Vancomycin-resistant Enterococcus faecium (VREfm) is a frequent cause of nosocomial outbreaks. In the second half of 2015, a sharp increase in the incidence of VREfm was observed at our university medical center.
KEYWORDS: hospital epidemiology, transmission routes, vancomycin resistance, whole-genome sequencing
ABSTRACT
Vancomycin-resistant Enterococcus faecium (VREfm) is a frequent cause of nosocomial outbreaks. In the second half of 2015, a sharp increase in the incidence of VREfm was observed at our university medical center. Next-generation sequencing (NGS) was used to analyze the first isolates of VREfm recovered from patients between 2010 and 2016 (n = 773) in order to decipher epidemiological change, outbreak dynamics, and possible transmission routes. VREfm isolates were analyzed using whole-genome sequencing followed by sequence type extraction and phylogenetic analysis. We examined epidemiological data, room occupancy data, and patient transferals and calculated an intensity score for patient-to-patient contact. Phylogenetic analysis revealed the presence of 38 NGS clusters and 110 single clones. The increase of VREfm was caused mainly by the expansion of two newly introduced NGS clusters, comprising VanB-type strains determined by multilocus sequence typing (MLST) as sequence type 80 (ST80) and ST117. By combining phylogenetic information with epidemiological data, intrahospital transmission could be demonstrated, however to a lesser extent than initially expected based solely on epidemiological data. The outbreak clones were continuously imported from other hospitals, suggesting a change in the epidemiological situation at a regional scale. By tracking intrahospital patient transferals, two major axes could be identified that contributed to the spread of VREfm within the hospital. NGS-based outbreak analysis revealed a dramatic change in the local and regional epidemiology of VREfm, emphasizing the role of health care networks in the spread of VREfm.
INTRODUCTION
Enterococci are an important cause of hospital-acquired infections. They are responsible for 6.7% of intensive care unit (ICU)-acquired bloodstream infections and were among the top five causes of ICU-acquired pneumonia and urinary tract infection (UTI) episodes in Europe in 2016 (1). Particularly worrisome are infections caused by vancomycin-resistant Enterococcus faecium (VREfm), which have limited treatment options and have been associated with longer hospital stays and higher hospitalization costs (2) and mortality in some studies (3, 4). Therefore, the occurrence of VREfm in hospitals currently represents one of the major concerns in infection control. Patients colonized with VREfm can function as sources for further dissemination through the shedding of vancomycin-resistant enterococci (VRE) in the hospital environment. Rooms occupied by VREfm patients become quickly contaminated, while VREfm can persist on inanimate surfaces for months (5, 6). Subsequently, patients or contaminated surfaces can lead to the contamination of the hands, gloves, or gowns of health care workers, thus facilitating transmission to other patients via personnel (5, 7). A basic reproductive rate of 1.32 based on 10 outbreaks was recently reported, confirming the epidemic nature of VREfm (8). Outbreaks involving both VanA-type and VanB-type VREfm strains have been reported from various countries (9–14), including two smaller outbreaks involving our hospital in 2001 (15) and in 2004 to 2005 (16).
In Germany, an uneven geographical distribution of reported VREfm cases was noted, with significantly higher proportions of VREfm in the federal states in the middle of Germany than in the northern and southern parts of Germany, where detection of VREfm continued at a lower level (17). In line with these findings, the detection of VREfm remained a rare event until 2015 at our 1,500-bed university hospital located in the federal state of Baden-Wuerttemberg in southwest Germany. However, starting in October 2015, we observed a sharp increase in VREfm, which led to an outbreak involving 561 patients by the end of 2016.
In order to explain the outbreak and its dynamic, all VREfm first isolates recovered from January 2010 to December 2016 (n = 773) were subjected to whole-genome sequencing (WGS). We also gathered exhaustive epidemiological data. Combining the information, we aimed to (i) characterize the clones involved in the outbreak, (ii) determine the rate of intrahospital transmission, and (iii) elucidate the impact of VREfm introduction into the hospital, as well as intrahospital patient transfers, on the spread of VREfm on a single-patient basis.
To the best of our knowledge, this represents the first resolution of a large-scale VanB-type VREfm outbreak based on WGS combined with patient-to-patient contact data and patient transferal data.
RESULTS
Outbreak dynamics and strain selection.
Beginning in October 2015, we observed an increase in the number of patients colonized or infected with VREfm in our hospital, representing a complete change in the epidemiological situation. From October 2015 until December 2016 (outbreak period), 561 patients were newly identified as having been colonized or infected with VREfm, whereas only 235 patients with VREfm were detected from January 2010 until September 2015 (baseline period). In total, 560 and 213 VREfm isolates were available for further analysis from the outbreak and the baseline periods, respectively. The following results refer to these 773 VREfm first isolates, including 607 (78.5%) VanB-type and 166 (21.5%) VanA-type VREfm isolates.
To rule out that increased VREfm detection was solely attributable to enhanced screening efforts, we calculated the rate of VREfm infection in clinically relevant specimens (blood cultures, intraoperative samples, ascites, and aspirates). During the baseline period, 0.94 patients/month were newly identified with VREfm from these sites. This rate increased to 5.20 patients/month during the outbreak period, revealing that the overall burden of VREfm indeed increased drastically during this time frame.
Phylogenetic analysis.
