‐phase of cell cycle. The activated FGFR stimulates STAT‐1/3/5, which induces cell migration and invasion by regulating STAT pathway target gene expression. NFκB is the downstream molecule of FGFR–PI3K–AKT pathway, which modulates MMP‐7/9 expression to increase cancer cell migration
2.1. RAS–MAPK pathway
As a major substrate of FGFR kinase, FGFR substrate 2α (FRS2α) is constitutively associated with the receptor kinase and phospholipase Cγ1 (PLCγ1).18, 19 Activated FRS2α binds with adaptor protein growth factor receptor‐bound 2 (GRB2),20 and then son of sevenless (SOS) and adaptor protein GRB2‐associated binding protein 1 (GAB1) are recruited by GRB2. Recruited SOS activates RAS GTPase, which stimulates the activation of the mitogen‐activated protein kinase (MAPK) cascade.20 As a critical signalling pathway in eukaryotic cells, RAS–MAPK enables specific phosphorylation of gene‐regulatory proteins on serine and threonine residues, thereby affecting cell proliferation and differentiation by regulating gene expression patterns.21 FGF–FGFR system could also activate AKT and MAPK pathways in a FRS2α‐dependent manner.22 It has been reported that FGF‐1 stimulates the phosphorylation of p38 MAPK (MAPK14) as well as the c‐jun N‐terminal kinase (JNK)1/2 (MAPK8/9), which is implicated in the regulation of cell apoptosis and growth arrest.23, 24 FGF‐2 has a close relationship with PKC and Ca2+ 25. FGF‐1‐mediated Egr‐1 induction is impaired by the inhibition of MEK‐1/2.26 FGF‐16 enhances the proliferation of human ovarian adenocarcinoma cells SKOV‐3 and OAW‐42 through the activation of FGFR‐mediated intracellular MAPK pathway.27 FGF‐7 and FGF‐10 are also involved in the proliferation of ameloblastoma cells through the MAPK pathway.28 Overall, these findings suggest that FGF‐mediated intracellular signalling pathways may represent the common mechanisms regulating tumour cell proliferation.29
2.2. PI3K–AKT pathway
FGFRs can activate substrate FRS2α, which combines with GRB1/2 and forms a ternary complex to activate class I phosphatidylinositol‐3 kinase (PI3K) and AKT.30 Activated AKT kinase suppresses pro‐apoptotic effectors, such as the BCL‐2 antagonist of cell death (BAD), forkhead box class O (FOXO) transcription factors and caspases, thereby promoting cell survival.31, 32 In addition, AKT regulates cell cycle, protein synthesis, cell proliferation and differentiation by activating mTOR. A number of studies have indicated the role of FGF‐regulated PI3K–AKT signalling pathway in tumorigenesis.33, 34, 35 FGFR‐3 plays a causative role in urothelial cancer pathogenesis in PTEN‐deficient mice.36 Similarly, the translational activation of FGF‐10 by PTEN deletion is reversed by genetic disruption of the mTORC1 complex, leading to the prevention of skin tumorigenesis. In another study,37 it was demonstrated that FGF‐2 could regulate G2/M phase of cell cycle by MEK, PI3K and PKC‐activated FGFR–RAS–SRC pathway. Based on these results, it can be concluded that FGF–FGFR system is involved in the activation of PI3K pathway and plays critical roles in tumour development and progression.38
2.3. PLCγ–PKC and Ca2+ channels
Activated PLCγ1 catalyses the hydrolysis of the membrane phospholipid phosphatidylinositol‐4,5‐bisphosphate (PtdIns (4,5) P2) into diacylglycerol (DAG) and inositol‐1,4,5,‐trisphosphate (IP3).18 As second messengers, DAG and IP3 regulate different downstream pathway, respectively. IP3 combines with IP3 receptors which locate at endoplasmic reticulum (ER), resulting in opening calcium channels. Then, Ca2+ is released from ER and translocates into the cytoplasm, which leads to the accumulation of free Ca2+ in cytoplasmic matrix. Ca2+ activates a variety of Ca2+‐dependent proteins, such as CaM (calmodulin) and Cacineurin. Cacineurin can regulate transcription factor nuclear factor of activated T cells (NFAT) to promote cell migration. Ca2+ together with DAG stimulates cytosolic protein kinase C(PKC), which further activates its substrate myristoylated alanine‐rich C‐kinase substrate (MARCKS). It has been found that δ‐PKC‐mediated MARCKS phosphorylation is essential for cancer cell migration and adhesion.39 Activation of the FGFs stimulates PLCγ–PKC pathways, which might be involved in cell proliferation, cell survival and metastasis of tumour cells.40 Browaeys‐Poly et al.41 found that the disruption of PLCγ–AKT interaction accelerated entry into M‐phase of the mitotic cycle. This fact indicated that maintaining PLCγ–AKT interaction triggered by FGFRs might inhibit the cell cycle M‐phase entry.41 Furthermore, FGF‐2 increases N‐cadherin expression by regulating PLCγ–PKC and Src‐kinase pathways, which promotes cell–cell adhesion.42
2.4. STAT and NFκB pathway
The activated FGFRs also stimulate STAT (STAT‐1, STAT‐3 and STAT‐5) and NFκB pathway, two signalling molecules that regulate the expression of STAT pathway target gene and matrix metalloproteinase (MMP), respectively.43 FGF–FGFR signalling phosphorylates and activates NFκB through PI3K–AKT pathway, which might induce cancer cell invasion by stimulating MMP. For example, FGF‐1/3 is reported to promote tumour progression in colon cancer through ERK and MMP‐7 and induce MMP‐9 expression through the NFκB pathway.44 FGF–FGFR pathway also regulates cell migration, invasion and growth arrest by activating STATs.45
2.5. Tendentiousness of intracellular pathways activated by FGFs
Although these signalling pathways mentioned above are commonly activated by FGFs in most cell types, FGFs might have different tendentiousness in regulating these signalling pathways. RAS–MAPK pathway activated by FGFs appears to ubiquitously exist in all cell types, while the various activity of the other three pathways in response to FGFs depends on the cell types.46, 47 In a certain cell type, on one hand, a part of FGFs positively activate a signal transduction pathway, while other FGFs might not be involved in its activation. For instance, FGF‐2 has a close relationship with PLCγ–PKC and Ca2+, while FGF‐1 has no effect on translocation of PLCγ in ovarian granulosa cells.25 FGF‐2 and FGF‐4 rapidly increase AKT and ERK1/2 phosphorylation in bovine granulosa cells, whereas FGF‐10 appears not able to trigger typical FGF signalling pathways, such as PI3K–AKT.47 On the other hand, a FGF may have different correlation with various pathways in a certain cell type. For example, FGF‐1‐mediated Egr‐1 induction in mouse hippocampal neuronal cell is impaired by inhibition of MEK‐1/2, but not of PI3K.26 Another investigation supports that FGF‐1 regulates cardiogenesis primarily in a mouse embryonic stem cell through the signalling of PKC, but not MAPK.48
3. Aberrant expression of FGFs in tumour
It is a feature that FGFs and their receptors are overexpressed in different types of human cancers. For instance, compared with normal ovarian surface epithelium (OSE), FGF‐18 is overexpressed in serous ovarian tumours and modulates ovarian tumour aggressiveness as well as microenvironment by increasing production of oncogenic cytokines and chemokines.49 Overproduction of FGF‐23 from the causative tumours is the main cause of tumour‐induced rickets/osteomalacia (TIO), whereas FGF‐23 production in normal bone is suppressed.50 FGF‐1 and FGF‐6 are undetectable in normal prostate, but their increased expression is observed in prostate cancers.51 FGF‐2 significantly has a higher expression level in cancer tissue when compared with normal prostate.51
The ectopic overexpression of FGFs usually correlates with tumorigenesis as well as poor prognosis. For example, the aberrant activation of FGF‐1 signalling is not only implicated in tumorigenesis but also associated with tumour invasion and metastasis.52 FGF‐2 in tumour cells is an independent negative prognostic factor. The co‐expression of FGF‐2/VEGFR‐3 and FGFR‐1/PDGF‐B is strongly associated with poor survival in patients with non–small‐cell lung carcinoma (NSCLC).53 FGF‐8b promotes cell cycle progression through the G1 restriction point and regulates key proteins that take part in chromosomal segregation during mitosis and cytokinesis of breast cancer cells.54 Thus, it is essential to consider the regulation of FGFs in the development of tumour and cancer therapeutics.
