Chryseobacterium infections are uncommon, and previous studies have revealed that Chryseobacterium gleum is frequently misidentified as Chryseobacterium indologenes. We aimed to explore the differences in clinical manifestations and antimicrobial susceptibility patterns between C. gleum and C. indologenes.
KEYWORDS: Chryseobacterium gleum, Chryseobacterium indologenes, drug resistance, quinolone resistance-determining region
ABSTRACT
Chryseobacterium infections are uncommon, and previous studies have revealed that Chryseobacterium gleum is frequently misidentified as Chryseobacterium indologenes. We aimed to explore the differences in clinical manifestations and antimicrobial susceptibility patterns between C. gleum and C. indologenes. The database of a clinical microbiology laboratory was searched to identify patients with Chryseobacterium infections between 2005 and 2017. Species were reidentified using 16S rRNA gene sequencing, and patients with C. gleum and C. indologenes infections were included in the study. A total of 42 C. gleum and 84 C. indologenes isolates were collected from consecutive patients. A significant increase in C. indologenes incidence was observed. C. gleum was significantly more associated with bacteremia than C. indologenes. Patients with C. gleum infections had more comorbidities of malignancy and liver cirrhosis than those with C. indologenes infections. The overall case fatality rate was 19.8%. Independent risk factors for mortality were female sex and C. indologenes infection. These isolates were most susceptible to minocycline (73%), followed by trimethoprim-sulfamethoxazole (47.6%), tigecycline (34.1%), and levofloxacin (32.5%). C. gleum exhibited a significantly higher rate of susceptibility than C. indologenes to piperacillin, piperacillin-tazobactam, ceftazidime, tigecycline, and levofloxacin. Alterations in DNA gyrase subunit A were identified to be associated with fluoroquinolone resistance in C. indologenes. No nonsynonymous substitutions were observed in the quinolone resistance-determining regions (QRDRs) of C. gleum. Differences in epidemiology, clinical manifestations, and antimicrobial susceptibility patterns exist between C. gleum and C. indologenes. Additional investigations are needed to explore the significance of these differences.
INTRODUCTION
Chryseobacterium, which normally exists in soil, water, and plants, is a genus of aerobic, Gram-negative, non-spore-forming, nonmotile, and nonfermenting, rod-shaped bacteria (1). These microorganisms are common pathogens of plants and animals but infrequently cause human infections (1). There are currently more than 100 species in the genus Chryseobacterium (2), among which Chryseobacterium indologenes is the most common species associated with human infections, followed by Chryseobacterium gleum (3–12). Nonetheless, C. gleum infections are substantially less frequently reported than infections with C. indologenes (3–12).
Our previous studies demonstrated that C. indologenes could be accurately identified using traditional biochemically based phenotyping methods and matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) (13, 14). In contrast, C. gleum was not reliably identified using commercial biochemically based phenotyping identification or Vitek MS systems (bioMérieux, Marcy l’Etoile, France) (13, 14). Moreover, these microbial identification systems usually misidentified C. gleum as C. indologenes. Overall, accurate identification of Chryseobacterium species relies on 16S rRNA gene sequencing or the Bruker Biotyper MALDI-TOF MS system (Bruker Daltonics GmbH, Bremen, Germany) (14, 15). Therefore, previous studies describing the epidemiology, clinical manifestations, virulence, pathogenesis, and antimicrobial susceptibilities of C. indologenes might have substantial bias if the identification was based on inaccurate methods.
Moreover, because the species in previous reports might not have been correctly identified, the actual clinical and molecular manifestations of C. gleum and C. indologenes remain unknown. To explore the exact epidemiology, clinical manifestations, and antimicrobial susceptibility patterns of C. gleum and C. indologenes, we apply 16S rRNA gene sequencing in this study to identify Chryseobacterium species and then compare clinical characteristics and antimicrobial susceptibility patterns between C. gleum and C. indologenes isolates causing human infections. We also investigate the contributions of mutations in the quinolone resistance-determining regions (QRDRs) to fluoroquinolone resistance in clinical isolates of C. gleum and C. indologenes.
RESULTS
Species identification.
During the 13-year period of investigation, 156 nonduplicated Chryseobacterium isolates collected from consecutive patients were found in the database of the clinical microbiology laboratory. Seven isolates died after thawing, and 11 could not be identified to the species level using 16S rRNA gene sequencing. Ultimately, 138 isolates were identified to the species level, including 42 C. gleum isolates (type strain ATCC 35910T; GenBank accession no. GL379781), 84 C. indologenes isolates (type strain DSM 16777T; GenBank accession no. LN681561), 4 C. arthrosphaerae isolates (type strain CC-VM-7T; GenBank accession no. NR_116977), 2 C. culicis isolates (type strain R4-1AT; GenBank accession no. FN554975), 2 C. cucumeris isolates (type strain GSE06T; GenBank accession no. KX146463), 2 C. bernardetii isolates (type strain CDC G229T; GenBank accession no. JX100816), 1 C. artocarpi isolate (type strain UTM-3T; GenBank accession no. KF751867), and 1 C. daecheongense isolate (type strain CPW406T; GenBank accession no. AJ457206).
Site of isolation.
