We report two KPC-producing Citrobacter freundii isolates from unrelated patients. In one case, blaKPC-2 was harbored on a novel variant of a Tn4401 transposon of an IncN plasmid conjugated together with a coresident IncA plasmid, whereas in the other one, blaKPC-3 was on a Tn4401a transposon located on an IncX3-IncA self-conjugative plasmid fusion.
KEYWORDS: Citrobacter freundii, IncA, IncN, IncX3, KPC, Tn4401a, Tn4401i
ABSTRACT
We report two KPC-producing Citrobacter freundii isolates from unrelated patients. In one case, blaKPC-2 was harbored on a novel variant of a Tn4401 transposon of an IncN plasmid conjugated together with a coresident IncA plasmid, whereas in the other one, blaKPC-3 was on a Tn4401a transposon located on an IncX3-IncA self-conjugative plasmid fusion. The interplay among plasmids carrying blaKPC and the coresident IncA plasmids offers new information on plasmids coresident within clinically relevant enterobacteria.
INTRODUCTION
Carbapenem-resistant Enterobacteriaceae is emerging as a global threat, especially due to the production of carbapenemase enzymes (1). The most frequent carbapenemases in clinical isolates of human origin are the class A KPC enzymes, which spread mostly in Klebsiella pneumoniae but also at lower frequency in other Enterobacteriaceae, such as Citrobacter freundii (2, 3). C. freundii is considered a low-risk pathogen in clinical settings but can act, as in the two examples described here, as a silent reservoir of relevant resistance genes.
Two strains of C. freundii, designated AA535 and AA593, were isolated during February to March 2016 from a screening rectal swab of a patient recovered at the S. Agostino-Estense-Baggiovara Hospital, Modena, Italy, and from a urine sample of a resident in the long-term care facility of the same geographic area, respectively. The two patients did not have any apparent epidemiological link.
Identification of the bacterial species was performed using the Vitek 2 system (bioMérieux, Marcy l’Etoile, France). Antimicrobial susceptibility was assessed by reference broth microdilution, and MICs were interpreted according to EUCAST 2019 breakpoints (http://www.eucast.org). The two strains were genotyped by XbaI digestion and pulsed-field gel electrophoresis (PFGE) (4) and by multilocus sequence typing (MLST) (5). The two isolates exhibited different PFGE patterns (data not shown), and the MLST profiles, assigned by the PubMLST database (https://pubmlst.org/cfreundii/), resulted in ST19 and ST46 for strains AA593 and AA535, respectively. Replicon content was detected by PCR-based replicon typing (PBRT 2.0 kit, DIATHEVA) (6). PBRT detected A/C and N replicons in strain AA535 and A/C and X3 replicons in strain AA593.
Conjugation experiments for strains AA535 and AA593 were performed by using streptomycin-resistant Escherichia coli K-12 J62 (pro−, his−, trp−, lac−, Smr) as recipient, selecting transconjugants on 100 μg/ml ampicillin plus 150 μg/ml streptomycin and on 6 μg/ml meropenem plus 150 μg/ml streptomycin. Ten transconjugant colonies from each experiment were identified as E. coli by matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) using MALDI Biotyper software (Bruker Daltonics, Bremen, Germany). PBRT 2.0 kit detected A/C and N replicons in all the selected AA535 transconjugants and A/C and X3 replicons in all AA593 transconjugants.
Plasmids were extracted from donors by use of the PureYield Plasmid Midiprep kit (Promega) and transformed into chemically competent E. coli DH5α cells (Invitrogen). Transformants were selected on Luria-Bertani agar plates containing 100 μg/ml ampicillin (Sigma). Transformation of strain AA535 provided two types of transformants, positive to A/C and N replicons, respectively. Presence of the blaKPC gene was detected by PCR using the universal primers KPC_FU_1_8 5′-GTGCAGCTCATTCAAGGG-3′ and KPC_RU_1_8 5′-GCCAATCAACAAACTGCTG-3′. The blaKPC-2 gene was identified in the IncN-positive transformants. Transformation with plasmid DNA of strain AA593 yielded one type of transformant, positive by PBRT for both A/C and X3 replicons and to the blaKPC-3 gene.