4. FGFs in the generation of new blood vessels and lymphangiogenesis
Numerous studies show that angiogenesis is one of the early events in the malignant transformation. The balance between endogenous activators and inhibitors of angiogenesis delicately maintains a normal quiescent vasculature to sustain homoeostasis. Disturbance of this balance causes pathogenic angiogenesis. FGF and vascular endothelial growth factor (VEGF) are two angiogenic factors produced by tumours to stimulate angiogenesis, both of them have been reported to be correlated with tumour growth, progression and metastasis55, 56 (Fig. 3). VEGFs which belong to the platelet‐derived growth factor super gene family play central roles in the modulation of angiogenesis and lymphangiogenesis.57, 58 In addition, FGFs are commonly reported to modulate angiogenesis through a variety of approaches. FGF‐1 and FGF‐2 are considered as major angiogenic factors among FGFs, which have been discovered currently.59 FGF‐1 induces angiogenesis in the chicken chorioallantoic membrane (CAM) through AKT–PKB signalling.60 FGF‐1 participating in vascular remodelling in endometriotic angiogenesis contributes to vascular wall formation and migration of endothelial cells (ECs) and vascular smooth muscle cells (VSMCs).61 FGF‐2 could stimulate angiogenesis via a VEGFR‐3‐independent pathway.50 FGF‐2 priming enhances the angiogenic potential of implanted tissue‐engineered constructs through the secretion of both hepatocyte growth factor (HGF) and VEGF.62 In addition, interleukin‐1β (IL‐1β) induces the expression of FGF‐2 in chondrocytes through ROS–AMPK–p38–NF‐κB signalling pathway, which subsequently increases endothelial progenitor cell (EPC) angiogenesis.63 FGFs also have been justified to play crucial roles in driving angiogenesis so that the formation of new blood vessels could assist in “feeding” cancer.64 There is a growing body of evidence implicating FGF‐1 and FGF‐2 in active angiogenesis and rapid tumour growth. For instance, cardiac‐specific overexpression of FGF‐1 contributes to angiogenesis‐independent cardioprotection.65 Active FGF‐2‐NDY1/EZH2‐miR‐101‐EZH2 axis is described to induce cell proliferation, migration and angiogenesis in bladder cancer.66
Figure 3.
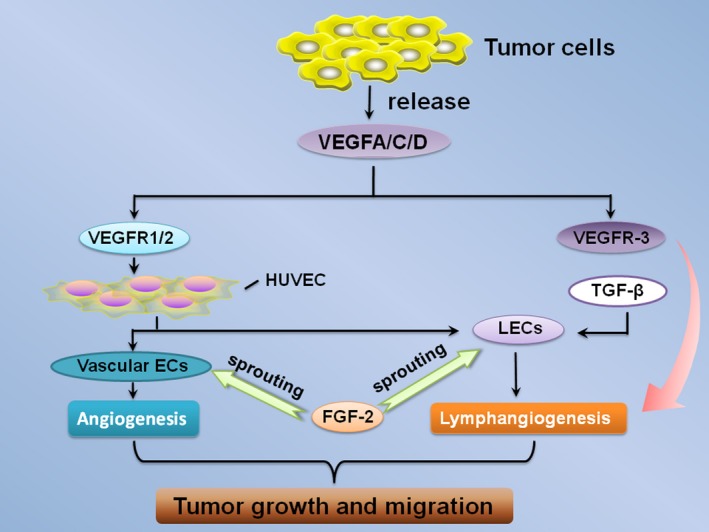
FGF‐2 in tumour angiogenesis and lymphangiogenesis. VEGF‐A/C/D and FGF‐2 are commonly expressed in various tumour tissues and their expression levels have been correlated with tumour growth, progression, and metastasis. The receptors of VEGF‐A, VEGFR‐1 (Flt‐1) and VEGFR‐2 (KDR/Flk‐1) could induce proliferation, migration and vessel formation of HUVECs.57, 66 Furthermore, the activation of VEGF‐C/VEGFR‐3 signalling results in lymphangiogenesis, and VEGFR‐3‐induced tip formation is a prerequisite for FGF‐2‐stimulated lymphangiogenesis. Moreover, significant new insights such as transforming growth factor β (TGF‐β) regulate the growth and remodelling of lymphatic vessels. FGF‐2 stimulates the sprouting of vascular ECs and LECs (lymphatic endothelial cells) which are isolated from human umbilical vein ECs (HUVECs). Vascular ECs and LECs independently induces angiogenesis and lymphangiogenesis
Lymphangiogenesis and the remodelling of existing lymphatics accomplish the generation of new lymphatic vessels.67 Lymphangiogenesis, which is similar to tumour angiogenesis in the molecular control, is distinctly an early step in lymphatic metastasis.67, 68, 69 In addition to the central role of VEGF‐C–VEGFR‐3 signalling in lymphangiogenesis,70 FGF‐2 may also be responsible for the growth and remodelling of lymphatic vasculature.70 The study by Cao et al.71 showed that FGF‐2 and VEGF‐C collaboratively facilitate corneal angiogenesis and lymphangiogenesis and independently stimulate lymphatic vascular endothelial cell (LEC) proliferation and migration. It has been found that FGF‐2 may simultaneously provoke lymphangiogenesis in different locations of the cornea through differential expression of VEGF ligands.72 According to a report, siRNA‐mediated FGFR‐1 knockdown abolishes FGF‐2‐mediated LEC proliferation.73
Evidence shows that FGFs are involved in tumour development by directly and indirectly regulating tumour angiogenesis.74 In addition, FGFs can also act in a paracrine manner on tumour lymphatics by facilitating the expression of prolymph angiogenic molecules.75 In general, FGFs might be the mediator or interact with other signal molecules such as VEGFs in most of cancers to promote angiogenesis and lymphangiogenesis. Thus, targeting FGFs may be a potential strategy to impair tumour progression.
