This study aimed to investigate the genetic characteristics, antibiotic resistance patterns, and novel mechanisms involved in fluoroquinolone (FQ) resistance in commensal Escherichia coli isolates. The E. coli isolates were recovered from a previous clinical study and subjected to antimicrobial susceptibility testing and molecular typing.
KEYWORDS: Escherichia coli, fluoroquinolones, multidrug resistance, resistance mechanisms
ABSTRACT
This study aimed to investigate the genetic characteristics, antibiotic resistance patterns, and novel mechanisms involved in fluoroquinolone (FQ) resistance in commensal Escherichia coli isolates. The E. coli isolates were recovered from a previous clinical study and subjected to antimicrobial susceptibility testing and molecular typing. Known mechanisms of FQ resistance (target site mutations, plasmid-mediated quinolone resistance [PMQR] genes, relative expression levels of efflux pumps and porins) were detected using DNA sequencing of PCR products and real-time quantitative PCR. Whole-genome shotgun sequencing was performed on 11 representative strains to screen for single nucleotide polymorphisms (SNPs). The function of a key SNP (A1541G) was investigated by site-directed mutagenesis and allelic exchange. The results showed that long-term enrofloxacin treatment selected multidrug-resistant (MDR) E. coli isolates in the chicken gut and that these E. coli isolates had diverse genetic backgrounds. Multiple genetic alterations, including double mutations on GyrA (S83L and D87N), a single mutation on ParC (S80I) and ParE (S458E), activation of efflux pumps, and the presence of the QnrS1 protein, contributed to the high-level FQ resistance (enrofloxacin MIC [MICENR] ≥ 128 μg/ml), while the relatively low-level FQ resistance (MICENR = 8 or 16 μg/ml) was commonly mediated by decreased expression of the porin OmpF, besides enhancement of the efflux pumps. No significant relationship was observed between resistance mechanisms and virulence genes. Introduction of the A1541G mutation on aegA was able to increase FQ susceptibility by 2-fold. This study contributes to a better understanding of the development of MDR and the differences underlying the mechanisms of high-level and low-level FQ resistance in E. coli.
INTRODUCTION
Antimicrobial resistance is an ancient natural phenomenon that predates the use of antibiotics in human and veterinary medicine (1, 2). Nevertheless, the clinical application of antibiotics accelerates the selection of antimicrobial resistance in pathogens and/or commensals (3, 4). Fluoroquinolones (FQs) comprise many critically important synthetic antimicrobial agents (5, 6) and have been intensively used in veterinary medicine for the treatment of salmonellosis and colibacillosis in broilers (7, 8). FQ usage may have various impacts on the intestinal microbiota (9). Oral administration of FQs might kill or inhibit susceptible pathogens but might simultaneously influence the composition and structure of the intestinal microbiota (10, 11). Most importantly, resistant pathogens and commensals may be selected under the antibiotic pressure (12, 13). Several studies have reported the dose-effect and time-effect relationships between FQ usage and the selection of bacterial resistance (14–17).
Over the past 3 decades, FQ resistance in pathogens and commensals from different animal species has been prevalent worldwide (18–22). In the United States, FDA withdrew approval for the usage of enrofloxacin in poultry in 2005 due to the risk of FQ-resistant Campylobacter (23). However, later studies still reported the detection of FQ-resistant strains from broiler samples in the United States (24). FQ resistance in poultry was even observed in Australia, which has never authorized the usage of enrofloxacin (25). It is proposed that the emergence and persistence of bacterial resistance to FQs might be due to (i) a low dosage of antibiotic residues that maintains the selective pressure, (ii) coselection by nonquinolone antibiotics, and (iii) low or no fitness cost in the FQ-resistant bacteria.
Several mechanisms have been found to mediate FQ resistance, including a mutation(s) at the drug binding sites (26), decreased accumulation of FQs in cells (27–29), and plasmid-mediated quinolone resistance (PMQR) genes [qnr family, aac(6′)-Ib-cr, oqxAB, and qepA genes] (30, 31). Although the contributions of different mechanisms to phenotypic FQ resistance have been extensively studied in vitro (32, 33), the mechanisms underlying in vivo-selected FQ resistance are not fully understood.
Our previous studies found that long-term enrofloxacin treatment of Salmonella infection in chicken selected FQ-resistant coliforms in the gut (34). However, the virulence and resistance mechanisms were not fully investigated. The aim of this study was to determine the genetic characteristics, the antibiotic resistance patterns, and the differences underlying the high-level and low-level FQ resistance mechanisms of these in vivo-selected Escherichia coli isolates.
RESULTS
Antimicrobial resistance phenotypes of E. coli strains selected by enrofloxacin.
A total of 1,048 strains of coliforms were recovered from 400 fecal samples, which were collected during the whole sampling period. Using biochemical tests and 16S rRNA gene sequencing, 330 strains of coliforms were identified to be E. coli, and they were isolated from agar plates without enrofloxacin (n = 85) and agar plates with enrofloxacin at concentrations of 0.125 mg/liter (n = 63), 0.25 mg/liter (n = 87), and 2 mg/liter (n = 95). Of all the identified E. coli isolates, 115 isolates were recovered from the middle-dosage group (4 mg/kg of body weight [b.w.]) on days 46 and 49; the other 215 isolates originated from the high-dosage group (100 mg/kg of b.w.) from days 35 to 49. However, no E. coli isolates were detected in the nonmedicated group or the low-dosage group (0.1 mg/kg of b.w.).
