Since the introduction of pneumococcal conjugate vaccines (PCVs), an increase in the incidence of disease attributable to serotype 15A-ST63 (sequence type 63) pneumococci has been observed in many regions worldwide. We conducted a nationwide pediatric pneumococcal infection surveillance study between 2012 and 2014 in Japan.
KEYWORDS: 15A, Japan, PCV13, PCV7, ST63, ST9084, Streptococcus pneumoniae, cefotaxime, multidrug resistance, serotype shift
ABSTRACT
Since the introduction of pneumococcal conjugate vaccines (PCVs), an increase in the incidence of disease attributable to serotype 15A-ST63 (sequence type 63) pneumococci has been observed in many regions worldwide. We conducted a nationwide pediatric pneumococcal infection surveillance study between 2012 and 2014 in Japan. In the surveillance study, we detected multidrug-resistant serotype 15A-CC63 (clonal complex 63) strains (resistant to macrolides, penicillin, cefotaxime, and meropenem); in this study, we analyzed these resistant isolates to determine the dynamics and mechanism of resistance using whole-genome sequencing. In most of the penicillin-, cefotaxime-, and meropenem-resistant strains, recombination occurred in the pbp2x region, resulting in the acquisition of cefotaxime resistance in addition to penicillin and meropenem resistance. In the multidrug-resistant serotype 15A-CC63 strains, we identified a specific clone with ST9084, and all of the isolates were recovered from the Yamaguchi prefecture in Japan. All of the serotype 15A-ST9084 isolates had a novel pbp2x type (pbp2x-JP3) that was inserted by recombination events. The conserved amino acid motif profiles of pbp1a, pbp2b, and pbp2x of the strains were identical to those of serotype 19A-ST320. A Bayesian analysis-based date estimation suggested that this clone emerged in approximately 2002 before the introduction of the PCV in Japan. This clone should be monitored because serotype 15A is not contained in the currently used 13-valent PCV (PCV13), and it was resistant to beta-lactams, which are often used in a clinical setting.
INTRODUCTION
Streptococcus pneumoniae is a common bacterial pathogen that causes infections such as pneumonia, otitis media, occult bacteremia, and meningitis. Global estimates suggest that pneumococcus was responsible for 826,000 deaths among children under 5 years of age in 2000 (1). To prevent infectious pneumococcal disease, 7-, 10-, and 13-valent pneumococcal conjugate vaccines (PCVs) that target a subset of pneumococcal serotypes have been introduced in many countries, which has resulted in a dramatic decrease in the number of invasive pneumococcal disease (IPD) cases. However, the identification of cases of non-PCV serotype pneumococcus has increased (2–8). One of the increasing non-PCV serotypes is serotype 15A, and most of these isolates belonged to sequence type 63 (ST63) (9–17). The serotype 15A-ST63 strain was submitted as Sweden15A-25 in the Pneumococcal Molecular Epidemiology Network (PMEN) and is highly resistant to macrolides (http://web1.sph.emory.edu/PMEN/) (18).
In Japan, the 7-valent PCV (PCV7) was licensed in February 2010 and used on a voluntary basis until April 2013. During this period, the estimated rates of PCV7 vaccination in children under 5 years of age increased from <10% in 2010 to 80% to 90% in 2012 (2). In April 2013, PCV7 was approved as a routine vaccine in Japan, and the vaccine was switched to PCV13 in October 2013. We conducted a nationwide pediatric pneumococcal infection surveillance study in Japan during the PCV13 era between 2012 and 2014 (19). According to the results of the surveillance study, the incidence of meropenem-resistant (MEM-R) pneumococci increased during the surveillance period, which was primarily caused by an increased incidence of isolates of the meropenem-resistant serotypes 15A and 35B. Previous studies demonstrated that penicillin-binding protein (PBP) typing using the pbp1a, pbp2b, and pbp2x transpeptidase domain sequences is useful for predicting beta-lactam resistance (3, 20, 21). The relationship between the three PBP sequences and beta-lactam susceptibility has been reported by the CDC (22). Using this typing methodology, we demonstrated that the meropenem resistance in serotype 15A-ST63 strains emerged due to recombination events overlapping the pbp1a region, which caused an insertion of the meropenem resistance-related pbp1a type 13 (pbp1a-13) sequence in Japan (23). This pbp1a-13 sequence was primarily identified in serotype 19A-ST320 isolates in the United States, which are well-known highly multidrug-resistant clones (3, 24). Thus, a detailed understanding of the dynamics of pneumococcal strains, especially antimicrobial-resistant clones, can be gained using the PBP typing method in combination with multilocus sequence typing (MLST).
In the treatment of such macrolide-, penicillin-, and meropenem-resistant serotype 15A-ST63 pneumococcal infections, cefotaxime (CTX) is effective in most cases and represents a key drug. Most of the Japanese serotype 15A-ST63 isolates with penicillin and meropenem resistance analyzed in a previous study were susceptible to cefotaxime (MIC, ≤0.5 μg/ml). In this study, we demonstrated the emergence of cefotaxime resistance, in addition to macrolide, penicillin, and meropenem resistance, in serotype 15A isolates in Japan. In addition, we demonstrated the prevalence of the multi-beta-lactam-resistant (MBLR) serotype 15A-ST9084 (single-locus variant [SLV] of ST63) clone in one prefecture in Japan. We analyzed the isolates using whole-genome sequencing and present here their genetic characteristics.
RESULTS
All 86 serotype 15A isolates analyzed in this study were penicillin G resistant (PG-R).
Among the 86 serotype 15A isolates, 62 were of ST63, 10 were of ST9084, and 14 were of other STs (Table 1). Of the 10 ST9084 isolates, 2 were assigned to ST10244, and 2 others were assigned to a new sequence type in a previous study (19). Among the 24 MBLR isolates, 9 were of ST63, 10 were of ST9084, and the other 4 isolates were of other STs.
TABLE 1.
Serotype 15A isolates from Japan in 2012 to 2014 and MICs for meropenem and cefotaximea
| Sequence type |
No. of MEM-R isolates (n = 10) with MIC for CTX (μg/ml) of: |
No. of MEM-IR isolates (n = 54) with MIC for CTX (μg/ml) of: |
No. of MEM-S isolates (n = 22) with MIC for CTX (μg/ml) of: |
||||||
|---|---|---|---|---|---|---|---|---|---|
| ≤0.5 | 1.0 | ≥2.0 | ≤0.5 | 1.0 | ≥2.0 | ≤0.5 | 1.0 | ≥2.0 | |
| 63 | 3 | 3 | 1 | 32 | 4 | 1 | 16 | 1 | 1 |
| 9084 | 0 | 0 | 2 | 0 | 0 | 8 | 0 | 0 | 0 |
| Other | 0 | 1 | 0 | 5 | 2 | 2 | 4 | 0 | 0 |
MEM-R, meropenem resistant (MIC of ≥1.0 μg/ml); MEM-IR, meropenem intermediate resistant (MIC of 0.5 μg/ml); MEM-S, meropenem susceptible (MIC of ≤0.25 μg/ml); CTX, cefotaxime.
