During the intraerythrocytic asexual cycle malaria parasites acquire nutrients and other solutes through a broad selectivity channel localized at the membrane of the infected erythrocyte termed the plasmodial surface anion channel (PSAC). The protein product of the Plasmodium falciparum clonally variant clag3.1 and clag3.2 genes determines PSAC activity.
KEYWORDS: malaria, Plasmodium falciparum, drug resistance, epigenetics, plasmodium surface anion channel, clag3
ABSTRACT
During the intraerythrocytic asexual cycle malaria parasites acquire nutrients and other solutes through a broad selectivity channel localized at the membrane of the infected erythrocyte termed the plasmodial surface anion channel (PSAC). The protein product of the Plasmodium falciparum clonally variant clag3.1 and clag3.2 genes determines PSAC activity. Switches in the expression of clag3 genes, which are regulated by epigenetic mechanisms, are associated with changes in PSAC-dependent permeability that can result in resistance to compounds toxic for the parasite, such as blasticidin S. Here, we investigated whether other antimalarial drugs require CLAG3 to reach their intracellular target and consequently are prone to parasite resistance by epigenetic mechanisms. We found that the bis-thiazolium salts T3 (also known as albitiazolium) and T16 require the product of clag3 genes to enter infected erythrocytes. P. falciparum populations can develop resistance to these compounds via the selection of parasites with dramatically reduced expression of both genes. However, other compounds previously demonstrated or predicted to enter infected erythrocytes through transport pathways absent from noninfected erythrocytes, such as fosmidomycin, doxycycline, azithromycin, lumefantrine, or pentamidine, do not require expression of clag3 genes for their antimalarial activity. This suggests that they use alternative CLAG3-independent routes to access parasites. Our results demonstrate that P. falciparum can develop resistance to diverse antimalarial compounds by epigenetic changes in the expression of clag3 genes. This is of concern for drug development efforts because drug resistance by epigenetic mechanisms can arise quickly, even during the course of a single infection.
INTRODUCTION
Malaria is a major public health problem that affects half of the world’s population. Plasmodium falciparum is the predominant species in Africa and the most deadly form of the parasite. It is responsible for half a million deaths every year, mostly among children and pregnant women (1). While chemotherapy is the main tool used for malaria control, P. falciparum has developed resistance to all antimalarial drugs, including artemisinin combination therapies (ACTs), which are the current frontline treatment (2, 3). Therefore, the appearance and spread of drug resistant parasites is a major obstacle to malaria control and elimination efforts and urges the discovery of new effective compounds to treat infections.
Most of the known mechanisms by which Plasmodium falciparum parasites develop resistance to antimalarial drugs are related to changes in the genome, such as single nucleotide polymorphisms (SNPs) or gene amplifications (4). SNPs can occur in parasite genes encoding the enzymes targeted by the drug, reducing the drug affinity as in the case of mutations in the P. falciparum dihydrofolate reductase (pfdhfr) and dihydropteroate synthase (pfdhps) genes (5, 6). Mutations associated with resistance can also occur in parasite-encoded transporters causing the active extrusion of the drug out of its site of action, as in the case of mutations in the chloroquine resistance transporter (pfcrt) gene associated with the efflux of chloroquine out of the digestive vacuole (7). Likewise, genetic amplifications can increase the expression of the target gene, or the expression of genes encoding transporters, as in the case of amplification of the multidrug resistance protein 1 (pfmdr1) gene that leads to accumulation of mefloquine in the digestive vacuole away from its predicted target (8).
Transport activities present in infected erythrocytes that mediate the uptake of solutes unable to enter uninfected erythrocytes are collectively referred to as new permeation pathways (NPPs) (9–11). The plasmodial surface anion channel (PSAC) has been proposed to be the single channel responsible for NPPs. This broad selectivity channel, localized at the membrane of the infected erythrocyte, is essential for the uptake of nutrients and several other solutes (12–16). The protein product of P. falciparum clag3.1 and clag3.2 genes, part of the five-member clag family that encodes the CLAG/RhopH1 component of the RhopH complex (17), plays a key role for the activity of the PSAC (16, 18–20). Other members of the RhopH complex, RhopH2 and RhopH3, are also necessary for PSAC activity (21–23). The structure of the PSAC has not yet been determined, but protease sensitivity assays and experiments with various transgenic parasite lines suggest that CLAG3s (and possibly also RhopH2 and RhopH3) may participate directly in the formation of the channel rather than only activating a channel formed by other proteins (20, 22, 24). The RhopH complex is initially expressed at the schizont stage and localized in the rhoptries (25). About 20 h after reinvasion, it is transported to the red cell membrane, where it determines PSAC activity (22, 23, 26). The sequence of clag3.1 and clag3.2 genes is 95% identical. These genes display clonally variant and mutually exclusive expression, such that commonly only one of the two clag3 genes is expressed at a time (27). The latter property was observed in culture-adapted parasite lines of different genetic backgrounds (18, 19, 28, 29) and has been later confirmed in uncomplicated human malaria infections (30), although mutual exclusion is not strict (31).
Recently, an epigenetic mechanism of drug resistance involving changes in the expression of clag3 genes was described in P. falciparum (32, 33). Previous studies demonstrated that blasticidin S and leupeptin require PSAC for their transport across the membrane of infected erythrocytes and that P. falciparum resistance to these compounds is associated with changes in PSAC function (13, 34, 35). Later, we and others showed that changes in PSAC-mediated transport of blasticidin S were associated with switches in clag3 genes expression regulated at the epigenetic level (32, 33). Resistance to low blasticidin S concentrations involved the selection of parasites that switched from clag3.2 to clag3.1 expression, whereas resistance to high concentrations of the drug was acquired by the selection of parasites with both clag3 genes simultaneously silenced (32). In all cases, clag3 silencing is mediated by heterochromatin (31). The pattern of clag3 expression in the selected parasites is transmitted to the next generations by epigenetic mechanisms even when the drug is no longer present. However, simultaneous silencing of the two genes poses a fitness cost for the parasite and in the absence of selection it is progressively reverted.
Whether other antimalarial drugs require the product of clag3 genes to reach their intracellular targets and consequently are prone to parasite resistance by this epigenetic mechanism is not known. Most antimalarials are small hydrophobic compounds that can diffuse through lipid membranes and do not require specific channels to enter infected erythrocytes (36). However, large hydrophilic compounds such as blasticidin S and leupeptin require facilitated uptake through PSAC. Drug physicochemical parameters such as molecular size and hydrophobicity indexes, e.g., the logP value, can be used to predict which antimalarial drugs require PSAC-facilitated transport (36, 37). However, while such in silico predictions are informative, only experimental validation can determine which drugs are actually prone to parasite resistance by epigenetic silencing of clag3 genes.
To address this question, we compared the 50% inhibitory concentrations (IC50s) of selected antimalarial compounds between the blasticidin S-selected 10G-0.6-2 line, which has both clag3 genes silenced and thus shows deficient PSAC transport, and the parental 10G line, which predominantly expresses clag3.2 (32). In addition, we selected parasites with some of the drugs and monitored switches in the expression of clag3 genes during selection. We also investigated PSAC transport in CLAG3-defficient parasites using the reporter compound 5-aminolevulinic acid (5-ALA).
RESULTS
The 10G-0.6-2 line is a valid tool to investigate transport via CLAG3-containing PSAC.