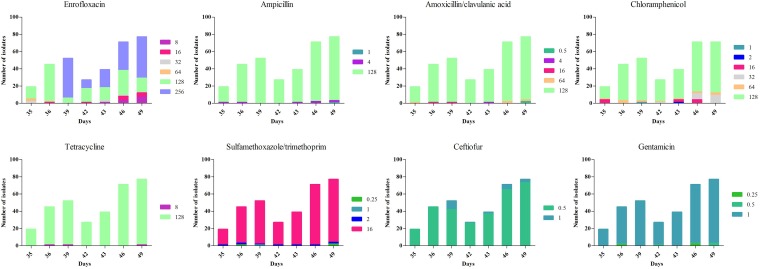
For all 330 isolates identified as E. coli, high rates of resistance to amoxicillin-clavulanic acid (98.5%), ampicillin (97.6%), sulfamethoxazole-trimethoprim (96.1%), tetracycline (99.1%), and chloramphenicol (96.1%) were observed (Fig. 1; see also Fig. S2 in the supplemental material). However, all the isolates were susceptible to gentamicin and ceftiofur. The rate of E. coli, which was resistant to six kinds of antibiotics, was 92.7%. The MICs of enrofloxacin against the E. coli isolates ranged from 8 to 256 μg/ml (Fig. 1 and S2). Analysis of the drug susceptibility testing results showed that enrofloxacin treatment of Salmonella infection in chicken induced multidrug resistance (MDR) in the commensal E. coli isolates in the chicken gut.
FIG 1.
Distributions of the MICs of enrofloxacin, amoxicillin-clavulanic acid, ampicillin, sulfamethoxazole-trimethoprim, gentamicin, tetracycline, chloramphenicol, and ceftiofur for the E. coli isolates with time.
Phylogenetic group, fingerprinting profiles, and virulence genes of E. coli.
According to the phylogenetic grouping result, group B1 was the most prevalent and accounted for 92.4% (305/330) of the total isolates, while the other 7.6% (25/330) of the strains belonged to phylogenetic group E (Table 1).
TABLE 1.
Eighteen subgroups of E. coli isolates classified by genetic characterizations, susceptibility to enrofloxacin, and resistance mechanisms
| Phylogenetic type | ERIC-PCR type | Presence of virulence genes |
Presence of qnrS |
ompF expression levela |
MICENR
(μg/ml) |
No. of isolates | Representative strain no. | ||
|---|---|---|---|---|---|---|---|---|---|
| iss | traT | iroN | |||||||
| E | B | − | − | − | − | ↓ | 8 | 3 | E283 |
| B | − | − | − | − | ↓ | 16 | 7 | E50 | |
| B | − | − | + | − | ↓ | 8 | 7 | E146 | |
| B | − | − | + | − | ↓ | 16 | 4 | E119 | |
| B | − | + | + | − | ↓ | 16 | 1 | E239 | |
| C | + | + | + | + | ↓ | 128 | 1 | E241 | |
| C | + | + | + | + | ↓ | 256 | 2 | E211 | |
| B1 | A | + | + | + | − | ↓ | 32 | 5 | E10 |
| A | + | + | + | − | ↓ | 64 | 2 | E1 | |
| A | + | + | + | − | ↓ | 256 | 1 | E176 | |
| C | + | + | + | − | ↓ | 8 | 1 | E255 | |
| C | + | + | + | − | ↓ | 64 | 2 | E9 | |
| C | + | + | + | − | ↓ | 128 | 3 | E157 | |
| C | + | + | + | + | ↓ | 128 | 135 | E2 | |
| C | + | + | + | + | ↓ | 256 | 150 | E65 | |
| C | − | + | + | + | ↓ | 256 | 4 | E115 | |
| C | + | − | − | + | ↓ | 128 | 1 | E163 | |
| C | − | + | − | + | ↓ | 256 | 1 | E291 | |
The downward-pointing arrows mean decreased expression of ompF.
The 330 strains of E. coli could be classified into three clusters (clusters A, B, and C), as defined by the enterobacterial repetitive intergenic consensus PCR (ERIC-PCR)-based fingerprinting profiles. Cluster C was the most prevalent, comprising 90.9% (300/330) of the total strains, followed in prevalence by cluster B (22/330) and cluster A (8/330) (Table 1). All 22 isolates in cluster B belonged to phylogenetic group E, and the 8 isolates in cluster A belonged to phylogenetic group B1, while the 297 strains in cluster C belonged to phylogenetic group B1 and the other 3 isolates in cluster C belonged to phylogenetic group E.
The virulence genes fimH, hlyF, and sitA were detected in all 330 E. coli isolates (100%). The iss (91.8%), traT (93.3%), and iroN (96.4%) genes were observed in the majority of the total isolates (Table 1). However, the tsh, iucA, ibeA, cdtB, and cvaC genes were not found in any of the E. coli strains.
Fluoroquinolone resistance genes in the selected E. coli isolates.
Double mutations on GyrA (S83L and D87N) and a single mutation on ParC (S80I) were observed in all 330 FQ-resistant E. coli isolates. No mutations were found on GyrB. In addition, a single mutation on ParE (S458E) was detected in 302 E. coli strains, which showed high-level resistance to enrofloxacin (enrofloxacin MIC [MICENR] ≥ 128 μg/ml). qnrS1 was the only PMQR element detected, and it was found in 294 (89.1%) high-level-resistant isolates. The qnrS1 gene could be successfully transferred to strain EC600 by conjugation, and the FQ susceptibility of the transconjugants (MICENR = 8 μg/ml) decreased 16-fold compared to that of the recipient strain, EC600 (MICENR = 0.5 μg/ml).
Contributions of efflux pumps and porins to fluoroquinolone resistance.