The 10 serotype 15A-ST9084 isolates were collected from different cases in the Yamaguchi prefecture between May 2012 and November 2014 (see Fig. S1 in the supplemental material). The 10 cases were diagnosed in three different cities. All of the cases except for one were diagnosed as having otitis media, and the pathogens were recovered from the middle-ear fluid. The remaining case was diagnosed as having pneumococcal meningitis, and the isolate was recovered from cerebrospinal fluid.
Whole-genome sequencing statistics.
Sequencing statistics are shown in Data Sets S1 and S2 in the supplemental material. The 52 isolate genomes that were newly tested in this study were sequenced at an average depth of 60.37 (standard deviation [SD], ±19.06) and an average coverage of 98.72% (SD, ±0.38%) using the 2,078,953-bp S. pneumoniae G54 chromosome as a reference sequence (GenBank accession no. NC_011072.1). The average number of contigs and the average N50 (base pairs) value for these isolates were 70.3 (SD, ±5.9) and 53,180 (SD, ±5,298), respectively.
Phylogenomics.
To determine the relationship between Japanese serotype 15A-CC63 isolates and foreign isolates, we created a mapping-based phylogenomic tree using serotype 15A-CC63 isolates from Japan, the United Kingdom, and the United States. In the tree, 82 of 86 Japanese serotype 15A-CC63 isolates were included in the same Japanese isolate-specific clade (Fig. S2). In addition, an MBLR serotype 15A-ST9084 isolate-specific clade was generated within the Japanese serotype 15A-CC63 isolate-specific clade; this result implied that the ST9084 lineage originated from the Japanese ST63 lineage. The tree created using only Japanese serotype 15A-CC63 isolates also revealed a serotype 15A-ST9084-specific subclade (subclade CTXR) that was included in a clade that specifically contained serotype 15A-CC63 isolates from the Yamaguchi prefecture (clade Y) (Fig. 1).
FIG 1.
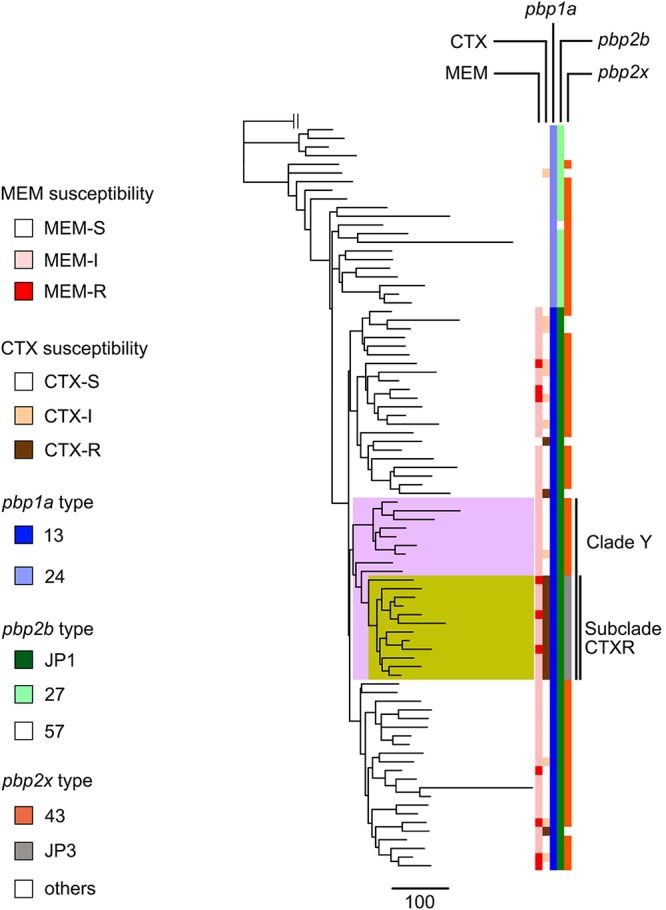
Phylogenetic tree, susceptibilities to cefotaxime and meropenem, and PBP profiles for all serotype 15A-CC63 isolates derived from Japan between 2012 and 2014. Clade Y is highlighted in pink, and subclade CTXR is highlighted in yellow. Subclade CTXR consists of all serotype 15A-ST9084 isolates and one isolate each of serotype 15A-ST63 and serotype 15A-ST9644. The phylogenetic tree was created by Gubbins using Streptococcus pneumoniae G54 as an outgroup isolate. MEM, meropenem; CTX, cefotaxime.
Prediction of recombination sites potentially responsible for additional cefotaxime resistance.
Using all the clade Y isolates, we predicted the recombination sites that could cause the additional cefotaxime resistance observed in the serotype 15A-ST9084 lineage.
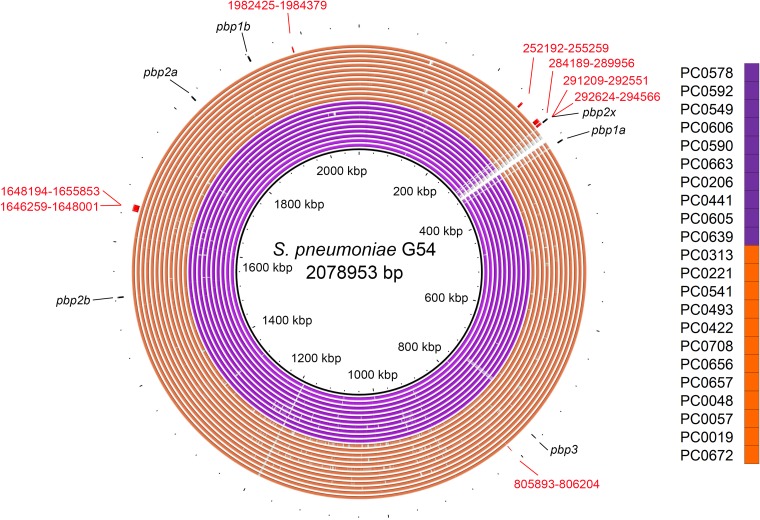
The predicted recombination sites present in all subclade CTXR serotype 15A-ST9084 isolates included eight sites, with one presenting a 1,943-nucleotide overlap of the pbp2x region of S. pneumoniae G54 (from positions 292624 to 294566 of the reference sequence) (Fig. 2 and Fig. S3 and S4) that seemed to be responsible for the additional cefotaxime resistance. The genes that included all of the recombination sites are listed in Table S1 in the supplemental material.
FIG 2.