5. FGFs in tumour invasion and metastasis
The development of cancer can be generally divided into two stages: invasion and metastasis of tumour cells.76 FGFs could mediate PLCγ–PKC and Ca2+ pathway to promote cell invasion.77 FGFs also play a role in tumour invasion and metastasis by interacting with other signalling molecules.78 For example, FGF‐2 participates in melanoma progression and cooperates with Thrombospondin‐1 (TSP‐1) in determining melanoma invasion and metastasis.79 Abolished nuclear FGF‐2 and FGFR‐1 inhibit pancreatic stellate cells (PSCs) invasion.80 FGF‐8 promotes colorectal cancer growth and metastasis by activating YAP1.81 FGF‐10 has also been reported to promote migration and invasion in pancreatic cancer cells.15 Thus, it can be concluded that FGFs could be the inducer of tumour invasion and metastasis.
Acquisition of the EMT phenotype of tumour cells not only enhances their invasion potentials but also promotes their capacity of metastasis.82 It is suggested that FGFs might induce EMT through downstream signalling pathways including RAS–MAPK–AKT–PI3K–mTOR and PLCγ–PKC, thus enhancing tumour cell metastasis.83 For instance, FGF‐10 may induce EMT through RAS–MAPK and AKT–PI3K–mTOR pathways.84 Additionally, TGF‐β and FGF‐2 may cooperate with each other and regulate EMT in various kinds of cells. TGF‐β1 induces EMT through Smad pathway85 and stimulates the isoform switching of FGF receptors, leading to the epithelial–myofibroblastic transition (EMyoT) by inactivating the MEK‐ERK pathway, thus causing the cells to be sensitive to FGF‐2.86 FGF‐2 disturbs EMyoT by reactivating the MEK‐ERK pathway and subsequently enhances EMT through the formation of MEK‐ERK‐dependent complexes87 (Fig. 4). TGF‐β1 and FGF‐2 can stimulate the EMT of HERS cells which is reversed by the MEK1/2 inhibitor U0126, suggesting that TGF‐β1 and FGF‐2 induce the EMT of HERS cells through a MAPK/ERK‐dependent signalling pathway.88 In addition, FGF‐2 alone could induce EMT in colon cancer cells.89 However, FGFs may facilitate EMT by increasing the expression of various mesenchymal factors and reducing the expression of epithelial markers.20 FGF‐16 has been reported to regulate the expression of MMP‐2, MMP‐9, SNAI1 and CDH1, which facilitates cell migration.27 FGF‐9 can be associated with EMT and metastasis by increasing the expression of N‐cadherin and VEGF‐A in prostate cancer cells.90 All these investigations demonstrate that FGFs could induce EMT to promote tumour invasion and metastasis.
Figure 4.
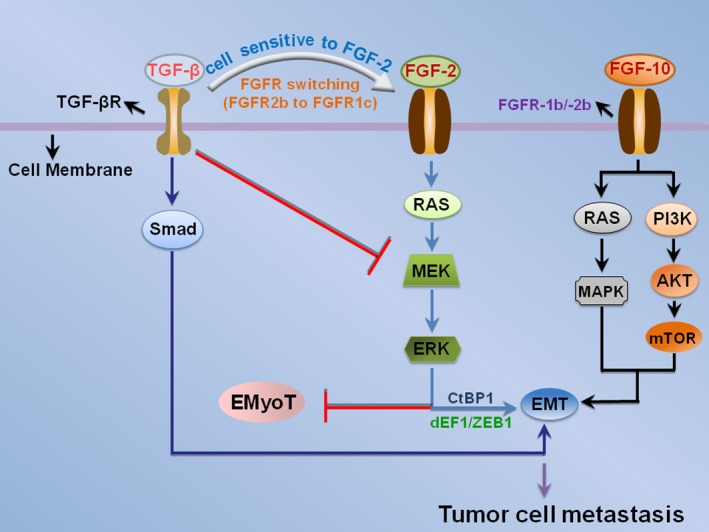
The interaction between FGF and TGF‐β acts on EMT. TGF‐β induces EMT and the isoform switching of FGF receptors (FGFR2b to FGFR1c), causing the cells to be sensitive to FGF‐2. In this context, epithelial–myofibroblastic transition (EMyoT) is induced through the inactivation of MEK‐ERK pathway. In the presence of FGF‐2, FGF‐2 perturbed EMyoT by reactivating MEK–ERK pathway and subsequently enhanced EMT through the formation of dEF1–ZEB1–CtBP1 complexes. FGF‐10 may also induce EMT by activating RAS–MAPK and AKT–PI3K–mTOR pathways
6. Targeting FGF–FGFR system for cancer therapy
Previous studies have reported that FGFs and FGFRs are significantly overexpressed in various kinds of cancers,91, 92, 93 and it is demonstrated that FGFs are involved in the regulation of cancer cell proliferation, survival and migration.94 Inhibitors and antibodies directly targeting different FGF ligands have shown therapeutic promise in different tumours.95, 96 Recently, several studies in vivo and in vitro have suggested that blocking FGF pathways could reduce the proliferation of tumour cells and inhibit metastasis.97 FGF‐19 is significantly overexpressed in hepatocellular carcinoma (HCC), and the introduction of FGF‐19 siRNA is able to reduce proliferation and increase apoptosis in HCC.98 In addition, the neutralizing antibody of FGF‐19 treatment significantly suppresses the growth of established colon cancer tumours in vivo.99 According to Schulze's study,100 RNAi‐mediated FGF‐binding protein (FGF‐BP) knockdown is integrated in the inhibition of colon carcinoma cell proliferation and induction of apoptosis alterations in redox status. FGF‐Trap distinctly abolishes FGF‐2‐stimulated activation of FGFs signalling and potently inhibits tumour growth and angiogenesis.101
Since mutations and amplifications of FGFRs are found in a range of cancers with some of the most striking clinical findings relating to their contribution to pathogenesis and progression of cancers,102, 103 FGFR‐targeted agents are currently being investigated in clinical studies for the treatment of cancer.104, 105, 106 For instance, cediranib, which is a multi‐tyrosine kinase inhibitor targeting FGFR and is in phase II evaluation, has proved as a monotherapy for recurrent or persistent endometrial cancer well tolerated.107 There are other inhibitors of FGFR, such as TKI258 and lucitanib. Phase I study shows that TKI258 is tolerable and has antitumour activity in renal cell carcinoma (RCC).108 Lucitanib is a confirmed and promising drug of treating advanced solid tumours in phase I/IIa study.109 In addition, the activity of AZD‐4547, a novel and potent FGFR kinase inhibitor in CRC cells, is correlated with the FGFR‐1/2 expression levels and inhibits CRC cell growth in vitro.110 The new non‐ATP competitive FGFR‐1 inhibitors A114 and A117 reveal significant anti‐tumour activity both in vitro and in vivo via targeting FGFR‐1.111 Although FGFR‐targeted inhibitors efficiently suppress tumour, there are still challenges. For example, TKI‐mediated durable clinical responses may be impacted by intrinsic tumour resistance.112 Furthermore, the median duration response of nintedanib (a inhibitor of FGFR) is only 4 months, and its function might be resisted by tumour through up‐regulating FGF signalling.113
A research shows that specific FGFR‐4‐targeted antibodies decreases the tumorigenic and invasive capabilities of colorectal cancer cells by reducing the expressions of Snail, Twist and TGF‐β and increasing the expression of E‐cadherin.114 Ablation of FGFR‐4 results in a net inhibitory effect on mammary tumour progression.115 However, FGFR‐4 deletion does not lead to an embryonic lethal phenotype, suggesting the possibility that its inhibition in cancer therapy might not cause grave adverse effects.116, 117 FGFR‐4‐mediated hormonal effects of several FGF ligands may also constitute a tissue‐protective tumour suppressor activity in liver.117 These results suggest that blocking FGF receptors might act different roles compared with FGF ligands in various carcinomas. Therefore, a systemic therapy that targets FGFs and FGFRs may be a potential strategy to inhibit tumour progression and metastatic spread.
