Candida albicans is known for its ability to form biofilms, which are communities of microorganisms embedded in an extracellular matrix developing on different surfaces. Biofilms are highly tolerant to antifungal therapy.
KEYWORDS: Candida albicans, antifungal tolerance, biofilms, persistence
ABSTRACT
Candida albicans is known for its ability to form biofilms, which are communities of microorganisms embedded in an extracellular matrix developing on different surfaces. Biofilms are highly tolerant to antifungal therapy. This phenomenon has been partially explained by the appearance of so-called persister cells, phenotypic variants of wild-type cells, capable of surviving very high concentrations of antimicrobial agents. Persister cells in C. albicans were found exceptionally in biofilms, while none were detected in planktonic cultures of this fungus. Yet, this topic remains controversial, as others could not observe persister cells in biofilms formed by the C. albicans SC5314 laboratory strain. Due to ambiguous data in the literature, this work aimed to reevaluate the presence of persister cells in C. albicans biofilms. We demonstrated that the isolation of C. albicans “persister cells” as described previously was likely to be the result of the survival of biofilm cells that were not reached by the antifungal. We tested biofilms of SC5314 and its derivatives, as well as 95 clinical isolates, using an improved protocol, demonstrating that persister cells are not a characteristic trait of C. albicans biofilms. Although some clinical isolates are able to yield survivors upon the antifungal treatment of biofilms, this phenomenon is rather stochastic and inconsistent.
INTRODUCTION
The yeast Candida albicans is a commensal of humans but also one of the most prevalent fungal pathogens, responsible for superficial infections as well as life-threatening systemic infections (1). C. albicans is recognized for its ability to form biofilms that are most frequently associated with nosocomial infections, particularly in immunocompromised patients.
C. albicans biofilms are communities of microorganisms with a complex structure composed of different cell types embedded in an extracellular matrix (2–4). They develop on different types of surfaces, either living or inert, and are characterized by their high tolerance to antifungals. Their high antifungal tolerance can result from the properties of the extracellular matrix that can serve as a trap for drug molecules (5–7). An additional source of antifungal tolerance has been proposed to result from the occurrence in biofilms of so-called persister cells, a subpopulation of phenotypic variants of wild-type cells capable of surviving concentrations of antimicrobial agents well above the MIC (8). Persister cells were first described in bacterial cultures as a drug-tolerant subpopulation that upon removal of the antimicrobial agent gave rise to a new population of susceptible cells (9). Persisters are known to be genetically identical to the rest of the population; thus, persistence is a noninherited trait (10–12).
In the clinical setting, persisters are usually associated with relapse of infections and with the development of chronic infections. For bacterial persisters, several mechanisms and pathways involved in their development have been described (13).
In 2006, LaFleur et al. presented the first report of persister cells in biofilms of C. albicans, which could contribute to biofilm tolerance to antifungals (8). In their paper, the authors reported that C. albicans exhibits a biphasic killing curve when exposed to antifungals such as amphotericin B (AMB), chlorhexidine, or a combination of both. This phenomenon is explained by the presence of a multidrug-tolerant subpopulation of persister cells within a biofilm, while planktonic cultures of C. albicans were found to be devoid of persisters. Notably, the experiments for this study were performed using an in vitro biofilm model of C. albicans developed in polystyrene 96-well plates. Following this work and relying on the protocol for persister cells isolation described therein (8), persister cells in C. albicans biofilms were described by a few other groups (14–16). However, later work by Al-Dhaheri and Douglas showed that not all Candida species and strains were able to form persister cells in laboratory-grown biofilms (17). This was in particular the case for C. albicans strain SC5314 (18), the parental strain of almost all C. albicans strains used for functional genomics and molecular genetics studies. Unlike in the previously mentioned papers (8, 14–16), the protocol Al-Dhaheri and Douglas (17) used for persister isolation involved growing biofilms on silicone discs, followed by their immersion into an antifungal solution. As the topic of C. albicans persister cells remains controversial, the main objective of this work was to reevaluate their occurrence in in vitro-grown C. albicans biofilms.
RESULTS AND DISCUSSION
In this work, we aimed to study the occurrence of persister cells in C. albicans biofilms. We applied the protocol published by LaFleur and colleagues, growing the biofilms in RPMI medium and in a 96-well plate format (8). We set up the protocol with 3 C. albicans prototroph strains, namely SC5314, CEC369, and CEC4664; CEC369 and CEC4664 are prototroph derivatives of BWP17 (19) and SN76 (20), respectively. BWP17 and SN76 are independent auxotroph derivatives of SC5314.
We encountered a technical problem at the biofilm recovery step, usually performed by scraping the cells in 1× phosphate-buffered saline (PBS) and vortexing prior to plating (8, 14, 16, 21). In our hands, the cells could not be properly resuspended and plated, as clumps of the biofilms would usually remain stranded inside the tips. Consequently, the CFU numbers obtained were highly varied for all samples, making any further analysis and comparison impossible (data not shown).