Genomic similarities between Streptococcus pneumoniae G54 (GenBank accession no. NC_011072.1) and multi-beta-lactam-resistant serotype 15A-ST9084-specific recombination sites predicted by Gubbins. The specific recombination sites show the predicted recombination sites identified within all subclade CTXR isolates (orange rings) that were not identified within any remaining clade Y isolates (purple rings). Colored segments indicate >95% similarity, and gray segments indicate >90% similarity, as indicated by a BLAST comparison between each isolate genome and Streptococcus pneumoniae G54. Red numbers indicate the sequence coordinates of the recombination sites using Streptococcus pneumoniae G54. Black arrows indicate each penicillin-binding protein region.
PBP profile and three conserved amino acid motifs. (i) pbp1a.
Sixty-five isolates, including all 24 MBLR isolates, presented pbp1a-13, which was identified as the cause of meropenem resistance in serotype 15A-ST63 strains in Japan in a previous study (23) (Table 2 and Fig. 1). The other 21 isolates contained pbp1a-24, which was identified in Sweden15A-25 (PMEN15A-25). Most of the isolates with pbp1a-24 were PCG-R (21/21 isolates), cefotaxime susceptible (CTX-S) (19/21 isolates), and meropenem susceptible (MEM-S) (21/21 isolates). The three conserved amino acid motifs of pbp1a-13 were 370SSMK, 466SSN, and 557KTG (Table S2). The sequences of the PBP transpeptidase domains that were not reported in the CDC database but were identified among the Japanese isolates are listed in Data Set S3 in the supplemental material.
TABLE 2.
PBP profile of each antimicrobial susceptibility patterna
| MEM susceptibility |
CTX susceptibility |
pbp1a type:pbp2b type:pbp2x type (no. of strains) |
|---|---|---|
| MEM-S | CTX-S | 24:27:43 (15), 24:27:28 (3), 24:27:JP1 (1), 24:57:43 (1) |
| CTX-IR | 24:27:112 (1) | |
| CTX-R | 13:JP1:JP19 (1) | |
| MEM-IR or -R | CTX-S | 13:JP1:43 (40) |
| CTX-IR | 13:JP1:43 (8), 13:JP1:JP2 (2) | |
| CTX-R | 13:JP1:JP3 (12), 13:JP1:147 (1), 13:JP1:JP6 (1) | |
MEM-R, meropenem resistant; MEM-IR, meropenem intermediate resistant; MEM-S, meropenem susceptible; CTX-R, cefotaxime resistant; CTX-IR, cefotaxime intermediate resistant; CTX-S, cefotaxime susceptible.
(ii) pbp2b.
Sixty-five isolates presented pbp2b-JP1 with pbp1a-13, and they included all 24 MBLR serotype 15A isolates. The other 21 isolates, except for one, contained pbp2b-27 (Table 2 and Fig. 1). This pbp2b-JP1 isolate was also identified in the meropenem-intermediate-resistant (MEM-IR) or MEM-R serotype 15A-ST63 strains in Japan in a previous study (23). Of the 65 isolates with pbp2b-JP1, 40 were CTX-S. The three conserved amino acid motifs of pbp2b-JP1 were 391SVVK, 448SSNA, and 620KTG (Table S3).
(iii) pbp2x.
The pbp2x types of the 24 MBLR serotype 15A isolates were more diverse than those of the other two PBPs and consisted of five different types. The most common pbp2x type was pbp2x-JP3 (n = 12), followed by pbp2x-43 (n = 8) and pbp2x-JP2 (n = 2) (Table 2 and Fig. 1). Ten of the 12 isolates with pbp2x-JP3 showed an MIC of 4 μg/ml for CTX; in contrast, all 8 isolates with pbp2x-43 showed an MIC of 1 μg/ml. All of the serotype 15A-ST9084 isolates contained pbp2x-JP3. In the reference pbp2x type database, pbp2x-33 was closest to pbp2x-JP3, which possessed 2 amino acid (aa) sequence mutations. Although all MBLR serotype 15A isolates contained the 337SAFK or SAMK motif, all 13 isolates with the SAFK motif were CTX-R; in contrast, 10 of 11 isolates with SAMK were CTX-IR (Table S4). In addition, all MBLR serotype 15A isolates had the 395SSN and 547KSG motifs.
Other resistance genes and pilus prevalence.
All of the Japanese serotype 15A-CC63 isolates contained the tetM and ermB genes and were negative for the ermTR, tetO, mefA, and mefE genes; the folA mutation; and the folP insertion. In addition, all Japanese serotype 15A-CC63 isolates lacked the pilus determinants PI-1 and PI-2 (Data Set S2).
Genome comparison via core-genome analysis.
A core-genome analysis was performed, and we identified one MBLR serotype 15A-ST9084 isolate-specific gene that was submitted as a pneumococcal hypothetical protein under accession no. WP_087671931.1 in the NCBI database (see the supplemental material).
Dating the origin of the multi-beta-lactam-resistant serotype 15A-ST9084 clone.
The date of origin for the subclade CTXR lineage was estimated using BEAST, which predicted a most recent lineage origination date (year) of 2001.8 and a 95% highest posterior density (HPD) interval between 1964.8 and 2010.8 (Fig. S5).
Variations in the serotype 15A-CC63 cps loci.
We obtained 15,432 bp of aligned cps locus sequences and created a maximum likelihood tree. There were variations in this region even among serotype 15A-CC63 isolates (Fig. S6); only 4 of 86 isolates from Japan and one PMEN15A-25 isolate contained the same sequence. Six of 10 serotype 15A-ST9084 isolates were clustered; all isolates in the cluster were from the Yamaguchi prefecture.
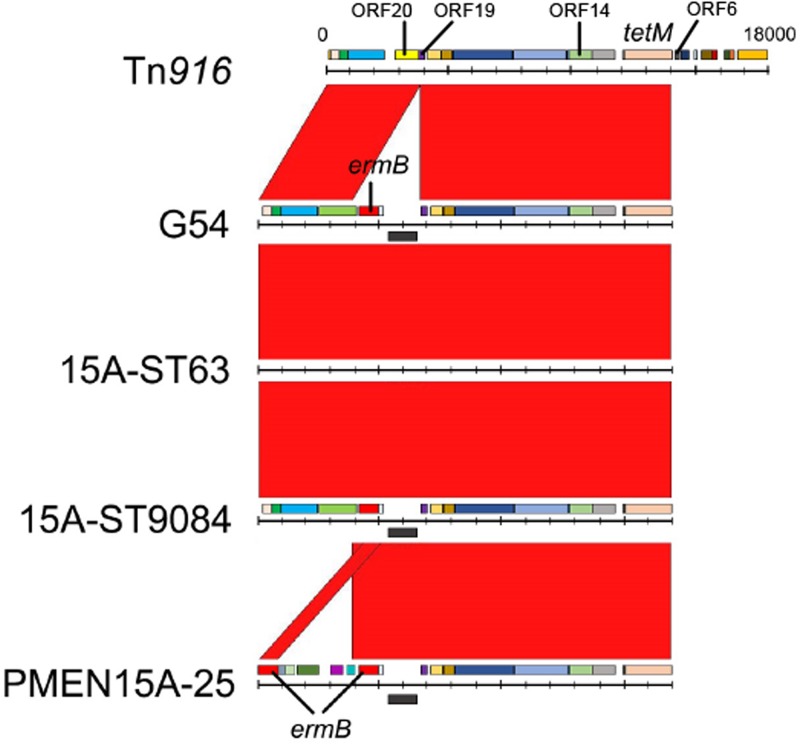
Structure of the Tn916-like integrative conjugative element (ICE).