7. Conclusion
Previous studies have provided plenty of evidence to support the fact that FGFs linked with FGFRs can activate their downstream crucial signalling pathways, including phospholipase Cγ (PLCγ), PI3K–AKT pathway, RAS–MAPK pathway, and STAT and NFκB pathway. We have described the signal transduction mechanism of these four pathways clearly in this review. In fact, these signalling pathways mentioned above are classic pathways of a majority of FGFs. However, FGFs might have different tendentiousness in regulating these signalling pathways between various cell types. Close link exists between FGF–FGFR system and tumour progression. With further comprehension of the molecular mechanisms of FGFs signalling and the development of more specific agents targeting FGFs and FGFRs, it is anticipated that an improved understanding of FGFs family and better anti‐cancer therapies that modulate FGF–FGFR signalling will emerge.
Conflict of interest
The authors declare no conflict of interest.
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (81302205 and 81402245), the Key Fund Project of Sichuan Provincial Department of Education (15ZA0250), and the University Innovation Research and Training Program of Chengdu Medical College (CXJS201412).
References
- 1. Sandhu DS, Baichoo E, Roberts LR. Fibroblast growth factor signaling in liver carcinogenesis. Hepatology. 2014;59:1166–1173. [DOI] [PubMed] [Google Scholar]
- 2. Li X, Wang C, Xiao J, McKeehan WL, Wang F. Fibroblast growth factors, old kids on the new block. Semin Cell Dev Biol. 2016;53:155–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Korc M, Friesel RE. The role of fibroblast growth factors in tumor growth. Curr Cancer Drug Targets. 2009;9:639–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Tran TA, Leong HS, Pavia‐Jimenez A, Fedyshyn S, Yang J, Kucejova B, et al. FGFR‐Dependent and ‐Independent Paracrine Signaling by Sunitinib‐Resistant RCC. Mol Cell Biol. 2016;36:1836–1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dalet A, Vigneron N, Stroobant V, Hanada K, Van den Eynde BJ. Splicing of distant peptide fragments occurs in the proteasome by transpeptidation and produces the spliced antigenic peptide derived from fibroblast growth factor‐5. J Immunol. 2010;184:3016–3024. [DOI] [PubMed] [Google Scholar]
- 6. Shimizu O, Yasumitsu T, Shiratsuchi H, Oka S, Watanabe T, Saito T, et al. Immunolocalization of FGF‐2, ‐7, ‐8, ‐10 and FGFR‐1‐4 during regeneration of the rat submandibular gland. J Mol Histol. 2015;46:421–429. [DOI] [PubMed] [Google Scholar]
- 7. Tong L, Zhou J, Rong L, Seeley EJ, Pan J, Zhu X, et al. Fibroblast Growth Factor‐10 (FGF‐10) Mobilizes Lung‐resident Mesenchymal Stem Cells and Protects Against Acute Lung Injury. Sci Rep. 2016;6:21642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yin F, Du R, Kuang W, Yang G, Wang H, Deng F, et al. Characterization of the viral fibroblast growth factor homolog of Helicoverpa armigera single nucleopolyhedrovirus. Virol Sin. 2016;31:240–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Goetz R, Mohammadi M. Exploring mechanisms of FGF signalling through the lens of structural biology. Nat Rev Mol Cell Biol. 2013;14:166–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhang J, Li Y. Therapeutic uses of FGFs. Semin Cell Dev Biol. 2016;53:144–154. [DOI] [PubMed] [Google Scholar]
- 11. Zhang F, Yu L, Lin X, Cheng P, He L, Li X, et al. Minireview: Roles of Fibroblast Growth Factors 19 and 21 in Metabolic Regulation and Chronic Diseases. Mol Endocrinol. 2015;29:1400–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nies VJ, Sancar G, Liu W, van Zutphen T, Struik D, Yu RT, et al. Fibroblast Growth Factor Signaling in Metabolic Regulation. Front Endocrinol (Lausanne). 2015;6:193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Degirolamo C, Sabba C, Moschetta A. Therapeutic potential of the endocrine fibroblast growth factors FGF19, FGF21 and FGF23. Nat Rev Drug Discov. 2016;15:51–69. [DOI] [PubMed] [Google Scholar]
- 14. Gallego‐Escuredo JM, Domingo P, Gutierrez Mdel M, Mateo MG, Cabeza MC, Fontanet A, et al. Reduced levels of serum FGF19 and impaired expression of receptors for endocrine FGFs in adipose tissue from HIV‐infected patients. J Acquir Immune Defic Syndr. 2012;61:527–534. [DOI] [PubMed] [Google Scholar]
- 15. Itoh N, Ohta H. Fgf10: a paracrine‐signaling molecule in development, disease, and regenerative medicine. Curr Mol Med. 2014;14:504–509. [DOI] [PubMed] [Google Scholar]
- 16. Hart KC, Robertson SC, Donoghue DJ. Identification of tyrosine residues in constitutively activated fibroblast growth factor receptor 3 involved in mitogenesis, Stat activation, and phosphatidylinositol 3‐kinase activation. Mol Biol Cell. 2001;12:931–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Katoh M, Nakagama H. FGF receptors: cancer biology and therapeutics. Med Res Rev. 2014;34:280–300. [DOI] [PubMed] [Google Scholar]
- 18. Carpenter G, Ji Q. Phospholipase C‐gamma as a signal‐transducing element. Exp Cell Res. 1999;253:15–24. [DOI] [PubMed] [Google Scholar]
- 19. Gotoh N. Regulation of growth factor signaling by FRS2 family docking/scaffold adaptor proteins. Cancer Sci. 2008;99:1319–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kouhara H, Hadari YR, Spivak‐Kroizman T, Schilling J, Bar‐Sagi D, Lax I, et al. A lipid‐anchored Grb2‐binding protein that links FGF‐receptor activation to the Ras/MAPK signaling pathway. Cell. 1997;89:693–702. [DOI] [PubMed] [Google Scholar]