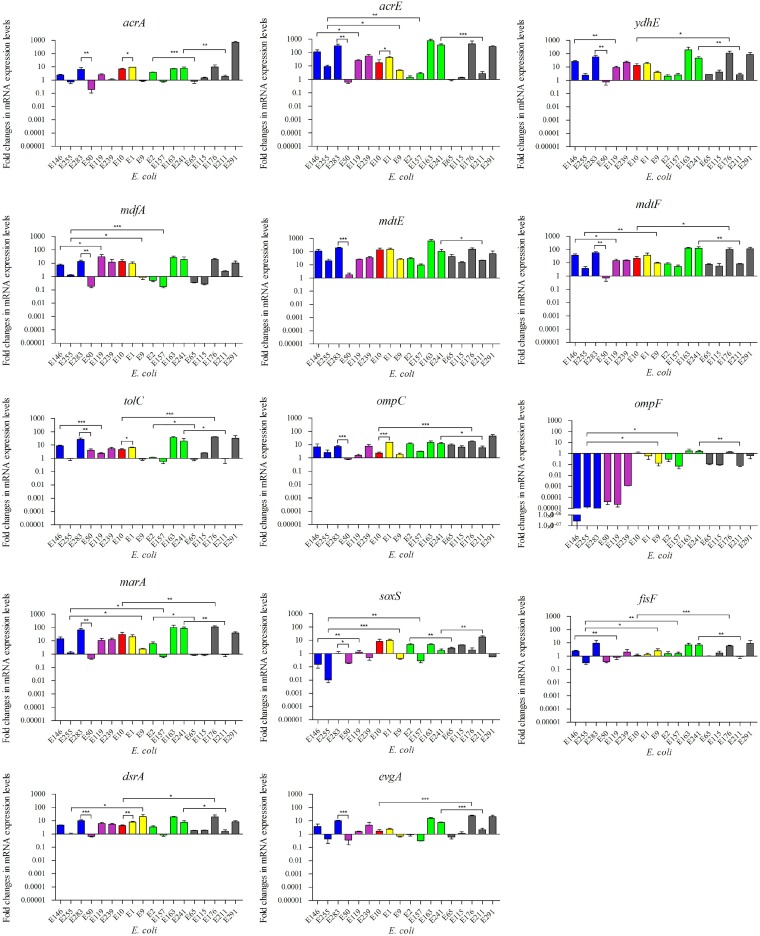
In comparison to their expression levels in FQ-susceptible strain ATCC 25922, the relative expression levels of the seven tested efflux pump-encoding genes were increased in most of the 18 representative FQ-resistant strains, as follows: acrA, 0.2- to 716.6-fold; acrE, 0.6- to 749.0-fold; tolC, 0.6- to 41.7-fold; mdtE, 1.8- to 629.0-fold; mdtF, 0.7- to 130.2-fold; mdfA, 0.2- to 30.5-fold; and ydhE, 0.7- to 191.8-fold (Fig. 2). For porin-encoding gene ompF, the expression levels were remarkably decreased (2.8 × 10−7- to 1.1 × 10−3-fold) in six low-level-resistant strains (MICENR = 8 or 16 μg/ml), while for high-level-resistant isolates (MICENR = 32 to 256 μg/ml), the changes in the expression level of ompF were not so obvious (0.08- to 1.73-fold) (Fig. 2). The expression of the other porin-encoding gene, ompC, was increased in 17 strains (1.5- to 46.1-fold) and slightly reduced in 1 isolate (0.8-fold) (Fig. 2). The relative expression levels of the five regulators were as follows: marA, 0.5- to 112.2-fold; soxS, 0.01- to 17.5-fold; fisF, 0.3- to 9.3-fold; dsrA, 0.7- to 29.5-fold; and evgA, 0.4- to 24.6-fold).
FIG 2.
Relative expression levels of efflux pumps, porins, and regulators. The different colors indicate the different MICs for the strains (blue, MIC = 8 mg/liter; purple, MIC = 16 mg/liter; red, MIC = 32 mg/liter; yellow, MIC = 64 mg/liter; green, MIC = 128 mg/liter; gray, MIC = 256 mg/liter). *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Role of an SNP (A1541G) on fluoroquinolone susceptibility in E. coli.
A total of 82,142 single nucleotide polymorphisms (SNPs) were identified among the 11 sequenced E. coli strains and the reference strain (E. coli CVM N37067PS) (Fig. S1). For all the SNPs that caused nonsynonymous mutations, there were 8 SNPs present in all 11 sequenced FQ-resistant strains. When the sequences were aligned to the reference genomes of two FQ-susceptible E. coli strains (ATCC 25922 and K-12 strain MG1655), only the SNP S80I was identified (Fig. S1). The mutation S80I was located on the DNA topoisomerase IV subunit ParC, and it had been reported to be the target site of FQs. On the other hand, 4 SNPs were present in the 7 high-level-resistant strains but absent in the 4 low-level-resistant strains. After aligning of the sequences to the reference genomes of E. coli ATCC 25922 and K-12 MG1655, only the key SNP A1541G was identified (Fig. S1). This A1541G mutation is located on the aegA gene, which encodes an oxidoreductase and has the functions of metal ion binding to clusters of 4 iron and 4 sulfur atoms. Functional assessment of the A1541G mutation was performed by site-directed mutagenesis and allelic exchange in the genome of E. coli ATCC 25922. The enrofloxacin MIC of wild-type strain ATCC 25922 was 0.015 μg/ml. After introduction of the A1541G mutation into ATCC 25922, the enrofloxacin MIC of the mutant strain was decreased to 0.0075 μg/ml.
DISCUSSION
The present results and our previous studies show that enrofloxacin treatment of Salmonella infection in chicken selects for multidrug-resistant E. coli isolates in the chicken gut and that these E. coli isolates have diverse genetic backgrounds. Our results are similar to previously published data which showed that coresistance to FQs, β-lactams, and tetracycline occurs in the commensal E. coli isolates of healthy chickens after the chickens are orally treated with enrofloxacin (35) or amoxicillin (36). The multidrug resistance could also be induced under stepwise selection with antimicrobial agents (e.g., norfloxacin) (37) and disinfectants (e.g., organic solvent, pine oil, and triclosan) (38). These data indicate that antimicrobial usage patterns may not always correlate with resistance prevalence (36, 39). The coselection of multidrug resistance may be due to the genetic linkage of different resistance genes on the same plasmid (39–41), upregulation of the AcrAB-TolC efflux pump and downregulation of porins (42, 43), or introduction of the A87G mutation on GyrA (44).