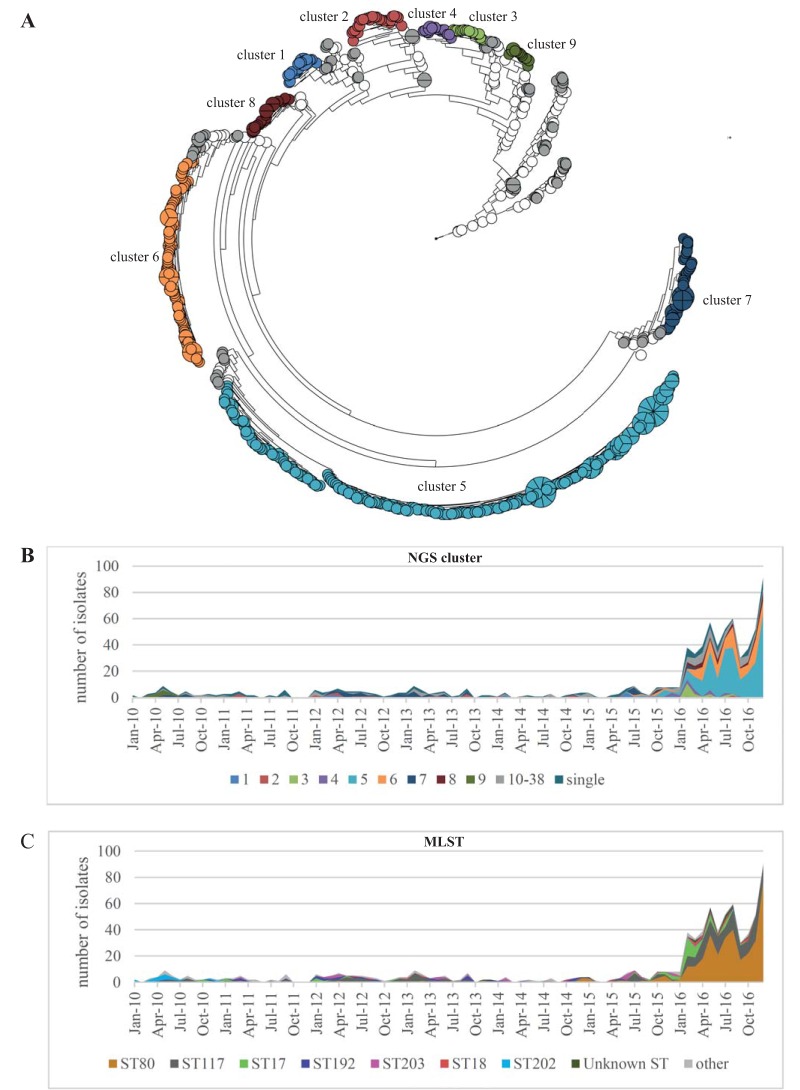
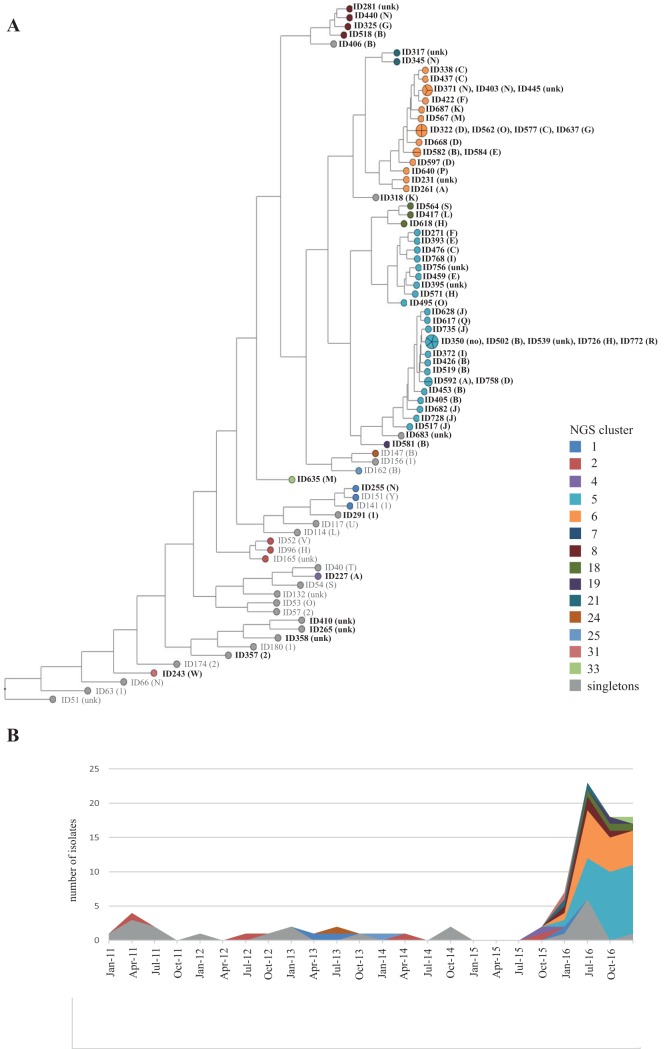
In total, the first VRE isolate of 773 hospitalized patients was subjected to whole-genome sequencing. A phylogeny was calculated based on pairwise similarity values (unweighted pair group method using average linkages [UPGMA]) and is displayed in Fig. 1A. The strains were grouped in next-generation sequencing (NGS) clusters defined as strains with a maximum single nucleotide polymorphism (SNP) difference of 13 SNPs (see Fig. S1 in the supplemental material). This approach allowed us to group the strains in 38 different NGS clusters, with cluster 5 (n = 275) and cluster 6 (n = 106) being the largest (Table S1). The subdivision of the NGS clusters resembled to a great extent the different clades observed in the maximum likelihood phylogeny based on core genome SNPs (Fig. S2). Interestingly, the increase observed from October 2015 onwards was mainly due to the expansion of the NGS clusters 5 and 6, which were observed in our patient population for the first time only shortly before this date (Fig. 1B). Since typing is often still based on multilocus sequence typing (MLST), the sequence types (STs) were extracted from the WGS sequencing data. The four largest were ST80 (n = 346), ST117 (n = 200), ST17 (n = 54), and ST192 (n = 38) (Fig. 1C and Table S1). Core genome phylogenies of ST80 and ST117 showed that both STs comprised genetically diverse strains in which homologous recombination had occurred to various extents (Fig.S3 and S4). For example, ST80 comprised strains of cluster 5 (vanB) and strains of cluster 8 (vanB), strains from smaller NGS clusters, and single isolates (Fig. S3). ST117 included the majority of cluster 6 and cluster 7 strains but also smaller NGS clusters and singletons (Fig. S4).
FIG 1.
(A) Phylogeny of 773 VREfm study isolates based on pairwise comparison of core genome SNP similarities and clustering using unweighted pair group method using average linkages (UPGMA). The branch length is shown on a logarithmic scale. In total, 38 NGS clusters were defined based on SNP differences of the isolates. NGS clusters 1 to 9 comprised more than 10 isolates each and are displayed in different colors, whereas the NGS clusters 10 to 38 with fewer than 10 isolates are displayed in gray. White circles represent singletons. (B and C) Temporal distribution of NGS clusters and MLST of the isolates over time. The increase of VREfm cases during the outbreak was mainly due to the expansion of isolates of NGS clusters 5 and 6. Interestingly, NGS cluster 5 and cluster 6 were not present in our hospital until June and July 2016, respectively, shortly before the outbreak started.
Intrahospital expansion of the predominant NGS clusters.