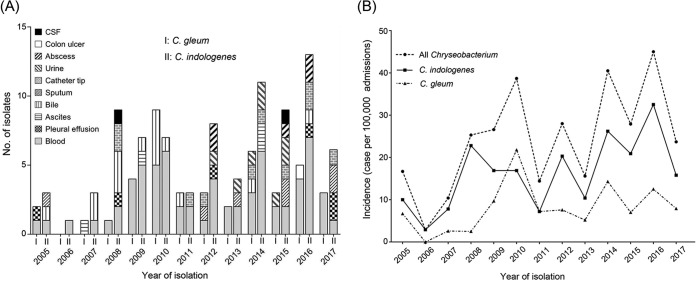
Among the 126 isolates of C. gleum and C. indologenes, 68 (54%) were collected from blood, 15 (11.9%) were from bile, 10 (7.9%) were from the tip of a central venous catheter, and 8 (6.3%) were from urine (Fig. 1A). C. gleum was significantly more prone to be isolated from blood than C. indologenes (66.7% for C. gleum versus 47.6% for C. indologenes; odds ratio [OR], 2.2; 95% confidence interval [CI], 1.02 to 4.76; P = 0.043). There was no statistically significant difference between the prevalences of these two species regarding the other sites of isolation.
FIG 1.
(A) Isolation sites of C. gleum and C. indologenes from 2005 to 2017. Blood was the most common site of isolation, followed by bile, the tip of a central venous catheter, and urine. (B) The incidence of Chryseobacterium from 2005 to 2017. A significant increase in the incidence of all Chryseobacterium isolates (chi-square for trend; χ2 = 4.1; P = 0.043) and C. indologenes (χ2 = 4.32; P = 0.038) was recognized. The incidence of C. gleum did not significantly increase during the study period (χ2 = 1.7; P = 0.193).
Incidence of infection.
The number of Chryseobacterium isolates and the incidence (number of cases per 100,000 admissions) distributed by the different years are shown in Fig. 1B. A significant increase in the incidence of Chryseobacterium infection was observed during the study period (P = 0.043). When the incidence of each species was calculated independently, a significant year-over-year increase was observed for C. indologenes (P = 0.038) but not for C. gleum (P = 0.193).
Clinical characteristics of Chryseobacterium infections.
Of the 126 consecutive patients with Chryseobacterium infections, male predominance (68.3%) was observed (Table 1). The median age was 59.5 years (range, 13 to 91 years; mean ± standard deviation, 59 ± 19.2 years). The majority (81.7%) of patients were found to have at least one comorbidity, with the most common being cardiovascular diseases (46.8%), followed by diabetes mellitus (38.1%), malignancy (33.3%), and liver cirrhosis (15.9%). Health care-associated infection accounted for 99.2%. One-third of the patients presented with shock, and 35.7% of patients were admitted to intensive care units. Empirical antibiotics administered included β-lactams (52.4%), β-lactam/lactamase inhibitors (46%), fluoroquinolones (50%), carbapenems (11.1%), aminoglycosides (14.3%), tigecycline (9.5%), and trimethoprim-sulfamethoxazole (7.1%), either singly or in combination. Inappropriate empirical antimicrobial therapy was recognized in 88.9% of patients. The overall mortality rate of those with Chryseobacterium infections was 19.8%.
TABLE 1.
Comparison of demographic characteristics, clinical information, and outcomes of patients with C. gleum and C. indologenes infections
| Parameter | Value for the parameter |
OR (95% CI)a | P value | ||
|---|---|---|---|---|---|
| Total group (n = 126) | C. gleum (n = 42) | C. indologenes (n = 84) | |||
| Sex (no. of patients [%]) | |||||
| Male | 86 (68.3) | 27 (64.3) | 59 (70.2) | 0.76 (0.35–1.67) | 0.499 |
| Female | 40 (31.7) | 15 (35.7) | 25 (29.8) | 1.31 (0.6–2.88) | 0.499 |
| Age (yr) | |||||
| Range | 13–91 | 20–91 | 13–90 | ||
| Median | 59.5 | 58.5 | 62 | ||
| Mean ± standard deviation | 59 ± 19.2 | 58.8 ± 16.5 | 59.1 ± 20.4 | 0.913 | |
| Comorbidity (no. of patients [%]) | 103 (81.7) | 36 (85.7) | 67 (79.8) | 1.52 (0.55–4.2) | 0.415 |
| Diabetes mellitus | 48 (38.1) | 13 (31) | 35 (41.7) | 0.63 (0.29–1.38) | 0.243 |
| Cardiovascular disease | 59 (46.8) | 17 (40.5) | 42 (50) | 0.68 (0.32–1.44) | 0.313 |
| End-stage renal disease (no. of patients [%]) | 5 (4) | 1 (2.4) | 4 (4.8) | 0.49 (0.05–4.51) | 0.664 |
| Malignancy | 42 (33.3) | 21 (50) | 21 (25) | 3 (1.37–6.55) | 0.005 |
| Liver cirrhosis | 20 (15.9) | 13 (31) | 7 (8.3) | 4.93 (1.79–13.58) | 0.001 |
| Chronic obstructive pulmonary disease | 10 (7.9) | 2 (4.8) | 8 (9.5) | 0.48 (0.1–2.34) | 0.494 |
| Type of infection acquisition (no. of patients [%]) | |||||
| Community-acquired infection | 1 (0.8) | 0 | 1 (1.2) | 0.999 | |
| Health care-associated infection | 125 (99.2) | 42 (100) | 83 (98.8) | 1.51 (1.33–1.71) | 0.999 |
| Laboratory data | |||||
| White blood cell count (cells/mm3) | 14,039 ± 39,343 | 9,578 ± 5,697 | 16,270 ± 47,959 | 0.37 | |
| Hemoglobin (g/dl) | 10.4 ± 2.3 | 10.8 ± 2.1 | 10.2 ± 2.4 | 0.187 | |
| Platelet count (×103 cells/mm3) | 198,629 ± 120,124 | 212,905 ± 108,507 | 191,492 ± 125,542 | 0.348 | |
| Serum creatinine (mg/dl) | 1.7 ± 1.5 | 1.6 ± 1.7 | 1.8 ± 1.4 | 0.487 | |
| Shock (no. of patients [%]) | 42 (33.3) | 6 (14.3) | 36 (42.9) | 0.22 (0.09–0.58) | 0.001 |
| Admission to intensive care unit (no. of patients [%]) | 45 (35.7) | 5 (11.9) | 40 (47.6) | 0.15 (0.05–0.42) | <0.001 |
| Inappropriate empirical antimicrobial therapy (no. of patients [%]) | 112 (88.9) | 38 (90.5) | 74 (88.1) | 1.28 (0.38–4.37) | 0.688 |
| Outcome | |||||
| Case fatality (no. [%]) | 25 (19.8) | 4 (9.5) | 21 (25) | 0.32 (0.1–0.99) | 0.04 |
OR, odds ratio; CI, confidence interval.
