Abstract
Objectives: MicroRNAs (miRNAs) are small functional RNAs that regulate mRNAs for degradation or translational suppression. In the present study, we aimed to reveal functional importance of miRNA‐494 (miR‐494) in A549 human lung cancer cells.
Materials and methods: We established A549 cells that constitutively expressed miR‐494. Next, we sought to investigate insulin‐like growth factor 2 mRNA‐binding protein 1 (IGF2BP1) mRNA as an miR‐494 target. For this, we constructed a reporter plasmid bearing potential miR‐494 binding sequences derived from the 3′‐untranslated region (3′‐UTR) of IGF2BP1 mRNA in the 3′‐UTR of the luciferase gene.
Results: Through comparison between miR‐494 expressing cells and control cells, we revealed that miR‐494 suppressed cell proliferation and colony forming activity, and induced senescence. Reporter activity was inhibited by miR‐494. In addition, IGF2BP1 mRNA levels were down‐regulated in A549 cells that constitutively expressed miR‐494. IGF2BP1 has been shown to bind and suppress IGF2 mRNA, and this could be a reason why IGF2BP1 can regulate cell function. Therefore, we analysed IGF2 mRNA levels and revealed that IGF2 was up‐regulated in A549 cells that constitutively expressed miR‐494. Finally, elevated IGF2 mRNA levels in A549 cells that constitutively expressed miR‐494 were suppressed to basal level by an miR‐494 inhibitor.
Conclusions: Taken together, IGF2BP1 and its downstream target IGF2 could be a crucial axis for miR‐494 in regulation of the destiny of A549 cells.
Introduction
In human cells, approximately 2% of the genome is transcribed by mRNAs and these are processed for translation to produce proteins. On the other hand, importance of the remaining large component of the genome has not been addressed for many years. Recent intensive OMICS studies have revealed that the whole body of the genome could be transcribed and non‐coding RNAs have been considered as crucial regulators in various cellular processes (1, 2). Among non‐coding RNAs, microRNAs (miRNAs), 18–25 nucleotides long, form base pairing with the 3′‐untranslated region (UTR) of their target mRNA, and this could lead to mRNA degradation and translational suppression. Currently, more than 700 types of human miRNA have been identified. One third of all genes is thought to be regulated by miRNAs as one miRNA is predicted to have approximately 200 kinds of target mRNA (3).
MicroRNAs have been shown to be profoundly involved in pathogenesis of human diseases such as cancer and metabolic diseases. Indeed, some miRNAs have been shown to act in the way of oncogenes or tumour suppressor genes, based on their expression levels, in certain cancers (4). To address upstream regulators of miRNA expression, we have searched for miRNAs whose expression levels were commonly or uniquely regulated by MAP (mitogen‐activated protein) kinase pathways, including oncogenic RAS, c‐Jun and E2F family transcription factors (5). Among miRNAs we identified, human miRNA‐494 (hereafter referred to as miR‐494, official symbol MIR494; hsa‐mir‐494) maps to chromosome 14q32.31, and has been shown to be up‐regulated in retinoblastoma (6); however, functional importance of miR‐494 in the context of cell proliferation and differentiation remains to be investigated.
In the present study, we have focused on analysing the target gene and cellular phenotype regulated by miR‐494, in human A549 lung cancer cells. Interestingly, insulin‐like growth factor 2 mRNA‐binding protein 1 (IGF2BP1), which we identified as one of the miR‐494 targets by in silico analysis, has been shown to be implicated in regulation of development of carcinogenesis by binding to pivotal mRNAs, such as IGF2 and c‐Myc (7, 8). To date, IGF2BP1 has been shown to be down‐regulated by let‐7 in A549 cells, and this regulation could account for suppression of cell proliferation (9). Here we report that miR‐494 induces senescence and suppresses cell proliferation in A549 cells through affecting IGF2BP1 and its downstream target, IGF2 mRNA level.
Materials and methods
Cell culture and RNA
Human A549 lung cancer cells were maintained in Earle’s modified Eagle’s medium (MEM) (Invitrogen, Carlsbad, CA, USA) supplemented with 1% non‐essential amino acids, 1% antibiotic‐antimycotic and 10% foetal bovine serum in a 5% CO2 incubator at 37 °C. Human miR‐494, 5′‐UGAAACAUACACGGGAAACCUC‐3′, was synthesized as double stranded RNA (miScript miRNA Mimic Cat. No. MSY0002816, Qiagen, Valencia, CA, USA). As control RNA, Stealth RNA interference Negative Control Duplexes (High GC Duplex, Invitrogen) was used. The miR‐494 inhibitor, anti‐hsa‐miR‐494 miScript miRNA inhibitor, was purchased from Qiagen (Cat. No. MIN0002816). miR‐494 or miR‐494 inhibitor was transfected into the cells using Lipofectamine 2000 (Invitrogen).