We previously showed that the 10G-0.6-2 parasite line, derived from the 10G line selected with a high concentration of blasticidin S, shows dramatically reduced expression of the two clag3 genes. The clonally variant clag2 gene is silenced in both the 10G-0.6-2 and the parental 10G lines, whereas the not clonally variant clag8 and clag9 genes are expressed in both lines (32). Silencing of clag3 genes in 10G-0.6-2 results in reduced permeability to structurally diverse compounds, such as blasticidin S, sorbitol, and the canonical amino acid l-alanine (32). To further demonstrate that the 10G-0.6-2 line is a valid tool to identify drugs that require CLAG3 to cross the membrane of the infected erythrocyte, we conducted immunofluorescence assays (IFAs) on 10G-derived lines selected with different concentrations of blasticidin S (32) using an anti-CLAG3 antibody that recognizes both CLAG3.1 and CLAG3.2 (19). By restricting the analysis only to schizonts positive for the mature schizont marker AMA1, we could unambiguously identify parasites in which absence of CLAG3 signal was attributable to epigenetic silencing rather than to parasite stage. We found that essentially all mature schizonts in the parental 10G line express CLAG3 (either CLAG3.1 or CLAG3.2), whereas the proportions of CLAG3-negative mature schizonts were 12% in 10G-0.4, 74% in 10G-0.6, and 98% in 10G-0.6-2 cultures (selected with 0.4, 0.6, and 2 μg/ml of blasticidin S, respectively) (Fig. 1). These experiments at the single cell level show that the vast majority of parasites in 10G-0.6-2 cultures do not express CLAG3 proteins at detectable levels, validating the 10G-06-2 line as an appropriate tool to identify compounds that require CLAG3 to enter the cell.
FIG 1.
CLAG3 expression in parasite cultures selected with different concentrations of blasticidin S. (A) IFA analysis of mature schizont-infected erythrocytes with anti-CLAG3 and anti-AMA1 antibodies in blasticidin S-selected lines and the parental 10G line. DAPI was used to mark the parasite nuclei. Anti-AMA1 antibodies were used to identify schizonts sufficiently mature for CLAG3-expression. (B) Proportion of AMA1-positive parasites that were negative for CLAG3 fluorescence in each parasite line. The results are averages of two independent biological replicates, along with the standard deviations (SD).
Parasites with deficient PSAC transport due to silencing of clag3 genes are resistant to bis-thiazolium salts.
To identify antimalarial compounds that require the product of clag3 genes for efficient transport across the membrane of infected erythrocytes, we selected drugs to be tested based on two criteria. First, we included drugs of clinical relevance for which physicochemical properties suggest that they may not enter infected erythrocytes by passive membrane diffusion, i.e., doxycycline, azithromycin and lumefantrine (36). Second, we included drugs for which there is previous evidence for uptake through NPPs, i.e., fosmidomycin (38), pentamidine (39), and the bis-thiazolium salts T3 (also known as albitiazolium) and T16 (40, 41). As positive controls we included drugs for which there is already clear evidence of CLAG3-dependent uptake through PSAC, i.e., blasticidin S and leupeptin (32, 33). For the selected compounds, we compared the IC50 between the 10G-0.6-2 line and the parental 10G line.
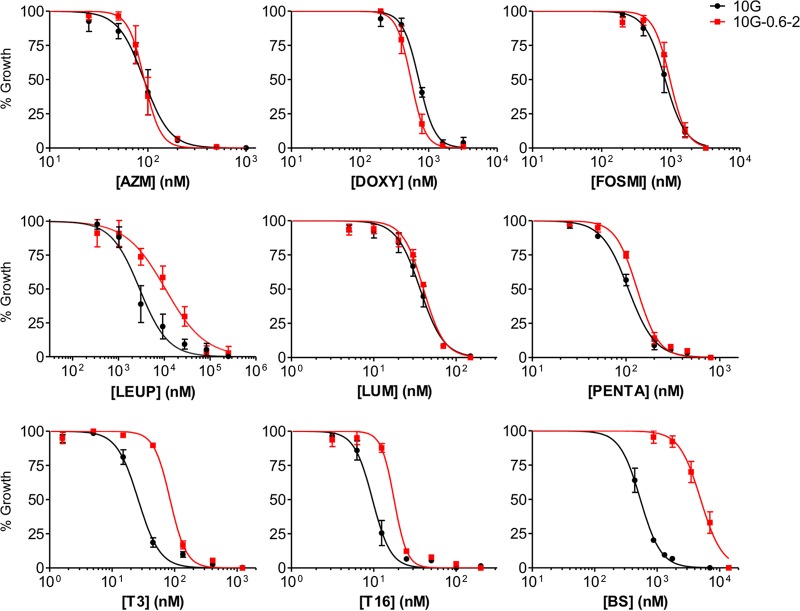
Our criteria to consider that the uptake of a drug is impaired by the absence of CLAG3 was a 1.5-fold increase in the IC50 in 10G-0.6-2 versus 10G, plus a statistically significant difference (P < 0.05). The former criterion was used because we consider that differences of lower magnitude are unlikely to have a major biological significance. Using these criteria, dose-response curves revealed that the 10G-0.6-2 line shows lower sensitivity to blasticidin S, T3, T16, and leupeptin than the 10G line (Fig. 2 and Table 1). These results support the idea that T3 and T16 require the expression of clag3 genes for their uptake, in addition to leupeptin and blasticidin S for which this was previously demonstrated (13, 32–34). The IC50 fold increases in 10G-0.6-2 compared to 10G were 3.3 for T3 and 1.8 for T16, which is lower than the 4.6- and 9.5-fold increases observed for leupeptin and blasticidin S, respectively (Table 1). On the other hand, we did not obtain evidence for clag3 genes playing a role in the uptake of the other compounds tested, since the difference in IC50s between 10G and 10G-0.6-2 cultures was not statistically significant and/or was of very low magnitude (Fig. 2 and Table 1).
FIG 2.
Drug dose-response curves for parasite lines 10G and 10G-0.6-2. The susceptibility to azithromycin (AZM), doxycycline (DOXY), fosmidomycin (FOSMI), leupeptin (LEUP), lumefantrine (LUM), pentamidine (PENTA), T3, T16, and blasticidin S (BS) were compared between the parasite lines 10G (predominantly expressing clag3.2) and 10G-0.6-2 (both clag3 genes silenced). Values are averages of three independent experiments, each performed in triplicate wells, along with the SD.
TABLE 1.
IC50s of different drugs in the 10G and 10G-0.6-2 linesa
| Drug | IC50 (nM) |
Fold change | P | |
|---|---|---|---|---|
| 10G | 10G-0.6-2 | |||
| Azithromycin | 95 (22) | 93 (19) | 1.0 | 0.899 |
| Doxycycline | 718 (11) | 563 (121) | 0.8 | 0.091 |
| Fosmidomycin | 841 (219) | 985 (184) | 1.2 | 0.434 |
| Leupeptin | 1,753 (1) | 8,010 (7) | 4.6 | 0.039* |
| Lumefantrine | 36 (6) | 40 (2) | 1.1 | 0.394 |
| Pentamidine | 106 (8) | 132 (9) | 1.2 | 0.019* |
| T3 | 26 (5) | 85 (8) | 3.3 | 0.003* |
| T16 | 10 (2) | 18 (1) | 1.8 | 0.002* |
| Blasticidin S | 530 (96) | 5,060 (1,045) | 9.5 | 0.002* |
IC50 values show the averages of three independent experiments (Fig. 2). Standard deviations are given in parentheses. P values were calculated using a two-tailed unpaired t test. Significant differences were determined between 10G and 10G-0.6-2 after applying the Benjamini-Hochberg correction for multiple testing, with a false discovery rate of 0.1, and are indicated by an asterisk (*).
clag3 expression patterns after selection with different drugs.
We and others have previously shown that adaptation to grow in the presence of blasticidin S is associated with selection of parasites with altered clag3 expression patterns, validating the involvement of these genes in the transport of the drug (30, 32, 33). Here, we investigated whether sublethal concentrations of other drugs can also select parasites with specific clag3 expression patterns. In these experiments we tested drugs to which 10G-0.6-2 is less sensitive than 10G (Fig. 2). In addition, doxycycline and fosmidomycin were included in spite of not showing differences between 10G and 10G-0.6-2 because previous studies suggested that they may require NPPs for their transport into infected erythrocytes (36, 38). We considered that selection experiments may be more sensitive than IC50 comparisons to detect the involvement of CLAG3 in their transport.