Among the 86 serotype 15A-CC63 isolates from Japan, the ermB gene was inserted between open reading frame 19 (ORF19) and ORF20 of Tn916, as in Tn6002 (25); 6 of the 86 isolates had a 40-amino-acid deletion in ORF14 that was annotated as iap (Fig. 3). PMEN15A-25 had the same ermB insertion as the serotype 15A-CC63 isolates in Japan; however, the components upstream of the ermB 3′ end were different from those of serotype 15A-CC63 isolates from Japan, and PMEN15A-25 contained another ermB gene, such as in Tn6003. All analyzed isolates, including PMEN15A-25, lacked the sequence from ORF6 to int of Tn916.
FIG 3.
Comparison of the Tn916-like integrative conjugative elements (ICE) of serotype 15A isolates and the S. pneumoniae G54 strain. Red bands between the sequences indicate BLASTN matches. The ermB element was inserted between ORF19 and ORF20 of Tn916. The ICE structures were homologous among the G54, serotype 15A-ST63, and serotype 15A-ST9084 strains; however, the PMEN15A-25 strain had different insertions at the same positions as the other strains.
DISCUSSION
Although the incidence of invasive pneumococcal infections dramatically decreased after the introduction of PCVs (2, 7, 8, 26, 27), the emergence and spread of resistant clones have been problematic (3, 23, 28–31). In particular, bacterial resistance to penicillin, cephalosporins, and carbapenems may affect the outcomes of pneumococcal infections because these antimicrobials are commonly used in many clinical settings. In the PCV13 era, increases in serotype 15A-ST63 pneumococcal infections and colonization have been observed in many countries (9–17, 19), and only isolates derived from Japan were previously shown to be resistant to meropenem (23). In addition, although most of these serotype 15A-CC63 isolates were susceptible to cefotaxime, we realized the emergence of additional cefotaxime-resistant isolates in this lineage; hence, we aimed to elucidate the mechanism of this resistance in this study.
We analyzed 24 MBLR (resistant to penicillin, cefotaxime, and meropenem) serotype 15A-CC63 isolates and another 62 serotype 15A-CC63 isolates using whole-genome sequencing. The results demonstrated that the additional cefotaxime resistance correlated with recombination events in the pbp2x region, and most of the MBLR isolates had a pbp2x type other than pbp2x-43, which was present mainly in CTX-S isolates. Of note, the isolates with pbp2x-JP3 showed higher MIC values for cefotaxime than those with the other pbp2x type, and most of these isolates belonged to ST9084.
All of the serotype 15A-ST9084 (SLV of ST63) isolates were MBLR, and this clone appears to have originated from the prevalent serotype 15A-ST63 clone according to results of the phylogenomic analysis. In the two phylogenic trees, one was created using global serotype 15A-CC63 isolates, and the other was created using only Japanese isolates. The serotype 15A-ST9084 isolates clustered exclusively and were included with the Japanese clusters. The identification of Japan-specific pbp2b-JP1 and pbp2x-JP3 in the serotype 15A-ST9084 strains may support this finding.
These serotype 15A-ST9084 isolates were derived from different cases between 2012 and 2014 in three cities in the Yamaguchi prefecture in Japan. The ages of the patients and dates of the cases indicated that these cases were not caused by an outbreak event but rather were caused by small outbreaks and sporadic incidents; therefore, this MBLR lineage may have already spread within the Yamaguchi prefecture. In addition, we are concerned about the future spread of this clone throughout Japan because the currently used PCV13 does not contain serotype 15A. A previous study in Japan demonstrated that the number of pediatric infection cases attributable to serotype 15A increased during the period between 2014 and 2016 (32). Moreover, resistance to multiple drugs (penicillin, cefotaxime, meropenem, and macrolides) may allow the clone to spread rapidly, as previously observed in the United States for a multidrug-resistant serotype 19A-CC320/271 clone (24).
The recombination site predictions in this study demonstrated that the additional cefotaxime resistance in serotype 15A-CC63, especially in the serotype 15A-ST9084 lineage, was caused by recombination events in the pbp2x region. We believe that developing a greater understanding of the mechanism of recombination events that cause beta-lactam resistance, serotype switching, and increases/decreases in fitness cost, etc., is very important for the control of pneumococcal infections. With regard to antibiotic resistance, recombination events that occur in the PBP regions (especially in the pbp1a, pbp2b, and pbp2x regions) are involved in beta-lactam resistance (33). Although the direct influence of the pneumococcal environment, such as antibiotic pressure and vaccine pressure, etc., on the recombination events is not well known, recent antibiotic use and pediatric serotypes (i.e., serotypes commonly found in children) were suggested as risk factors for the development of resistance among IPD cases (34–36). S. pneumoniae is naturally transformable by the uptake and chromosomal integration of DNA from other pneumococcal strains and other related nonpathogenic mitis group species. Therefore, we believe that future studies on pneumococcal genetic ecology, including the mechanisms underlying genetic evolution, should focus on both species.
With respect to conserved amino acid motifs in the PBP transpeptidase domains, all MBLR serotype 15A-ST9084 isolates had the 337SAFK motif in pbp2x-43. Although this 337SAFK motif was identified in only the MBLR serotype 15A-ST9084 isolates in this study, we observed the same sequence in the multidrug-resistant serotype 19A-ST320 isolates from a previous U.S. data set (3). Comparing the other pbp1a, pbp2b, and pbp2x motifs between serotype 15A-ST9084 isolates in Japan and serotype 19A-ST320 isolates in the United States, all eight motifs also had the same sequences, suggesting that the basic mechanism of cephalosporin resistance in these two clones is identical. However, this match must be coincidental because these strains are not closely related based on the phylogenetic tree reported in a previous study (23), and the PBPs are associated with certain distances in the whole-genome sequence.
Estimation of the date of origin for the most recent common ancestor (MRCA) of the MBLR serotype 15A-ST9084 clone showed that the clone emerged in approximately 2002, prior to the introduction of PCVs. This finding may explain the dynamics of the resistant clone, as the clone developed naturally by chance in the pre-PCV era and subsequently spread in response to PCV pressure and/or other factors, such as antimicrobial use and changes in lifestyle (including child care systems). Interestingly, Obolski et al. and Lehtinen et al. demonstrated that vaccination impacts antibiotic resistance frequencies in pneumococci by removing competition from susceptible strains and prolonging the duration of carriage of resistant isolates (37, 38). The results of these studies clearly support the results of this study.