In this study, no counterparts of the susceptible or resistant E. coli isolates were detected before drug administration. Sequencing of the 16S rRNA of the intestinal microbiota of specific-pathogen-free (SPF) chickens also confirmed that the number of E. coli isolates at the initial phase was low (10). All 330 E. coli isolates with low to high levels of resistance to FQs were selected from the middle-dosage group (4 mg/kg of b.w.) and high-dosage group (100 mg/kg of b.w.) at the later phase of the experiment. To understand the development or emergence of antimicrobial resistance in commensal E. coli isolates under the selective pressure of enrofloxacin, the SPF chickens were raised in separate isolators to rule out the possibility of exogenous contamination and the transmission of resistance genes. Molecular typing results showed that the 330 isolates of E. coli had diverse genetic backgrounds. Among those, more than 285 E. coli isolates were classified into phylogenetic group B1 and ERIC-PCR cluster C (Table 1), indicating a wide dissemination of a unique clone of E. coli in the gut of chickens under the selective pressure of enrofloxacin.
For FQ resistance, we determined each known mechanism and illustrated the differences underlying the mechanisms of high-level and low-level FQ resistance. It is reported that an individual mechanism usually confers only a small reduction in susceptibility to FQs, and to reach the clinical resistance breakpoint, several genetic alterations need to occur in a stepwise fashion (33, 45, 46). Singh et al. reported that early low-level FQ resistance was conferred by acrAB overexpression and that it preceded the high-level FQ resistance development mediated by a target site mutation(s) (33). Our results were consistent with the results of those studies, with a difference being that the decreased expression of ompF instead of activation of efflux pumps played a significant role in low-level FQ resistance. The OmpF porin is a major outer membrane channel for the entry of quinolones (29, 47, 48), and several studies have reported that a reduction in ompF expression could confer only low-level FQ resistance but that it was a stepping stone to higher-level resistance (29, 49).
In comparison with the low-level resistance, the in vivo-selected high-level FQ resistance was most likely due to QnrS1 and an extra mutation consisting of S458E on ParE. As one of the PMQR determinants, qnr homologues are found on the chromosomes of many Gram-negative bacteria, mediate a modest reduction in FQ susceptibility, and facilitate the selection of higher-level FQ-resistant mutants (30, 50). qnr is also located on a plasmid and can transfer to other bacterial species through horizontal gene transfer (51, 52). Machuca et al. found that the acquisition of qnrS1 in E. coli ATCC 25922 with triple mutations (S83L, D87N, ΔmarR) significantly enhances its fitness and maintains its high-level FQ resistance (53). A recent study showed that recombination of qnrS onto the chromosome of E. coli sufficiently confers clinical resistance to ciprofloxacin without a fitness cost (51).
Using whole-genome sequencing and site-directed mutagenesis, we screened for a key SNP (A1541G) on the gene aegA, which was able to increase FQ susceptibility by 2-fold in E. coli ATCC 25922. AegA is an oxidoreductase and has the function of metal ion binding (54). It has been reported that metal ions play important roles in quinolone action by forming a water-metal ion interaction that bridges the drug to the topoisomerase (5, 55, 56). Considering that this SNP was present only in the 7 high-level-resistant isolates but absent in the 4 low-level-resistant strains, we predict that this SNP may have some association with FQ resistance. The results showed that introduction of the A1541G substitution caused a small increase in the susceptibility of E. coli ATCC 25922 to enrofloxacin, which might have been due to a reduction of bacterial survival ability that resulted from changes in cellular physiology or metabolic pathways. The presence of A1541G only in the high-level-resistant isolates indicates that there may be a small fitness cost that accompanies acquisition of in vivo-selected high-level FQ resistance, which needs further investigations in the future.
Conclusions.
In conclusion, these data show that enrofloxacin treatment of Salmonella infection in chicken selected for multidrug-resistant commensal E. coli isolates in the gut. The decreased expression of ompF played a key role in low-level FQ resistance, while qnrS1 encoded a protective protein and an extra mutation on ParE contributed to the high-level FQ resistance. In addition, the A1541G substitution on aegA was able to increase FQ susceptibility by 2-fold in E. coli ATCC 25922. A better understanding of the evolvement of MDR and mechanisms of FQ resistance would be useful for developing effective strategies to combat the problem of antimicrobial resistance.
MATERIALS AND METHODS
Studied isolates.
A collection of E. coli isolates which were recovered from a previous clinical efficacy study were used in this work (34). Briefly, 20 specific-pathogen-free (SPF) chicks were orally challenged with Salmonella enterica serovar Typhimurium CVCC541 at 4 days of age, and then the infected chickens were randomly allotted into four groups (n = 5) and gavaged with enrofloxacin at different doses (100, 4, 0.1, and 0 mg/kg of b.w.) for three rounds of 7 days of treatment following 7 days of withdrawal from the age of 8 days on. Cloacal swab specimens were collected from each chicken when they were 3, 7, 8, 11, 14, 15, 18, 21, 22, 25, 28, 29, 32, 35, 36, 39, 42, 43, 46, and 49 days old and spread onto MacConkey agar plates containing 0, 0.125, 0.25, and 2 mg/liter of enrofloxacin. For each chicken, two colonies of coliforms were randomly picked from each plate at each time point. The identification of E. coli was performed by biochemical tests and 16S rRNA sequencing (34). All the experimental procedures in this study were performed according to the guidelines of the laboratory care and use committee in Hubei Province, China. The study was approved by Animal Ethics Committee of Huazhong Agricultural University and the Animal Care Center (hzauch 2014-002) and Hubei Science and Technology Agency in China (SYXK 2013-0044). All the animals were monitored throughout the study for any adverse effect signs.
Antimicrobial susceptibility testing.
All the confirmed E. coli isolates were subjected to antimicrobial susceptibility testing using the agar dilution method, and the results were interpreted according to NCCLS guidelines (2002). The susceptibilities of the isolates to eight classes of antimicrobial agents were tested: enrofloxacin (0.0075 to 256 μg/ml), amoxicillin-clavulanic acid (0.5/0.25 to 128/64 μg/ml), ampicillin (1 to 128 μg/ml), sulfamethoxazole-trimethoprim (0.25/4.75 to 16/304 μg/ml), gentamicin (0.125 to 128 μg/ml), tetracycline (0.25 to 128 μg/ml), chloramphenicol (1 to 128 μg/ml), and ceftiofur (0.125 to 128 μg/ml). E. coli ATCC 25922 was used as a quality control strain.