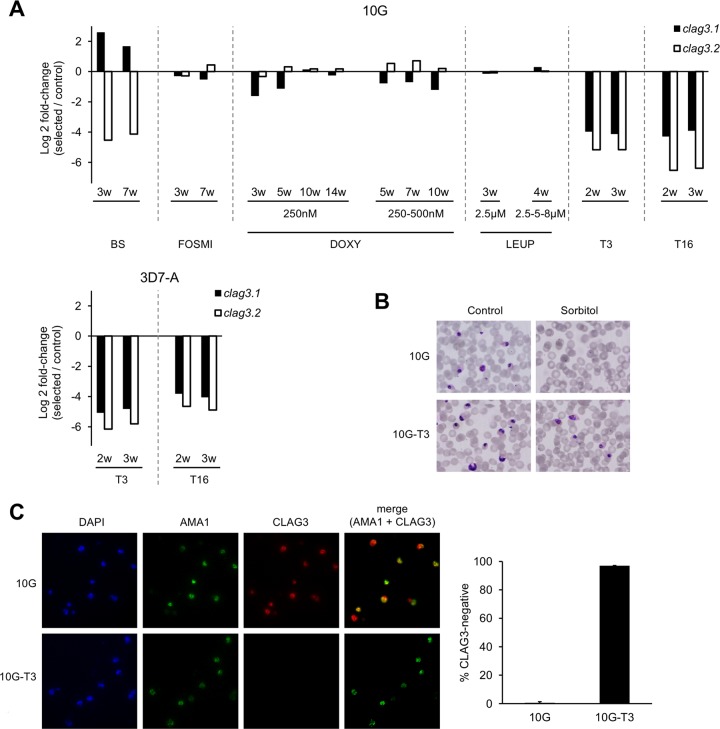
We selected 10G cultures with the drugs at concentrations ranging from (approximately) the IC50 to the IC80. We used these relatively low concentrations because we predicted that this may facilitate adaptation by selection of parasites expressing clag3.1 or clag3.2, if expression of one or the other paralog was associated with differential transport of the drug as in the case of blasticidin S (32). However, for T3 and T16 we observed that toxicity is much higher in the second and subsequent cycles than in the first cycle, such that selection with this range of drug concentrations killed the cultures after only a few cycles. We determined the approximate IC50s of T3 and T16 over two cycles and found that they were ∼10 times lower than in a one-cycle assay; we used T3 at 2.86 nM and T16 at 0.6 nM for selection. For doxycycline, which shows a delayed death effect, we also selected cultures with a concentration of the drug based on the IC50 in a two-cycle assay. Cultures were selected until we obtained evidence for adaptation (see Materials and Methods and Fig. S1) or for a maximum of 14 weeks. 10G cultures selected with blasticidin S (650 nM, corresponding to 0.3 μg/ml, a concentration that inhibits growth by ∼60%) were used as a positive control. These cultures switched from predominant clag3.2 to clag3.1 expression after only 3 weeks of selection (80-fold increase in the clag3.1/clag3.2 ratio), which is consistent with previous results (32) (Fig. 3A).
FIG 3.
Changes in CLAG3 expression in cultures selected with different drugs. (A) Changes in the transcript levels of clag3.1 and clag3.2 in cultures selected with blasticidin S (BS), fosmidomycin (FOSMI), doxycycline (DOXY), leupeptin (LEUP), T3, and T16 compared to unselected control cultures maintained in parallel. The 10G or 3D7-A lines were used for these experiments, as indicated. Cultures selected with DOXY were maintained with the drug at 250 nM for 14 weeks or at 250 nM for 3 weeks and then at 500 nM for 7 additional weeks. Cultures selected with LEUP were first selected at 2.5 μM for 2 weeks and then maintained one additional week at the same concentration (for a total of 3 weeks at 2.5 μM) or one additional week at 5 μM and one week at 8 μM (2.5-5-8 μM bars). Transcript levels are normalized against rhoph2, which has temporal expression dynamics similar to clag3 genes along the asexual cycle. Values are the log2 of the normalized expression fold change in drug-selected cultures versus cultures maintained in parallel in the absence of drug. Zero indicates the same expression in selected and control cultures, whereas positive values reflect an increase of expression in drug-selected cultures and negative values reflect reduced expression in drug-selected cultures. Individual values are the average of reactions performed in triplicate, but for each drug selection the result of independent biological samples collected at different times (indicated in weeks) is presented. (B) Resistance of late-stage parasites (pigmented trophozoites and schizonts) to treatment with sorbitol in 10G cultures selected with T3 (10G-T3) or unselected 10G cultures maintained in parallel (10G). “Control” are the same cultures before sorbitol treatment. (C) IFA analysis of mature schizont-infected erythrocytes with anti-CLAG3 and anti-AMA1 antibodies in T3-selected (10G-T3) or unselected 10G cultures. DAPI marks parasite nuclei. Anti-AMA1 antibodies were used to identify schizonts sufficiently mature for CLAG3 expression. The proportion of AMA1-positive infected erythrocytes that are negative for CLAG3 in 10G-T3 or unselected 10G cultures is shown. Values in the bar chart are averages of two independent biological replicates, along with the SD.
Cultures selected with T3 or T16 showed a prominent decrease in the expression levels of both clag3 genes after only 2 weeks (Fig. 3A and Fig. S2). This was observed in selection experiments performed with the parasite lines 10G and also 3D7-A, a previously described stock of the P. falciparum clonal line 3D7, from which the 10G subclone was derived (27, 42). As expected from this result, in cultures selected with T3 the vast majority of parasites were resistant to sorbitol lysis (Fig. 3B) and negative for CLAG3 expression by IFA (Fig. 3C), similar to the blasticidin S-selected 10G-0.6-2 line. Together with the increased IC50 values observed for these drugs in the 10G-0.6-2 line, these results demonstrate that the uptake of T3 and T16 by infected erythrocytes requires CLAG3. As in the case of blasticidin S-selected cultures (32, 33), clag3 silencing was reversible, and removal of the drug resulted in a progressive selection of parasites that express clag3 (Fig. S3). This is probably attributable to the fitness cost associated with simultaneous silencing of the two clag3 genes (32). We also selected cultures with lower concentrations of T3, but we did not obtain evidence for any major alteration in the clag3.1/clag3.2 ratio that would reflect selection of parasites expressing one or the other clag3 gene (Fig. S4).
On the other hand, 10G cultures selected with fosmidomycin, doxycycline, and leupeptin did not show any clear alteration in the expression of clag3 genes compared to control 10G cultures maintained in parallel without drug (Fig. 3A).
Parasites with dramatically reduced levels of CLAG3 expression still acquire some compounds through the NPPs.
Previous reports demonstrated that the uptake of fosmidomycin or pentamidine depends on NPPs (38, 39), and the uptake of other compounds such as doxycycline, lumefantrine, and azithromycin is strongly predicted to also require NPPs (36). Therefore, the results of the selection experiments with these drugs and especially the similar IC50s between 10G and 10G-0.6-2 were somehow unexpected. To test the possibility that the transport of specific compounds via NPPs is active in the 10G-0.6-2 line in spite of its dramatically reduced levels of CLAG3, we compared the uptake of 5-ALA between the 10G-0.6-2 line and the parental 10G. Inside the infected erythrocyte, 5-ALA is processed to the fluorescent compound protoporphyrin IX (PPIX) that can be visualized by microscopy. 5-ALA has been previously observed to enter infected erythrocytes through NPPs and its uptake requires RhopH3 (a component of the RhopH complex), which indicates that it uses the PSAC (23, 43). We observed that the uptake of 5-ALA was impaired in RhopH3-defficient parasites, as previously reported (23). In contrast, essentially all mature parasite-infected erythrocytes acquired the compound in the 10G, 10G-0.6-2, and T3-selected lines (Fig. 4). This result indicates that CLAG3 is not essential for the acquisition of some specific PSAC substrates such as 5-ALA. This is in contrast to the PSAC-mediated transport of blasticidin S, leupeptin, sorbitol, l-alanine, T3, or T16, which appears to depend more strongly on CLAG3 expression.