Tn916-like ICE analysis demonstrated that all serotype 15A-CC63 isolates in Japan had Tn6002-like ICEs; this result was consistent with data from a previous publication by Santoro et al. that demonstrated that S. pneumoniae G54, the predicted ancestor of the serotype 15A-ST63 strain, had a defective Tn6002-like ICE (39). Although our analysis was based on assembled contigs, which resulted in a lack of data concerning the downstream sequences of tetM, all Tn6002-like ICEs identified in serotype 15A-CC63 isolates in Japan seemed to lack genes from ORF6 to int of Tn6002. Interestingly, the PMEN15A-25 isolate that was recovered in 1998 in Portugal had a Tn916-like ICE different from that of the Japanese isolates (i.e., a defective Tn6003-like ICE); however, PMEN15A-25 also lacked the sequences downstream of tetM. Considering that S. pneumoniae G54 was isolated in 1991 before the PMEN15A-25 strain was recovered, the ermB insertion region of S. pneumoniae G54 may be replaced by that of PMEN15A-25; thus, the defective Tn6003-like ICE was inserted in the PMEN15A-25 isolate.
In this study, we identified a PBP motif associated with cefotaxime resistance in an emerging lineage of MBLR serotype 15A-ST9084 strains in Japan. Although the incidence of this strain was identified in only one prefecture in Japan, additional and careful monitoring is warranted due to its high level of resistance. To perform multifaceted surveillance, the global PBP profile database should be enhanced.
MATERIALS AND METHODS
Patient data and bacterial isolates.
In a previous nationwide prospective surveillance study of pediatric invasive pneumococcal disease (IPD) (n = 343) and non-IPD (n = 286) isolates collected from January 2012 to December 2014 from 154 Japanese medical institutions, 87 of 92 serotype 15A isolates belonged to MLST clonal complex 63 (CC63) (19). We analyzed all 87 isolates except one, which did not grow from the stock. Patient data were collected during the surveillance period using a standardized questionnaire, as described previously.
Antimicrobial susceptibility definition.
Based on 2008 Clinical and Laboratory Standards Institute (CLSI) guidelines (52), we performed susceptibility testing for penicillin, cefotaxime, meropenem, and erythromycin using the broth microdilution method, as described previously (19). Penicillin G-susceptible (PG-S) and penicillin G-resistant (PG-R) isolates were defined as having MICs of ≤0.06 and ≥0.12 mg/liter, respectively. Cefotaxime-susceptible (CTX-S), cefotaxime-intermediate-resistant (CTX-IR), and cefotaxime-resistant (CTX-R) isolates were defined as having MICs of ≤0.5, 1.0, and ≥2.0 mg/liter, respectively. Meropenem-susceptible (MEM-S), meropenem-intermediate-resistant (MEM-IR), and meropenem-resistant (MEM-R) isolates were defined as having MICs of ≤0.25, 0.5, and ≥1.0 mg/liter, respectively. Multi-beta-lactam resistance (MBLR) was defined as PG-R, CTX-R or -IR, and MEM-R or -IR.
Whole-genome sequencing.
Of the 86 serotype 15A-CC63 isolates described above, 34 were analyzed in our previous study using whole-genome sequencing (23). Therefore, we obtained additional whole-genome sequencing data for the 52 remaining isolates in this study. Total genomic DNA was extracted, and sequence libraries were prepared using a QIAamp DNA minikit (Qiagen, Hilden, Germany) and a Nextera XT DNA library preparation kit (Illumina, San Diego, CA, USA). We multiplexed and sequenced the samples on an Illumina NextSeq system for 300 cycles (2- by 150-bp paired-end reads). Isolates with N50 values of <20,000 were excluded from subsequent analysis. The resulting sequences were assembled using SPAdes v3.12.0 (40), followed by MLST (http://pubmlst.org/spneumoniae/) using BLAST+ v2.6.0 (41).
PBP typing, antimicrobial resistance genes, and pilus detection.
We allocated PBP transpeptidase domain type numbers to the extracted pbp1a, pbp2b, and pbp2x transpeptidase domain sequences of the examined isolates. The type numbers originated from the CDC’s PBP types, which were reported in previous studies (3, 20–22). PBP types that were not previously reported in the CDC database (22) were numbered with the prefix “JP,” such as in pbp1a-JP1. Three conserved amino acid motifs (SXXK, SXN, and KT/SG) of the three PBPs were identified according to the R6 amino acid sequence (GenBank accession no. NC_003098.1). In addition, we identified the presence of the ermB, ermTR, mefA, mefE, tetM, tetO, rrgA1 (pilin 1), and pitB1 (pilin 2) genes and searched for mutations within the folA and folP genes in the assembled contigs (3).
Phylogenetic analysis.
To predict recombination sites and create a phylogenetic tree, we used Genealogies Unbiased by Recombinations in Nucleotide Sequences (Gubbins) v1.4.6 (42), which identifies recombination sites and constructs a phylogenetic tree using an algorithm that iteratively identifies loci containing increased densities of base substitutions while constructing a phylogenetic tree based on putative point mutations outside the recombination regions. S. pneumoniae G54 (GenBank accession no. NC_011072.1) was used as a reference sequence because a previous study suggested that this strain is closely related to serotype 15A-ST63 isolates (43). To elucidate the relationships among the serotype 15A-CC63 lineages in Japan, the United Kingdom, and the United States, we downloaded the foreign isolates’ whole-genome read data from the Sequence Read Archive (SRA) database (https://www.ncbi.nlm.nih.gov/sra/) (see Data Set S2 in the supplemental material) (44, 45) and created a phylogenetic tree using all of the isolates. With regard to the foreign isolates, serotypes and STs were examined by extracting the corresponding genetic regions with BLAST+ v2.6.0, as previously described (23). Following this process, we created additional trees, one tree using all the data for the Japanese isolates and another tree using isolates included in the serotype 15A-ST9084-specific clade, to predict recombination sites that could cause cefotaxime resistance. The genes identified at the recombination sites were annotated using Prokka v1.12 (46), and the protein sequences were searched in the NCBI database using ACT v18.0.0 (47).
Dating the origin of the multi-beta-lactam-resistant serotype 15A-ST9084 clone.
The date of the most recent common ancestor (MRCA) of the MBLR serotype 15A-ST9084 clone was estimated using BEAST v1.10.4 with isolate alignment data obtained from the read data (48). In this analysis, we used serotype 15A-ST9084-specific clade isolates. The tree was used to analyze the final maximum likelihood tree and the alignment of base substitutions occurring outside putative recombination regions using an uncorrelated relaxed-clock model. The tree was calibrated using the isolation dates (month and year) of the strains. The coalescent exponential population was used as the tree prior, and the length of the chain value was set such that all output values had an effective sample size of >200.