Plasmid construction
To construct an expression vector for miR‐494 (hereafter referred to as Green‐494), the genomic region containing miR‐494 stem‐loop sequence was amplified using polymerase chain reaction (PCR), and was inserted downstream of ZsGreen (Zoanthus sp. green fluorescent protein) gene of pmR‐ZsGreen1 (Takara Bio, Otsu, Shiga, Japan). For this, human genomic DNA (Promega, Madison, WI, USA) was used as template DNA, and primers were designed based on the sequence of the genomic clone (GenBank Accession No. AC208187) as follows:
5′‐GAAGATCTACGTCTGGTCTACCCAGTGC‐3′ and 5′‐GGGGTACCACCGAGAGTGGAGCCGGCAA‐3′, in which the restriction enzyme sites are underlined.
To construct the pGL3‐Promoter‐IGF2BP1 reporter plasmid, miR‐494 target sites predicted in the 3′‐UTR of IGF2BP1 mRNA were synthesized as oligonucleotides and inserted into the 3′‐UTR of the luciferase gene of pGL3‐Promoter (Promega). Primers used were 5′‐GCTCTAGA GAATTCCTTTCCCGTGAAGGTTGTTTCAGCCACAAACCACTTCATTTTGCTGTTTCAATCTAGAGC‐3′ and 5′‐GCTCTAGATTGAAACAGCAAAATGAAGTGGTTTGTGGCTGAAACAACCTTCACGGGAAAGGAATTC TCTAGAGC‐3′, in which Xba1 sites are underlined and EcoRI sites are indicated in italic script. Similarly, the pGL3‐Promoter‐JUND reporter plasmid was constructed. Primers used were 5′‐GCTCTAGA GAATTCGATTCTGCCCTATTTATGTTTCTTCTAGAGC‐3′ and 5′‐GCTCTAGAAGAAACATAAATAGGGCAGAATCGAATTC TCTAGAGC‐3′. After annealing these primers, double stranded oligonucleotides were digested with XbaI, and inserted into the XbaI‐digested pGL3‐Promoter. Confirmation of oligonucleotide insertion was achieved with EcoRI digestion, and then sequencing was performed with an RV4 primer. The pGL3‐Promoter‐494 has been reported previously (5).
Screening for an miR‐494 stable transformant
pmR‐ZsGreen1 or Green‐494 was transfected into A549 cells using Lipofectamine Plus reagent (Invitrogen). After 48 h transfection, cells were replated, and after 24 h, 800 μg/ml of G418 (Invitrogen) was added to the medium.
Luciferase assay
The protocol for luciferase assay was performed as previously reported (5). An attractene reagent (Qiagen) was used when plasmid DNA and miRNA‐494 or the miRNA‐494 inhibitor was co‐transfected into the cells. Briefly, 0.4 μg of plasmid DNA and 6 pmol (0.3 μl of 20 μm stock) of RNA together with 2 μl of Attractene reagent were incubated in 60 μl of MEM for 15 min at room temperature, then the mixture was added directly to 5 × 104 cells/well, of 24‐well plates.
Senescence assay
β‐galactosidase activity was analysed using a senescence detection kit (BioVision, Mountain View, CA, USA). Briefly, 2 × 105 cells/well in 6‐well plates were fixed in fixative solution for 10 min, cells were washed twice in phosphate‐buffered saline; then staining solution mix containing X‐gal was added to the cells which were then incubated for 24 h at 37 °C. Blue‐stained cells were counted in five separate areas (each area >30 cells).
Colony formation and the WST‐1 assay
For the colony formation assay, cells were inoculated into 96‐well plates at 1 cell/well, then cultured for 7 days. 5 × 102 cells/well were inoculated into 24‐well plates, and after 24, 48, 72 and 96 h, a WST‐1 reagent (Roche, Basel, Switzerland) was added to each well and incubated for 1 h at 37 °C. Absorbance at 450 nm was measured by subtracting the reference value at 600 nm.
Reverse transcription (RT)‐PCR and Taqman quantitative real‐time RT‐PCR
Total RNA (500 ng) was extracted using an RNeasy Mini kit (Qiagen) and was treated with deoxyribonuclease I (Invitrogen), in accordance with the manufacturer’s instructions. cDNA was synthesized with a High‐capacity cDNA Reverse Transcription kit (Applied Biosystems, Foster City, CA, USA); PCR reaction mixture (20 μl) contained 1 × buffer, 200 μm of dNTPs, 400 nm of primers, 1 mm MgSO4, 1 unit of KOD plus DNA polymerase (Toyobo, Osaka, Japan) and one‐twentieth part of synthesized cDNA. Hot‐start PCR was performed as follows; 94 °C for 3 min, following multiple steps of 94 °C for 15 s, 60 °C for 30 s and 68 °C for 30 s, then the final step of 68 °C for 3 min. Primers for detection of IGF2 mRNA were as follows; 5′‐CAGCCCGACTAGCAGTCTAG‐3′ and 5′‐GGCCAGGATGGTTAGTGGCC‐3′. PCR products were separated in 1.0–1.5 % agarose gel and visualized under ultraviolet irradiation. Gel images were quantified with Quantity One (Bio‐Rad Laboratories, Hercules, CA, USA). Values of quantified band images were normalized with GAPDH (glyceraldehyde‐3‐phosphate dehydrogenase), and background value was subtracted.