FIG 4.
Uptake of 5-ALA by erythrocytes infected with CLAG3-defficient parasites. Analysis was performed with the CLAG3-defficient 10G-0.6-2 and T3-selected 10G lines, the parental 10G control line, and the previously described 5F5 transgenic line treated with rapamycin to induce the deletion of rhoph3 exons 4 to 6 (5F5-RAPA) or treated in parallel with DMSO solvent (5F5-CTRL). The uptake of 5-ALA and its subsequent conversion to PPIX in infected erythrocytes was visualized by fluorescence microscopy. Parasite nuclei were stained with Hoechst. The bar chart shows the proportion of PPIX fluorescence-positive cells among pigmented parasite-infected erythrocytes. Values are averages of three independent biological replicates, along with the SD.
DISCUSSION
We and others have previously shown that epigenetic changes in the expression of clag3 genes modify the permeability of the infected erythrocyte membrane through alterations in PSAC function, and that these changes can confer resistance to compounds such as blasticidin S and leupeptin (13, 32–35, 44). Here, we investigated whether other compounds with antimalarial activity require the product of clag3 genes to enter the infected erythrocyte and are thus prone to parasite resistance due to epigenetic changes in clag3 genes expression. For this aim, we compared the IC50 for several compounds with antimalarial activity between the parental 10G line and the 10G-0.6-2 line, which expresses dramatically reduced levels of clag3 genes. Of note, the clonally variant gene clag2, which has also been implicated in infected erythrocyte permeability, is silenced in both the 10G and the 10G-0.6-2 lines (27, 32). This excludes changes in the expression of this gene as a confounding factor for the results obtained. In addition, we selected cultures with some of the drugs for several weeks and measured clag3.1 and clag3.2 transcript levels to investigate whether the drugs select for parasites with altered clag3 expression. Both approaches led to the identification of T3 and T16 as new antimalarial compounds that require CLAG3 for their uptake.
We characterized the 10G-0.6-2 line by IFA using anti-CLAG3 antibodies and found that ∼98% of the parasites in the population do not express CLAG3 at detectable levels, which is consistent with the previously described high level of resistance to sorbitol and blasticidin S and dramatically reduced clag3 transcript levels in this parasite line (32). However, we also characterized PSAC functionality in this parasite line using 5-ALA, a compound that specifically enters infected erythrocytes and is then converted to fluorescent PPIX. The uptake of 5-ALA is inhibited by furosemide (a PSAC/NPPs inhibitor) or by conditional depletion of RhopH3 (23, 43), which demonstrates that it is a PSAC substrate that can be used as a convenient reporter for PSAC activity. Surprisingly, we observed PPIX fluorescence indicative of transport through PSAC in the 10G-0.6-2 line, despite silencing of both clag3 genes. Although we cannot exclude the possibility that undetected quantitative differences may occur in the transport of 5-ALA between control and CLAG3-defficient parasites, these results suggest that the transport of some compounds into infected erythrocytes may be mediated by CLAG3-independent PSAC, possibly involving other CLAGs such as CLAG8 or CLAG9. These genes appear to be expressed by all parasites, as they have not been found to show clonally variant expression or to carry epigenetic marks of silencing (45–48). We hypothesize that PSAC involving CLAG8 or CLAG9 may ensure transport of some solutes even when total PSAC transport is highly reduced due to silencing of both clag3 genes. The PSAC formed in the absence of CLAG3 appears to be able to mediate the uptake of compounds such as 5-ALA, but its ability to transport other compounds such as blasticidin S, sorbitol, l-alanine, T3, or T16 is severely impaired. Altogether, these results indicate that the 10G-0.6-2 line provides an appropriate tool to measure drug uptake in the absence of CLAG3 and CLAG2. However, NPPs likely mediated by PSAC formed in the absence of CLAG3 are active in this parasite line, resulting in the uptake of some solutes. Considering that variant expression and epigenetic silencing has only been reported for CLAG3s and CLAG2, only drugs that show lower activity on the 10G-0.6-2 line are prone to parasite resistance by epigenetic silencing of clag genes. Drugs for which uptake may require PSAC but independently of the presence of CLAG3 are unlikely to develop parasite resistance by this mechanism.
Our results demonstrate that in addition to blasticidin S, leupeptin, sorbitol and l-alanine (19, 32, 33), T3 and T16 also require the product of clag3 genes to enter infected erythrocytes. The bis-thiazolium salts T3 and T16 are choline analogs that inhibit the synthesis of phosphatidylcholine. They are able to cure malaria infections in vivo in mice, primates, and humans, but their clinical development was discontinued because of rapid drug clearance in children (49). T3 concentrates massively in infected erythrocytes in an energy-dependent and saturable process, which underlies its antiplasmodial activity at low nanomolar concentrations (50). We observed a clear increase in the IC50 values for T3 and T16 in 10G-0.6-2 compared to 10G, which together with silencing of both clag3 genes after only 2 weeks of selection with these drugs demonstrates that P. falciparum can develop resistance to them by changes in the expression of clag3 genes (30, 32). The concentration of T3 and T16 that was used in the selection experiments was below the IC50 of the drugs, which explains the difference with previous reports concluding that choline analogs are likely not prone to drug resistance (50). Resistance to drugs at low concentrations is physiologically relevant because low drug concentrations can be encountered by parasites at some points during a treatment course. However, in spite of selecting parasites with T3 and T16 at low concentrations, we observed silencing of both clag3 genes, a pattern that in the case of blasticidin S was only observed when using high concentrations (≫IC50) of the drug. Although resistance to blasticidin S at low concentration can be acquired by selection of parasites that switched from expression of one clag3 gene to expression of the other, which does not pose a fitness cost, this was not observed for T3. Thus, our experiments did not identify additional compounds for which there are differences in sensitivity (likely reflecting differences in transport efficiency) between parasites that express clag3.1 and parasites that express clag3.2.
The IC50 increase for T3 and T16 in the 10G-0.6-2 line relative to 10G is moderate compared to the increase observed for blasticidin S (∼2- to ∼3-fold versus ∼10-fold). This is consistent with previous reports showing that the transport of T3/T16 into the infected erythrocytes is highly reduced, but not eliminated, by treatment with the NPP inhibitor furosemide. The authors concluded that transport of these compounds occurs mainly through PSAC/NPPs, but residual transport (∼15%) is nonsaturable and continues to occur even in the presence of high concentrations of the inhibitor (40, 41). Residual transport of T3/T16 through membrane diffusion or endogenous transporters may explain the relatively modest differences in IC50 values between 10G and 10G-0.6-2 cultures.
A previous study analyzing the effect of T4, a bis-thiazolium compound structurally related with T3 and T16, did not detect significant transcriptional changes in 3D7 cultures after 30 min to 36 h of exposure to the drug (51). The apparent discrepancy with our results reflects the very different experimental approaches used: while the previous study analyzed changes in mRNA levels soon after drug exposure to explore the occurrence of directed protective transcriptional responses, we studied adaptations at the transcriptional level after several cycles of selection with the drug. Our approach revealed changes in the expression of clag3 genes linked to adaptation, indicating that selection of parasites with specific expression patterns of clonally variant genes can occur in the development of resistance to compounds to which the parasite is unable to mount a directed transcriptional response. Of note, natural selection of parasites with specific transcriptional patterns is the basis of bet-hedging adaptive strategies in P. falciparum (47, 48).