Genome comparison by core genome analysis.
To identify the genes that were specific to the MBLR serotype 15A-ST9084 isolates, we performed pangenome and core genome analyses using get_homologues (49). To prepare the input data, the assembled contigs of each isolate were annotated using Prokka v1.12 (46). The base sequences of the identified genes that were specific to the MBLR serotype 15A-ST9084 isolates were searched in the NCBI database using BLAST+ v2.6.0.
cps locus and Tn916-like integrative conjugative element comparison.
A phylogenetic tree was constructed for the serotype 15A cps locus using RAxML v8.2.10 (50) by mapping short reads of each isolate to previously reported reference sequences (51). To clarify the structure of the Tn916-like integrative conjugative element (ICE), the corresponding regions were extracted from the de novo strain assemblies. The sequences were compared using ACT v18.0.0 (47) with standard parameters in BLAST+ v2.6.0.
Details of the genomic analysis process are described in the supplemental material.
Data availability.
The accession numbers of the sequence data are available in Data Set S1 in the supplemental material.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Toshiaki Ihara for his substantial contribution to the Pneumocatch surveillance study. We are also grateful to the members of the Pneumocatch surveillance study group.
Bin Chang was supported by research funding (16fk0108311j0403) from the Japan Agency for Medical Research and Development. Takao Fujisawa was supported by a research grant through his institution from Pfizer Inc. Yutaka Ito was supported by a research grant through his institution from Daiichi-Sankyo, MSD, and the Japan Society for the Promotion of Science (17K10023). Yutaka Ito and Satoshi Nakano were supported by research funding from Pfizer for the surveillance study.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.02579-18.
REFERENCES
- 1.O’Brien KL, Wolfson LJ, Watt JP, Henkle E, Deloria-Knoll M, McCall N, Lee E, Mulholland K, Levine OS, Cherian T. 2009. Burden of disease caused by Streptococcus pneumoniae in children younger than 5 years: global estimates. Lancet 374:893–902. doi: 10.1016/S0140-6736(09)61204-6. [DOI] [PubMed] [Google Scholar]
- 2.Chiba N, Morozumi M, Shouji M, Wajima T, Iwata S, Ubukata K. 2014. Changes in capsule and drug resistance of pneumococci after introduction of PCV7, Japan, 2010–2013. Emerg Infect Dis 20:1132–1139. doi: 10.3201/eid2007.131485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Metcalf BJ, Gertz RE Jr, Gladstone RA, Walker H, Sherwood LK, Jackson D, Li Z, Law C, Hawkins PA, Chochua S, Sheth M, Rayamajhi N, Bentley SD, Kim L, Whitney CG, McGee L, Beall B. 2016. Strain features and distributions in pneumococci from children with invasive disease before and after 13 valent conjugate vaccine implementation in the United States. Clin Microbiol Infect 22:60.e9–60.e29. doi: 10.1016/j.cmi.2015.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Song JY, Nahm MH, Moseley MA. 2013. Clinical implications of pneumococcal serotypes: invasive disease potential, clinical presentations, and antibiotic resistance. J Korean Med Sci 28:4–15. doi: 10.3346/jkms.2013.28.1.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Suga S, Chang B, Asada K, Akeda H, Nishi J, Okada K, Wakiguchi H, Maeda A, Oda M, Ishiwada N, Saitoh A, Oishi T, Hosoya M, Togashi T, Oishi K, Ihara T. 2015. Nationwide population-based surveillance of invasive pneumococcal disease in Japanese children: effects of the seven-valent pneumococcal conjugate vaccine. Vaccine 33:6054–6060. doi: 10.1016/j.vaccine.2015.07.069. [DOI] [PubMed] [Google Scholar]
- 6.Schroeder MR, Chancey ST, Thomas S, Kuo WH, Satola SW, Farley MM, Stephens DS. 2017. A population-based assessment of the impact of 7- and 13-valent pneumococcal conjugate vaccines on macrolide-resistant invasive pneumococcal disease: emergence and decline of Streptococcus pneumoniae serotype 19A (CC320) with dual macrolide resistance mechanisms. Clin Infect Dis 65:990–998. doi: 10.1093/cid/cix446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Camilli R, D’Ambrosio F, Del Grosso M, Pimentel de Araujo F, Caporali MG, Del Manso M, Gherardi G, D’Ancona F, Pantosti A. 2017. Impact of pneumococcal conjugate vaccine (PCV7 and PCV13) on pneumococcal invasive diseases in Italian children and insight into evolution of pneumococcal population structure. Vaccine 35:4587–4593. doi: 10.1016/j.vaccine.2017.07.010. [DOI] [PubMed] [Google Scholar]
- 8.Waight PA, Andrews NJ, Ladhani NJ, Sheppard CL, Slack MP, Miller E. 2015. Effect of the 13-valent pneumococcal conjugate vaccine on invasive pneumococcal disease in England and Wales 4 years after its introduction: an observational cohort study. Lancet Infect Dis 15:629. doi: 10.1016/S1473-3099(15)00028-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duvvuri VR, Deng X, Teatero S, Memari N, Athey T, Fittipaldi N, Gubbay JB. 2016. Population structure and drug resistance patterns of emerging non-PCV-13 Streptococcus pneumoniae serotypes 22F, 15A, and 8 isolated from adults in Ontario, Canada. Infect Genet Evol 42:1–8. doi: 10.1016/j.meegid.2016.04.007. [DOI] [PubMed] [Google Scholar]
- 10.van der Linden M, Perniciaro S, Imöhl M. 2015. Increase of serotypes 15A and 23B in IPD in Germany in the PCV13 vaccination era. BMC Infect Dis 15:207. doi: 10.1186/s12879-015-0941-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sheppard C, Fry NK, Mushtaq S, Woodford N, Reynolds R, Janes R, Pike R, Hill R, Kimuli M, Staves P, Doumith M, Harrison T, Livermore DM. 2016. Rise of multidrug-resistant non-vaccine serotype 15A Streptococcus pneumoniae in the United Kingdom, 2001 to 2014. Euro Surveill 21(50):pii=30423. doi: 10.2807/1560-7917.ES.2016.21.50.30423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chi HC, Hsieh YC, Tsai MH, Lee CH, Kuo KC, Huang CT, Huang YC. 2018. Impact of pneumococcal conjugate vaccine in children on the serotypic epidemiology of adult invasive pneumococcal diseases in Taiwan. J Microbiol Immunol Infect 51:332–336. doi: 10.1016/j.jmii.2016.08.009. [DOI] [PubMed] [Google Scholar]