We decided to test alternative approaches to circumvent the stickiness of biofilms. Resuspending cells in 20% glycerol-1× PBS for plating helped reduce stickiness but did not improve consistency (data not shown). We hypothesized that EDTA might reduce the adherence of biofilms by binding bivalent cations that are required for the activity of cell surface adhesins (22). Thus, we attempted applying 20% glycerol with a range of EDTA concentrations (0, 50, and 100 mM) for plating. One hundred microliters of EDTA solutions of different concentrations was added to biofilms and left for 10 min at room temperature prior to biofilm disruption by scraping and vortexing. None of the applied EDTA solutions allowed abolishment of stickiness. Additionally, colonies growing on yeast-peptone-dextrose (YPD) agar exhibited a wrinkled morphology, most probably linked to the toxicity of EDTA (23). Finally, we tried adding Tween 20 (0.05%) to PBS. Tween 20 eradicated the problems of stickiness and poor disruption and improved the recovery of cells from the biofilms (Fig. 1). The effect on cell viability was tested using a planktonic culture of SC5314 that was washed and plated on YPD agar using PBS and PBS-Tween 20 solutions. No impact on viability was observed (data not shown). Thus, in the experiments described below, biofilms were resuspended in a 0.05% Tween 20-1× PBS solution.
FIG 1.
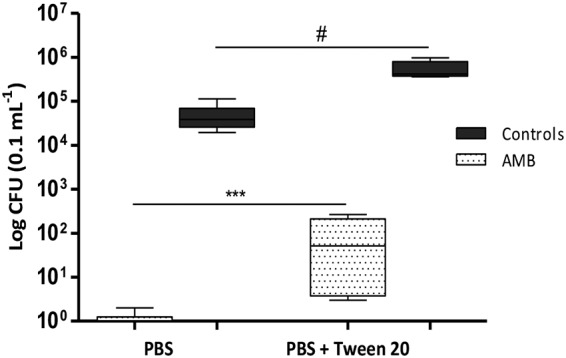
Effect of Tween 20 on the recovery of CFU from C. albicans SC5314 biofilms. C. albicans SC5314 was allowed to form biofilms in 100 μl RPMI medium in a 96-well plate according to the protocol adapted from LaFleur et al. (8). Error bars indicate the standard deviation (SD) of the results from 6 biological replicates generated from 2 independent experiments. #, nonsignificant difference; ***, significant difference (P = 0.0007; unpaired t test was applied to compare data sets).
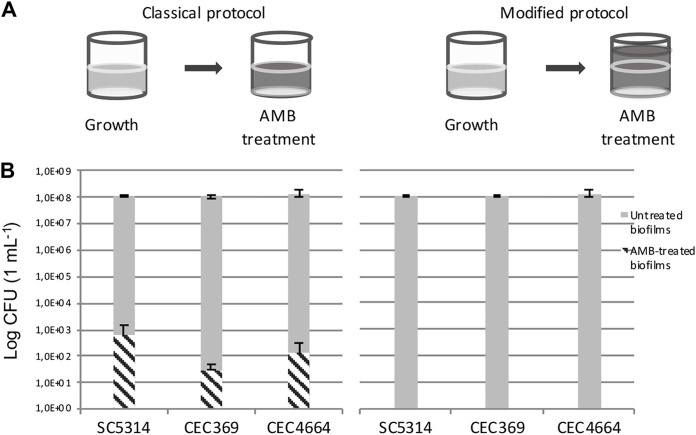
However, even after this modification, the ratios of cells that survived AMB treatment were still inconsistent between repeats. According to LaFleur and colleagues, the ratios of C. albicans persister cells in biofilms vary from 0.1% to 2% for different strains, notably from 0.05 to 0.1% for strain CAI4, a derivative of C. albicans SC5314 (8). Our values hardly ever exceeded 0.01% persisters per biofilm, even after improving the recovery protocol, thus bordering with statistical error. We reasoned that increasing the surface of a biofilm and changing the growth medium could improve persister yields, and we decided to test the protocol described in reference 14, applying the modifications that were mentioned previously. However, the problems of inconsistency and low ratios of persisters remained (Fig. 2).
FIG 2.
Schemes of the protocols (A) and levels of persisters (B) obtained from biofilms grown using modified protocol from Li et al. (14). Biofilms were grown in 1 ml of GHAUM medium in 24-well plates before application of either 1 ml of AMB solution (left) or 3 ml of AMB solution (right). Ratios of surviving cells are as follows: SC5314, 5.6 × 10−4%; CEC369, 2.6 × 10−5%; and CEC4664, 9.4 × 10−5%. Error bars indicate the SD of the results from 6 biological replicates generated from 2 independent experiments.