- 21. Cargnello M, Roux PP. Activation and function of the MAPKs and their substrates, the MAPK‐activated protein kinases. Microbiol Mol Biol Rev. 2011;75:50–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhang J, Liu J, Liu L, McKeehan WL, Wang F. The fibroblast growth factor signaling axis controls cardiac stem cell differentiation through regulating autophagy. Autophagy. 2012;8:690–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chaffee BR, Hoang TV, Leonard MR, Bruney DG, Wagner BD, Dowd JR, et al. FGFR and PTEN signaling interact during lens development to regulate cell survival. Dev Biol. 2016;410:150–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Raju R, Palapetta SM, Sandhya VK, Sahu A, Alipoor A, Balakrishnan L. A Network Map of FGF‐1/FGFR Signaling System. J Signal Transduct. 2014;2014:962962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lin J, Jia Y, Zeng W, Mi Y, Zhang C. Basic FGF promotes proliferation of ovarian granulosa cells in the laying chickens via FGFR1 and PKC pathway. Reprod Domest Anim. 2012;47:135–142. [DOI] [PubMed] [Google Scholar]
- 26. Benz AH, Shajari M, Peruzki N, Dehghani F, Maronde E. Early growth response‐1 induction by fibroblast growth factor‐1 via increase of mitogen‐activated protein kinase and inhibition of protein kinase B in hippocampal neurons. Br J Pharmacol. 2010;160:1621–1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Basu M, Mukhopadhyay S, Chatterjee U, Roy SS. FGF16 promotes invasive behavior of SKOV‐3 ovarian cancer cells through activation of mitogen‐activated protein kinase (MAPK) signaling pathway. J Biol Chem. 2014;289:1415–1428. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 28. Nakao Y, Mitsuyasu T, Kawano S, Nakamura N, Kanda S, Nakamura S. Fibroblast growth factors 7 and 10 are involved in ameloblastoma proliferation via the mitogen‐activated protein kinase pathway. Int J Oncol. 2013;43:1377–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Murphy T, Hori S, Sewell J, Gnanapragasam VJ. Expression and functional role of negative signalling regulators in tumour development and progression. Int J Cancer. 2010;127:2491–2499. [DOI] [PubMed] [Google Scholar]
- 30. Lamothe B, Yamada M, Schaeper U, Birchmeier W, Lax I, Schlessinger J. The docking protein Gab1 is an essential component of an indirect mechanism for fibroblast growth factor stimulation of the phosphatidylinositol 3‐kinase/Akt antiapoptotic pathway. Mol Cell Biol. 2004;24:5657–5666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Brunet A, Bonni A, Zigmond MJ, Lin MZ, Juo P, Hu LS, et al. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell. 1999;96:857–868. [DOI] [PubMed] [Google Scholar]
- 32. Datta SR, Dudek H, Tao X, Masters S, Fu H, Gotoh Y, et al. Akt phosphorylation of BAD couples survival signals to the cell‐intrinsic death machinery. Cell. 1997;91:231–241. [DOI] [PubMed] [Google Scholar]
- 33. Chen GJ, Weylie B, Hu C, Zhu J, Forough R. FGFR1/PI3K/AKT signaling pathway is a novel target for antiangiogenic effects of the cancer drug fumagillin (TNP‐470). J Cell Biochem. 2007;101:1492–1504. [DOI] [PubMed] [Google Scholar]
- 34. Di Serio C, Doria L, Pellerito S, Prudovsky I, Micucci I, Massi D, et al. The release of fibroblast growth factor‐1 from melanoma cells requires copper ions and is mediated by phosphatidylinositol 3‐kinase/Akt intracellular signaling pathway. Cancer Lett. 2008;267:67–74. [DOI] [PubMed] [Google Scholar]
- 35. Vandermoere F, El Yazidi‐Belkoura I, Adriaenssens E, Lemoine J, Hondermarck H. The antiapoptotic effect of fibroblast growth factor‐2 is mediated through nuclear factor‐kappaB activation induced via interaction between Akt and IkappaB kinase‐beta in breast cancer cells. Oncogene. 2005;24:5482–5491. [DOI] [PubMed] [Google Scholar]
- 36. Foth M, Ahmad I, van Rhijn BW, van der Kwast T, Bergman AM, King L, et al. Fibroblast growth factor receptor 3 activation plays a causative role in urothelial cancer pathogenesis in cooperation with Pten loss in mice. J Pathol. 2014;233:148–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Salotti J, Dias MH, Koga MM, Armelin HA. Fibroblast growth factor 2 causes G2/M cell cycle arrest in ras‐driven tumor cells through a Src‐dependent pathway. PLoS ONE. 2013;8:e72582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ghayad SE, Cohen PA. Inhibitors of the PI3K/Akt/mTOR pathway: new hope for breast cancer patients. Recent Pat Anticancer Drug Discov. 2010;5:29–57. [DOI] [PubMed] [Google Scholar]
- 39. Sheats MK, Sung EJ, Adler KB, Jones SL. In Vitro Neutrophil Migration Requires Protein Kinase C‐Delta (delta‐PKC)‐Mediated Myristoylated Alanine‐Rich C‐Kinase Substrate (MARCKS) Phosphorylation. Inflammation. 2015;38:1126–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Salazar L, Kashiwada T, Krejci P, Muchowski P, Donoghue D, Wilcox WR, et al. A novel interaction between fibroblast growth factor receptor 3 and the p85 subunit of phosphoinositide 3‐kinase: activation‐dependent regulation of ERK by p85 in multiple myeloma cells. Hum Mol Genet. 2009;18:1951–1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Browaeys‐Poly E, Perdereau D, Lescuyer A, Burnol AF, Cailliau K. Akt interaction with PLC(gamma) regulates the G(2)/M transition triggered by FGF receptors from MDA‐MB‐231 breast cancer cells. Anticancer Res. 2009;29:4965–4969. [PubMed] [Google Scholar]
- 42. Debiais F, Lemonnier J, Hay E, Delannoy P, Caverzasio J, Marie PJ. Fibroblast growth factor‐2 (FGF‐2) increases N‐cadherin expression through protein kinase C and Src‐kinase pathways in human calvaria osteoblasts. J Cell Biochem. 2001;81:68–81. [DOI] [PubMed] [Google Scholar]
- 43. Yang X, Qiao D, Meyer K, Friedl A. Signal transducers and activators of transcription mediate fibroblast growth factor‐induced vascular endothelial morphogenesis. Cancer Res. 2009;69:1668–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bai YP, Shang K, Chen H, Ding F, Wang Z, Liang C, et al. FGF‐1/‐3/FGFR4 signaling in cancer‐associated fibroblasts promotes tumor progression in colon cancer through Erk and MMP‐7. Cancer Sci. 2015;106:1278–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ornitz DM, Itoh N. The Fibroblast Growth Factor signaling pathway. Wiley Interdiscip Rev Dev Biol. 2015;4:215–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Dailey L, Ambrosetti D, Mansukhani A, Basilico C. Mechanisms underlying differential responses to FGF signaling. Cytokine Growth Factor Rev. 2005;16:233–247. [DOI] [PubMed] [Google Scholar]