We did not observe major differences in the IC50 for doxycycline, fosmidomycin, azithromycin, lumefantrine, or pentamidine between the 10G and 10G-0.6-2 lines. However, there is robust previous data for fosmidomycin and pentamidine indicating that these drugs require NPPs for their entry into infected erythrocytes: their uptake is abrogated by NPPs inhibitors, and they concentrate massively inside the infected erythrocytes (38, 39). Hence, the most plausible explanation for the similar sensitivity to these compounds between the 10G-0.6-2 and 10G lines is that their uptake occurs via CLAG3-independent NPPs, as in the case of the PSAC substrate 5-ALA. We consider it likely that PSAC involving nonvariantly expressed CLAGs, which we predict to mediate 5-ALA uptake in CLAG3-defficient lines, also mediates the uptake of drugs such as fosmidomycin and pentamidine in these lines. However, we cannot fully exclude the possibility that the residual expression of CLAG3 in 10G-0.6-2, which is undistinguishable from background signal in IFA experiments, is sufficient to mediate transport of these compounds. In any case, given that doxycycline, fosmidomycin, lumefantrine, azithromycin, and pentamidine can access and kill parasites expressing dramatically reduced levels of clag3 genes, it is unlikely that P. falciparum can develop resistance to these compounds by selection of parasite subpopulations with altered clag3 expression as in the case of blasticidin S, T3, or T16.
Altogether, we identified T3 and T16 as antimalarials that require CLAG3-containing PSAC to enter the infected erythrocyte, and thus these drugs are prone to parasite resistance by epigenetic changes in the expression of clag3 genes. This is an important concern because resistance acquired at the epigenetic level can arise quickly during the course of a single infection and is easily reversible, providing the parasite with a level of plasticity toward susceptible drugs that would promptly render them ineffective. On the other hand, our results suggest that other compounds known or predicted to require NPPs for their uptake by infected erythrocytes may use PSAC involving CLAG8 or CLAG9, or other channels different from PSAC. To determine which drugs require any form of PSAC to access infected erythrocytes, experiments similar to the ones presented here could be performed using conditional KO lines for rhoph2 or rhoph3 (21–23). However, even if these experiments identified additional drugs that require PSAC for their uptake, these drugs would not be prone to the resistance mechanism regulated at the epigenetic level studied here if they show normal transport in the 10G-0.6-2 line.
MATERIALS AND METHODS
Parasite cultures.
The 3D7-A stock of the clonal P. falciparum line 3D7 and the 10G subclone of 3D7-A have been previously described and characterized (27, 42, 52). The 10G-0.4 and 10G-0.6 lines were generated by selection of the 10G line with blasticidin S at 0.4 and 0.6 μg/ml, respectively, whereas the 10G-0.6-2 line was generated by sequential selection with 0.6 and 2 μg/ml (32). The 5F5 inducible rhoph3 disruption line has been previously described (23). Parasites were cultured in O erythrocytes at a 3% hematocrit with inactivated human serum under standard culture conditions, except for selection experiments with T3 and their controls that were performed with B+ erythrocytes and media containing Albumax II instead of serum. The 10G-0.6-2 parasite line was regularly cultured under blasticidin S pressure (2 μg/ml) to maintain silencing of both clag3 genes (32). For synchronization, we used treatment with 5% sorbitol, except for the 10G-0.6-2 line, the 10G line selected with T3 or T16, and the unselected 10G controls analyzed in parallel that were synchronized with l-proline as previously described (31). To prepare RNA for transcriptional analysis, cultures were harvested when the majority of parasites were at the schizont stage, i.e., when clag3 genes are expressed, and a small proportion of schizonts had already burst.
IFAs.
IFAs were performed on Percoll-purified parasites at the mature schizont stage because at this stage CLAG3 yields a strong and unambiguous signal in rhoptries, whereas detection at other stages in which the protein is only present in the surface of infected erythrocytes is less clear. Air-dried smears were fixed for 10 min with 1% paraformaldehyde and permeabilized for 10 min with 0.1% Triton X-100 in phosphate-buffered saline (PBS). Smears were incubated with rabbit anti-3D7 AMA1 (1:2,000; kindly provided by Robin F. Anders, La Trobe University, Australia) (53) and mouse anti-CLAG3 (1:2,000; from mouse 167#2, kindly provided by Sanjay A. Desai, NIAID-NIH) (19) polyclonal antibodies. We used secondary anti-rabbit antibodies conjugated with Alexa Fluor 488 (Life Technologies, A-11034) and anti-mouse antibodies conjugated with Alexa Fluor 546 (Life Technologies, A-10036). Nuclei were stained with DAPI (4′,6′-diamidino-2-phenylindole). Preparations were observed under a confocal Leica TCS-SP5 microscope with LAS-AF image acquisition software and processed using ImageJ. AMA1, which is expressed later than CLAG3 during intraerythrocytic development and does not show variant expression, was used to identify parasites that were mature enough for CLAG3 expression. This enabled us to distinguish between parasites that did not express CLAG3 because they were at a too early stage of development from parasites in which both clag3 genes were simultaneously silenced at the epigenetic level. The analysis of CLAG3 expression was restricted to AMA1-positive parasites: the proportion of schizonts that have both CLAG3s silenced was determined by counting >100 AMA1-postitive mature schizonts in each replicate experiment.
Drugs.
Blasticidin S, lumefantrine, doxycycline, fosmidomycin, leupeptin, and azithromycin were purchased from Sigma-Aldrich (reference numbers 15205, L5420, D9891, F8682, L8511, and PZ0007, respectively). T3, T16, and pentamidine were kindly provided by Henri J. Vial (CNRS, Montpellier, France). Stock solutions for each drug were prepared as follows: azithromycin and pentamidine in dimethyl sulfoxide (DMSO) at 10 mM, blasticidin S in H2O at 10 mM, doxycycline in methanol at 20 mM, fosmidomycin in PBS at 1 mM, T3 and T16 in H2O at 10 mM, and lumefantrine at 1 mM in a 1:1:1 volume mix of Tween 80, ethanol, and linoleic acid (prepared from a 5 mM stock in DMSO).
Growth inhibition assays.
To determine the IC50 of the different compounds in 10G and 10G-0.6-2 cultures, we used a previously described SYBR Green-based assay (54) with some adjustments. We incubated parasites in triplicate wells for 96 h for experiments with blasticidin S, leupeptin, lumefantrine, fosmidomycin, pentamidine, T3, and T16, which kill the parasites in the first cycle after drug administration, or for 144 h for drugs that produce a delayed death effect (55), killing parasites only at the second cycle after adding the drug (doxycycline and azithromycin). Initial parasitemias were adjusted for each incubation time and parasite strain in order to prevent the culture from collapsing, while ensuring a sufficiently high final parasitemia (in the absence of drug) for accurate determination of the inhibition levels. The parasitemia of synchronized ring-stage cultures was determined by light microscopy and adjusted to 0.1 to 0.2 or 0.05% for 96- and 144-h experiments with the 10G line, respectively, and to 0.2 to 0.5% or 0.1% for 96- and 144-h assays with 10G-0.6-2, respectively. In experiments with 10G-0.6-2 cultures, which were regularly maintained under blasticidin S pressure, blasticidin S was removed 5 h before starting the assay. After 96 or 144 h of incubation, parasites were exposed to one freeze-thaw cycle, and 100 μl of lysis buffer (54) with SYBR Green 5× was added per well. Plates were incubated at 37°C in the dark for 3 h, and then fluorescence was measured on a plate reader (Victor X3; Perkin-Elmer) with excitation and emission wavelength bands centered at 485 and 535 nm, respectively. After log transforming the drug concentrations, the data were fit to sigmoidal dose-response curves using GraphPad Prism (version 5), setting the maximum to 100 and the minimum to 0. IC50 values were compared between 10G and 10G-0.6-2 using a two-tailed t test for unpaired data (Stata, version 12). We applied the Benjamini-Hochberg multiple testing correction of P values (56), with a false discovery rate of 0.1.