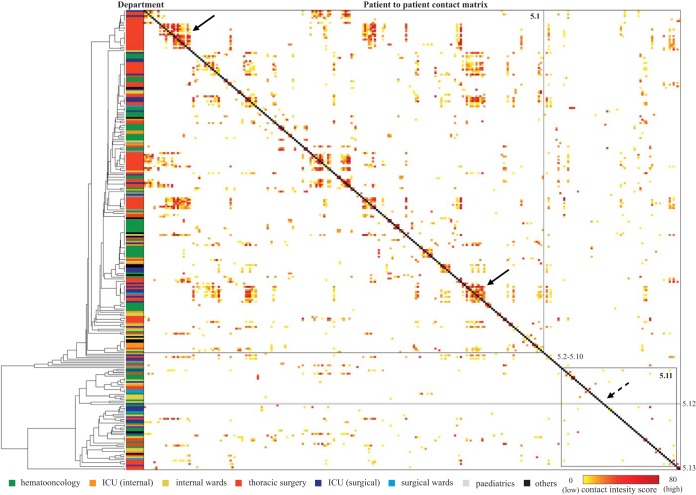
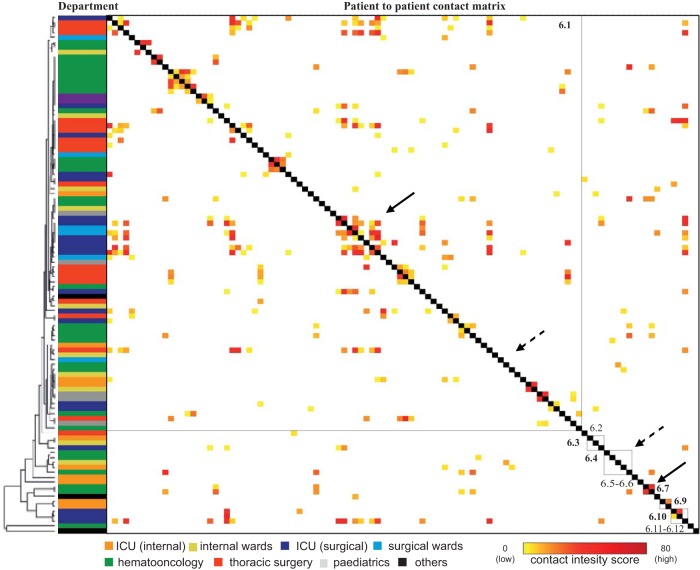
In order to get a more detailed picture of the transmission routes and the dynamics at the different locations in our hospital, we accessed phylogenetic data obtained by clustering based on the core genome SNP matrix of the major outbreak clusters (clusters 3, 5, 6, and 8). Within each cluster a SNP cutoff value (<7 SNPs) was determined based on the UPGMA analysis in order to define subclusters (Table S2). The genetic data were combined with a patient-to-patient contact matrix of each of the major outbreak clusters. The contact intensity score included exposure time and quality. The higher the score, the closer and longer the indications were of patient-to-patient contact (Fig. 2 and 3 and Fig. S4). The following observations were made. (i) The different NGS clusters and subclusters were found on different wards throughout the hospital. The different clusters were not restricted to certain geographical areas of the hospital. (ii) Parallel to this, genetic relatedness and score for epidemiological contact intensity in some cases indicated intrahospital transmission, for example, in clusters 5.1 (Fig. 2, solid arrows), 6.1, 6.7 (Fig. 3, solid arrows) 3.1, and 8.7 (Fig. S5). (iii) Patients were colonized with VREfm strains belonging to the outbreak cluster but with only a slight epidemiological link or no epidemiological link at all (Fig. 2 and 3, dashed arrows). (iv) In addition, sporadic, single-subcluster strains with no epidemiological link were detected. Despite the fact that cases of intrahospital transmission had most likely occurred in some instances, the proportion of patients with no or little epidemiological link was higher than we had expected. This led us to investigate the transmission dynamics in more detail in a defined patient subpopulation.
FIG 2.
Phylogeny of NGS cluster 5 in combination with the epidemiological links of the corresponding patients. The phylogenetic tree is based on the specific NGS cluster core genome. The department in which VREfm was initially detected and the epidemiological links between all patients of the NGS cluster are displayed. Strains were divided into 13 subclusters (black right angles and gray lines), with clusters 5.1 and 5.13 being the largest. Epidemiological links of all patients were calculated based on room occupancy data, and the contact intensity score was color-coded for each patient pair in the matrix. The contact intensity score of each patient with another is shown on the x and y axes in the same order (patient 1 to patient 275). For orientation, the dark gray middle line represents the symmetry axis of the matrix. Strains of cluster 5 and the different subclusters were present in the majority of hospital departments, and numerous links between the patients existed. Phylogenetically closely related isolates with strong epidemiological links (clustering of high contact intensity scores around the gray middle line) suggested intrahospital transmission (e.g., cluster 5.1). However, for several patients, an epidemiological link could not be established (e.g., cluster 5.11, marked with dashed arrows).
FIG 3.
Phylogeny of NGS cluster 6 in combination with the epidemiological links of the corresponding patients. The phylogenetic tree is based on the specific NGS cluster core genome. Epidemiological links of all patients were calculated based on room occupancy data, and the contact intensity score was color-coded for each patient pair in the matrix. The contact intensity score of each patient with another is shown on the x and y axes in the same order (patient 1 patient to 106). While closely related isolates with strong epidemiological links (clustering of high-contact intensity scores around the gray middle line) suggested intrahospital transmission (cluster 6.1 and 6.7, marked with solid arrows), for several patients an epidemiological link could not be established (e.g., 6.1, lower part; 6.4, marked with dashed arrows).
Transmission dynamics in a defined patient subpopulation.
The combination of epidemiological data and typing information was used to reconstruct the transmission routes for the hemato-oncology department, described in detail below. The rationale for selection of this patient population was that a weekly VRE screening program using VRE chromogenic agar had been implemented before the outbreak in January 2015 and had not been changed since then. Detailed analysis of the 24 months from January 2015 to December 2016 showed that the overall screening rate remained at a stable level, while the rate of hemato-oncology patients colonized with VRE infection increased from 0.3% in the first quarter of 2015 to 10.4% in the last quarter of 2016 for all hemato-oncology patients admitted during the respective quarter (Table S3). The number of VREfm isolates from blood cultures remained low, with a total of 3 cases in the 24 months. The blood culture VREfm isolates (n = 3) belonged to the same NGS cluster as the initial screening isolate. In one of these cases, no prior VREfm colonization was noted, and the blood culture isolate was the first detection of VREfm in the patient.