Short-read sequence data were obtained from AA535 and AA593 parental strains using Illumina MiSeq next-generation sequencer (Illumina Inc., CA). De novo assembly was performed with SPAdes 3.10 and A5 MiSeq software (http://cab.spbu.ru/software/spades/) using the default SPAdes parameters (7, 8). Resistance and replicon content were determined by using default threshold parameters in ResFinder (https://cge.cbs.dtu.dk/services/ResFinder/ [9]) and PlasmidFinder (https://cge.cbs.dtu.dk/services/PlasmidFinder/ [10]), respectively. Assembled genomes were also tested by BLASTN against the IncA (FJ705807), IncN (AY046276), and IncX3 (JN247852) plasmid reference sequences, as suggested by PlasmidFinder results. Therefore, IncA (previously IncA/C1, renamed by Ambrose et al. [11]) and IncN plasmid sequences were identified in the AA535 genome in one (144,490-bp) and two (33,378- and 15,559-bp) large scaffolds, respectively. The AA535 genome was also positive for Col440 I and Col440I small plasmids (9,532 and 1,736 bp). IncA and IncX3 plasmid sequences were identified in five large scaffolds (12,876, 57,440, 45,859, 13,171, and 52,783 bp) in the AA593 genome. In addition, IncQ1 was detected within this genome. Complete plasmid sequence assembly was obtained by checking pair-end overlaps and performing PCR-based gap closure, using plasmids from both donors and transconjugants as DNA templates. Plasmid sequences were annotated by BLASTP (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE=Proteins).
The pIBAC_IncA plasmid (MH594477) of strain AA535 was 145,294 bp in size and carried a class I integron with aac(6')-Ib-cr5, blaOXA-1, and catB3 gene cassettes. It showed 99% nucleotide identity and 90% coverage with pIncAC-KP4898 (KY882285) (12), a blaVIM-1-positive plasmid described in a K. pneumoniae from Naples, Italy. The copresence of pIBAC_IncA and IncN plasmids was identified in most of the AA535 transconjugants by PBRT; presence of the blaKPC gene was confirmed by PCR in the same strains. An integration of the IS3 family element (ISEhe3) was observed between the traW and traU genes, which were not disrupted by this integration.
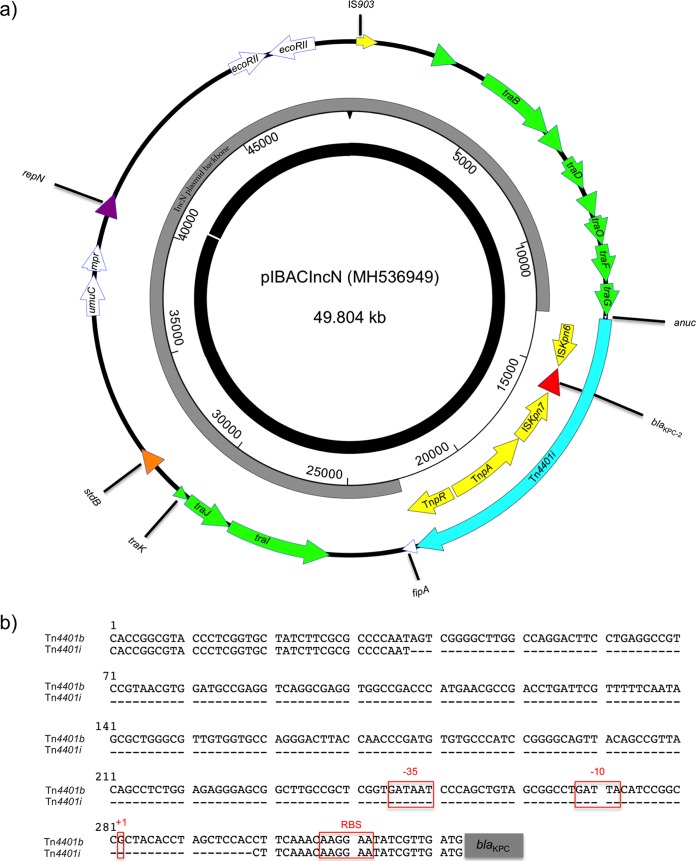
The pIBAC_IncN plasmid (MH536949) of AA535 was 49,804 bp in size and showed 99% nucleotide similarity and 95% coverage with pKp148 PINH-4900 (KX062091), recovered from a K. pneumoniae strain from an urban river in Brazil. The IncN plasmid carried a blaKPC-2 gene on a novel variant of the Tn4401 transposon designated Tn4401i, integrated within the nuc gene (Fig. 1). This transposon had the same genetic environment surrounding the blaKPC-2 gene as Tn4401b, except for a deletion of 260 bp upstream of blaKPC-2 (Fig. 1). The IncN sequence type was determined to be ST15 at the pMLST (https://cge.cbs.dtu.dk/services/pMLST/). ST15 IncN plasmids were described as being efficient shuttles between various species and clones in a study conducted on KPC-producing Enterobacteriaceae isolates in Netanya, Israel (13).
FIG 1.