Drug selection experiments.
To select cultures with doxycycline, fosmidomycin, T3, T16, leupeptin, or blasticidin S, the drugs were initially applied to cultures at the ring stage. Cultures were maintained under sublethal drug concentrations (determined in preliminary experiments) for a maximum of 14 weeks, or until clear evidence of adaptation to the drug was observed, i.e., an increase in the growth rate in the presence of the drug compared to the initial growth rate when the drug was first added (see Fig. S1 in the supplemental material). Unselected cultures were maintained in parallel. In all selection experiments, parasites were harvested for RNA extraction every 3 to 5 weeks.
Transcriptional analysis.
For RNA purification, erythrocyte pellets were collected in TRIzol (Invitrogen), and phase separation was conducted according to the manufacturer’s instructions. RNA was purified from the aqueous phase using an RNeasy minikit (Qiagen) as previously described (30) and reverse transcribed using a reverse transcription system (Promega). To exclude gDNA contamination, parallel reactions were performed in the absence of reverse transcriptase. cDNAs were analyzed by quantitative PCR in triplicate wells using Power SYBR Green master mix (Applied Biosystems) in a StepOnePlus or a 7900HT Fast real-time PCR system (Applied Biosystems). clag3.1 and clag3.2 expression values, in arbitrary units, were calculated using the standard curve method as previously described (30). The primers used have been previously described (30).
Analysis of erythrocyte membrane permeability by 5-ALA uptake.
The 5F5 inducible rhoph3 disruption line was treated with rapamycin at the cycle prior to testing 5-ALA uptake to induce deletion of exons 4 to 6 (23) or treated in parallel with DMSO as a control. 5-ALA uptake was determined as previously described (23, 43), with minor modifications. In brief, cultures of synchronous ring-stage parasites were incubated overnight in normal RPMI-based parasite culture medium with Albumax II supplemented with 200 μM 5-ALA (Sigma-Aldrich, catalog no. A3785). After uptake by infected erythrocytes, 5-ALA is converted to PPIX. Just prior to analysis, parasite nuclei were stained with 2 μg/ml Hoechst for 10 min at 37°C and washed with PBS. Samples were placed in an eight-well chamber slide for live cell fluorescence microscopy analysis using the Fluorescence Imaging System Leica AF6000. Images were captured with the same acquisition settings for all samples, so that signal intensities are directly comparable. Images were analyzed using ImageJ software.
Supplementary Material
ACKNOWLEDGMENTS
We thank Sanjay A. Desai (National Institutes of Health) for kindly providing mouse anti-CLAG3 polyclonal antibody; Robin F. Anders (La Trobe, Australia) for the rabbit anti-3D7 AMA1 polyclonal antibody; Henri J. Vial and Sharon Wein (CNRS, Montpellier, France) for providing T3, T16, and pentamidine; and Emma Sherling and Mike Blackman for the P. falciparum conditional rhoph3 deletion line 5F5.
This study was supported by the Spanish Ministry of Economy and Competitiveness through the Agencia Estatal de Investigación, cofunded by the European Regional Development Fund (ERDF/FEDER), European Union (SAF2013-43601-R and SAF2016-76190-R to A.C.); the Secretary for Universities and Research, Department of Economy and Knowledge, Government of Catalonia (2014 SGR 485 to A.C.); the Institute of Tropical Medicine, Antwerp (funding to A.R.-U.). ISGlobal is a member of the CERCA Program, Government of Catalonia. ITM and ISGlobal are members of the Trans Global Health–Erasmus Mundus Joint Doctorate Program, European Union (scholarship to S.M.-M.). E.T.-F. is supported by a fellowship from the Spanish Ministry of Economy and Competitiveness (BES-2014-067901), cofunded by the European Social Fund (ESF), European Commission. A.K.P. is supported by a fellowship from the Secretary for Universities and Research, Catalan Government (FI_B 00373), cofunded by the European Social Fund (ESF), European Commission. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.00052-19.
REFERENCES
- 1.World Health Organization. 2016. Global malaria programme, World Malaria Report 2016. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 2.Ashley EA, Dhorda M, Fairhurst RM, Amaratunga C, Lim P, Suon S, Sreng S, Anderson JM, Mao S, Sam B, Sopha C, Chuor CM, Nguon C, Sovannaroth S, Pukrittayakamee S, Jittamala P, Chotivanich K, Chutasmit K, Suchatsoonthorn C, Runcharoen R, Hien TT, Thuy-Nhien NT, Thanh NV, Phu NH, Htut Y, Han K-T, Aye KH, Mokuolu OA, Olaosebikan RR, Folaranmi OO, Mayxay M, Khanthavong M, Hongvanthong B, Newton PN, Onyamboko MA, Fanello CI, Tshefu AK, Mishra N, Valecha N, Phyo AP, Nosten F, Yi P, Tripura R, Borrmann S, Bashraheil M, Peshu J, Faiz MA, Ghose A, Hossain MA, Samad R, Rahman MR, Hasan MM, Islam A, Miotto O, Amato R, MacInnis B, Stalker J, Kwiatkowski DP, Bozdech Z, Jeeyapant A, Cheah PY, Sakulthaew T, Chalk J, Intharabut B, Silamut K, Lee SJ, Vihokhern B, Kunasol C, Imwong M, Tarning J, Taylor WJ, Yeung S, Woodrow CJ, Flegg JA, Das D, Smith J, Venkatesan M, Plowe CV, Stepniewska K, Guerin PJ, Dondorp AM, Day NP, White NJ. 2014. Spread of artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med 371:411–423. doi: 10.1056/NEJMoa1314981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amaratunga C, Lim P, Suon S, Sreng S, Mao S, Sopha C, Sam B, Dek D, Try V, Amato R, Blessborn D, Song L, Tullo GS, Fay MP, Anderson JM, Tarning J, Fairhurst RM. 2016. Dihydroartemisinin-piperaquine resistance in Plasmodium falciparum malaria in Cambodia: a multisite prospective cohort study. Lancet Infect Dis 16:357–365. doi: 10.1016/S1473-3099(15)00487-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Petersen I, Eastman R, Lanzer M. 2011. Drug-resistant malaria: molecular mechanisms and implications for public health. FEBS Lett 585:1551–1562. doi: 10.1016/j.febslet.2011.04.042. [DOI] [PubMed] [Google Scholar]
- 5.Peterson DS, Walliker D, Wellems TE. 1988. Evidence that a point mutation in dihydrofolate reductase-thymidylate synthase confers resistance to pyrimethamine in falciparum malaria. Proc Natl Acad Sci U S A 85:9114–9118. doi: 10.1073/pnas.85.23.9114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Triglia T, Menting JG, Wilson C, Cowman AF. 1997. Mutations in dihydropteroate synthase are responsible for sulfone and sulfonamide resistance in Plasmodium falciparum. Proc Natl Acad Sci U S A 94:13944–13949. doi: 10.1073/pnas.94.25.13944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lakshmanan V, Bray PG, Verdier-Pinard D, Johnson DJ, Horrocks P, Muhle RA, Alakpa GE, Hughes RH, Ward SA, Krogstad DJ, Sidhu AB, Fidock DA. 2005. A critical role for PfCRT K76T in Plasmodium falciparum verapamil-reversible chloroquine resistance. EMBO J 24:2294–2305. doi: 10.1038/sj.emboj.7600681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barnes DA, Foote SJ, Galatis D, Kemp DJ, Cowman AF. 1992. Selection for high-level chloroquine resistance results in deamplification of the pfmdr1 gene and increased sensitivity to mefloquine in Plasmodium falciparum. EMBO J 11:3067–3075. doi: 10.1002/j.1460-2075.1992.tb05378.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elford BC, Haynes JD, Chulay JD, Wilson RJM. 1985. Selective stage-specific changes in the permeability to small hydrophilic solutes of human erythrocytes infected with Plasmodium falciparum. Mol and Biochem Parasitol 16:43–60. doi: 10.1016/0166-6851(85)90048-9. [DOI] [PubMed] [Google Scholar]