- 47. Jiang Z, Price CA. Differential actions of fibroblast growth factors on intracellular pathways and target gene expression in bovine ovarian granulosa cells. Reproduction. 2012;144:625–632. [DOI] [PubMed] [Google Scholar]
- 48. Lin HY, Lee DC, Wang HD, Chi YH, Chiu IM. Activation of FGF1B Promoter and FGF1 Are Involved in Cardiogenesis Through the Signaling of PKC, but Not MAPK. Stem Cells Dev. 2015;24:2853–2863. [DOI] [PubMed] [Google Scholar]
- 49. Wei W, Mok SC, Oliva E, Kim SH, Mohapatra G, Birrer MJ. FGF18 as a prognostic and therapeutic biomarker in ovarian cancer. J Clin Invest. 2013;123:4435–4448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Fukumoto S. Diagnostic Modalities for FGF23‐Producing Tumors in Patients with Tumor‐Induced Osteomalacia. Endocrinol Metab (Seoul). 2014;29:136–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kwabi‐Addo B, Ozen M, Ittmann M. The role of fibroblast growth factors and their receptors in prostate cancer. Endocr Relat Cancer. 2004;11:709–724. [DOI] [PubMed] [Google Scholar]
- 52. Yang X, Friedl A. A positive feedback loop between prolactin and STAT5 promotes angiogenesis. Adv Exp Med Biol. 2015;846:265–280. [DOI] [PubMed] [Google Scholar]
- 53. Donnem T, Al‐Shibli K, Al‐Saad S, Busund LT, Bremnes RM. Prognostic impact of fibroblast growth factor 2 in non‐small cell lung cancer: coexpression with VEGFR‐3 and PDGF‐B predicts poor survival. J Thorac Oncol. 2009;4:578–585. [DOI] [PubMed] [Google Scholar]
- 54. Nilsson EM, Brokken LJ, Narvi E, Kallio MJ, Harkonen PL. Identification of fibroblast growth factor‐8b target genes associated with early and late cell cycle events in breast cancer cells. Mol Cell Endocrinol. 2012;358:104–115. [DOI] [PubMed] [Google Scholar]
- 55. Nguyen M, Watanabe H, Budson AE, Richie JP, Folkman J. Elevated levels of the angiogenic peptide basic fibroblast growth factor in urine of bladder cancer patients. J Natl Cancer Inst. 1993;85:241–242. [DOI] [PubMed] [Google Scholar]
- 56. Van der Auwera I, Cao Y, Tille JC, Pepper MS, Jackson DG, Fox SB, et al. First international consensus on the methodology of lymphangiogenesis quantification in solid human tumours. Br J Cancer. 2006;95:1611–1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Shibuya M. Vascular endothelial growth factor and its receptor system: physiological functions in angiogenesis and pathological roles in various diseases. J Biochem. 2013;153:13–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Yamamizu K, Hamada Y, Narita M. kappa Opioid receptor ligands regulate angiogenesis in development and in tumours. Br J Pharmacol. 2015;172:268–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Gerwins P, Skoldenberg E, Claesson‐Welsh L. Function of fibroblast growth factors and vascular endothelial growth factors and their receptors in angiogenesis. Crit Rev Oncol Hematol. 2000;34:185–194. [DOI] [PubMed] [Google Scholar]
- 60. Forough R, Weylie B, Patel C, Ambrus S, Singh US, Zhu J. Role of AKT/PKB signaling in fibroblast growth factor‐1 (FGF‐1)‐induced angiogenesis in the chicken chorioallantoic membrane (CAM). J Cell Biochem. 2005;94:109–116. [DOI] [PubMed] [Google Scholar]
- 61. Hayrabedyan S, Kyurkchiev S, Kehayov I. FGF‐1 and S100A13 possibly contribute to angiogenesis in endometriosis. J Reprod Immunol. 2005;67:87–101. [DOI] [PubMed] [Google Scholar]
- 62. Gorin C, Rochefort GY, Bascetin R, Ying H, Lesieur J, Sadoine J, et al. Priming Dental Pulp Stem Cells With Fibroblast Growth Factor‐2 Increases Angiogenesis of Implanted Tissue‐Engineered Constructs Through Hepatocyte Growth Factor and Vascular Endothelial Growth Factor Secretion. Stem Cells Transl Med. 2016;5:392–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Chien SY, Huang CY, Tsai CH, Wang SW, Lin YM, Tang CH. Interleukin‐1beta induces fibroblast growth factor 2 expression and subsequently promotes endothelial progenitor cell angiogenesis in chondrocytes. Clin Sci (Lond). 2016;130:667–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Gavalas NG, Liontos M, Trachana SP, Bagratuni T, Arapinis C, Liacos C, et al. Angiogenesis‐related pathways in the pathogenesis of ovarian cancer. Int J Mol Sci. 2013;14:15885–15909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Buehler A, Martire A, Strohm C, Wolfram S, Fernandez B, Palmen M, et al. Angiogenesis‐independent cardioprotection in FGF‐1 transgenic mice. Cardiovasc Res. 2002;55:768–777. [DOI] [PubMed] [Google Scholar]
- 66. Huang SW, Lien JC, Kuo SC, Huang TF. Antiangiogenic mechanisms of PJ‐8, a novel inhibitor of vascular endothelial growth factor receptor signaling. Carcinogenesis. 2012;33:1022–1030. [DOI] [PubMed] [Google Scholar]
- 67. Stacker SA, Williams SP, Karnezis T, Shayan R, Fox SB, Achen MG. Lymphangiogenesis and lymphatic vessel remodelling in cancer. Nat Rev Cancer. 2014;14:159–172. [DOI] [PubMed] [Google Scholar]
- 68. Loffredo S, Staiano RI, Granata F, Genovese A, Marone G. Immune cells as a source and target of angiogenic and lymphangiogenic factors. Chem Immunol Allergy. 2014;99:15–36. [DOI] [PubMed] [Google Scholar]
- 69. Qiu X, Yao S, Zhang S. Advances in the research on lymphangiogenesis in carcinoma tissues (Review). Oncol Lett. 2010;1:579–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Coso S, Bovay E, Petrova TV. Pressing the right buttons: signaling in lymphangiogenesis. Blood. 2014;123:2614–2624. [DOI] [PubMed] [Google Scholar]
- 71. Cao R, Ji H, Feng N, Zhang Y, Yang X, Andersson P, et al. Collaborative interplay between FGF‐2 and VEGF‐C promotes lymphangiogenesis and metastasis. Proc Natl Acad Sci USA. 2012;109:15894–15899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Hajrasouliha AR, Sadrai Z, Chauhan SK, Dana R. b‐FGF induces corneal blood and lymphatic vessel growth in a spatially distinct pattern. Cornea. 2012;31:804–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Kazenwadel J, Secker GA, Betterman KL, Harvey NL. In vitro assays using primary embryonic mouse lymphatic endothelial cells uncover key roles for FGFR1 signalling in lymphangiogenesis. PLoS ONE. 2012;7:e40497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Rousseau B, Larrieu‐Lahargue F, Javerzat S, Guilhem‐Ducleon F, Beermann F, Bikfalvi A. The tyrp1‐Tag/tyrp1‐FGFR1‐DN bigenic mouse: a model for selective inhibition of tumor development, angiogenesis, and invasion into the neural tissue by blockade of fibroblast growth factor receptor activity. Cancer Res. 2004;64:2490–2495. [DOI] [PubMed] [Google Scholar]