In all protocols described previously, the volumes of the medium and solutions used for biofilm growth, washing, and AMB treatment were identical. Upon careful observation, we noticed that C. albicans cells form a dense rim at the border of the air and liquid phases as a result of agitation during growth. Treating a biofilm with the exact same volume of antifungal and growth medium under static conditions thus could result in cells from the rim escaping treatment. We decided to increase the volume of the applied antifungal solution (filling wells to the top), and, to our surprise, this change in the protocol led to a complete eradication of persisters for the laboratory strain SC5314 and its derivatives. Reproducibly, we did not obtain any persisters after applying this change for all strains for biofilms grown in both RPMI and GHAUM (Sabouraud dextrose [SD] supplemented with 2% glucose and 1 mg/ml histidine, 1 mg/ml arginine, 0.02 mg/ml uridine, and 2 mg/ml methionine) media. Thus, the volume of the antifungal applied in the original protocols for persister isolation was skewing the results. Increasing the volume of antifungal eliminated this bias, resulting in a complete eradication of any survivors after the antifungal treatment.
In our work, we used a modified protocol for persister cell isolation with a starting cell suspension of an optical density at 600 nm (OD600) of 0.3 used for biofilm growth instead of 0.1 as described in the original protocols (8, 14). To assess the impact of the initial cell number used for seeding biofilms on persister cells’ appearance, we tested our protocol for SC5314 using cell suspensions at an OD600 of 0.1, 0.3, and 0.5 for seeding. Regardless of the initial biomass, persister cells did not form in SC5314 biofilms grown either in RPMI or GHAUM medium (data not shown).
These results made us question the very existence of persister cells in C. albicans biofilms. Previously, Al-Dhaheri and Douglas showed that not all strains of C. albicans can form persister cells (17). Particularly, in their hands, SC5314 biofilms lost all viability after exposure to 30 μg/ml AMB. However, biofilms of another clinical isolate, GDH2346, appeared to contain a small proportion (0.01%) of cells that survived 100 μg/ml AMB treatment. These authors used a different in vitro model for assessing persistence, as they grew biofilms on silicone discs that were transferred to a new well filled with an antifungal solution. This prevented the escape of any cells from the antifungal treatment. Thus, our modified protocol for the treatment of biofilms formed in 96-well or 24-well plates corroborated the results obtained by the Douglas group for C. albicans strain SC5314 (17).
Since the clinical isolate GDH2346 could give rise to survivors (17), we could not exclude that persisters could emerge in biofilms of different C. albicans isolates. Additionally, in 2010, LaFleur and colleagues isolated and described C. albicans strains from patients with long-term oral infection that gave yield to increased levels of persisters (up to 8.9%) (21). These were called hip mutants, by analogy with the high-persister strains previously described for bacteria (24, 25). Although hip mutants were identified using a protocol that showed limitations in our hands, we hypothesized that some C. albicans clinical isolates could generally be more prone to form persisters than others (namely SC5314). To test this assumption, we tested 95 clinical isolates (see Table S1 in the supplemental material) for their ability to form biofilms and for the occurrence of persister cells following AMB treatment. In a first round of experiments, biofilms were treated with a 64 μg/ml AMB solution. Only 38 isolates (39.6%) displayed survivors (notably, never exceeding a rate of 0.02%). According to the generally accepted concept of persistence (10), the frequency of persisters’ appearance is independent of the increase in antibiotic concentration. Thus, in a second round of experiments, biofilms were developed for these 38 isolates and treated with a 100 μg/ml AMB solution. Notably, only 7 isolates out of these 38 displayed survivors when grown with 100 μg/ml AMB (CEC3622, CEC3668, CEC3669, CEC4514, CEC4521, CEC5317, CEC5318). These 7 strains, together with 4 other isolates randomly picked in the remaining 31 strains (CEC712, CEC3708, CEC3711, and CEC5316), were tested seven more times with 100 μg/ml AMB. In most cases, these strains did not yield persister cells (Fig. 3); however, 7 strains (CEC3622, CEC3669, CEC4514, CEC4521, CEC5316, CEC5317, and CEC5318) gave rise to small numbers of survivors in 1 to 4 of the experiments (Fig. 3), with the survival rate never exceeding 9.1 × 10−4% per biofilm (for CEC3622). This could be explained either by the stochastic nature of persistence as a phenomenon or by technical errors during the experiment.
FIG 3.
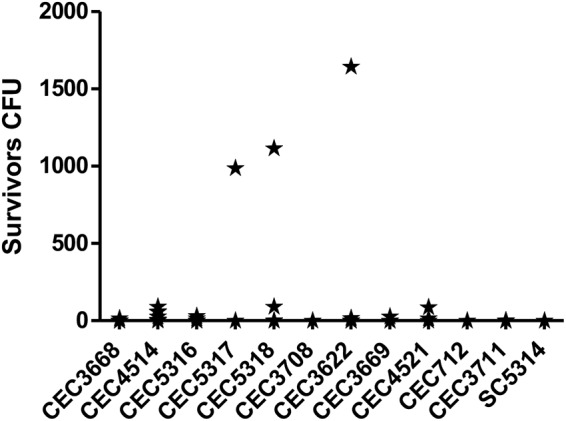
Analysis of persister cell formation in 11 clinical isolates. Biofilms were grown in 1 ml of GHAUM medium in 24-well plates and treated with 3 ml of AMB solution (modified protocol from Li et al. [14]). The values obtained from 7 biofilms were used to draw the graph.