- 13.Cilveti R, Olmo M, Perez-Jove J, Picazo JJ, Arimany JL, Mora E, Perez-Porcuna TM, Aguilar I, Alonso A, Molina F, Del Amo M, Mendez C. 2017. Epidemiology of otitis media with spontaneous perforation of the tympanic membrane in young children and association with bacterial nasopharyngeal carriage, recurrences and pneumococcal vaccination in Catalonia, Spain—the prospective HERMES study. PLoS One 12:e0170316. doi: 10.1371/journal.pone.0170316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Devine VT, Cleary DW, Jefferies JM, Anderson R, Morris DE, Tuck AC, Gladstone RA, O’Doherty G, Kuruparan P, Bentley SD, Faust SN, Clarke SC. 2017. The rise and fall of pneumococcal serotypes carried in the PCV era. Vaccine 35:1293–1298. doi: 10.1016/j.vaccine.2017.01.035. [DOI] [PubMed] [Google Scholar]
- 15.Kaur R, Casey JR, Pichichero ME. 2016. Emerging Streptococcus pneumoniae strains colonizing the nasopharynx in children after 13-valent pneumococcal conjugate vaccination in comparison to the 7-valent era, 2006-2015. Pediatr Infect Dis J 35:901–906. doi: 10.1097/INF.0000000000001206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horacio AN, Silva-Costa C, Lopes JP, Ramirez M, Melo-Cristino J. 2016. Serotype 3 remains the leading cause of invasive pneumococcal disease in adults in Portugal (2012-2014) despite continued reductions in other 13-valent conjugate vaccine serotypes. Front Microbiol 7:1616. doi: 10.3389/fmicb.2016.01616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Soysal A, Karabağ-Yılmaz E, Kepenekli E, Karaaslan A, Cagan E, Atıcı S, Atınkanat-Gelmez G, Boran P, Merdan S, Hasdemir U, Söyletir G, Bakır M. 2016. The impact of a pneumococcal conjugate vaccination program on the nasopharyngeal carriage, serotype distribution and antimicrobial resistance of Streptococcus pneumoniae among healthy children in Turkey. Vaccine 34:3894–3900. doi: 10.1016/j.vaccine.2016.05.043. [DOI] [PubMed] [Google Scholar]
- 18.Sa-Leao R, Tomasz A, de Lencastre H. 2001. Multilocus sequence typing of Streptococcus pneumoniae clones with unusual drug resistance patterns: genetic backgrounds and relatedness to other epidemic clones. J Infect Dis 184:1206–1210. doi: 10.1086/323663. [DOI] [PubMed] [Google Scholar]
- 19.Nakano S, Fujisawa T, Ito Y, Chang B, Suga S, Noguchi T, Yamamoto M, Matsumura Y, Nagao M, Takakura S, Ohnishi M, Ihara T, Ichiyama S. 2016. Serotypes, antimicrobial susceptibility, and molecular epidemiology of invasive and non-invasive Streptococcus pneumoniae isolates in paediatric patients after the introduction of 13-valent conjugate vaccine in a nationwide surveillance study conducted in Japan in 2012-2014. Vaccine 34:67–76. doi: 10.1016/j.vaccine.2015.11.015. [DOI] [PubMed] [Google Scholar]
- 20.Metcalf BJ, Chochua S, Gertz RE Jr, Li Z, Walker H, Tran T, Hawkins PA, Glennen A, Lynfield R, Li Y, McGee L, Beall B, Active Bacterial Core Surveillance Team. 2016. Using whole genome sequencing to identify resistance determinants and predict antimicrobial resistance phenotypes for year 2015 invasive pneumococcal disease isolates recovered in the United States. Clin Microbiol Infect 22:1002.e1–1002.e8. doi: 10.1016/j.cmi.2016.08.001. [DOI] [PubMed] [Google Scholar]
- 21.Li Y, Metcalf BJ, Chochua S, Li Z, Gertz RE Jr, Walker H, Hawkins PA, Tran T, Whitney CG, McGee L, Beall BW. 2016. Penicillin-binding protein transpeptidase signatures for tracking and predicting beta-lactam resistance levels in Streptococcus pneumoniae. mBio 7:e00756-16. doi: 10.1128/mBio.00756-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.CDC. 2018. Minimum inhibitory concentrations (MICs) for β-lactam antibiotics predicted by penicillin binding protein gene types. CDC, Atlanta, GA: https://www.cdc.gov/streplab/pneumococcus/mic.html. [Google Scholar]
- 23.Nakano S, Fujisawa T, Ito Y, Chang B, Matsumura Y, Yamamoto M, Nagao M, Suga S, Ohnishi M, Ichiyama S. 2018. Spread of meropenem-resistant Streptococcus pneumoniae serotype 15A-ST63 clone in Japan, 2012-2014. Emerg Infect Dis 24:275–283. doi: 10.3201/eid2402.171268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beall BW, Gertz RE, Hulkower RL, Whitney CG, Moore MR, Brueggemann AB. 2011. Shifting genetic structure of invasive serotype 19A pneumococci in the United States. J Infect Dis 203:1360–1368. doi: 10.1093/infdis/jir052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cochetti I, Tili E, Mingoia M, Varaldo PE, Montanari MP. 2008. erm(B)-carrying elements in tetracycline-resistant pneumococci and correspondence between Tn1545 and Tn6003. Antimicrob Agents Chemother 52:1285–1290. doi: 10.1128/AAC.01457-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luca DL, Kwong JC, Chu A, Sander B, O’Reilly R, McGeer AJ, Bloom DE. 2018. Impact of pneumococcal vaccination on pneumonia hospitalizations and related costs in Ontario: a population-based ecological study. Clin Infect Dis 66:541–547. doi: 10.1093/cid/cix850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mackenzie GA, Hill PC, Sahito SM, Jeffries DJ, Hossain I, Bottomley C, Uchendu U, Ameh D, Ndiaye M, Osuorah CD, Adeyemi O, Pathirana J, Olatunji Y, Abatan B, Ahameefula E, Muhammad BS, Fombah AE, Saha D, Mackenzie R, Plumb I, Akano A, Ebruke B, Ideh RC, Kuti B, Githua P, Olutunde E, Ofordile O, Green E, Usuf E, Badji H, Ikumapayi UNA, Manjang A, Salaudeen R, Nsekpong ED, Jarju S, Antonio M, Sambou S, Ceesay L, Lowe-Jallow Y, Sowe D, Jasseh M, Mulholland K, Knoll M, Levine OS, Howie SR, Adegbola RA, Greenwood BM, Corrah T. 2017. Impact of the introduction of pneumococcal conjugate vaccination on pneumonia in The Gambia: population-based surveillance and case-control studies. Lancet Infect Dis 17:965–973. doi: 10.1016/S1473-3099(17)30321-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Neves FPG, Cardoso NT, Souza ARV, Snyder RE, Marlow MM, Pinto TCA, Teixeira LM, Riley LW. 2018. Population structure of Streptococcus pneumoniae colonizing children before and after universal use of pneumococcal conjugate vaccines in Brazil: emergence and expansion of the MDR serotype 6C-CC386 lineage. J Antimicrob Chemother 73:1206–1212. doi: 10.1093/jac/dky001. [DOI] [PubMed] [Google Scholar]