Molecular typing.
DNA extracts of the E. coli isolates were prepared using an AxyPrep bacterial genomic DNA miniprep kit (Axygen Scientific, Inc., CA, USA) following the manufacturer’s instructions. The E. coli strains were assigned to different groups by a multiplex PCR-based phylogenetic typing method (57) and an enterobacterial repetitive intergenic consensus PCR (ERIC-PCR) fingerprinting method (58).
All the identified E. coli isolates were analyzed for 11 potential virulence genes (59). Three multiplex PCR assays were performed to detect different virulence genes simultaneously: (i) tsh, iss, and iucA; (ii) hlyF and sitA; and (iii) ibeA, traT, cdtB, iroN, and cvaC.
Detection of target gene mutations and PMQR genes.
The quinolone resistance-determining regions of the gyrA, gyrB, parC, and parE genes were amplified from all E. coli isolates by PCR (60). The PCR products were sequenced, and the sequences were compared to the sequence of the genome of wild-type strain Escherichia coli ATCC 25922 by BLAST analysis for identification of target gene mutations. The presence of the PMQR genes [qnrA, qnrB, qnrS, oqxAB, qepA, and aac(6′)-Ib] was determined as described previously (61). The transferability of the detected PMQR genes was tested by conjugation using E. coli strain EC600 (rifampin resistant) as the recipient strain. The transconjugants were screened on Luria-Bertani (LB) agar plates supplemented with rifampin (500 μg/ml) plus enrofloxacin (1 μg/ml). The clonal relatedness between the recipient strain and the transconjugants was confirmed by ERIC-PCR fingerprinting. The acquisition of PMQR genes in the transconjugants was confirmed by PCR.
Relative expression levels of genes encoding efflux pumps, porins, and regulators.
All the E. coli isolates were classified into 18 subgroups according to their genetic characteristics of phylogenetic group, ERIC-PCR type, virulence genotype, MICENR, and PMQR genes (Table 1). One representative strain of E. coli was chosen from each subgroup for subsequent detection. Reverse transcription followed by real-time quantitative PCR (RT-qPCR) was performed on the 18 representative FQ-resistant E. coli isolates and a reference strain, E. coli ATCC 25922, to determine the expression levels of the selected efflux pump-encoding genes (acrA, mdfA, ydhE, acrE, tolC, mdtE, and mdtF), regulators (marA, soxS, fisF, dsrA, and evgA), and porin protein-encoding genes (ompC and ompF) according to the methods described by Vinué et al. (50). The expression levels of each tested gene were compared to those of the housekeeping gene mdh in the 18 FQ-resistant isolates and E. coli ATCC 25922.
DNA sequencing.
According to the results of the relative expression levels of the efflux pumps, porins, and regulators, 11 representative strains of E. coli were further chosen for whole-genome shotgun sequencing. Whole-genome sequencing was performed on an Illumina MiSeq platform (250 PE; Illumina) using the paired-end mode (2 × 250), and the entire data were analyzed by Shanghai Personalbio Biotechnology (Shanghai, China). The sequencing data were mapped to the published genome sequence of E. coli CVM N37067PS to identify single nucleotide polymorphisms (SNPs). The SNPs, that were present in all 11 isolates or that were present in the 7 high-level-resistant isolates but not in the 4 low-level-resistant isolates were screened and then confirmed by PCR and conventional Sanger sequencing. Primers (see Table S1 in the supplemental material) targeting the regions containing the predicted SNPs were designed based on the reference genome of E. coli CVM N37067PS. The confirmed SNPs in the FQ-resistant isolates were compared to the whole-genome sequences of two FQ-susceptible strains (E. coli ATCC 25922 and K-12 MG1655) by BLAST analysis to screen for key SNPs which might have a relationship with FQ resistance (Fig. S1).
Site-directed mutagenesis.
A key SNP (A1541G) was identified through the screening process (Fig. S1). The A1541G codon mutation leads to the K514R amino acid substitution on the AegA protein. To investigate the role of the key SNP (A1541G) in FQ resistance, a homologous recombination method was employed to construct the isogenic mutation (A1541G) on the genome of FQ-susceptible strain E. coli ATCC 25922. The upstream and downstream homologous arms of wild-type aegA (aegAw) and the kanamycin resistance (kan) cassette [pmdT-TKarmA-TKarmB-Kan-ef1a-BFP-WPRE-poly(A)] were ligated to the suicide plasmid pRE112 to construct recombinant plasmid pRE112-kan. After conjugative transfer of pRE112-kan from donor strain χ7213 to recipient strain ATCC 25922, the wild-type aegAw gene on the genome of E. coli ATCC 25922 was replaced by the kan cassette. Using sacB sucrose-negative screening and two-step exchange homologous recombination, the strain ATCC 25922-kan was constructed. Then, the whole fragment of the wild-type aegAw with upstream and downstream homologous arms was connected to pMD18-T. Site-directed mutagenesis of the key SNP (A1541G) was constructed on plasmid pMD18-T-aegAw using a TaKaRa MutanBEST kit. Then, the mutant aegA (aegAm) gene fragment with upstream and downstream homologous arms was ligated to pRE112 to construct a recombinant suicide plasmid, pRE112-aegAm. pRE112-aegAm was transferred to ATCC 25922-kan by conjugation, and the kan cassette was replaced by aegAm, so that mutant strain ATCC 25922-aegAm was constructed. Then, the susceptibilities of both the parent strain (ATCC 25922) and the mutant strain (ATCC 25922-aegAm) to enrofloxacin were detected.
Statistical analysis.
All assays were performed in triplicate and repeated at least three times on different days. Student’s t tests were performed to examine the differences between two groups, and a P value of <0.05 was set as the significance level.
Supplementary Material
ACKNOWLEDGMENT
This work was supported by the National Basic Research Program of China (2013CB127200).
The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
We have no conflicts of interest to declare.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.01824-18.