We tested up to 30 randomly picked colonies for three isolates (CEC3622, CEC4514, and CEC5316) on yeast-nitrogen-glucose (YNG) medium containing 10 μg/ml AMB. None of the tested colonies were able to grow in the presence of amphotericin B (data not shown), proving that their survival was not a result of AMB resistance development.
With an improved protocol in our hands, we decided to test other Candida species for their ability to form persister cells in biofilms. Previously, Al-Dhaheri and Douglas (17) reported that clinical isolates of Candida krusei (Glasgow strain) and Candida parapsilosis (AAHB 4479) developed persister cells in biofilms (approximately 0.001% and 0.07%, respectively) upon treatment with 100 μg/ml AMB. We selected 3 clinical isolates of Candida tropicalis (CEC5296, CEC5297, and CEC5298) and 3 of C. parapsilosis (CEC5299, CEC5300, and CEC5301) from our lab collection to test with our protocol. One of the C. tropicalis strains (CEC5298) and the 3 selected C. parapsilosis strains were unable to grow as biofilms and were excluded from the study. C. tropicalis CEC5296 and CEC5297 formed proper biofilms, with a small fraction of persisters ranging from 2 × 10−5 to 6.4 × 10−3% and 2.3 × 10−7 to 2.6 × 10−4%, respectively (data not shown). Such low values are comparable to the survival rates we observed for some of the C. albicans clinical isolates tested in this study. As before, we cannot exclude that these survivors are persister cells arising within C. tropicalis biofilms or that they are the consequence of a technical error during the experiment.
Since 1944, when Bigger first described persister cells in Staphylococcus spp. (9), many advances have been made in exploring this phenomenon, especially in bacteria. It is known that microbial cultures growing in vivo can sometimes be very difficult to eradicate completely by an antibiotic treatment, causing relapses or development of chronic infections in patients. From an evolutionary point of view, a small pool of cells with the same genotype as the rest of the population but differing in their ability to tolerate stress, including drug treatment, provides a form of insurance to the population.
The phenomenon of persistence has not only been described for bacteria, but also in other types of pathogens, and it has been proposed that persister cells significantly contributed to the recalcitrance of C. albicans biofilms to antifungal treatments (26–28).
C. albicans persister cells were first described in 2006 (8), and since then, only a few reports, sometimes contradictory, have been presented. In our study, we explored standard protocols to obtain persisters and showed that their proportion in biofilms formed by different C. albicans strains has been overestimated. Only Al-Dhaheri and Douglas did not detect persisters in SC5314 biofilms (17). In their study, biofilms were grown on silicon discs that were transferred in antifungal solutions for treatment. In contrast, the other published experiments were performed using 96-well plates and RPMI medium or 24-well plates and SD-based medium, while keeping the incubation volumes constant throughout the experiment (8, 14–16). In this study, we modified the protocols growing biofilms at the bottom of 96- or 24-well plates (8, 14–16) by increasing the volume of antifungal. This change led to the eradication of biofilms, indicating that previously detected “persisters” were likely the result of survival of cells that were not reached by the antifungal. Our results corroborate the findings of Al-Dhaheri and Douglas (17).
Notably, these authors were able to detect some persisters in biofilms of a clinical isolate (17), but the ratio obtained was much lower (0.01%) than the numbers published by others (8, 14). Although some of the clinical isolates of C. albicans and C. tropicalis tested in our study were occasionally able to yield survivors after the treatment of biofilms with AMB, this phenomenon was rather inconsistent, pointing either to the stochastic nature of persistence itself or to another skew in the protocol while carrying out particular experiments.
At this time, we cannot completely exclude the possibility of persistence in all C. albicans strains, though with the described protocol, we managed to disprove their presence for 92 C. albicans strains out of 98. It is important to stress that our results reflect only the behavior of C. albicans biofilms grown in vitro; we cannot rule out that in the context of the host, persister cells could appear and contribute to the general resistance and dissemination of C. albicans.
MATERIALS AND METHODS
Strains and growth conditions.
In this study, we used 3 reference strains (listed in Table 1) and a set of 95 C. albicans (Table 2), 3 C. tropicalis, and 3 C. parapsilosis clinical isolates.
TABLE 1.
C. albicans reference strains used in this study
| C. albicans strain | Genotype | Reference or source |
|---|---|---|
| SC5314 | 18 | |
| CEC369 | ura3::λimm434/ura3::λimm434 ARG4/arg4::hisG HIS1/his1Δ::hisG RPS1/RPS1::CIp10 | 31 |
| CEC4664 | ura3Δ::λimm434/ura3Δ::λimm434 iro1Δ::λimm434/iro1Δ::λimm434 ADH1/adh1::PTDH3-carTA::SAT1 arg4Δ/ARG4 his1Δ::hisG/HIS1 RPS1/RPS1::CIp10 | Lab collection |
TABLE 2.