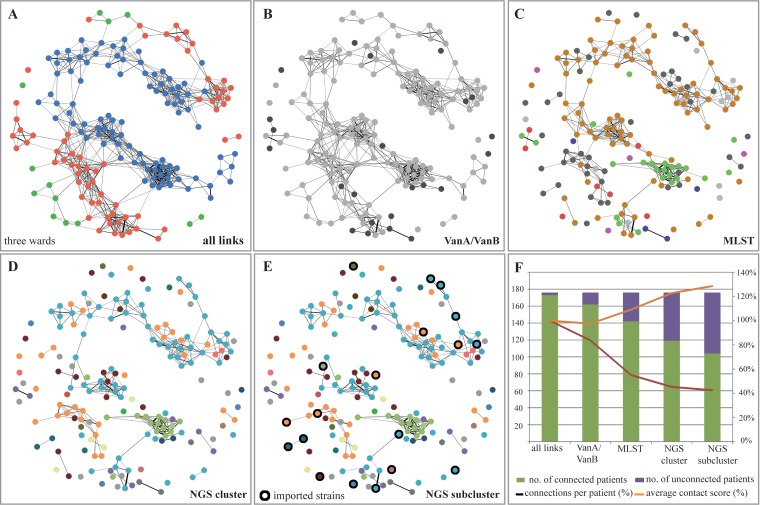
Epidemiological data demonstrated that various links existed between the patients (Fig. 4A). These links could be reduced by more detailed typing, thus reducing the possible transmission routes and allowing a more detailed picture to emerge of the evolution of the outbreak (Fig. 4B to E). At the same time, the epidemiological score for contact intensity increased, making transmission more likely (Fig. 4F) and leading to an increase in patients for whom we could not establish any epidemiological links to other patients (Fig. 4E). Reviewing the history of these patients identified strains that were most likely imported in our hospital (Fig. 4E, black circles).
FIG 4.
Overview of epidemiological links and possible transmission routes depending on the depth of typing. The epidemiological links between all VRE patients of one department comprising three wards were scored, and the network of patient contacts is displayed. The connection line thickness represents the contact intensity score between two patients. (A) All links between the patients of the three wards are shown. (B and C) The patient contact network is displayed integrating the information of the type of van gene, whereas the MLST information was integrated into the contact network. (D and E) Integration of NGS cluster information and subcluster information further reduced the number of epidemiological links and possible transmissions. In addition, strains imported into the hospital are marked with a black ring in panel E. (F) Summary of patient contacts and the strength of the epidemiological link. More detailed resolution of the typing method limited the potential epidemiological transmissions and led to more patients for whom no epidemiological link could be established. At the same time, epidemiological contact intensity scores increased, thus making transmission more likely.
Influx of VREfm into our hospital.
In order to gather more information on the strains imported into our hospital, we identified isolates of patients screened positive for VREfm within 72 h after admission and with no prior hospitalization in our hospital within the previous 5 years (n = 88). Surprisingly, the imported strains after October 2015 resembled the distribution of NGS clusters observed during the outbreak, with the majority of imported strains belonging to clusters 5, 6, and 8 (Fig. 5). In contrast, the imported strains before October 2015 were more diverse, belonging to smaller NGS clusters or representing single clones. The majority of patients (n = 65) were transferred from smaller local hospitals within the federal state of Baden-Wuerttemberg, while 5 patients were transferred from hospitals within Germany, and 4 were from hospitals abroad. For one patient, no prior contact to our hospital could be discovered, while for the remaining 14 patients, no data could be obtained. Strains belonging to the two major NGS clusters 5 and 6 were imported from several hospitals, indicating that in these hospitals more than one VREfm clone was also present. This clearly indicates that the expansion of certain strains is not restricted to a single hospital but, rather, encompasses a health care network.
FIG 5.
(A) Phylogenetic maximum likelihood tree of imported strains. The NGS cluster information, as well as the most likely origin of the VREfm, is displayed. Patient identification (ID) numbers are indicated along with (in parentheses) whether patients were transferred from a local hospital (A to Z), from a hospital within Germany (1), or from any other hospital (2). If no information was available, the isolate is marked as unknown (unk). Most of the imported isolates belonged to NGS clusters 5 and 6. However, smaller clusters (e.g., cluster 2) and single isolates have also been observed. (B) Timeline of VREfm isolates imported in the hospital. As within the hospital, a sharp increase in the import of VREfm isolates was noted. Interestingly, the composition of NGS clusters of the imported strains, observed from October 2015 onwards, was also dominated by NGS clusters 5 and 6.
Patient movements.
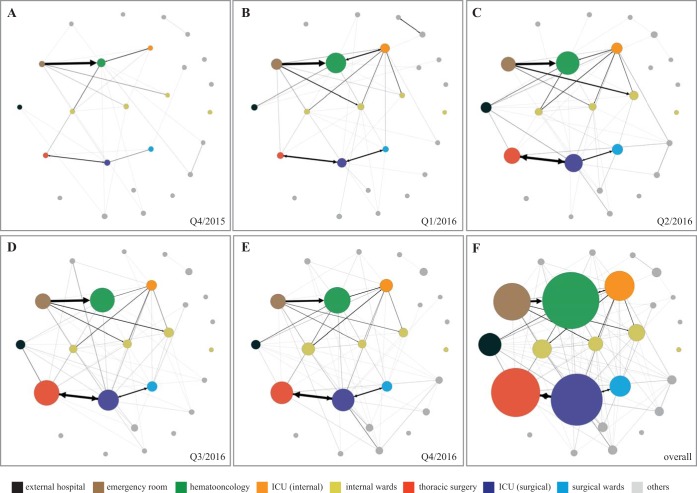
Figure 6 illustrates the movements of patients from external hospitals or within our hospital departments who were VREfm positive or became VREfm positive within 14 days after being transferred. In the beginning, not many transfers occurred of VREfm patients between the different departments or from external hospitals. The number of movements increased over time, and more VREfm patients were present in the different wards, as illustrated by the size of the dots in Fig. 6. With increasing numbers of VREfm patients being transferred, two major axes were observed. The first axis involved the emergency room, internal wards, and the internal ICU. Thoracic surgery, the surgical ICU, and surgical wards formed the second axis. Additionally, an increasing influx of VREfm patients could be demonstrated over time (Fig. 6A to E, black circles).
FIG 6.