Comparison of C. gleum and C. indologenes infections.
When comparing patients with C. gleum and C. indologenes infections, we found no significant differences in the age of onset, sex, or presence of diabetes mellitus, cardiovascular disease, end-stage renal disease, and chronic obstructive pulmonary disease. However, the prevalence of malignancy (OR, 3; 95% CI, 1.37 to 6.55; P = 0.005) and of liver cirrhosis (OR, 4.93; 95% CI, 1.79 to 13.58; P = 0.001) was significantly higher in patients with C. gleum infections than in those with C. indologenes infections. There was no significant difference in inappropriate empirical antimicrobial therapies between these two groups. Patients with C. gleum infections had a significantly lower risk of shock (OR, 0.22; 95% CI, 0.09 to 0.58; P = 0.001), rate of admission to an intensive care unit (OR, 0.15; 95% CI, 0.05 to 0.42; P < 0.001), and case fatality rate (OR, 0.32; 95% CI, 0.1 to 0.99; P = 0.04) than those with C. indologenes infections (Table 1).
Factors associated with mortality.
Univariate analysis showed that female patients generally and all patients with C. indologenes infection had higher mortality rates. Moreover, multivariate logistic regression analysis revealed that female sex (adjusted OR, 5.34; 95% CI, 2.04 to 13.97; P = 0.001) and C. indologenes infection (adjusted OR, 4.01; 95% CI, 1.2 to 13.38; P = 0.024) were independent risk factors for mortality in these Chryseobacterium-infected patients (Table 2).
TABLE 2.
Factors associated with mortality in patients with Chryseobacterium infections
| Factor | Outcome (no. of patients [%]) |
Univariate analysis |
Multivariate analysis |
|||
|---|---|---|---|---|---|---|
| Death (n = 25) | Survival (n = 101) | OR (95% CI)a | P value | OR (95% CI) | P value | |
| Isolate | ||||||
| C. gleum | 4 (16) | 38 (37.6) | 0.32 (0.1–0.99) | 0.04 | 0.25 (0.08–0.83) | 0.024 |
| C. indologenes | 21 (84) | 63 (62.4) | 3.17 (1.01–9.93) | 0.04 | 4.01 (1.2–13.38) | 0.024 |
| Age of ≥65 yr | 13 (52) | 39 (38.6) | 1.72 (0.71–4.16) | 0.224 | 1.17 (0.38–3.66) | 0.785 |
| Gender | ||||||
| Male | 10 (40) | 76 (75.2) | 0.22 (0.09–0.55) | 0.001 | 0.19 (0.07–0.49) | 0.001 |
| Female | 15 (60) | 25 (24.8) | 4.56 (1.82–11.43) | 0.001 | 5.34 (2.04–13.97) | 0.001 |
| Underlying disease | ||||||
| Diabetes mellitus | 11 (44) | 37 (36.6) | 1.36 (0.56–3.3) | 0.497 | 0.66 (0.23–1.93) | 0.446 |
| Cardiovascular disease | 14 (56) | 45 (44.6) | 1.58 (0.66–3.83) | 0.305 | 0.86 (0.28–2.61) | 0.784 |
| End-stage renal disease | 2 (8) | 3 (3) | 2.84 (0.45–17.99) | 0.258 | 1.44 (0.17–12.21) | 0.74 |
| Malignancy | 5 (20) | 37 (36.6) | 0.43 (0.15–1.25) | 0.114 | 0.85 (0.25–2.85) | 0.787 |
| Liver cirrhosis | 2 (8) | 18 (17.8) | 0.4 (0.09–1.86) | 0.36 | 0.87 (0.15–5) | 0.876 |
| Chronic obstructive pulmonary disease | 3 (12) | 7 (6.9) | 1.83 (0.44–7.65) | 0.414 | 2.13 (0.43–10.48) | 0.352 |
| Inappropriate empirical antimicrobial therapy | 21 (84) | 91 (90.1) | 0.58 (0.17–2.02) | 0.385 | 0.53 (0.13–2.1) | 0.364 |
OR, odds ratio; CI, confidence interval.