Clinical isolates used in this study
| Name | Reference or source |
|---|---|
| CEC704 | 32 |
| CEC712 | 32 |
| CEC718 | 32 |
| CEC723 | 32 |
| CEC1289 | 33 |
| CEC1424 | 34 |
| CEC2018 | 35 |
| CEC2019 | 34 |
| CEC2020 | 36 |
| CEC2021 | 35 |
| CEC2022 | 37 |
| CEC2871 | 38 |
| CEC2876 | 38 |
| CEC3494 | 39 |
| CEC3533 | 37 |
| CEC3534 | 36 |
| CEC3535 | 36 |
| CEC3536 | 36 |
| CEC3540 | 32 |
| CEC3541 | 33 |
| CEC3544 | 39 |
| CEC3547 | 39 |
| CEC3548 | 39 |
| CEC3549 | 39 |
| CEC3550 | 40 |
| CEC3553 | 39 |
| CEC3555 | 33 |
| CEC3556 | 36 |
| CEC3560 | 34 |
| CEC3561 | 39 |
| CEC3596 | 33 |
| CEC3611 | 36 |
| CEC3614 | 36 |
| CEC3615 | 36 |
| CEC3621 | 32 |
| CEC3622 | 36 |
| CEC3623 | 32 |
| CEC3626 | 39 |
| CEC3627 | 36 |
| CEC3634 | 36 |
| CEC3637 | 34 |
| CEC3659 | 35 |
| CEC3662 | 35 |
| CEC3663 | 32 |
| CEC3664 | 35 |
| CEC3665 | 35 |
| CEC3668 | 35 |
| CEC3669 | 35 |
| CEC3672 | 35 |
| CEC3675 | 35 |
| CEC3681 | 35 |
| CEC3682 | 35 |
| CEC3685 | 35 |
| CEC3706 | 35 |
| CEC3708 | 35 |
| CEC3711 | 35 |
| CEC4035 | 32 |
| CEC4039 | 32 |
| CEC4103 | 41 |
| CEC4104 | 41 |
| CEC4106 | 41 |
| CEC4108 | 41 |
| CEC4256 | 42 |
| CEC4259 | 42 |
| CEC4481 | 32 |
| CEC4482 | 32 |
| CEC4485 | 32 |
| CEC4486 | 32 |
| CEC4487 | 32 |
| CEC4488 | 32 |
| CEC4489 | 32 |
| CEC4492 | 32 |
| CEC4494 | 32 |
| CEC4495 | Lab collection |
| CEC4496 | 32 |
| CEC4501 | Lab collection |
| CEC4504 | Lab collection |
| CEC4505 | Lab collection |
| CEC4511 | Lab collection |
| CEC4514 | Lab collection |
| CEC4515 | Lab collection |
| CEC4517 | Lab collection |
| CEC4521 | Lab collection |
| CEC4524 | Lab collection |
| CEC4526 | 32 |
| CEC4527 | Lab collection |
| CEC4547 | Lab collection |
| CEC4548 | Lab collection |
| CEC4549 | Lab collection |
| CEC4550 | Lab collection |
| CEC4552 | 32 |
| CEC5029 | 32 |
| CEC5316 | Lab collection |
| CEC5317 | Lab collection |
| CEC5318 | Lab collection |
Yeast precultures were grown overnight in YPD (1% yeast extract, 2% peptone, 2% glucose) agar with shaking at 30°C.
Biofilms were grown either in RPMI 1640 medium with l-glutamine (buffered with 50 mM HEPES), as described previously (8, 29), or in GHAUM medium (SD supplemented with 2% glucose and 1 mg/ml histidine, 1 mg/ml arginine, 0.02 mg/ml uridine, and 2 mg/ml methionine [30]).
Resistance was checked on solid YNG (6,7 g/liter yeast nitrogen base without amino acids and with ammonium sulfate, 2% glucose, and 2% agar) medium supplemented with 10 μg/ml AMB.
Biofilm growth and persister cell isolation.
To assess persister cell appearance in biofilms, we used two protocols adapted either from LaFleur et al. (8) or Li et al. (14). The first protocol uses 96-well plates, and the biofilms are grown in RPMI medium. In the second protocol, the biofilms are grown in 24-well plates but using GHAUM medium instead of yeast nitrogen base (YNB) agar.
Biofilm growth.
Overnight cultures were washed in sterile 1× PBS and diluted in the corresponding medium to an OD600 of 0.3. Either 100 μl or 1 ml of cells in the 96-well plate or the 24-well plate, respectively, was allowed to adhere for 1.5 h without agitation. The nonadhered cells were then washed with 1× PBS, the same volume of fresh medium was added, plates were covered with a breathable seal, and biofilms were allowed to form for 48 h at 37°C with agitation (110 rpm), with a medium change after 24 h.