(a) Circular map of pIBAC_IncN plasmid compared with pKp148 (black circular box). Green arrows, conjugal transfer system of the plasmid; red arrow, blaKPC gene; yellow arrows, mobile elements; white arrows, hypothetical proteins; purple arrow, replication protein repN; orange arrow, stability protein stdB. Gray circular box represents the plasmid backbone, and transposon Tn4401i is represented by a blue arrow. (b) Alignment in the position of interest between the sequence of Tn4401i and that of Tn4401b. −35 and −10, putative promoter regions; +1, putative transcription starting point; RBS, ribosome-binding site.
The AA593 pIBAC_IncX3_IncA (MH594478) plasmid of 192,802 bp was a fusion of IncA and IncX3 plasmids. The occurrence of such an IncA-IncX3 plasmid fusion in the parental strain was ascertained by the results of the transformation experiments described here: a unique type of E. coli DH5α transformant, positive to both A/C and X3 replicons and to the blaKPC-3 gene, was obtained.
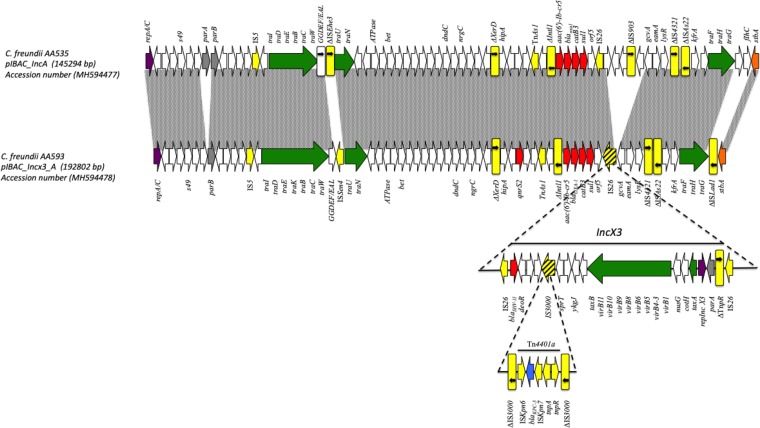
The blaKPC-3 gene was associated with a Tn4401a transposon flanked by truncated tnpA of IS3000 in a configuration very similar to the IncX3 plasmid pCfr-145 (KY659388), recently identified in several C. freundii in Italy (Fig. 2) (14). The IncA-fused portion of the pIBAC_IncX3_IncA plasmid showed 99% nucleotide identity and 92% coverage with the pIBAC_IncA plasmid of AA535, carrying the same class I integron with aac(6')-Ib-cr5, blaOXA-1, and catB3 gene cassettes but a different IS3 element (ISSen4) integrated within the traW and traU genes. The fusion of the IncA and IncX3 plasmids in pIBAAC539 may have been mediated by recombination between IS26 elements. The IncX3 plasmid scaffold was interrupted by two directly repeated IS26 elements located next to the blaSHV-11 gene, followed by the resistance region of the IncA plasmid. Moreover, hypothetical proteins and an IS903, which were located in the proximity of IS26 in the pIBAC_IncA plasmid of the AA535 scaffold, were lost in the fusion process in the pIBAC_IncX3_IncA plasmid of AA593, and a truncated ISLad1 was inserted just before the stbA stability gene, probably independent of the fusion process (Fig. 2).
FIG 2.
Linear map of pIBAC_IncA and pIBAC_IncX3_A. Arrows, direction of transcription of open reading frames (ORFs); rectangles, truncated ORFs. Replicons, partitioning genes, mobile elements, conjugal transfer genes, antibiotic resistance, blaKPC-3, and other remaining genes are designated by violet, gray, yellow, green, red, blue, and white, respectively. Gray shaded area in upper part shows similarity of both IncA plasmids. Lower part shows the IncX3 plasmid region of pIBAC_IncX3_A and its insertion/fusion point, which is shown as striped yellow arrows/rectangles in the IncX3 region.
In conclusion, both C. freundii strains had the same resident IncA plasmid. In strain AA593, it was permanently fused to IncX3 carrying the blaKPC-3 gene. In the other strain, despite the two plasmids being physically distinct in the donor and separately transferred in transformation experiments, they conjugated together, with the coresident IncN plasmid carrying blaKPC-2. The role of the companion IncA plasmids in these C. freundii isolates is not clear. The transfer locus of IncA plasmids were likely not functional while those of the IncX3 and IncN were conserved. IncA may have contributed to stabilization of the coresident IncX3 plasmid in one strain and used the IncN as a helper plasmid for conjugation in the other. The IncA plasmids may be normal residents of C. freundii and may favor adaptation, replication, and stability of the plasmids imported from other clinically relevant, carbapenemase-producing Enterobacteriaceae, favoring their spread. On the other hand, IncA plasmids may be symbionts of other coresident plasmids, using their conjugative properties to spread.
Accession number(s).
The nucleotide sequences of plasmids pIBAC_IncA, pIBAC_IncN, and pIBAC_IncX3_IncA have been deposited in GenBank, and the following accession numbers have been assigned, respectively: MH594477, MH536949, and MH594478.