(A to F) Movement of VREfm patients at different time points. The movement of patients who became positive within 14 days after the transfer is displayed. An increase in movements of VREfm patients between different departments is indicated by an increase in the thickness of the line between them. The increase in size of a department represents the number of VREfm patients in the department shown during the corresponding quarter. The black dot on the left-hand side represents external hospitals and unidirectional transfer of VREfm patients into our hospital. In the beginning, only few movements were detected (A), with an increasing dynamic over time (B to E). External transfer of VREfm patients became more frequent. In addition, two major axes of patient movements were identified within the hospital, comprising, on the one hand, emergency room, hemato-oncology, and internal ICU, and on the other, thoracic surgery, surgical ICU, and surgical wards. (F) Overview of VREfm patient movements. Q, quarter.
DISCUSSION
The prevalence of VREfm in Germany has evolved in recent years (17). Whereas the incidence of VREfm colonization or infection was low between 2010 and 2015 in our hospital, we have observed a dramatic increase in VanB-type strains, beginning in the last quarter of 2015. The most recent report from the German National Reference Center for Staphylococci and Enterococci described increasing requests for typing of VanB-type enterococci, indicating a higher occurrence of suspected transmission and local clusters of these strains. For the first time, the number of VanB-type isolates sent for analysis outnumbered VanA-type isolates (18), suggesting that VanB-type VREfm has become much more prevalent in Germany in recent years. Another study demonstrated the involvement of VanB-type ST192 VREfm in invasive infections in Germany (19). Even though, on a national level, systematic epidemiological information on the distribution of VREfm in hospitals is lacking, our data are in line with a changing epidemiology on a larger, nationwide scale.
The screening protocol and the sensitivity of the microbiological procedures could influence the number of VRE patients detected. During the course of the outbreak, the screening program was expanded to several wards in our hospital, and diagnostic procedures have been adjusted. These measures might have in part contributed to the increase in VREfm observed. However, in the hemato-oncology department a comprehensive screening program had been implemented by the beginning of 2015 without any changes in diagnostic procedures during the course of the outbreak. Here, we could show that the overall number of screened patients remained stable while the number of colonized patients clearly increased toward the end of 2015 (see Table S3 in the supplemental material). In addition, the number of patients with at least one occurrence of VRE infection in clinically relevant specimens (invasive isolates) increased from 9, 5, and 6 for the years 2013, 2014, and 2015, respectively, to 55 in the year 2016. This included detection of VRE throughout the stay in the hospital and not only the first isolate. The rise in numbers of patients with invasive VRE infection was detectable in all departments and most pronounced in surgical specialties, probably due to more invasive sampling. This underlines that the overall burden of VRE in the hospital increased not only in hematological patients but also in all patients and that this was not attributable to changes in the VRE screening policy.
Potential factors influencing their spread could include the exposure of large numbers of persons to a yet unknown source in the community, e.g., through food, water, or contaminated hospital environments. Other possible explanations could involve strain-specific traits of these clones, such as those enabling more efficient colonization of the gastrointestinal system of humans and animals or those strengthening their persistence in the environment. Gut-commensal E. faecium strains have been shown to be an important reservoir for traits that can be transferred to successful clinical E. faecium strains (20). Therefore, another scenario worth considering is that successfully colonizing vancomycin-susceptible enterococci acquired vanB-harboring genetic elements, resulting in the increase of VanB-type VREfm. This was shown for VanA-type VREfm in a study of 495 isolates from Denmark that demonstrated the spread of a vanA plasmid into different sequence-type backgrounds of E. faecium (21). Further studies are needed to address these questions, preferably with the application of long-range read sequencing technology to characterize the genetic environment, plasmids, and transposons without the bias and insecurity of the assembly of short-read sequencing data.
A recent report describes the value of genome sequencing of (mainly VanA-type) VREfm bloodstream isolates for elucidating transmission routes (22). Our study also shows that combining WGS and epidemiological data provides us with a powerful tool for gaining insights into the dynamic of VREfm outbreaks and transmission routes within the hospital. A clustering approach based on a core genome SNP similarity matrix was chosen in order to define NGS clusters and subclusters. The advantage of this approach compared to maximum likelihood phylogeny is that we acquire the ability to establish SNP cutoffs for the cluster definitions. However, criteria or definitions used for determining SNP cutoff values are lacking to date. Therefore, we based the cutoff selection on the SNP distance between each isolate and each other (Fig. S1). The NGS clusters defined based on the cutoff resembled, to a great extent, the distribution of the isolates based on a core genome maximum likelihood phylogeny (Fig. S2) and the maximum likelihood phylogeny of the different lineages of the MLSTs (Fig.S3 and S4). This agreement increased our confidence in the cutoff selected based on the SNP distance distribution. Likewise, only limited data are available regarding the impact of different transmission routes for VREfm in the hospital setting. A recent systematic review described the major transmission routes as being the hands of health care workers, the contamination of the environment, and patient-to-patient transmission (23). In order to account for all these modes in our study, we established an epidemiological contact intensity score to quantify the possibility of transmission of a clone between two patients. The highest value was assigned for patients sharing a room since all modes of transmission might have occurred, followed by a consecutive stay in the same room where a room-specific reservoir and personnel might have served as vehicles of transmission and a stay on the same ward with a potential contact to a more general reservoir or personnel. Due to the lack of comprehensive valid data on the impact of the different transmission modes, we used this score solely in a descriptive way. The calculation of the transmission routes was not based on the value of the score, and no intensity contact score cutoff was determined.
Whereas outbreaks of VanB-type VREfm have been described before (24–26), this study reports, to the best of our knowledge, the largest outbreak with this organism so far, comprising more than 500 isolates and dominated by a VanB-type ST80 VREfm. In this study, we developed an approach to combine NGS data with epidemiological data from 773 patients, allowing the elucidation of the outbreak dynamic on a single-patient basis. We could demonstrate that the definition of NGS clusters and subclusters leads to a much more detailed outbreak resolution than MLST, resulting in the observation of many small transmission clusters and the elimination of a large number of possible transmission events. Interestingly, we identified 88 patients who were positive for VRE upon admission to our hospital. Phylogenetic analysis revealed that these induced strains also included isolates of the two predominant clusters 5 and 6, thus indicating that the network of the health care system is important to take into consideration. However, of course the opposite might have happened as well such that patients who became colonized during their stay in our hospital contributed to the spread of the VRE into other hospitals. In order to be able to combat the spread of multidrug-resistant (MDR) bacteria, systematic data collection on patient transferals is essential. These data could be collected in national databases or, if such databases do not exist, might be collected from health insurance companies. In addition, systematic surveillance is essential, including harmonized regional screening procedures and outbreak management and reporting of invasive infections, as well as collection and typing of these strains. These strategies would enable data integration to decipher a thoroughly comprehensive picture of the mode and form the basis to establish efficient infectious control measures.