- 29.Ktari S, Jmal I, Mroua M, Maalej S, Ben Ayed NE, Mnif B, Rhimi F, Hammami A. 2017. Serotype distribution and antibiotic susceptibility of Streptococcus pneumoniae strains in the south of Tunisia: a five-year study (2012-2016) of pediatric and adult populations. Int J Infect Dis 65:110–115. doi: 10.1016/j.ijid.2017.10.015. [DOI] [PubMed] [Google Scholar]
- 30.Chaguza C, Cornick JE, Andam CP, Gladstone RA, Alaerts M, Musicha P, Peno C, Bar-Zeev N, Kamng’ona AW, Kiran AM, Msefula CL, McGee L, Breiman RF, Kadioglu A, French N, Heyderman RS, Hanage WP, Bentley SD, Everett DB. 2017. Population genetic structure, antibiotic resistance, capsule switching and evolution of invasive pneumococci before conjugate vaccination in Malawi. Vaccine 35:4594–4602. doi: 10.1016/j.vaccine.2017.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmitz J, van der Linden M, Al-Lahham A, Levina N, Pletz MW, Imöhl M. 2017. Fluoroquinolone resistance in Streptococcus pneumoniae isolates in Germany from 2004–2005 to 2014–2015. Int J Med Microbiol 307:216–222. doi: 10.1016/j.ijmm.2017.04.003. [DOI] [PubMed] [Google Scholar]
- 32.Ubukata K, Takata M, Morozumi M, Chiba N, Wajima T, Hanada S, Shouji M, Sakuma M, Iwata S. 2018. Effects of pneumococcal conjugate vaccine on genotypic penicillin resistance and serotype changes, Japan, 2010-2017. Emerg Infect Dis 24:2010–2020. doi: 10.3201/eid2411.180326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hakenbeck R, Bruckner R, Denapaite D, Maurer P. 2012. Molecular mechanisms of beta-lactam resistance in Streptococcus pneumoniae. Future Microbiol 7:395–410. doi: 10.2217/fmb.12.2. [DOI] [PubMed] [Google Scholar]
- 34.Dowell SF, Schwartz B. 1997. Resistant pneumococci: protecting patients through judicious use of antibiotics. Am Fam Physician 55:1647–1654, 1657–1658. [PubMed] [Google Scholar]
- 35.Hofmann J, Cetron MS, Farley MM, Baughman WS, Facklam RR, Elliott JA, Deaver KA, Breiman RF. 1995. The prevalence of drug-resistant Streptococcus pneumoniae in Atlanta. N Engl J Med 333:481–486. doi: 10.1056/NEJM199508243330803. [DOI] [PubMed] [Google Scholar]
- 36.Ruhe JJ, Hasbun R. 2003. Streptococcus pneumoniae bacteremia: duration of previous antibiotic use and association with penicillin resistance. Clin Infect Dis 36:1132–1138. doi: 10.1086/374556. [DOI] [PubMed] [Google Scholar]
- 37.Obolski U, Lourenco J, Thompson C, Thompson R, Gori A, Gupta S. 2018. Vaccination can drive an increase in frequencies of antibiotic resistance among nonvaccine serotypes of Streptococcus pneumoniae. Proc Natl Acad Sci U S A 115:3102–3107. doi: 10.1073/pnas.1718712115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lehtinen S, Blanquart F, Croucher NJ, Turner P, Lipsitch M, Fraser C. 2017. Evolution of antibiotic resistance is linked to any genetic mechanism affecting bacterial duration of carriage. Proc Natl Acad Sci U S A 114:1075–1080. doi: 10.1073/pnas.1617849114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Santoro F, Vianna ME, Roberts AP. 2014. Variation on a theme; an overview of the Tn916/Tn1545 family of mobile genetic elements in the oral and nasopharyngeal streptococci. Front Microbiol 5:535. doi: 10.3389/fmicb.2014.00535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 42.Croucher NJ, Page AJ, Connor TR, Delaney AJ, Keane JA, Bentley SD, Parkhill J, Harris SR. 2015. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res 43:e15. doi: 10.1093/nar/gku1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dopazo J, Mendoza A, Herrero J, Caldara F, Humbert Y, Friedli L, Guerrier M, Grand-Schenk E, Gandin C, de Francesco M, Polissi A, Buell G, Feger G, García E, Peitsch M, García-Bustos JF. 2001. Annotated draft genomic sequence from a Streptococcus pneumoniae type 19F clinical isolate. Microb Drug Resist 7:99–125. doi: 10.1089/10766290152044995. [DOI] [PubMed] [Google Scholar]
- 44.Kapatai G, Sheppard CL, Al-Shahib A, Litt DJ, Underwood AP, Harrison TG, Fry NK. 2016. Whole genome sequencing of Streptococcus pneumoniae: development, evaluation and verification of targets for serogroup and serotype prediction using an automated pipeline. PeerJ 4:e2477. doi: 10.7717/peerj.2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Andam CP, Mitchell PK, Callendrello A, Chang Q, Corander J, Chaguza C, McGee L, Beall BW, Hanage WP. 2017. Genomic epidemiology of penicillin-nonsusceptible pneumococci with nonvaccine serotypes causing invasive disease in the United States. J Clin Microbiol 55:1104–1115. doi: 10.1128/JCM.02453-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 47.Carver T, Berriman M, Tivey A, Patel C, Bohme U, Barrell BG, Parkhill J, Rajandream MA. 2008. Artemis and ACT: viewing, annotating and comparing sequences stored in a relational database. Bioinformatics 24:2672–2676. doi: 10.1093/bioinformatics/btn529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Drummond AJ, Rambaut A. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol 7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Contreras-Moreira B, Vinuesa P. 2013. GET_HOMOLOGUES, a versatile software package for scalable and robust microbial pangenome analysis. Appl Environ Microbiol 79:7696–7701. doi: 10.1128/AEM.02411-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bentley SD, Aanensen DM, Mavroidi A, Saunders D, Rabbinowitsch E, Collins M, Donohoe K, Harris D, Murphy L, Quail MA, Samuel G, Skovsted IC, Kaltoft MS, Barrell B, Reeves PR, Parkhill J, Spratt BG. 2006. Genetic analysis of the capsular biosynthetic locus from all 90 pneumococcal serotypes. PLoS Genet 2:e31. doi: 10.1371/journal.pgen.0020031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Clinical and Laboratory Standards Institute. 2008. Performance standards for antimicrobial susceptibility testing; 18th informational supplement CLSI document M100-S18. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession numbers of the sequence data are available in Data Set S1 in the supplemental material.