Antifungal treatment.
Media were carefully aspirated from the 48-h-old biofilms without disrupting the biofilm structure. Biofilms were washed once with either 100 μl or 1 ml of 1× PBS, respectively, and treated with a 100 μg/ml AMB solution in either RPMI or GHAUM medium for 24 h at 37°C, statically. AMB solutions were prepared from an 8 mg/ml stock in dimethyl sulfoxide (DMSO) so that the final concentration of DMSO in a working solution did not exceed 1.25%. For control biofilms, a corresponding amount of DMSO was added to the medium instead of the antifungal solution.
This step was either performed using the same volumes of antifungal solution as for biofilm growth as described by LaFleur et al. (8) or Li et al. (14) or the volume of antifungal was increased to fill the well to the top (350 μl and 3 ml for 96- and 24-well plates, respectively).
Clinical isolates were first treated with a 64 μg/ml AMB solution. Strains giving rise to colonies were then tested 5 times with 100 μg/ml AMB.
Plating.
Upon 24 h of antifungal treatment, AMB solution was aspirated, and biofilms were washed twice with 1× PBS prior to plating on YPD agar plates. Biofilms were resuspended in 1× PBS–0.05% Tween 20. For the AMB-treated samples, the whole biofilms were plated. For control biofilms, serial dilutions were performed to allow CFU counting. CFU were counted after incubating the plates at 30°C for 48 h.
ACKNOWLEDGMENTS
Iryna Denega is part of the Pasteur–Paris University (PPU) International Ph.D. program and of Ecole Doctorale BioSPC.
This project has received funding from the European Union's Horizon 2020 research and innovation program under the Marie Sklodowska-Curie grant agreement 665807, and from the Institut Carnot Pasteur Microbes & Santé. This work has been supported by grants from the French Government’s Investissement d’Avenir Program (Laboratoire d’Excellence Integrative Biology of Emerging Infectious Diseases, grant ANR-10-LABX-62-IBEID) to C.D.
REFERENCES
- 1.Sardi JCO, Scorzoni L, Bernardi T, Fusco-Almeida AM, Mendes Giannini MJS. 2013. Candida species: current epidemiology, pathogenicity, biofilm formation, natural antifungal products and new therapeutic options. J Med Microbiol 62:10–24. doi: 10.1099/jmm.0.045054-0. [DOI] [PubMed] [Google Scholar]
- 2.Ramage G, Saville SP, Thomas DP, López-Ribot JL. 2005. Candida biofilms: an update. Eukaryot Cell 4:633–638. doi: 10.1128/EC.4.4.633-638.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nobile CJ, Johnson AD. 2015. Candida albicans biofilms and human disease. Annu Rev Microbiol 69:71–92. doi: 10.1146/annurev-micro-091014-104330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chandra J, Kuhn DM, Mukherjee PK, Hoyer LL, McCormick T, Ghannoum MA. 2001. Biofilm formation by the fungal pathogen Candida albicans: development, architecture, and drug resistance. J Bacteriol 183:5385–5394. doi: 10.1128/JB.183.18.5385-5394.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nett JE, Crawford K, Marchillo K, Andes DR. 2010. Role of Fks1p and matrix glucan in Candida albicans biofilm resistance to an echinocandin, pyrimidine, and polyene. Antimicrob Agents Chemother 54:3505–3508. doi: 10.1128/AAC.00227-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nett JE, Sanchez H, Cain MT, Andes DR. 2010. Genetic basis of Candida biofilm resistance due to drug sequestering matrix glucan. J Infect Dis 202:171–175. doi: 10.1086/651200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nett JE, Sanchez H, Cain MT, Ross KM, Andes DR. 2011. Interface of Candida albicans biofilm matrix-associated drug resistance and cell wall integrity regulation. Eukaryot Cell 10:1660–1669. doi: 10.1128/EC.05126-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.LaFleur MD, Kumamoto CA, Lewis K. 2006. Candida albicans bofilms produce antifungal-tolerant persister cells. Antimicrob Agents Chemother 50:3839–3846. doi: 10.1128/AAC.00684-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bigger J. 1944. Treatment of staphylococcal infections with penicillin by intermittent sterilisation. Lancet 244:497–500. doi: 10.1016/S0140-6736(00)74210-3. [DOI] [Google Scholar]
- 10.Brauner A, Fridman O, Gefen O, Balaban NQ. 2016. Distinguishing between resistance, tolerance and persistence to antibiotic treatment. Nat Rev Microbiol 14:320–330. doi: 10.1038/nrmicro.2016.34. [DOI] [PubMed] [Google Scholar]