Antimicrobial susceptibility testing.
The MICs and susceptibilities of Chryseobacterium bacteria are presented in Table 3. The MICs of β-lactams, β-lactam/β-lactam inhibitors, carbapenems, and aminoglycosides were generally high. Furthermore, almost all Chryseobacterium bacteria showed little susceptibility to aminoglycosides, carbapenems, aztreonam, and tetracycline. These isolates were most susceptible to minocycline (73%), followed by susceptibility to trimethoprim-sulfamethoxazole (47.6%), tigecycline (34.1%), levofloxacin (32.5%), piperacillin-tazobactam (19.8%), and piperacillin (19%).
TABLE 3.
The MIC and susceptibility of Chryseobacterium in this study
| Drug | All Chryseobacterium isolates (n = 126) |
C. gleum (n = 42) |
C. indologenes (n = 84) |
OR (95% CI)b | P valuec | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| MIC50 | MIC90 | S (no. [%])a | MIC50 | MIC90 | S (no. [%]) | MIC50 | MIC90 | S (no. [%]) | |||
| Piperacillin | 64 | >64 | 24 (19) | 64 | >64 | 14 (33.3) | >64 | >64 | 10 (11.9) | 3.7 (1.47–9.29) | 0.004 |
| Piperacillin-tazobactam | 128/4 | >128/4 | 25 (19.8) | 64/4 | 128/4 | 14 (33.3) | 128/4 | >128/4 | 11 (13.1) | 3.32 (1.35–8.18) | 0.007 |
| Ticarcillin-clavulanic acid | >64/2 | >64/2 | 0 | >64/2 | >64/2 | 0 | >64/2 | >64/2 | 0 | ||
| Ceftazidime | >16 | >16 | 17 (13.5) | >16 | >16 | 10 (23.8) | >16 | >16 | 7 (8.3) | 3.44 (1.2–9.83) | 0.017 |
| Cefepime | >32 | >32 | 22 (17.5) | 32 | >32 | 8 (19) | >32 | >32 | 14 (16.7) | 1.18 (0.45–3.07) | 0.74 |
| Ceftriaxone | >32 | >32 | 0 | >32 | >32 | 0 | >32 | >32 | 0 | ||
| Aztreonam | >16 | >16 | 0 | >16 | >16 | 0 | >16 | >16 | 0 | ||
| Imipenem | >8 | >8 | 0 | >8 | >8 | 1 (2.4) | >8 | >8 | 0 | 0.333 | |
| Meropenem | >8 | >8 | 0 | >8 | >8 | 0 | >8 | >8 | 0 | ||
| Gentamicin | >8 | >8 | 0 | >8 | >8 | 0 | >8 | >8 | 0 | ||
| Tobramycin | >8 | >8 | 0 | >8 | >8 | 0 | >8 | >8 | 0 | ||
| Amikacin | >32 | >32 | 2 (1.6) | >32 | >32 | 0 | >32 | >32 | 2 (2.4) | 0.552 | |
| Tetracycline | >8 | >8 | 0 | >8 | >8 | 0 | >8 | >8 | 0 | ||
| Minocycline | 2 | 8 | 92 (73) | 2 | 8 | 35 (83.3) | 4 | 8 | 57 (67.9) | 2.41 (0.95–6.13) | 0.065 |
| Tigecycline | 4 | >8 | 43 (34.1) | 2 | 8 | 22 (52.4) | 4 | >8 | 21 (25) | 3.3 (1.51–7.21) | 0.002 |
| Ciprofloxacin | >2 | >2 | 23 (18.3) | 2 | >2 | 9 (21.4) | >2 | >2 | 14 (16.7) | 1.36 (0.54–3.47) | 0.514 |
| Levofloxacin | 8 | >8 | 41 (32.5) | <1 | 8 | 25 (59.5) | >8 | >8 | 16 (19) | 6.25 (2.75–14.22) | <0.001 |
| Trimethoprim-sulfamethoxazole | 4/76 | >4/76 | 60 (47.6) | <2/38 | >4/76 | 24 (57.1) | 4/76 | >4/76 | 36 (42.9) | 1.78 (0.84–3.76) | 0.13 |
S, susceptible.
OR, odd ratio; CI, confidence interval.
Comparison of the susceptibility rates between C. gleum and C. indologenes.
Analysis of the MICs and susceptibilities of each species revealed that the MIC50 values of minocycline, tigecycline, ciprofloxacin, levofloxacin, and trimethoprim-sulfamethoxazole for C. gleum were generally lower than those for C. indologenes. C. gleum was most susceptible to minocycline (83.3%), levofloxacin (59.5%), trimethoprim-sulfamethoxazole (57.1%), and tigecycline (52.4%). C. indologenes was also most susceptible to minocycline (67.9%), followed by susceptibility to trimethoprim-sulfamethoxazole (42.9%), tigecycline (25%), and levofloxacin (19%). C. gleum expressed a significantly higher susceptibility rate than C. indologenes to piperacillin (OR, 3.7; 95% CI, 1.47 to 9.29; P = 0.004), piperacillin-tazobactam (OR, 3.32; 95% CI, 1.35 to 8.18; P = 0.007), ceftazidime (OR, 3.44; 95% CI, 1.2 to 9.83; P = 0.017), tigecycline (OR, 3.3; 95% CI, 1.51 to 7.21; P = 0.002), and levofloxacin (OR, 6.25; 95% CI, 2.75 to 14.22; P < 0.001).