- 75. Larrieu‐Lahargue F, Welm AL, Bouchecareilh M, Alitalo K, Li DY, Bikfalvi A, et al. Blocking Fibroblast Growth Factor receptor signaling inhibits tumor growth, lymphangiogenesis, and metastasis. PLoS ONE. 2012;7:e39540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Mukaida N, Sasaki S. Fibroblasts, an inconspicuous but essential player in colon cancer development and progression. World J Gastroenterol, 2016;22:5301–5316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Wang ZG, Wang Y, Huang Y, Lu Q, Zheng L, Hu D, et al. bFGF regulates autophagy and ubiquitinated protein accumulation induced by myocardial ischemia/reperfusion via the activation of the PI3K/Akt/mTOR pathway. Sci Rep. 2015;5:9287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Venero Galanternik M, Kramer KL, Piotrowski T. Heparan Sulfate Proteoglycans Regulate Fgf Signaling and Cell Polarity during Collective Cell Migration. Cell Rep. 2015; pii: S2211‐1247(14)01096‐1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Borsotti P, Ghilardi C, Ostano P, Silini A, Dossi R, Pinessi D, et al. Thrombospondin‐1 is part of a Slug‐independent motility and metastatic program in cutaneous melanoma, in association with VEGFR‐1 and FGF‐2. Pigment Cell Melanoma Res. 2015;28:73–81. [DOI] [PubMed] [Google Scholar]
- 80. Coleman SJ, Chioni AM, Ghallab M, Anderson RK, Lemoine NR, Kocher HM, et al. Nuclear translocation of FGFR1 and FGF2 in pancreatic stellate cells facilitates pancreatic cancer cell invasion. EMBO Mol Med. 2014;6:467–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Liu R, Huang S, Lei Y, Zhang T, Wang K, Liu B, et al. FGF8 promotes colorectal cancer growth and metastasis by activating YAP1. Oncotarget. 2015;6:935–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Liu H, Zhang X, Li J, Sun B, Qian H, Yin Z. The biological and clinical importance of epithelial‐mesenchymal transition in circulating tumor cells. J Cancer Res Clin Oncol. 2015;141:189–201. [DOI] [PubMed] [Google Scholar]
- 83. Xu W, Yang Z, Lu N. A new role for the PI3K/Akt signaling pathway in the epithelial‐mesenchymal transition. Cell Adh Migr. 2015;9:317–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Abolhassani A, Riazi GH, Azizi E, Amanpour S, Muhammadnejad S, Haddadi M, et al. FGF10: Type III Epithelial Mesenchymal Transition and Invasion in Breast Cancer Cell Lines. J Cancer. 2014;5:537–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Ramos C, Becerril C, Montano M, Garcia‐De‐Alba C, Ramirez R, Checa M, et al. FGF‐1 reverts epithelial‐mesenchymal transition induced by TGF‐{beta}1 through MAPK/ERK kinase pathway. Am J Physiol Lung Cell Mol Physiol. 2010;299:L222–L231. [DOI] [PubMed] [Google Scholar]
- 86. Shirakihara T, Horiguchi K, Miyazawa K, Ehata S, Shibata T, Morita I, et al. TGF‐beta regulates isoform switching of FGF receptors and epithelial‐mesenchymal transition. EMBO J. 2011;30:783–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Horiguchi K, Shirakihara T, Nakano A, Imamura T, Miyazono K, Saitoh M. Role of Ras signaling in the induction of snail by transforming growth factor‐beta. J Biol Chem. 2009;284:245–253. [DOI] [PubMed] [Google Scholar]
- 88. Chen J, Chen G, Yan Z, Guo Y, Yu M, Feng L, et al. TGF‐beta1 and FGF2 stimulate the epithelial‐mesenchymal transition of HERS cells through a MEK‐dependent mechanism. J Cell Physiol. 2014;229:1647–1659. [DOI] [PubMed] [Google Scholar]
- 89. Sakuma K, Aoki M, Kannagi R. Transcription factors c‐Myc and CDX2 mediate E‐selectin ligand expression in colon cancer cells undergoing EGF/bFGF‐induced epithelial‐mesenchymal transition. Proc Natl Acad Sci U S A. 2012;109:7776–7781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Teishima J, Yano S, Shoji K, Hayashi T, Goto K, Kitano H, et al. Accumulation of FGF9 in prostate cancer correlates with epithelial‐to‐mesenchymal transition and induction of VEGF‐A expression. Anticancer Res. 2014;34:695–700. [PubMed] [Google Scholar]
- 91. Williams SV, Hurst CD, Knowles MA. Oncogenic FGFR3 gene fusions in bladder cancer. Hum Mol Genet. 2013;22:795–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Valencia T, Joseph A, Kachroo N, Darby S, Meakin S, Gnanapragasam VJ. Role and expression of FRS2 and FRS3 in prostate cancer. BMC Cancer. 2011;11:484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Moussavi M, Moshgabadi N, Fazli L, Leblanc E, Zhang K, Jia W, et al. Fibroblast growth factor and ornithine decarboxylase 5'UTRs enable preferential expression in human prostate cancer cells and in prostate tumors of PTEN(‐/‐) transgenic mice. Cancer Gene Ther. 2012;19:19–29. [DOI] [PubMed] [Google Scholar]
- 94. Helsten T, Schwaederle M, Kurzrock R. Fibroblast growth factor receptor signaling in hereditary and neoplastic disease: biologic and clinical implications. Cancer Metastasis Rev. 2015;34:479–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Lien IC, Horng LY, Hsu PL, Wu CL, Sung HC, Wu RT. Internal ribosome entry site of bFGF is the target of thalidomide for IMiDs development in multiple myeloma. Genes Cancer. 2014;5:127–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Rosli SN, Shintani T, Toratani S, Usui E, Okamoto T. 1alpha,25(OH)(2)D(3) inhibits FGF‐2 release from oral squamous cell carcinoma cells through down‐regulation of HBp17/FGFBP‐1. In Vitro Cell Dev Biol Anim. 2014;50:802–806. [DOI] [PubMed] [Google Scholar]