Our study presents the following conclusions: (i) the increase observed in VREfm was due to the expansion of two major clusters that were present throughout the hospital; (ii) patient-to-patient transmission had occurred, but to a lesser extent than suggested by the epidemiological data alone; (iii) continuous reintroduction of the two major NGS cluster strains from local hospitals was detected; and (iv) patient transferals play a significant role in distributing VREfm within the hospital and between different health care providers. Our findings emphasize the importance of establishing network strategies as means of understanding and combating the spread of these antibiotic-resistant organisms. In particular, sharing whole-genome sequencing data and metadata on a real-time basis could enable monitoring and comparison of the epidemiological situation and transmissions on a global scale.
MATERIALS AND METHODS
Strain selection.
Overall, VREfm was detected in 797 patients during the 7-year study period (January 2010 until December 2016). From these patients, 773 initial VREfm isolates were available for further analysis. This included 614 screening isolates and 159 isolates from clinical specimens.
Bacteriological procedures.
Screening cultures (rectal swabs and stool samples) obtained between 2010 and February 2016 were examined for the presence of VRE by plating the specimens on CNA agar (Becton, Dickinson and Company, France) with a vancomycin disk (30 μg) placed in the first fraction and interpreted after incubation for 48 h at 35°C (27). In January 2015, the chromID VRE agar (bioMérieux, Marcy l'Etoile, France) was introduced for VRE screening from the hemato-oncology ward and in March 2016 for all screening specimens. Identification of isolates was achieved by linear matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry (AXIMA Assurance; [bioMérieux SA, France] or Microflex LT [Bruker Daltonics, Germany]) and Vitek 2 system identification (bioMérieux, Marcy l’Etoile, France). Antimicrobial susceptibility testing was performed using the Vitek 2 system (bioMérieux SA, France) and interpreted according to the guidelines of the European Committee on Antimicrobial Susceptibility Testing (EUCAST). Vancomycin-resistant isolates and all Enterococcus species recovered from the chromID VRE agar (including vancomycin-susceptible isolates) were subjected to molecular vanA and vanB gene detection as described previously (28).
Whole-genome sequencing.
Genomic DNA was extracted using an UltraClean Microbial DNA isolation kit (Mo Bio Laboratories, Inc., Carlsbad, CA, USA) and sheared by Covaris M220 (Covaris, Woburn, MA, USA) to obtain 550-bp fragments. DNA libraries were prepared with a TruSeqNano DNA LT kit (Illumina, San Diego, CA, USA), followed by analysis of barcoded libraries on a QIAxcel Advanced Instrument (Qiagen, Hilden, Germany). All libraries were sequenced on an Illumina MiSeq (Illumina, San Diego, CA, USA) with v2 chemistry (2 by 250 bp) or an Illumina Nextseq (Illumina, San Diego, CA, USA) with v2 chemistry (2 by 150 bp), depending on sequencing capacities.
WGS data analysis.
(i) Assembly, core genome calculation, maximum likelihood phylogeny, and homologous recombination. Sequencing reads were assembled using the A5 pipeline (version 20140604) and SPAdes (version 3.7.0) (29, 30). High-quality reads were generated by applying Trimmomatic (31). The core genomes of all strains and clusters were generated by Spine (version 0.1.2) using the default parameter setting except that segment length was adjusted to 1,000 bp (32). Prophage regions were investigated and removed using PHAST (33). For core-genome single nucleotide polymorphism (SNP) calling, a customized python pipeline including GATK (version 3.2-2) and SAMtools (version 0.1.19) (34, 35) was applied, followed by calculation of a maximum likelihood tree using RAxML (version 8.2.6) with a general time-reversible (GTR) substitution model and gamma distribution of rates undergoing 1,000 bootstraps (36) and with visualization by FigTree (version 1.4.2). In order to take homologous recombination into account within the major MLSTs, a maximum likelihood phylogeny was calculated using Gubbins (version 2.1.0) (37).
(ii) MLST sequence types and unweighted pair-grouping (UPGMA) SNP similarity matrix. MLST sequence types of all isolates were extracted from the assembled sequences with the BioNumerics, version 7.6, software using the MLST definition for Enterococcus faecium comprising seven gene loci (pstS, purK, adk, gyd, ddl, atpA, and gdh). MLST sequence types could be assigned to 731 (94.6%) strains. An SNP-based phylogeny was calculated for all 773 VREfm strains using BioNumerics, version 7.6, software (Applied Maths, Sint-Martens-Latem, Belgium). In a first step, the high-quality sequencing reads were mapped to the previously calculated core genome (see above) using the default settings. Next, SNPs were determined using the predefined strict SNP filtering settings. Clustering was performed by applying the UPGMA algorithm to the SNP similarity matrix.
(iii) Definition of NGS clusters and subclusters. NGS clusters were then defined from the SNP similarity matrix. To determine the SNP cutoff for definition of NGS clusters, the SNP distance from each VREfm isolate to each other was determined and plotted in a diagram (see Fig. S1 in the supplemental material). An isolate was assigned to a cluster if the SNP difference to at least one cluster member was less than 13 bp. To increase resolution within the clusters, core genomes were then recalculated for each NGS cluster using the cluster member genomes only (Table S1). This allowed the designation of subclusters, in which member isolates differed from at least one other member isolate by a maximum of six SNPs (22). Subclusters were generated only for NGS clusters with more than eight isolates. For NGS clusters comprising less than eight strains, it was assumed that all isolates also belonged to the same subcluster.
Epidemiological data.
Room occupancy data were electronically available for each patient and for each day from January 2011 through December 2016. These data were used to calculate a summarized contact intensity score for each pair of VREfm patients to determine and quantify the risk of transmission; a score of 10 was attributed to each day the patients occupied the same room at the same time. A score of 3 was given for each day the patients were on the same ward (but not in the same room) at the same time. A score of 5 was counted for each day a future VREfm patient was located in the same room that was previously occupied (but not longer than 28 days ago) by a patient with VREfm infection. Only occupancy days before VREfm detection in the second patient were considered to avoid skewing the contact score by cohort isolation.