Mutations in the QRDRs.
No nonsynonymous substitutions were observed in the QRDRs of GyrA, GyrB, ParC, or ParE in any isolate of C. gleum. However, amino acid alternations were recognized at positions 83 and 87 in GyrA and at position 66 in ParC of C. indologenes (Table 4). Four different amino acids were identified at position 83 in GyrA of C. indologenes, namely, serine (n = 24), phenylalanine (n = 4), valine (n = 12), and tyrosine (n = 44). Isolates of C. indologenes with serine at position 83 in GyrA were significantly more susceptible to fluoroquinolones (P < 0.001); in contrast, isolates with tyrosine at position 83 in GyrA were less susceptible to fluoroquinolones (P < 0.001). There were also four different amino acids at position 87 in GyrA of C. indologenes. Isolates with aspartate at position 87 in GyrA were significantly associated with levofloxacin (P < 0.001) and ciprofloxacin (P = 0.001) susceptibility, but those with tyrosine at position 87 in GyrA were significantly less susceptible to levofloxacin (P < 0.01) and ciprofloxacin (P = 0.018). Despite the amino acid alteration at position 66 in ParC of C. indologenes, no significant difference in the fluoroquinolone susceptibility was observed.
TABLE 4.
Amino acid alternations in the quinolone resistance-determining regions of C. indologenes
| Amino acid residue | Levofloxacin resistance |
Ciprofloxacin resistance |
||||||
|---|---|---|---|---|---|---|---|---|
| No. (%) of susceptible isolates (n = 16) | No. (%) of nonsusceptible isolates (n = 68) | OR (95% CI)a | P value | No. (%) of susceptible isolates (n =14) | No. (%) of nonsusceptible isolates (n = 70) | OR (95% CI) | P value | |
| Position 83 of GyrA | ||||||||
| Serine | 13 (81.3) | 11 (16.2) | 22.46 (5.47–92.12) | <0.001 | 13 (92.9) | 11 (15.7) | 69.73 (8.26–588.73) | <0.001 |
| Phenylalanine | 0 | 4 (5.9) | 0.999 | 0 | 4 (5.7) | 0.999 | ||
| Valine | 1 (6.3) | 11 (16.2) | 0.345 (0.04–2.89) | 0.447 | 0 | 12 (17.1) | 0.203 | |
| Tyrosine | 2 (12.5) | 42 (61.8) | 0.09 (0.2–0.42) | <0.001 | 1 (7.1) | 43 (61.4) | 0.05 (0.01–0.39) | <0.001 |
| Position 87 of GyrA | ||||||||
| Aspartate | 15 (93.8) | 29 (42.6) | 20.17 (2.52–161.55) | <0.001 | 13 (92.9) | 31 (44.3) | 16.36 (2.03–131.95) | 0.001 |
| Glycine | 1 (6.3) | 18 (26.5) | 0.19 (0.02–1.5) | 0.104 | 1 (7.1) | 18 (25.7) | 0.22 (0.03–1.82) | 0.174 |
| Histidine | 0 | 1 (1.5) | 0.999 | 0 | 1 (1.4) | 0.999 | ||
| Tyrosine | 0 | 20 (29.4) | 0.01 | 0 | 20 (28.6) | 0.018 | ||
| Position 66 of ParC | ||||||||
| Aspartate | 1 (6.3) | 3 (4.4) | 1.44 (0.14–14.87) | 0.578 | 1 (7.1) | 3 (4.3) | 1.72 (0.17–17.83) | 0.525 |
| Valine | 15 (93.8) | 65 (95.6) | 0.69 (0.07–7.13) | 0.578 | 13 (92.9) | 67 (95.7) | 0.58 (0.06–6.04) | 0.525 |
OR, odds ratio; CI, confidence interval.
DISCUSSION
As stated above, Chryseobacterium infections in humans, particularly C. gleum infections, are rarely reported. By using 16S rRNA gene sequencing as a reference method, we have identified 42 C. gleum and 84 C. indologenes isolates as well as 12 isolates of six uncommon Chryseobacterium species over the past 13 years. We reported the clinical features of six rarely observed Chryseobacterium species in our previous study (14). It is noteworthy that C. gleum accounted for approximately one-third of all Chryseobacterium bacteria isolated from clinical specimens, suggesting that the incidence of C. gleum infections in humans is substantially underestimated.
According to previous studies, Chryseobacterium commonly causes bacteremia, catheter-related infection, and pneumonia, and these infections are usually hospital acquired (3–11). Indeed, a previous study found that 47% (17/36) of C. indologenes-infected patients developed bacteremia (7), and in a study of 215 Chryseobacterium isolates (4), 64.2% and 18.1% were obtained from sputum and blood, respectively. Our current study also showed that patients infected with Chryseobacterium, particularly C. gleum, are prone to bacteremia.
Patients with Chryseobacterium infections usually have comorbidities, such as cardiovascular disease, malignancy, chronic kidney disease, diabetes mellitus, and chronic obstructive pulmonary disease (3–11). In our study, one or more comorbidities were recognized in 81.7% of Chryseobacterium-infected patients. Furthermore, patients infected with C. gleum had a significantly higher prevalence of malignancy and liver cirrhosis than those with C. indologenes infections. The host-pathogen interaction between humans and Chryseobacterium requires further investigation.