- 11.Lewis K. 2007. Persister cells, dormancy and infectious disease. Nat Rev Microbiol 5:48–56. doi: 10.1038/nrmicro1557. [DOI] [PubMed] [Google Scholar]
- 12.Lewis K. 2010. Persister cells. Annu Rev Microbiol 64:357–372. doi: 10.1146/annurev.micro.112408.134306. [DOI] [PubMed] [Google Scholar]
- 13.Harms A, Maisonneuve E, Gerdes K. 2016. Mechanisms of bacterial persistence during stress and antibiotic exposure. Science 354:aaf4268. doi: 10.1126/science.aaf4268. [DOI] [PubMed] [Google Scholar]
- 14.Li P, Seneviratne CJ, Alpi E, Vizcaino JA, Jin L. 2015. Delicate metabolic control and coordinated stress response critically determine antifungal tolerance of Candida albicans biofilm persisters. Antimicrob Agents Chemother 59:6101–6112. doi: 10.1128/AAC.00543-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Truong T, Zeng G, Qingsong L, Kwang LT, Tong C, Chan FY, Wang Y, Seneviratne CJ. 2016. Comparative ploidy proteomics of Candida albicans biofilms unraveled the role of the AHP1 gene in the biofilm persistence against amphotericin B. Mol Cell Proteomics 15:3488–3500. doi: 10.1074/mcp.M116.061523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun J, Li Z, Chu H, Guo J, Jiang G, Qi Q. 2016. Candida albicans amphotericin B-tolerant persister formation is cosely related to surface adhesion. Mycopathologia 181:41–49. doi: 10.1007/s11046-015-9894-1. [DOI] [PubMed] [Google Scholar]
- 17.Al-Dhaheri RS, Douglas LJ. 2008. Absence of amphotericin B-tolerant persister cells in biofilms of some Candida species. Antimicrob Agents Chemother 52:1884–1887. doi: 10.1128/AAC.01473-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gillum AM, Tsay EY, Kirsch DR. 1984. Isolation of the Candida albicans gene for orotidine-5′-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Mol Gen Genet 198:179–182. doi: 10.1007/BF00328721. [DOI] [PubMed] [Google Scholar]
- 19.Wilson RB, Davis D, Mitchell AP. 1999. Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J Bacteriol 181:1868–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Noble SM, Johnson AD. 2005. Strains and strategies for large-scale gene deletion studies of the diploid human fungal pathogen Candida albicans. Eukaryot Cell 4:298–309. doi: 10.1128/EC.4.2.298-309.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.LaFleur MD, Qi Q, Lewis K. 2010. Patients with long-term oral carriage harbor high-persister mutants of Candida albicans. Antimicrob Agents Chemother 54:39–44. doi: 10.1128/AAC.00860-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klotz SA, Rutten MJ, Smith RL, Babcock SR, Cunningham MD. 1993. Adherence of Candida albicans to immobilized extracellular matrix proteins is mediated by calcium-dependent surface glycoproteins. Microb Pathog 14:133–147. doi: 10.1006/mpat.1993.1014. [DOI] [PubMed] [Google Scholar]
- 23.Chudzik B, Malm A, Rautar B, Polz-Dacewicz M. 2007. In vitro inhibitory activity of EDTA against planktonic and adherent cells of Candida sp. Ann Microbiol 57:115. doi: 10.1007/BF03175059. [DOI] [Google Scholar]
- 24.Moyed HS, Bertrand KP. 1983. hipA, a newly recognized gene of Escherichia coli K-12 that affects frequency of persistence after inhibition of murein synthesis. J Bacteriol 155:768–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wolfson JS, Hooper DC, McHugh GL, Bozza MA, Swartz MN. 1990. Mutants of Escherichia coli K-12 exhibiting reduced killing by both quinolone and beta-lactam antimicrobial agents. Antimicrob Agents Chemother 34:1938–1943. doi: 10.1128/AAC.34.10.1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Borghi E, Borgo F, Morace G. 2016. Fungal biofilms: update on resistance, p 37–47. In Imbert C. (ed), Fungal biofilms and related infections: advances in microbiology, infectious diseases and public health, volume 3 (advances in medicine and biology book 931). Springer, Cham, Switzerland. [DOI] [PubMed] [Google Scholar]
- 27.Ramage G, Rajendran R, Sherry L, Williams C. 2012. Fungal biofilm resistance. Int J Microbiol 2012:528521. doi: 10.1155/2012/528521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mathé L, Van Dijck P. 2013. Recent insights into Candida albicans biofilm resistance mechanisms. Curr Genet 59:251–264. doi: 10.1007/s00294-013-0400-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.De Brucker K, De Cremer K, Cammue BPA, Thevissen K. 2016. Protocol for determination of the persister subpopulation in Candida albicans biofilms. Methods Mol Biol 1333:67–72. doi: 10.1007/978-1-4939-2854-5_6. [DOI] [PubMed] [Google Scholar]