- 97. Pu D, Hou M. Advanced research of fibroblast growth factor receptor in non‐small cell lung cancer. Zhongguo Fei Ai Za Zhi. 2013;16:609–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Miura S, Mitsuhashi N, Shimizu H, Kimura F, Yoshidome H, Otsuka M, et al. Fibroblast growth factor 19 expression correlates with tumor progression and poorer prognosis of hepatocellular carcinoma. BMC Cancer. 2012;12:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Desnoyers LR, Pai R, Ferrando RE, Hotzel K, Le T, Ross J, et al. Targeting FGF19 inhibits tumor growth in colon cancer xenograft and FGF19 transgenic hepatocellular carcinoma models. Oncogene. 2008;27:85–97. [DOI] [PubMed] [Google Scholar]
- 100. Schulze D, Plohmann P, Hobel S, Aigner A. Anti‐tumor effects of fibroblast growth factor‐binding protein (FGF‐BP) knockdown in colon carcinoma. Mol Cancer. 2011;10:144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Li D, Wei X, Xie K, Chen K, Li J, Fang J. A novel decoy receptor fusion protein for FGF‐2 potently inhibits tumour growth. Br J Cancer. 2014;111:68–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Fearon AE, Gould CR, Grose RP. FGFR signalling in women's cancers. Int J Biochem Cell Biol. 2013;45:2832–2842. [DOI] [PubMed] [Google Scholar]
- 103. Cappelletti MR, Gnetti L, Santini D, Spada D, Fox SB, Generali D. Would the combination of everolimus with endocrine‐therapy help in FGFR2 positive serous endometrial cancer? Oncoscience. 2015;2:567–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. di Martino E, Tomlinson DC, Knowles MA. A Decade of FGF Receptor Research in Bladder Cancer: Past, Present, and Future Challenges. Adv Urol. 2012;2012:429213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Ronca R, Giacomini A, Rusnati M, Presta M. The potential of fibroblast growth factor/fibroblast growth factor receptor signaling as a therapeutic target in tumor angiogenesis. Expert Opin Ther Targets. 2015;19:1361–1377. [DOI] [PubMed] [Google Scholar]
- 106. Ling L, Tan SK, Goh TH, Cheung E, Nurcombe V, van Wijnen AJ, et al. Targeting the heparin‐binding domain of fibroblast growth factor receptor 1 as a potential cancer therapy. Mol Cancer. 2015;14:136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Bender D, Sill MW, Lankes HA, Reyes HD, Darus CJ, Delmore JE, et al. A phase II evaluation of cediranib in the treatment of recurrent or persistent endometrial cancer: An NRG Oncology/Gynecologic Oncology Group study. Gynecol Oncol. 2015;138:507–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Angevin E, Lopez‐Martin JA, Lin CC, Gschwend JE, Harzstark A, Castellano D, et al. Phase I study of dovitinib (TKI258), an oral FGFR, VEGFR, and PDGFR inhibitor, in advanced or metastatic renal cell carcinoma. Clin Cancer Res. 2013;19:1257–1268. [DOI] [PubMed] [Google Scholar]
- 109. Soria JC, DeBraud F, Bahleda R, Adamo B, Andre F, Dienstmann R, et al. Phase I/IIa study evaluating the safety, efficacy, pharmacokinetics, and pharmacodynamics of lucitanib in advanced solid tumors. Ann Oncol. 2014;25:2244–2251. [DOI] [PubMed] [Google Scholar]
- 110. Yao TJ, Zhu JH, Peng DF, Cui Z, Zhang C, Lu PH. AZD‐4547 exerts potent cytostatic and cytotoxic activities against fibroblast growth factor receptor (FGFR)‐expressing colorectal cancer cells. Tumour Biol. 2015;36:5641–5648. [DOI] [PubMed] [Google Scholar]
- 111. Wang Y, Cai Y, Ji J, Liu Z, Zhao C, Zhao Y, et al. Discovery and identification of new non‐ATP competitive FGFR1 inhibitors with therapeutic potential on non‐small‐cell lung cancer. Cancer Lett. 2014;344:82–89. [DOI] [PubMed] [Google Scholar]
- 112. Ho TH, Liu XD, Huang Y, Warneke CL, Johnson MM, Hoang A, et al. The impact of FGFR1 and FRS2alpha expression on sorafenib treatment in metastatic renal cell carcinoma. BMC Cancer. 2015;15:304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Norden AD, Schiff D, Ahluwalia MS, Lesser GJ, Nayak L, Lee EQ, et al. Phase II trial of triple tyrosine kinase receptor inhibitor nintedanib in recurrent high‐grade gliomas. J Neurooncol. 2015;121:297–302. [DOI] [PubMed] [Google Scholar]
- 114. Peláz‐García A, Barderas R, Torres S, Hernández‐Varas P, Teixidó J, Bonilla F, et al. FGFR4 role in epithelial‐mesenchymal transition and its therapeutic value in colorectal cancer. PLoS ONE. 2013;8:e63695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Luo Y, Yang C, Ye M, Jin C, Abbruzzese JL, Lee MH, et al. Deficiency of metabolic regulator FGFR4 delays breast cancer progression through systemic and microenvironmental metabolic alterations. Cancer Metab. 2013;1:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Li H, Martin A, David V, Quarles LD. Compound deletion of Fgfr3 and Fgfr4 partially rescues the Hyp mouse phenotype. Am J Physiol Endocrinol Metab. 2011;300:E508–E517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Heinzle C, Erdem Z, Paur J, Grasl‐Kraupp B, Holzmann K, Grusch M, et al. Is fibroblast growth factor receptor 4 a suitable target of cancer therapy? Curr Pharm Des. 2014;20:2881–2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Coutu DL, Galipeau J. Roles of FGF signaling in stem cell self‐renewal, senescence and aging. Aging (Albany NY). 2011;3:920–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Miki T, Bottaro DP, Fleming TP, Smith CL, Burgess WH, Chan AM, et al. Determination of ligand‐binding specificity by alternative splicing: two distinct growth factor receptors encoded by a single gene. Proc Natl Acad Sci U S A. 1992;89:246–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Orr‐Urtreger A, Bedford MT, Burakova T, Arman E, Zimmer Y, Yayon A, et al. Developmental localization of the splicing alternatives of fibroblast growth factor receptor‐2 (FGFR2). Dev Biol. 1993;158:475–486. [DOI] [PubMed] [Google Scholar]
- 121. Wuechner C, Nordqvist AC, Winterpacht A, Zabel B, Schalling M. Developmental expression of splicing variants of fibroblast growth factor receptor 3 (FGFR3) in mouse. Int J Dev Biol. 1996;40:1185–1188. [PubMed] [Google Scholar]
- 122. Chandrasekaran S, Deng H, Fang Y. PTEN deletion potentiates invasion of colorectal cancer spheroidal cells through 3D Matrigel. Integr Biol (Camb). 2015;7:324–334. [DOI] [PubMed] [Google Scholar]