Chryseobacterium infection had a high mortality rate, with a reported overall rate of 13.9% to 40.7% (3–8). Nevertheless, research to date has not employed an accurate microbial identification method to assess the mortality of patients with C. gleum and C. indologenes infections. In our study, the overall case fatality rate of patients with Chryseobacterium infections was 19.8%, with the rate of C. gleum infection being much lower than that of C. indologenes infection (9.5% versus 25%). After adjusting for confounding variables in the multivariate analysis model, we found C. indologenes infection to be an independent risk factor for mortality. However, the reason for the different mortality rates of patients infected with C. gleum and C. indologenes is unknown. Further studies to explore the possible factors, such as virulence or pathogen-host interaction, are warranted.
Information about the susceptibilities of Chryseobacterium to antimicrobial agents is limited. Previous studies have demonstrated that Chryseobacterium is usually resistant to ceftriaxone, aztreonam, carbapenems, and aminoglycosides but variably susceptible to piperacillin, piperacillin-tazobactam, ceftazidime, cefepime, minocycline, tigecycline, ciprofloxacin, levofloxacin, and trimethoprim-sulfamethoxazole (4–12). The study by Hsueh et al. (7) reported that 75% (27/36) of C. indologenes isolates obtained in 1992 to 1995 were susceptible to minocycline, 63.9% were susceptible to piperacillin, 41.7% were susceptible to ceftazidime, and 30.6% were susceptible to trimethoprim. Kirby et al. (12). reported that >85% of 20 C. indologenes isolates collected during 1997 to 2001 were susceptible to piperacillin, piperacillin-tazobactam, ceftazidime, cefepime, ciprofloxacin, levofloxacin, and trimethoprim-sulfamethoxazole. In addition, Chen et al. (4) evaluated the antimicrobial susceptibilities of 113 C. indologenes isolates obtained in 2004 to 2011 and found that 87.6% were susceptible to trimethoprim-sulfamethoxazole, 41.8% were susceptible to tigecycline, 34.4% were susceptible to levofloxacin, 31.6% were susceptible to ciprofloxacin, and 29.4% were susceptible to piperacillin-tazobactam. Regardless, the species of C. indologenes in these above-mentioned studies was identified using inaccurate methods. Consequently, these susceptibility results likely do not represent the genuine antimicrobial susceptibility patterns of C. indologenes.
In our study, all Chryseobacterium strains examined showed the highest susceptibilities to minocycline, trimethoprim-sulfamethoxazole, tigecycline, and levofloxacin. Interestingly, a significant difference in the susceptibility rates was found between C. gleum and C. indologenes, whereby C. gleum was significantly more susceptible to piperacillin, piperacillin-tazobactam, ceftazidime, tigecycline, and levofloxacin. Only a few studies to date have examined the antimicrobial susceptibility of Chryseobacterium using accurate identification methods (10, 11). For example, Lo and Chang (10) and Chang et al. (11) used 16S rRNA gene sequencing to identify Chryseobacterium species and reported the susceptibilities of 15 C. gleum and 34 C. indologenes strains as follows (in respective order): minocycline, 100% and 100%; trimethoprim-sulfamethoxazole, 93.3% and 52%; ciprofloxacin, 40% and 14.7%; piperacillin, 0 and 2.9%; and piperacillin-tazobactam, 0 and 2.9%. In their reports, the susceptibility rates of C. gleum to trimethoprim-sulfamethoxazole and ciprofloxacin were also much higher than those of C. indologenes.
Fluoroquinolones are a suggested empirical antimicrobial therapy for Chryseobacterium infection (6, 12). However, our study demonstrated that a large number of Chryseobacterium isolates were not susceptible to fluoroquinolones. Moreover, C. indologenes isolates were significantly less susceptible to fluoroquinolones than C. gleum isolates. The mechanisms for fluoroquinolone resistance include gene alterations in drug target enzymes (GyrA, GyrB, ParC, and ParE), alterations in drug permeation (efflux pumps), and the plasmid-encoded quinolone resistance protein Qnr (16). Gene mutations in drug target enzymes are the major mechanism for fluoroquinolone resistance in many microorganisms (16). However, there is no information concerning the mechanisms of fluoroquinolone resistance in Chryseobacterium. Our study revealed that several C. indologenes isolates possessed amino acid alternations in the QRDR of GyrA, and these nonsynonymous substitutions were significantly associated with fluoroquinolone resistance. But no nonsynonymous amino acid substitution was identified in the QRDRs of C. gleum. Other mechanisms, such as efflux pumps and Qnr, could be responsible for fluoroquinolone resistance in C. gleum.
Conclusions.
This study reveals the different epidemiologies, clinical manifestations, and antimicrobial susceptibility patterns between C. gleum and C. indologenes. In particular, C. gleum demonstrates a significantly lower case fatality rate than C. indologenes and a higher susceptibility rate to several antimicrobial agents. To better understand these life-threatening pathogens, additional studies are necessary to investigate the significance of these differences.
MATERIALS AND METHODS
Ethics.