- 10.Ginsburg H, Kutner S, Krugliak M, Cabantchik ZI. 1985. Characterization of permeation pathways appearing in the host membrane of Plasmodium falciparum infected red blood cells. Mol Biochem Parasitol 14:313–322. doi: 10.1016/0166-6851(85)90059-3. [DOI] [PubMed] [Google Scholar]
- 11.Neame KD, Homewood CA. 1975. Alterations in the permeability of mouse erythrocytes infected with the malaria parasite, Plasmodium berghei. Int J Parasitol 5:537–540. doi: 10.1016/0020-7519(75)90046-6. [DOI] [PubMed] [Google Scholar]
- 12.Desai SA, Bezrukov SM, Zimmerberg J. 2000. A voltage-dependent channel involved in nutrient uptake by red blood cells infected with the malaria parasite. Nature 406:1001–1005. doi: 10.1038/35023000. [DOI] [PubMed] [Google Scholar]
- 13.Hill DA, Pillai AD, Nawaz F, Hayton K, Doan L, Lisk G, Desai SA. 2007. A blasticidin S-resistant Plasmodium falciparum mutant with a defective plasmodial surface anion channel. Proc Natl Acad Sci U S A 104:1063–1068. doi: 10.1073/pnas.0610353104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alkhalil A, Cohn JV, Wagner MA, Cabrera JS, Rajapandi T, Desai SA. 2004. Plasmodium falciparum likely encodes the principal anion channel on infected human erythrocytes. Blood 104:4279–4286. doi: 10.1182/blood-2004-05-2047. [DOI] [PubMed] [Google Scholar]
- 15.Desai SA. 2014. Why do malaria parasites increase host erythrocyte permeability? Trends Parasitol 30:151–159. doi: 10.1016/j.pt.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Desai SA. 2012. Ion and nutrient uptake by malaria parasite-infected erythrocytes. Cell Microbiol 14:1003–1009. doi: 10.1111/j.1462-5822.2012.01790.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaneko O, Tsuboi T, Ling IT, Howell S, Shirano M, Tachibana M, Cao YM, Holder AA, Torii M. 2001. The high molecular mass rhoptry protein, RhopH1, is encoded by members of the clag multigene family in Plasmodium falciparum and Plasmodium yoelii. Mol Biochem Parasitol 118:223–231. doi: 10.1016/S0166-6851(01)00391-7. [DOI] [PubMed] [Google Scholar]
- 18.Pillai AD, Nguitragool W, Lyko B, Dolinta K, Butler MM, Nguyen ST, Peet NP, Bowlin TL, Desai SA. 2012. Solute restriction reveals an essential role for clag3-associated channels in malaria parasite nutrient acquisition. Mol Pharmacol 82:1104–1114. doi: 10.1124/mol.112.081224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nguitragool W, Bokhari AA, Pillai AD, Rayavara K, Sharma P, Turpin B, Aravind L, Desai SA. 2011. Malaria parasite clag3 genes determine channel-mediated nutrient uptake by infected red blood cells. Cell 145:665–677. doi: 10.1016/j.cell.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nguitragool W, Rayavara K, Desai SA. 2014. Proteolysis at a specific extracellular residue implicates integral membrane CLAG3 in malaria parasite nutrient channels. PLoS One 9:e93759. doi: 10.1371/journal.pone.0093759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Counihan NA, Chisholm SA, Bullen HE, Srivastava A, Sanders PR, Jonsdottir TK, Weiss GE, Ghosh S, Crabb BS, Creek DJ, Gilson PR, de Koning-Ward TF. 2017. Plasmodium falciparum parasites deploy RhopH2 into the host erythrocyte to obtain nutrients, grow, and replicate. Elife 6:e23217. doi: 10.7554/eLife.23217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ito D, Schureck MA, Desai SA. 2017. An essential dual-function complex mediates erythrocyte invasion and channel-mediated nutrient uptake in malaria parasites. Elife 6:e23485. doi: 10.7554/eLife.23485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sherling ES, Knuepfer E, Brzostowski JA, Miller LH, Blackman MJ, van Ooij C. 2017. The Plasmodium falciparum rhoptry protein RhopH3 plays essential roles in host cell invasion and nutrient uptake. Elife 6:e23239. doi: 10.7554/eLife.23239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gupta A, Balabaskaran-Nina P, Nguitragool W, Saggu GS, Schureck MA, Desai SA. 2018. CLAG3 self-associates in malaria parasites and quantitatively determines nutrient uptake channels at the host membrane. Mol Biol 9:e02293-17. doi: 10.1128/mBio.02293-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cooper JA, Ingram LT, Bushell GR, Fardoulys CA, Stenzel D, Schofield L, Saul AJ. 1988. The 140/130/105 kilodalton protein complex in the rhoptries of Plasmodium falciparum consists of discrete polypeptides. Mol Biochem Parasitol 29:251–260. doi: 10.1016/0166-6851(88)90080-1. [DOI] [PubMed] [Google Scholar]
- 26.Gupta A, Thiruvengadam G, Desai SA. 2015. The conserved clag multigene family of malaria parasites: essential roles in host-pathogen interaction. Drug Resist Updat 18:47–54. doi: 10.1016/j.drup.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cortes A, Carret C, Kaneko O, Yim Lim BY, Ivens A, Holder AA. 2007. Epigenetic silencing of Plasmodium falciparum genes linked to erythrocyte invasion. PLoS Pathog 3:e107. doi: 10.1371/journal.ppat.0030107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Comeaux CA, Coleman BI, Bei AK, Whitehurst N, Duraisingh MT. 2011. Functional analysis of epigenetic regulation of tandem RhopH1/clag genes reveals a role in Plasmodium falciparum growth. Mol Microbiol 80:378–390. doi: 10.1111/j.1365-2958.2011.07572.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Crowley VM, Rovira-Graells N, de Pouplana LR, Cortés A. 2011. Heterochromatin formation in bistable chromatin domains controls the epigenetic repression of clonally variant Plasmodium falciparum genes linked to erythrocyte invasion. Mol Microbiol 80:391–406. doi: 10.1111/j.1365-2958.2011.07574.x. [DOI] [PubMed] [Google Scholar]