REFERENCES
- 1.D'Costa VM, King CE, Kalan L, Morar M, Sung WWL, Schwarz C, Froese D, Zazula G, Calmels F, Debruyne R, Golding GB, Poinar HN, Wright GD. 2011. Antibiotic resistance is ancient. Nature 477:457–461. doi: 10.1038/nature10388. [DOI] [PubMed] [Google Scholar]
- 2.Wright GD, Poinar H. 2012. Antibiotic resistance is ancient: implications for drug discovery. Trends Microbiol 20:157–159. doi: 10.1016/j.tim.2012.01.002. [DOI] [PubMed] [Google Scholar]
- 3.Wegener HC. 2003. Antibiotics in animal feed and their role in resistance development. Curr Opin Microbiol 6:439–445. doi: 10.1016/j.mib.2003.09.009. [DOI] [PubMed] [Google Scholar]
- 4.Singer RS, Finch R, Wegener HC, Bywater R, Walters J, Lipsitch M. 2003. Antibiotic resistance—the interplay between antibiotic use in animals and human beings. Lancet Infect Dis 3:47–51. doi: 10.1016/S1473-3099(03)00490-0. [DOI] [PubMed] [Google Scholar]
- 5.Aldred KJ, Kerns RJ, Osheroff N. 2014. Mechanism of quinolone action and resistance. Biochemistry 53:1565–1574. doi: 10.1021/bi5000564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Correia S, Poeta P, Hebraud M, Capelo JL, Igrejas G. 2017. Mechanisms of quinolone action and resistance: where do we stand? J Med Microbiol 66:551–559. doi: 10.1099/jmm.0.000475. [DOI] [PubMed] [Google Scholar]
- 7.Landoni MF, Albarellos G. 2015. The use of antimicrobial agents in broiler chickens. Vet J 205:21–27. doi: 10.1016/j.tvjl.2015.04.016. [DOI] [PubMed] [Google Scholar]
- 8.Gouvea R, Dos SFF, Aquino MHCD, Pereira VLD. 2015. Fluoroquinolones in industrial poultry production, bacterial resistance and food residues: a review. Rev Bras Cienc Avic 17:1–10. doi: 10.1590/1516-635X17011-10. [DOI] [Google Scholar]
- 9.de Lastours V, Fantin B. 2015. Impact of fluoroquinolones on human microbiota. Focus on the emergence of antibiotic resistance. Future Microbiol 10:1241–1255. doi: 10.2217/fmb.15.40. [DOI] [PubMed] [Google Scholar]
- 10.Li J, Hao HH, Cheng GY, Liu CB, Ahmed S, Shabbir MAB, Hussain HI, Dai MH, Yuan ZH. 2017. Microbial shifts in the intestinal microbiota of Salmonella infected chickens in response to enrofloxacin. Front Microbiol 8:1711. doi: 10.3389/fmicb.2017.01711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen Y, Sun J, Liao XP, Shao Y, Li L, Fang LX, Liu YH. 2016. Impact of enrofloxacin and florfenicol therapy on the spread of OqxAB gene and intestinal microbiota in chickens. Vet Microbiol 192:1–9. doi: 10.1016/j.vetmic.2016.05.014. [DOI] [PubMed] [Google Scholar]
- 12.Redgrave LS, Sutton SB, Webber MA, Piddock LJV. 2014. Fluoroquinolone resistance: mechanisms, impact on bacteria, and role in evolutionary success. Trends Microbiol 22:438–445. doi: 10.1016/j.tim.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 13.Holmes AH, Moore LSP, Sundsfjord A, Steinbakk M, Regmi S, Karkey A, Guerin PJ, Piddock LJV. 2016. Understanding the mechanisms and drivers of antimicrobial resistance. Lancet 387:176–187. doi: 10.1016/S0140-6736(15)00473-0. [DOI] [PubMed] [Google Scholar]
- 14.Olofsson SK, Marcusson LL, Stromback A, Hughes D, Cars O. 2007. Dose-related selection of fluoroquinolone-resistant Escherichia coli. J Antimicrob Chemother 60:795–801. doi: 10.1093/jac/dkm265. [DOI] [PubMed] [Google Scholar]
- 15.Wiuff C, Lykkesfeldt J, Svendsen O, Aarestrup FM. 2003. The effects of oral and intramuscular administration and dose escalation of enrofloxacin on the selection of quinolone resistance among Salmonella and coliforms in pigs. Res Vet Sci 75:185–193. doi: 10.1016/S0034-5288(03)00112-7. [DOI] [PubMed] [Google Scholar]
- 16.Burow E, Simoneit C, Tenhagen BA, Kasbohrer A. 2014. Oral antimicrobials increase antimicrobial resistance in porcine E. coli—a systematic review. Prev Vet Med 113:364–375. doi: 10.1016/j.prevetmed.2013.12.007. [DOI] [PubMed] [Google Scholar]
- 17.Simoneit C, Burow E, Tenhagen BA, Kasbohrer A. 2015. Oral administration of antimicrobials increase antimicrobial resistance in E. coli from chicken—a systematic review. Prev Vet Med 118:1–7. doi: 10.1016/j.prevetmed.2014.11.010. [DOI] [PubMed] [Google Scholar]
- 18.de Jong A, Stephan B, Silley P. 2012. Fluoroquinolone resistance of Escherichia coli and Salmonella from healthy livestock and poultry in the EU. J Appl Microbiol 112:239–245. doi: 10.1111/j.1365-2672.2011.05193.x. [DOI] [PubMed] [Google Scholar]
- 19.Hooper DC, Jacoby GA. 2016. Topoisomerase inhibitors: fluoroquinolone mechanisms of action and resistance. Cold Spring Harb Perspect Med 6:a025320. doi: 10.1101/cshperspect.a025320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith JL, Fratamico PM. 2010. Fluoroquinolone resistance in Campylobacter. J Food Prot 73:1141–1152. doi: 10.4315/0362-028X-73.6.1141. [DOI] [PubMed] [Google Scholar]
- 21.Threlfall EJ, Frost JA, Rowe B. 1999. Fluoroquinolone resistance in salmonellas and campylobacters from humans. BMJ 318:943–944. doi: 10.1136/bmj.318.7188.943c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hooper DC, Jacoby GA. 2015. Mechanisms of drug resistance: quinolone resistance. Ann N Y Acad Sci 1354(1):12–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.FDA. 2005. Withdrawal of enrofloxacin for poultry. http://www.fda.gov/animalveterinary/safetyhealth/recallswithdrawals/ucm042004.htm.