ACKNOWLEDGMENTS
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
We have no conflicts to declare.
REFERENCES
- 1.Wu W, Espedido B, Feng Y, Zong Z. 2016. Citrobacter freundii carrying blaKPC-2 and blaNDM-1: characterization by whole genome sequencing. Sci Rep 6:30670. doi: 10.1038/srep30670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yao Y, Lazaro-Perona F, Falgenhauer L, Valverde A, Imirzalioglu C, Dominguez L, Cantón R, Mingorance J, Chakraborty T. 2017. Insights into a novel blaKPC-2-encoding IncP-6 plasmid reveal carbapenem-resistance circulation in several Enterobacteriaceae species from wastewater and a hospital source in Spain. Front Microbiol 8:1143. doi: 10.3389/fmicb.2017.01143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cuzon G, Naas T, Nordmann P. 2011. Functional characterization of Tn4401, a Tn3-based transposon involved in blaKPC gene mobilization. Antimicrob Agents Chemother 55:5370–5373. doi: 10.1128/AAC.05202-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Piazza A, Caltagirone M, Bitar I, Nucleo E, Spalla M, Fogato E, D’Angelo R, Pagani L, Migliavacca R. 2015. Emergence of Escherichia coli sequence type 131 (ST131) and ST3948 with KPC-2, KPC-3 and KPC-8 carbapenemases from a long-term care and rehabilitation facility (LTCRF) in northern Italy. Adv Exp Med Biol 901:77–89. doi: 10.1007/5584_2015_5017. [DOI] [PubMed] [Google Scholar]
- 5.Gaibani P, Ambretti S, Farruggia P, Bua G, Berlingeri A, Tamburini MV, Cordovana M, Guerra L, Mazzetti M, Roncarati G, Tenace C, Moro ML, Gagliotti C, Landini MP, Sambri V. 2013. Outbreak of Citrobacter freundii carrying VIM-1 in an Italian hospital, identified during the carbapenemases screening actions, June 2012. Int J Infect Dis 17:e714–e717. doi: 10.1016/j.ijid.2013.02.007. [DOI] [PubMed] [Google Scholar]
- 6.Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threlfall EJ. 2005. Identification of plasmids by PCR-based replicon typing. J Microbiol Methods 63:219–228. doi: 10.1016/j.mimet.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 7.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nurk S, Bankevich A, Antipov D, Gurevich A, Korobeynikov A, Lapidus A, Prjibelsky A, Pyshkin A, Sirotkin A, Sirotkin Y, Stepanauskas R, Mclean J, Lasken R, Clingenpeel SR, Woyke T, Tesler G, Alekseyev MA, Pevzner PA. 2013. Assembling genomes and mini-metagenomes from highly chimeric reads. J Comput Biol 20:714–739. doi: 10.1089/cmb.2013.0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV. 2012. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carattoli A, Zankari E, García-Fernández A, Larsen MV, Lund O, Villa L, Aarestrup FM, Hasman H. 2014. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother 58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ambrose SJ, Harmer CJ, Hall RM. 2018. Compatibility and entry exclusion of IncA and IncC plasmids revisited: IncA and IncC plasmids are compatible. Plasmid 96-97:7–12. doi: 10.1016/j.plasmid.2018.02.002. [DOI] [PubMed] [Google Scholar]
- 12.Esposito EP, Gaiarsa S, Franco MD, Crivaro V, Bernardo M, Cuccurullo S, Pennino F, Triassi M, Marone P, Sassera D, Zarrilli R. 2017. A novel IncA/C1 group conjugative plasmid, encoding VIM-1 metallo-beta-lactamase, mediates the acquisition of carbapenem resistance in ST104 Klebsiella pneumoniae isolates from neonates in the intensive care unit of V. Monaldi Hospital in Naples. Front Microbiol 8:2135. doi: 10.3389/fmicb.2017.02135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adler A, Khabra E, Paikin S, Carmeli Y. 2016. Dissemination of the blaKPC gene by clonal spread and horizontal gene transfer: comparative study of incidence and molecular mechanisms. J Antimicrob Chemother 71:2143–2146. doi: 10.1093/jac/dkw106. [DOI] [PubMed] [Google Scholar]
- 14.Venditti C, Fortini D, Villa L, Vulcano A, D’Arezzo S, Capone A, Petrosillo N, Nisii C, Carattoli A, Caro AD. 2017. Circulation of blaKPC-3-carrying IncX3 plasmids among Citrobacter freundii isolates in an Italian hospital. Antimicrob Agents Chemother 61:e00505-17. doi: 10.1128/AAC.00505-17. [DOI] [PMC free article] [PubMed] [Google Scholar]