This study was conducted in accordance with the Declaration of Helsinki and national and institutional standards. This study was approved by the Institutional Review Board of E-Da Hospital (EMRP-106-105). The need for patient informed consent was waived by the Institutional Review Board because the retrospective analysis of routine cultures posed no more than minimal risk of harm to the subjects.
Study setting and design.
This study was performed at a 1,000-bed university-affiliated medical center in Taiwan. Microbial isolates were routinely collected according to the clinical requirements of patients. The computer database of the clinical microbial laboratory was searched for cultures that yielded Chryseobacterium species between January 2005 and December 2017. The microbial laboratory used an API/ID32 phenotyping kit (bioMérieux) (2005 to 2013) and Vitek MS System (2014 to 2017) to identify clinical isolates. All isolates were stored as glycerol stocks at −80°C until use. The species of Chryseobacterium was reidentified using 16S rRNA gene sequencing, and patients infected with C. gleum and C. indologenes were included. Demographic information, underlying illnesses, clinical manifestations, and outcomes were collected from medical records.
Definition.
Empirical antimicrobial therapy was defined as inappropriate if the isolates were not susceptible to the prescribed antibiotics. Shock was defined as systolic pressure of <90 mm Hg, a reduction of 40 mm Hg in the systolic blood pressure from baseline, or a condition requiring inotropic agents to maintain blood pressure.
Species identification using 16S rRNA gene sequencing.
The frozen bacteria were thawed and subcultured on tryptic soy agar with 5% sheep blood (Becton, Dickinson, Sparks, MD, USA). The colony was then inoculated onto fresh tryptic soy agar with 5% sheep blood and grown overnight. Total DNA from freshly collected colonies was prepared using a Wizard Genomic DNA purification kit (Promega, Madison, WI, USA). The primers and protocols for amplification and sequencing of the 16S rRNA gene are listed in Table S1 in the supplemental material (17). The assembled sequences of 16S rRNA genes were submitted to the National Center for Biotechnology Information website for comparison with GenBank sequence databases using the Basic Local Alignment Search Tool (https://blast.ncbi.nlm.nih.gov/Blast.cgi). To determine similarity, 16S rRNA sequences among isolates were compared to those of type strains in the GenBank sequence databases. An isolate was considered to be a Chryseobacterium species match if it shared ≥99.5% 16S rRNA sequence identity with the respective type strain in GenBank (18).
Antimicrobial susceptibility testing.
The MICs of antibiotics were examined using the broth microdilution method of the Sensititre 96-well panels according to the manufacturer's instructions (Thermo Fisher Scientific/Trek Diagnostics Systems, Oakwood Village, OH, USA). Susceptibility was interpreted according to the MIC criteria for “other non-Enterobacteriaceae” in the Clinical and Laboratory Standards Institute (CLSI) guidelines (19). Because there are no interpretive susceptibility criteria for “other non-Enterobacteriaceae” to tigecycline in the CLSI guidelines, the tigecycline MIC was interpreted according to the Enterobacteriaceae susceptibility breakpoints of the U.S. Food and Drug Administration (susceptible MIC, ≤2 mg/liter; intermediate MIC, 4 mg/liter; resistant MIC, ≥8 mg/liter) (20).
Identification of mutations in the QRDRs.
The primers and PCR conditions for the amplification of the QRDRs in gyrA, gyrB, parC, and parE are listed in Table S1.
Data analysis.
SPSS software (version 24.0; IBM, Armonk, NY, USA) was used to analyze the data. Categorical variables were calculated using a chi-square test or Fisher’s exact test, as appropriate. All factors associated with mortality were computed via univariate analyses and with the logistic regression model of multivariate analysis using backward stepwise methods by the likelihood ratio. The significance of variables was calculated by the odds ratio (OR), 95% confidence interval (CI), and two-tailed P value. A factor was considered statistically significant at a P value of <0.05.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grant EDPJ106075 from E-Da Hospital and MOST 105-2314-B-214-008 and MOST 106-2314-B-214-009-MY2 from the Ministry of Science and Technology, Taiwan.
We report that we have no conflicts of interest.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.02256-18.