- 24.Love DC, Halden RU, Davis MF, Nachman KE. 2012. Feather meal: a previously unrecognized route for reentry into the food supply of multiple pharmaceuticals and personal care products (PPCPs). Environ Sci Technol 46:3795–3802. doi: 10.1021/es203970e (Erratum, 46:5631, doi:.) [DOI] [PubMed] [Google Scholar]
- 25.Ingram PR, Rogers BA, Sidjabat HE, Gibson JS, Inglis TJJ. 2013. Co-selection may explain high rates of ciprofloxacin non-susceptible Escherichia coli from retail poultry reared without prior fluoroquinolone exposure. J Med Microbiol 62:1743–1746. doi: 10.1099/jmm.0.062729-0. [DOI] [PubMed] [Google Scholar]
- 26.Hopkins KL, Davies RH, Threlfall EJ. 2005. Mechanisms of quinolone resistance in Escherichia coli and Salmonella: recent developments. Int J Antimicrob Agents 25:358–373. doi: 10.1016/j.ijantimicag.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 27.Blair JMA, Richmond GE, Piddock LJV. 2014. Multidrug efflux pumps in Gram-negative bacteria and their role in antibiotic resistance. Future Microbiol 9:1165–1177. doi: 10.2217/fmb.14.66. [DOI] [PubMed] [Google Scholar]
- 28.Alcalde-Rico M, Hernando-Amado S, Blanco P, Martinez JL. 2016. Multidrug efflux pumps at the crossroad between antibiotic resistance and bacterial virulence. Front Microbiol 7:1483. doi: 10.3389/fmicb.2016.01483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kishii R, Takei M. 2009. Relationship between the expression of ompF and quinolone resistance in Escherichia coli. J Infect Chemother 15:361–366. doi: 10.1007/s10156-009-0716-6. [DOI] [PubMed] [Google Scholar]
- 30.Strahilevitz J, Jacoby GA, Hooper DC, Robicsek A. 2009. Plasmid-mediated quinolone resistance: a multifaceted threat. Clin Microbiol Rev 22:664. doi: 10.1128/CMR.00016-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Robicsek A, Jacoby GA, Hooper DC. 2006. The worldwide emergence of plasmid-mediated quinolone resistance. Lancet Infect Dis 6:629–640. doi: 10.1016/S1473-3099(06)70599-0. [DOI] [PubMed] [Google Scholar]
- 32.Rushdy AA, Mabrouk MI, Abu-Sef FA, Kheiralla ZH, Mohamed Abdel-All S, Saleh NM. 2013. Contribution of different mechanisms to the resistance to fluoroquinolones in clinical isolates of Salmonella enterica. Braz J Infect Dis 17:431–437. doi: 10.1016/j.bjid.2012.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Singh R, Swick MC, Ledesma KR, Yang Z, Hu M, Zechiedrich L, Tam VH. 2012. Temporal interplay between efflux pumps and target mutations in development of antibiotic resistance in Escherichia coli. Antimicrob Agents Chemother 56:1680–1685. doi: 10.1128/AAC.05693-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li J, Hao H, Cheng G, Wang X, Ahmed S, Shabbir MAB, Liu Z, Dai M, Yuan Z. 2017. The effects of different enrofloxacin dosages on clinical efficacy and resistance development in chickens experimentally infected with Salmonella Typhimurium. Sci Rep 7:11676. doi: 10.1038/s41598-017-12294-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jurado S, Medina A, de la Fuente R, Ruiz-Santa-Quiteria JA, Orden JA. 2015. Resistance to non-quinolone antimicrobials in commensal Escherichia coli isolates from chickens treated orally with enrofloxacin. Jpn J Vet Res 63:195–200. [PubMed] [Google Scholar]
- 36.Jiménez-Belenguer A, Doménech E, Villagrá A, Fenollar A, Ferrús MA. 2016. Antimicrobial resistance of Escherichia coli isolated in newly-hatched chickens and effect of amoxicillin treatment during their growth. Avian Pathol 45:501–507. doi: 10.1080/03079457.2016.1168515. [DOI] [PubMed] [Google Scholar]
- 37.Lindgren PK, Marcusson LL, Sandvang D, Frimodt-Moller N, Hughes D. 2005. Biological cost of single and multiple norfloxacin resistance mutations in Escherichia coli implicated in urinary tract infections. Antimicrob Agents Chemother 49:2343–2351. doi: 10.1128/AAC.49.6.2343-2351.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Randall LP, Bagnall MC, Karatzas KA, Coldham NC, Piddock LJ, Woodward MJ. 2008. Fitness and dissemination of disinfectant-selected multiple-antibiotic-resistant (MAR) strains of Salmonella enterica serovar Typhimurium in chickens. J Antimicrob Chemother 61:156–162. doi: 10.1093/jac/dkm415. [DOI] [PubMed] [Google Scholar]
- 39.da Costa PM, Belo A, Goncalves J, Bernardo F. 2009. Field trial evaluating changes in prevalence and patterns of antimicrobial resistance among Escherichia coli and Enterococcus spp. isolated from growing broilers medicated with enrofloxacin, apramycin and amoxicillin. Vet Microbiol 139:284–292. doi: 10.1016/j.vetmic.2009.06.006. [DOI] [PubMed] [Google Scholar]
- 40.Baker-Austin C, Wright MS, Stepanauskas R, McArthur JV. 2006. Co-selection of antibiotic and metal resistance. Trends Microbiol 14:176–182. doi: 10.1016/j.tim.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 41.Canton R, Ruiz-Garbajosa P. 2011. Co-resistance: an opportunity for the bacteria and resistance genes. Curr Opin Pharmacol 11:477–485. doi: 10.1016/j.coph.2011.07.007. [DOI] [PubMed] [Google Scholar]
- 42.Koronakis V, Eswaran J, Hughes C. 2004. Structure and function of TolC: the bacterial exit duct for proteins and drugs. Annu Rev Biochem 73:467–489. doi: 10.1146/annurev.biochem.73.011303.074104. [DOI] [PubMed] [Google Scholar]