- 30.Mira-Martinez S, van Schuppen E, Amambua-Ngwa A, Bottieau E, Affara M, Van Esbroeck M, Vlieghe E, Guetens P, Rovira-Graells N, Gomez-Perez GP, Alonso PL, D’Alessandro U, Rosanas-Urgell A, Cortes A. 2017. Expression of the Plasmodium falciparum clonally variant clag3 genes in human infections. J Infect Dis 215:938–945. doi: 10.1093/infdis/jix053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rovira-Graells N, Crowley VM, Bancells C, Mira-Martínez S, Ribas de Pouplana L, Cortés A. 2015. Deciphering the principles that govern mutually exclusive expression of Plasmodium falciparum clag3 genes. Nucleic Acids Res 43:8243–8257. doi: 10.1093/nar/gkv730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mira-Martinez S, Rovira-Graells N, Crowley VM, Altenhofen LM, Llinas M, Cortes A. 2013. Epigenetic switches in clag3 genes mediate blasticidin S resistance in malaria parasites. Cell Microbiol 15:1913–1923. doi: 10.1111/cmi.12162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sharma P, Wollenberg K, Sellers M, Zainabadi K, Galinsky K, Moss E, Nguitragool W, Neafsey D, Desai SA. 2013. An epigenetic antimalarial resistance mechanism involving parasite genes linked to nutrient uptake. J Biol Chem 288:19429–19440. doi: 10.1074/jbc.M113.468371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lisk G, Pain M, Gluzman IY, Kambhampati S, Furuya T, Su XZ, Fay MP, Goldberg DE, Desai SA. 2008. Changes in the plasmodial surface anion channel reduce leupeptin uptake and can confer drug resistance in Plasmodium falciparum-infected erythrocytes. Antimicrob Agents Chemother 52:2346–2354. doi: 10.1128/AAC.00057-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hill DA, Desai SA. 2010. Malaria parasite mutants with altered erythrocyte permeability: a new drug resistance mechanism and important molecular tool. Future Microbiol 5:81–97. doi: 10.2217/fmb.09.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Basore K, Cheng Y, Kushwaha AK, Nguyen ST, Desai SA. 2015. How do antimalarial drugs reach their intracellular targets? Front Pharmacol 6:91. doi: 10.3389/fphar.2015.00091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lisk G, Pain M, Sellers M, Gurnev PA, Pillai AD, Bezrukov SM, Desai SA. 2010. Altered plasmodial surface anion channel activity and in vitro resistance to permeating antimalarial compounds. Biochim Biophys Acta 1798:1679–1688. doi: 10.1016/j.bbamem.2010.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baumeister S, Wiesner J, Reichenberg A, Hintz M, Bietz S, Harb OS, Roos DS, Kordes M, Friesen J, Matuschewski K, Lingelbach K, Jomaa H, Seeber F. 2011. Fosmidomycin uptake into Plasmodium and Babesia-infected erythrocytes is facilitated by parasite-induced new permeability pathways. PLoS One 6:e19334. doi: 10.1371/journal.pone.0019334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stead AM, Bray PG, Edwards IG, DeKoning HP, Elford BC, Stocks PA, Ward SA. 2001. Diamidine compounds: selective uptake and targeting in Plasmodium falciparum. Mol Pharmacol 59:1298–1306. doi: 10.1124/mol.59.5.1298. [DOI] [PubMed] [Google Scholar]
- 40.Biagini GA, Richier E, Bray PG, Calas M, Vial H, Ward SA. 2003. Heme binding contributes to antimalarial activity of bis-quaternary ammoniums. Antimicrob Agents Chemother 47:2584–2589. doi: 10.1128/AAC.47.8.2584-2589.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wein S, Maynadier M, Bordat Y, Perez J, Maheshwari S, Bette-Bobillo P, Tran Van Ba C, Penarete-Vargas D, Fraisse L, Cerdan R, Vial H. 2012. Transport and pharmacodynamics of albitiazolium, an antimalarial drug candidate. Br J Pharmacol 166:2263–2276. doi: 10.1111/j.1476-5381.2012.01966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cortes A, Benet A, Cooke BM, Barnwell JW, Reeder JC. 2004. Ability of Plasmodium falciparum to invade Southeast Asian ovalocytes varies between parasite lines. Blood 104:2961–2966. doi: 10.1182/blood-2004-06-2136. [DOI] [PubMed] [Google Scholar]
- 43.Sigala PA, Crowley JR, Henderson JP, Goldberg DE. 2015. Deconvoluting heme biosynthesis to target blood-stage malaria parasites. Elife 4:e09143. doi: 10.7554/eLife.09143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lisk G, Scott S, Solomon T, Pillai AD, Desai SA. 2007. Solute-inhibitor interactions in the plasmodial surface anion channel reveal complexities in the transport process. Mol Pharmacol 71:1241–1250. doi: 10.1124/mol.106.030734. [DOI] [PubMed] [Google Scholar]
- 45.Lopez-Rubio JJ, Mancio-Silva L, Scherf A. 2009. Genome-wide analysis of heterochromatin associates clonally variant gene regulation with perinuclear repressive centers in malaria parasites. Cell Host Microbe 5:179–190. doi: 10.1016/j.chom.2008.12.012. [DOI] [PubMed] [Google Scholar]
- 46.Flueck C, Bartfai R, Volz J, Niederwieser I, Salcedo-Amaya AM, Alako BT, Ehlgen F, Ralph SA, Cowman AF, Bozdech Z, Stunnenberg HG, Voss TS. 2009. Plasmodium falciparum heterochromatin protein 1 marks genomic loci linked to phenotypic variation of exported virulence factors. PLoS Pathog 5:e1000569. doi: 10.1371/journal.ppat.1000569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rovira-Graells N, Gupta AP, Planet E, Crowley VM, Mok S, Ribas de Pouplana L, Preiser PR, Bozdech Z, Cortes A. 2012. Transcriptional variation in the malaria parasite Plasmodium falciparum. Genome Res 22:925–938. doi: 10.1101/gr.129692.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cortes A, Deitsch KW. 2017. Malaria epigenetics. Cold Spring Harb Perspect Med 7:a025528. doi: 10.1101/cshperspect.a025528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wein S, Taudon N, Maynadier M, Tran Van Ba C, Margout D, Bordat Y, Fraisse L, Wengelnik K, Cerdan R, Bressolle-Gomeni F, Vial HJ. 2017. High accumulation and in vivo recycling of the new antimalarial albitiazolium lead to rapid parasite death. Antimicrob Agents Chemother 61:e00352-17. doi: 10.1128/AAC.00352-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wein S, Tran Van Ba C, Maynadier M, Bordat Y, Perez J, Peyrottes S, Fraisse L, Vial HJ. 2014. New insight into the mechanism of accumulation and intraerythrocytic compartmentation of albitiazolium, a new type of antimalarial. Antimicrob Agents Chemother 58:5519–5527. doi: 10.1128/AAC.00040-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Le Roch KG, Johnson JR, Ahiboh H, Chung DW, Prudhomme J, Plouffe D, Henson K, Zhou Y, Witola W, Yates JR, Mamoun CB, Winzeler EA, Vial H. 2008. A systematic approach to understand the mechanism of action of the bisthiazolium compound T4 on the human malaria parasite, Plasmodium falciparum. BMC Genomics 9:513. doi: 10.1186/1471-2164-9-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cortes A. 2005. A chimeric Plasmodium falciparum Pfnbp2b/Pfnbp2a gene originated during asexual growth. Int J Parasitol 35:125–130. doi: 10.1016/j.ijpara.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 53.Healer J, Murphy V, Hodder AN, Masciantonio R, Gemmill AW, Anders RF, Cowman AF, Batchelor A. 2004. Allelic polymorphisms in apical membrane antigen-1 are responsible for evasion of antibody-mediated inhibition in Plasmodium falciparum. Mol Microbiol 52:159–168. doi: 10.1111/j.1365-2958.2003.03974.x. [DOI] [PubMed] [Google Scholar]
- 54.Worldwide Antimalarial Resistance Network. 2011. In vitro module: Plasmodium falciparum drug sensitivity assay using SYBR Green I assay technique WWARN procedure. Worldwide Antimalarial Resistance Network, LOCATION. [Google Scholar]
- 55.Goodman CD, Pasaje CF, Kennedy K, McFadden GI, Ralph SA. 2016. Targeting protein translation in organelles of the Apicomplexa. Trends Parasitol 32:953–965. doi: 10.1016/j.pt.2016.09.011. [DOI] [PubMed] [Google Scholar]
- 56.Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol 57:289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.