- 30.Cabral V, Znaidi S, Walker LA, Martin-Yken H, Dague E, Legrand M, Lee K, Chauvel M, Firon A, Rossignol T, Richard ML, Munro CA, Bachellier-Bassi S, d’Enfert C. 2014. Targeted changes of the cell wall proteome influence Candida albicans ability to form single- and multi-strain biofilms. PLoS Pathog 10:e1004542. doi: 10.1371/journal.ppat.1004542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Enjalbert B, Rachini A, Vediyappan G, Pietrella D, Spaccapelo R, Vecchiarelli A, Brown AJP, d’Enfert C. 2009. A multifunctional, synthetic Gaussia princeps luciferase reporter for live imaging of Candida albicans infections. Infect Immun 77:4847–4858. doi: 10.1128/IAI.00223-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ropars J, Maufrais C, Diogo D, Marcet-Houben M, Perin A, Sertour N, Mosca K, Permal E, Laval G, Bouchier C, Ma L, Schwartz K, Voelz K, May RC, Poulain J, Battail C, Wincker P, Borman AM, Chowdhary A, Fan S, Kim SH, Pape PL, Romeo O, Shin JH, Gabaldon T, Sherlock G, Bougnoux M-E, d’Enfert C. 2018. Gene flow contributes to diversification of the major fungal pathogen Candida albicans. Nat Commun 9:2253. doi: 10.1038/s41467-018-04787-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bougnoux M-E, Morand S, d’Enfert C. 2002. Usefulness of multilocus sequence typing for characterization of clinical isolates of Candida albicans. J Clin Microbiol 40:1290–1297. doi: 10.1128/JCM.40.4.1290-1297.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Odds FC, Bougnoux M-E, Shaw DJ, Bain JM, Davidson AD, Diogo D, Jacobsen MD, Lecomte M, Li S-Y, Tavanti A, Maiden MCJ, Gow NAR, d’Enfert C. 2007. Molecular phylogenetics of Candida albicans. Eukaryot Cell 6:1041–1052. doi: 10.1128/EC.00041-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bougnoux M-E, Kac G, Aegerter P, d’Enfert C, Fagon J-Y, CandiRea Study Group. 2008. Candidemia and candiduria in critically ill patients admitted to intensive care units in France: incidence, molecular diversity, management and outcome. Intensive Care Med 34:292–299. doi: 10.1007/s00134-007-0865-y. [DOI] [PubMed] [Google Scholar]
- 36.Sdoudi K, Bougnoux M-E, Hamoumi RE, Diogo D, Mdaghri NE, d’Enfert C, Razki A. 2014. Phylogeny and diversity of Candida albicans vaginal isolates from three continents. Int J Curr Microbiol App Sci 3:471–480. [Google Scholar]
- 37.Schönherr FA, Sparber F, Kirchner FR, Guiducci E, Trautwein-Weidner K, Gladiator A, Sertour N, Hetzel U, Le GTT, Pavelka N, d'Enfert C, Bougnoux M-E, Corti CF, LeibundGut-Landmann S. 2017. The intraspecies diversity of C. albicans triggers qualitatively and temporally distinct host responses that determine the balance between commensalism and pathogenicity. Mucosal Immunol 10:1335–1350. doi: 10.1038/mi.2017.2. [DOI] [PubMed] [Google Scholar]
- 38.Shin JH, Bougnoux M-E, d'Enfert C, Kim SH, Moon C-J, Joo MY, Lee K, Kim M-N, Lee HS, Shin MG, Suh SP, Ryang DW. 2011. Genetic diversity among Korean Candida albicans bloodstream isolates: assessment by multilocus sequence typing and restriction endonuclease analysis of genomic DNA by use of BssHII. J Clin Microbiol 49:2572–2577. doi: 10.1128/JCM.02153-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bougnoux M-E, Diogo D, Francois N, Sendid B, Veirmeire S, Colombel JF, Bouchier C, Van Kruiningen H, d’Enfert C, Poulain D. 2006. Multilocus sequence typing reveals intrafamilial transmission and microevolutions of Candida albicans isolates from the human digestive tract. J Clin Microbiol 44:1810–1820. doi: 10.1128/JCM.44.5.1810-1820.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bougnoux M-E, Aanensen DM, Morand S, Théraud M, Spratt BG, d'Enfert C. 2004. Multilocus sequence typing of Candida albicans: strategies, data exchange and applications. Infect Genet Evol 4:243–252. doi: 10.1016/j.meegid.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 41.Dieng Y, Sow D, Ndiaye M, Guichet E, Faye B, Tine R, Lo A, Sylla K, Ndiaye M, Abiola A, Dieng T, Ndiaye JL, Le Pape P, Gaye O. 2012. Identification of three Candida africana strains in Senegal. J Mycol Med 22:335–340. doi: 10.1016/j.mycmed.2012.07.052. [DOI] [PubMed] [Google Scholar]
- 42.Garnaud C, Botterel F, Sertour N, Bougnoux M-E, Dannaoui E, Larrat S, Hennequin C, Guinea J, Cornet M, Maubon D. 2015. Next-generation sequencing offers new insights into the resistance of Candida spp. to echinocandins and azoles. J Antimicrob Chemother 70:2556–2565. doi: 10.1093/jac/dkv139. [DOI] [PubMed] [Google Scholar]