- 43.Li XZ, Plesiat P, Nikaido H. 2015. The challenge of efflux-mediated antibiotic resistance in Gram-negative bacteria. Clin Microbiol Rev 28:337–418. doi: 10.1128/CMR.00117-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Webber MA, Ricci V, Whitehead R, Patel M, Fookes M, Ivens A, Piddock LJ. 2013. Clinically relevant mutant DNA gyrase alters supercoiling, changes the transcriptome, and confers multidrug resistance. mBio 4:e00273-13. doi: 10.1128/mBio.00273-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marcusson LL, Frimodt-Møller N, Hughes D. 2009. Interplay in the selection of fluoroquinolone resistance and bacterial fitness. PLoS Pathog 5:e1000541. doi: 10.1371/journal.ppat.1000541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huseby DL, Pietsch F, Brandis G, Garoff L, Tegehall A, Hughes D. 2017. Mutation supply and relative fitness shape the genotypes of ciprofloxacin-resistant Escherichia coli. Mol Biol Evol 34:1029–1039. doi: 10.1093/molbev/msx052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chapman JS, Georgopapadakou NH. 1988. Routes of quinolone permeation in Escherichia coli. Antimicrob Agents Chemother 32:438–442. doi: 10.1128/AAC.32.4.438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Delcour AH. 2009. Outer membrane permeability and antibiotic resistance. Biochim Biophys Acta 1794:808–816. doi: 10.1016/j.bbapap.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Randall LP, Woodward MJ. 2002. The multiple antibiotic resistance (mar) locus and its significance. Res Vet Sci 72:87–93. doi: 10.1053/rvsc.2001.0537. [DOI] [PubMed] [Google Scholar]
- 50.Vinué L, Corcoran MA, Hooper DC, Jacoby GA. 2015. Mutations that enhance the ciprofloxacin resistance of Escherichia coli with qnrA1. Antimicrob Agents Chemother 60:1537–1545. doi: 10.1128/AAC.02167-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Garoff L, Yadav K, Hughes D. 2018. Increased expression of Qnr is sufficient to confer clinical resistance to ciprofloxacin in Escherichia coli. J Antimicrob Chemother 73:348–352. doi: 10.1093/jac/dkx375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hooper DC, Jacoby GA. 2015. Mechanisms of drug resistance: quinolone resistance. Ann N Y Acad Sci 1354:12–31. doi: 10.1111/nyas.12830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Machuca J, Briales A, Labrador G, Díaz-de-Alba P, López-Rojas R, Docobo-Pérez F, Martínez-Martínez L, Rodríguez-Baño J, Pachón ME, Pascual A, Rodríguez-Martínez J-M. 2014. Interplay between plasmid-mediated and chromosomal-mediated fluoroquinolone resistance and bacterial fitness in Escherichia coli. J Antimicrob Chemother 69:3203–3215. doi: 10.1093/jac/dku308. [DOI] [PubMed] [Google Scholar]
- 54.Cavicchioli R, Kolesnikow T, Chiang RC, Gunsalus RP. 1996. Characterization of the aegA locus of Escherichia coli: control of gene expression in response to anaerobiosis and nitrate. J Bacteriol 178:6968–6974. doi: 10.1128/jb.178.23.6968-6974.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sissi C, Perdona E, Domenici E, Feriani A, Howells AJ, Maxwell A, Palumbo M. 2001. Ciprofloxacin affects conformational equilibria of DNA gyrase A in the presence of magnesium ions. J Mol Biol 311:195–203. doi: 10.1006/jmbi.2001.4838. [DOI] [PubMed] [Google Scholar]
- 56.Wohlkonig A, Chan PF, Fosberry AP, Homes P, Huang JZ, Kranz M, Leydon VR, Miles TJ, Pearson ND, Perera RL, Shillings AJ, Gwynn MN, Bax BD. 2010. Structural basis of quinolone inhibition of type IIA topoisomerases and target-mediated resistance. Nat Struct Mol Biol 17:1152–1153. doi: 10.1038/nsmb.1892. [DOI] [PubMed] [Google Scholar]
- 57.Clermont O, Christenson JK, Denamur E, Gordon DM. 2013. The Clermont Escherichia coli phylo-typing method revisited: improvement of specificity and detection of new phylo-groups. Environ Microbiol Rep 5:58–65. doi: 10.1111/1758-2229.12019. [DOI] [PubMed] [Google Scholar]
- 58.Duan HY, Chai TJ, Liu JZ, Zhang XX, Qi CH, Gao J, Wang YL, Cai YM, Miao ZM, Yao ML, Schlenker G. 2009. Source identification of airborne Escherichia coli of swine house surroundings using ERIC-PCR and REP-PCR. Environ Res 109:511–517. doi: 10.1016/j.envres.2009.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang XM, Liao XP, Zhang WJ, Jiang HX, Sun J, Zhang MJ, He XF, Lao DX, Liu YH. 2010. Prevalence of serogroups, virulence genotypes, antimicrobial resistance, and phylogenetic background of avian pathogenic Escherichia coli in south of China. Foodborne Pathog Dis 7:1099–1106. doi: 10.1089/fpd.2010.0542. [DOI] [PubMed] [Google Scholar]
- 60.Everett MJ, Jin YF, Ricci V, Piddock LJ. 1996. Contributions of individual mechanisms to fluoroquinolone resistance in 36 Escherichia coli strains isolated from humans and animals. Antimicrob Agents Chemother 40:2380–2386. doi: 10.1128/AAC.40.10.2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Domokos J, Kristof K, Szabo D. 2016. Plasmid-mediated quinolone resistance among extended-spectrum beta-lactamase producing Enterobacteriaceae from bloodstream infections. Acta Microbiol Immunol Hung 63:313–323. doi: 10.1556/030.63.2016.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.