Screening program and infection control measures.
No hospital-wide screening protocol for VREfm detection was in place during the baseline period, with the exception of screening of contact patients. In 2015, a rectal screening for VREfm once weekly and upon admission was introduced in the hemato-oncology department. During the outbreak, rectal VREfm screening of risk patients (mainly transfers from other hospitals) was additionally implemented in the departments for thoracic surgery and visceral surgery as well as in the admission and intensive care units.
The study was conducted in accordance with the local ethics committee (Ethic Committee, Medical Faculty Tuebingen, no. 537/2017BO2).
Data availability.
The raw sequencing data were deposited at the European Nucleotide Archive under accession number PRJEB30772.
Supplementary Material
ACKNOWLEDGMENTS
We thank Nadine Hoffmann, Cornelia Lueth, and Baris Bader for expert technical assistance.
The study was partly funded by the Faculty of Medicine, Tuebingen, and the German Center for Infection Research (DZIF TTU 08.809). The funders had no role in the design, analysis, or interpretation and writing of the manuscript.
S.P. and J.L. designed the study and wrote the manuscript. S.P., J.L., L.S., P.O., L.T., T.N., D.D., W.V., M.M., I.A., and M.W. were involved in the generation of experimental data, clinical information, and bioinformatic data analysis.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.01978-18.
REFERENCES
- 1.European Centre for Disease Prevention and Control. 2016. Healthcare-associated infections acquired in intensive care units—annual epidemiological report 2016. https://ecdc.europa.eu/en/publications-data/infections-acquired-intensive-care-units-annual-report-2016.
- 2.Cheah AL, Spelman T, Liew D, Peel T, Howden BP, Spelman D, Grayson ML, Nation RL, Kong DC. 2013. Enterococcal bacteraemia: factors influencing mortality, length of stay and costs of hospitalization. Clin Microbiol Infect 19:E181–E189. doi: 10.1111/1469-0691.12132. [DOI] [PubMed] [Google Scholar]
- 3.Lode HM. 2009. Clinical impact of antibiotic-resistant Gram-positive pathogens. Clin Microbiol Infect 15:212–217. doi: 10.1111/j.1469-0691.2009.02738.x. [DOI] [PubMed] [Google Scholar]
- 4.DiazGranados CA, Zimmer SM, Klein M, Jernigan JA. 2005. Comparison of mortality associated with vancomycin-resistant and vancomycin-susceptible enterococcal bloodstream infections: a meta-analysis. Clin Infect Dis 41:327–333. doi: 10.1086/430909. [DOI] [PubMed] [Google Scholar]
- 5.Miller WR, Munita JM, Arias CA. 2014. Mechanisms of antibiotic resistance in enterococci. Expert Rev Anti Infect Ther 12:1221–1236. doi: 10.1586/14787210.2014.956092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kramer A, Schwebke I, Kampf G. 2006. How long do nosocomial pathogens persist on inanimate surfaces? A systematic review. BMC Infect Dis 6:130. doi: 10.1186/1471-2334-6-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Snyder GM, Thom KA, Furuno JP, Perencevich EN, Roghmann MC, Strauss SM, Netzer G, Harris AD. 2008. Detection of methicillin-resistant Staphylococcus aureus and vancomycin-resistant enterococci on the gowns and gloves of healthcare workers. Infect Control Hosp Epidemiol 29:583–589. doi: 10.1086/588701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Satilmis L, Vanhems P, Benet T. 2016. Outbreaks of vancomycin-resistant enterococci in hospital settings: a systematic review and calculation of the basic reproductive number. Infect Control Hosp Epidemiol 37:289–294. doi: 10.1017/ice.2015.301. [DOI] [PubMed] [Google Scholar]
- 9.Fournier S, Brossier F, Fortineau N, Gillaizeau F, Akpabie A, Aubry A, Barbut F, Chedhomme FX, Kassis-Chikhani N, Lucet JC, Robert J, Seytre D, Simon I, Vanjak D, Zahar JR, Brun-Buisson C, Jarlier V. 2012. Long-term control of vancomycin-resistant Enterococcus faecium at the scale of a large multihospital institution: a seven-year experience. Euro Surveill 17(30):pii=20229 https://www.eurosurveillance.org/content/10.2807/ese.17.30.20229-en. [PubMed] [Google Scholar]
- 10.Kurup A, Chlebicki MP, Ling ML, Koh TH, Tan KY, Lee LC, Howe KB. 2008. Control of a hospital-wide vancomycin-resistant Enterococci outbreak. Am J Infect Control 36:206–211. doi: 10.1016/j.ajic.2007.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peta M, Carretto E, Barbarini D, Zamperoni A, Carnevale L, Perversi L, Pagani M, Bonora MG, Fontana R, Marone P, Langer M. 2006. Outbreak of vancomycin-resistant Enterococcus spp. in an Italian general intensive care unit. Clin Microbiol Infect 12:163–169. doi: 10.1111/j.1469-0691.2005.01331.x. [DOI] [PubMed] [Google Scholar]
- 12.Tuon FF, Penteado-Filho SR, Camilotti J, van der Heijden IM, Costa SF. 2011. Outbreak of vancomycin-resistant Enterococcus in a renal transplant unit. Braz J Infect Dis 15:403–405. doi: 10.1016/S1413-8670(11)70216-1. [DOI] [PubMed] [Google Scholar]
- 13.Ergani-Ozcan A, Naas T, Baysan BO, Ogunc D, Inan D, Colak D, Nordmann P. 2008. Nosocomial outbreak of vancomycin-resistant Enterococcus faecium in a paediatric unit at a Turkish university hospital. J Antimicrob Chemother 61:1033–1039. doi: 10.1093/jac/dkn066. [DOI] [PubMed] [Google Scholar]
- 14.Pinholt M, Larner-Svensson H, Littauer P, Moser CE, Pedersen M, Lemming LE, Ejlertsen T, Sondergaard TS, Holzknecht BJ, Justesen US, Dzajic E, Olsen SS, Nielsen JB, Worning P, Hammerum AM, Westh H, Jakobsen L. 2015. Multiple hospital outbreaks of vanA Enterococcus faecium in Denmark, 2012-13, investigated by WGS, MLST and PFGE. J Antimicrob Chemother 70:2474–2482. doi: 10.1093/jac/dkv142. [DOI] [PubMed] [Google Scholar]