REFERENCES
- 1.Vandamme P, Bernardet JF, Segers P, Kersters K, Holmes B. 1994. Notes: new perspectives in the classification of the Flavobacteria: Description of Chryseobacterium gen. nov., Bergeyella gen. nov., and Empedobacter nom. rev. Int J Syst Evol Microbiol 44:827–831. doi: 10.1099/00207713-44-4-827. [DOI] [Google Scholar]
- 2.Parte AC. 2017. List of prokaryotic names with standing in nomenclature: Chryseobacterium. http://www.bacterio.net/chryseobacterium.html. [DOI] [PMC free article] [PubMed]
- 3.Cantero M, Parra LM, Muñez E, Iranzo R, Sánchez-Romero MI, Oteo J, Asensio A. 2018. A cluster of Chryseobacterium indologenes cases related to drainage water in intensive care units. Infect Control Hosp Epidemiol 39:997–999. doi: 10.1017/ice.2018.126. [DOI] [PubMed] [Google Scholar]
- 4.Chen FL, Wang GC, Teng SO, Ou TY, Yu FL, Lee WS. 2013. Clinical and epidemiological features of Chryseobacterium indologenes infections: analysis of 215 cases. J Microbiol Immunol Infect 46:425–432. doi: 10.1016/j.jmii.2012.08.007. [DOI] [PubMed] [Google Scholar]
- 5.Chou DW, Wu SL, Lee CT, Tai FT, Yu WL. 2011. Clinical characteristics, antimicrobial susceptibilities, and outcomes of patients with Chryseobacterium indologenes bacteremia in an intensive care unit. Jpn J Infect Dis 64:520–524. [PubMed] [Google Scholar]
- 6.Lin YT, Jeng YY, Lin ML, Yu KW, Wang FD, Liu CY. 2010. Clinical and microbiological characteristics of Chryseobacterium indologenes bacteremia. J Microbiol Immunol Infect 43:498–505. doi: 10.1016/S1684-1182(10)60077-1. [DOI] [PubMed] [Google Scholar]
- 7.Hsueh PR, Teng LJ, Yang PC, Ho SW, Hsieh WC, Luh KT. 1997. Increasing incidence of nosocomial Chryseobacterium indologenes infections in Taiwan. Eur J Clin Microbiol Infect Dis 16:568–574. doi: 10.1007/BF02447918. [DOI] [PubMed] [Google Scholar]
- 8.Hsueh PR, Teng LJ, Ho SW, Hsieh WC, Luh KT. 1996. Clinical and microbiological characteristics of Flavobacterium indologenes infections associated with indwelling devices. J Clin Microbiol 34:1908–1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hsueh PR, Hsiue TR, Wu JJ, Teng LJ, Ho SW, Hsieh WC, Luh KT. 1996. Flavobacterium indologenes bacteremia: clinical and microbiological characteristics. Clin Infect Dis 23:550–555. doi: 10.1093/clinids/23.3.550. [DOI] [PubMed] [Google Scholar]
- 10.Lo HH, Chang SM. 2014. Identification, characterization, and biofilm formation of clinical Chryseobacterium gleum isolates. Diagn Microbiol Infect Dis 79:298–302. doi: 10.1016/j.diagmicrobio.2014.01.027. [DOI] [PubMed] [Google Scholar]
- 11.Chang YC, Lo HH, Hsieh HY, Chang SM. 2015. Identification, epidemiological relatedness, and biofilm formation of clinical Chryseobacterium indologenes isolates from central Taiwan. J Microbiol Immunol Infect 48:559–564. doi: 10.1016/j.jmii.2014.04.004. [DOI] [PubMed] [Google Scholar]
- 12.Kirby JT, Sader HS, Walsh TR, Jones RN. 2004. Antimicrobial susceptibility and epidemiology of a worldwide collection of Chryseobacterium spp: report from the SENTRY Antimicrobial Surveillance Program (1997–2001). J Clin Microbiol 42:445–448. doi: 10.1128/JCM.42.1.445-448.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin JN, Lai CH, Yang CH, Huang YH, Lin HF, Lin HH. 2017. Comparison of four automated microbiology systems with 16S rRNA gene sequencing for identification of Chryseobacterium and Elizabethkingia species. Sci Rep 7:13824. doi: 10.1038/s41598-017-14244-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin JN, Teng SH, Lai CH, Yang CH, Huang YH, Lin HF, Lin HH. 2018. Comparison of the VITEK MS and Bruker matrix-assisted laser desorption ionization-time of flight mass spectrometry systems for identification of Chryseobacterium isolated from clinical specimens and report of uncommon Chryseobacterium infections in humans. J Clin Microbiol Aug 22:e00712-18. doi: 10.1128/JCM.00712-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holmes B, Steigerwalt AG, Nicholson AC. 2013. DNA-DNA hybridization study of strains of Chryseobacterium, Elizabethkingia and Empedobacter and of other usually indole-producing non-fermenters of CDC groups IIc, IIe, IIh and IIi, mostly from human clinical sources, and proposals of Chryseobacterium bernardetii sp. nov., Chryseobacterium carnis sp. nov., Chryseobacterium lactis sp. nov., Chryseobacterium nakagawai sp. nov. and Chryseobacterium taklimakanense comb. nov. Int J Syst Evol Microbiol 63:4639–4662. doi: 10.1099/ijs.0.054353-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jacoby GA. 2005. Mechanisms of resistance to quinolones. Clin Infect Dis 41:S120–S126. doi: 10.1086/428052. [DOI] [PubMed] [Google Scholar]
- 17.Hantsis-Zacharov E, Shakéd T, Senderovich Y, Halpern M. 2008. Chryseobacterium oranimense sp. nov., a psychrotolerant, proteolytic and lipolytic bacterium isolated from raw cow’s milk. Int J Syst Evol Microbiol 58:2635–2639. doi: 10.1099/ijs.0.65819-0. [DOI] [PubMed] [Google Scholar]
- 18.Janda JM, Abbott SL. 2007. 16S rRNA gene sequencing for bacterial identification in the diagnostic laboratory: pluses, perils, and pitfalls. J Clin Microbiol 45:2761–2764. doi: 10.1128/JCM.01228-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clinical and Laboratory Standards Institute. 2016. Performance standards for antimicrobial susceptibility testing: twenty-sixth informational supplement M100-S26. CLSI, Wayne, PA. [Google Scholar]
- 20.Kelesidis T, Karageorgopoulos DE, Kelesidis I, Falagas ME. 2008. Tigecycline for the treatment of multidrug-resistant Enterobacteriaceae: a systematic review of the evidence from microbiological and clinical studies. J Antimicrob Chemother 62:895–904. doi: 10.1093/jac/dkn311. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.