- 15.Borgmann S, Niklas DM, Klare I, Zabel LT, Buchenau P, Autenrieth IB, Heeg P. 2004. Two episodes of vancomycin-resistant Enterococcus faecium outbreaks caused by two genetically different clones in a newborn intensive care unit. Int J Hyg Environ Health 207:386–389. doi: 10.1078/1438-4639-00304. [DOI] [PubMed] [Google Scholar]
- 16.Sagel U, Schulte B, Heeg P, Borgmann S. 2008. Vancomycin-resistant enterococci outbreak, Germany, and calculation of outbreak start. Emerg Infect Dis 14:317–319. doi: 10.3201/eid1402.070752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gastmeier P, Schroder C, Behnke M, Meyer E, Geffers C. 2014. Dramatic increase in vancomycin-resistant enterococci in Germany. J Antimicrob Chemother 69:1660–1664. doi: 10.1093/jac/dku035. [DOI] [PubMed] [Google Scholar]
- 18.Klare IBJ, Koppe U, Abu Sin M, Eckmanns T, Werner G. 2017. Eigenschaften, Häufigkeit und Verbreitung von Vancomycin-resistenten Enterokokken (VRE) in Deutschland—update 2015/2016. Epidemiol Bull 46:519–527. [Google Scholar]
- 19.Bender JK, Kalmbach A, Fleige C, Klare I, Fuchs S, Werner G. 2016. Population structure and acquisition of the vanB resistance determinant in German clinical isolates of Enterococcus faecium ST192. Sci Rep 6:21847. doi: 10.1038/srep21847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de Been M, van Schaik W, Cheng L, Corander J, Willems RJ. 2013. Recent recombination events in the core genome are associated with adaptive evolution in Enterococcus faecium. Genome Biol Evol 5:1524–1535. doi: 10.1093/gbe/evt111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pinholt M, Gumpert H, Bayliss S, Nielsen JB, Vorobieva V, Pedersen M, Feil E, Worning P, Westh H. 2017. Genomic analysis of 495 vancomycin-resistant Enterococcus faecium reveals broad dissemination of a vanA plasmid in more than 19 clones from Copenhagen, Denmark. J Antimicrob Chemother 72:40–47. doi: 10.1093/jac/dkw360. [DOI] [PubMed] [Google Scholar]
- 22.Raven KE, Gouliouris T, Brodrick H, Coll F, Brown NM, Reynolds R, Reuter S, Torok ME, Parkhill J, Peacock SJ. 2017. Complex routes of nosocomial vancomycin-resistant Enterococcus faecium transmission revealed by genome sequencing. Clin Infect Dis 64:886–893. doi: 10.1093/cid/ciw872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ulrich N, Vonberg RP, Gastmeier P. 2017. Outbreaks caused by vancomycin-resistant Enterococcus faecium in hematology and oncology departments: a systematic review. Heliyon 3:e00473. doi: 10.1016/j.heliyon.2017.e00473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lister DM, Kotsanas D, Ballard SA, Howden BP, Carse E, Tan K, Scott C, Gillespie EE, Mahony AA, Doherty R, Korman TM, Johnson PD, Stuart RL. 2015. Outbreak of vanB vancomycin-resistant Enterococcus faecium colonization in a neonatal service. Am J Infect Control 43:1061–1065. doi: 10.1016/j.ajic.2015.05.047. [DOI] [PubMed] [Google Scholar]
- 25.Johnson PD, Ballard SA, Grabsch EA, Stinear TP, Seemann T, Young HL, Grayson ML, Howden BP. 2010. A sustained hospital outbreak of vancomycin-resistant Enterococcus faecium bacteremia due to emergence of vanB E. faecium sequence type 203. J Infect Dis 202:1278–1286. doi: 10.1086/656319. [DOI] [PubMed] [Google Scholar]
- 26.Donskey CJ, Schreiber JR, Jacobs MR, Shekar R, Salata RA, Gordon S, Whalen CC, Smith F, Rice LB. 1999. A polyclonal outbreak of predominantly VanB vancomycin-resistant enterococci in northeast Ohio. Northeast Ohio Vancomycin-Resistant Enterococcus Surveillance Program. Clin Infect Dis 29:573–579. [DOI] [PubMed] [Google Scholar]
- 27.European Committee on Antimicrobial Susceptibility Testing. 2010. Breakpoint tables for interpretation of MICs and zone diameters, version 1.0 2010. http://www.eucast.org/ast_of_bacteria/previous_versions_of_documents/.
- 28.Bell JM, Paton JC, Turnidge J. 1998. Emergence of vancomycin-resistant enterococci in Australia: phenotypic and genotypic characteristics of isolates. J Clin Microbiol 36:2187–2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Coil D, Jospin G, Darling AE. 2015. A5-miseq: an updated pipeline to assemble microbial genomes from Illumina MiSeq data. Bioinformatics 31:587–589. doi: 10.1093/bioinformatics/btu661. [DOI] [PubMed] [Google Scholar]
- 30.Nurk S, Bankevich A, Antipov D, Gurevich AA, Korobeynikov A, Lapidus A, Prjibelski AD, Pyshkin A, Sirotkin A, Sirotkin Y, Stepanauskas R, Clingenpeel SR, Woyke T, McLean JS, Lasken R, Tesler G, Alekseyev MA, Pevzner PA. 2013. Assembling single-cell genomes and mini-metagenomes from chimeric MDA products. J Comput Biol 20:714–737. doi: 10.1089/cmb.2013.0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ozer EA, Allen JP, Hauser AR. 2014. Characterization of the core and accessory genomes of Pseudomonas aeruginosa using bioinformatic tools Spine and AGEnt. BMC Genomics 15:737. doi: 10.1186/1471-2164-15-737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou Y, Liang Y, Lynch KH, Dennis JJ, Wishart DS. 2011. PHAST: a fast phage search tool. Nucleic Acids Res 39:W347–W352. doi: 10.1093/nar/gkr485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, Genome Project Data Processing S. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, Del Angel G, Levy-Moonshine A, Jordan T, Shakir K, Roazen D, Thibault J, Banks E, Garimella KV, Altshuler D, Gabriel S, DePristo MA. 2013. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics 43:11.10.1–33. doi: 10.1002/0471250953.bi1110s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Croucher NJ, Page AJ, Connor TR, Delaney AJ, Keane JA, Bentley SD, Parkhill J, Harris SR. 2015. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res 43:e15. doi: 10.1093/nar/gku1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw sequencing data were deposited at the European Nucleotide Archive under accession number PRJEB30772.