Total RNA was extracted and cDNA was synthesized as described above, then a Taqman Gene Expression Assay was performed according to the manufacturer’s protocol (Applied Biosystems). PCR mixtures included Taqman Gene Expression Assay Primer (Applied Biosystems) for human IGF2BP1 mRNA (Assay ID: Hs00198023_m1, Applied Biosystems) and TaqMan Universal PCR Mix (Applied Biosystems) in total reaction volume of 20 μl. Reactions were performed with the 7500 Standard program on a 7500 Fast Real‐Time PCR System (Applied Biosystems). Cycling parameters were 95 °C for 10 min then 40 cycles of 95 °C for 15 s, annealed/extended at 60 °C for 1 min. Cycle threshold (Ct) values, corresponding to PCR cycle number at which fluorescence emission reached a threshold above baseline emission, were determined, and mRNA expression values were calculated using GAPDH as endogenous control (Applied Biosystems) following comparative Ct (ΔΔCt) method. Details of miRNA detection have been described in the previous report (5). Briefly, RNA (100 ng) was extracted using a mirVana miRNA Isolation kit (Applied Biosystems) and was treated with deoxyribonuclease I (Invitrogen). The miR‐494 (Assay ID 002365) and mature miRNA control RNU6B (Assay ID 001093) were purchased from Applied Biosystems.
Results
Establishment and functional analysis of miR‐494 expressing A549 cells
To observe functional aspects of miR‐494 in A549 cells, we first constructed a pmR‐ZsGreen1 vector bearing the miR‐494 sequence (Green‐494), which could express GFP and miR‐494 simultaneously. To determine whether miR‐494 transcribed from Green‐494 was functional as a miRNA, we co‐expressed Green‐494 with pGL3‐Promoter‐494, in which the miR‐494 target sequence was inserted into the 3′‐UTR of the luciferase gene, in A549 cells. After 48 h co‐transfection, luciferase activity of the pGL3‐Promoter‐494 was clearly suppressed by miR‐494 derived from Green‐494 when compared to the control vector pmR‐ZsGreen1 (Fig. 1a). To establish constitutively miR‐494 expressing A549 cells, we transfected Green‐494 into A549 cells and selected them with G418, whose resistant clones were further checked using GFP expression, by fluorescence microscopy. Expression level of miR‐494 was also determined by Taqman real‐time RT‐PCR (Fig. 1b). Consequently, we obtained four clones that stably expressed miR‐494 (A549G494‐1 to ‐4) and one control group of cells bearing pmR‐ZsGreen1 in a stable manner (A549G). To test whether miR‐494 transcribed in A549G494‐1 to ‐4 cells was functional, we transfected the pGL3‐Promoter‐494 reporter into these clones and measured luciferase activity. In all of four clones stably expressing miR‐494, reporter activity was significantly suppressed when compared to control cells such as A549G and parental A549 cells (Fig. 1c). Taken together, the miR‐494 stably expressed in A549G494‐1 to ‐4 cells were functional sufficiently to down‐regulate the target mRNAs. Thus, we decided to use these clones for further functional assay.
Figure 1.
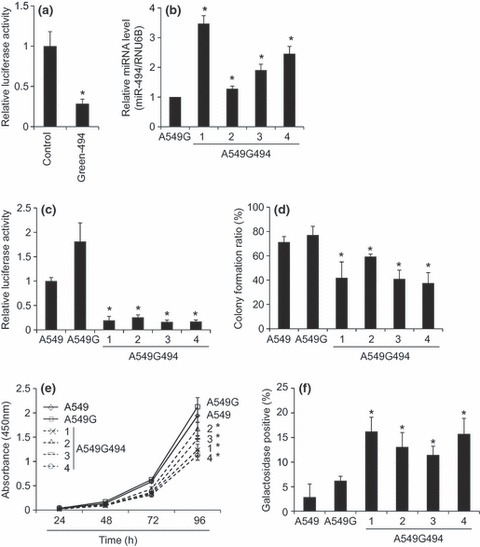
Functional characterization of A549 cells stably expressing miR‐494. (a) Verification of miR‐494 function expressed from the miR‐494 expression vector in A549 cells. Green‐494, an miR‐494 expression vector, or control pmR‐Zs‐Green1, was co‐transfected with pGL3‐Promoter‐494, which possesses the miR‐494 target sequence in the 3’‐UTR of the luciferase gene. After 48 h transfection, relative luciferase activities between control and Green‐494 were normalized by luciferase activities obtained from the pGL3‐Promoter set to 1. This means that luciferase activities obtained from pGL3‐Promoter‐494 reporter was divided by those obtained from pGL3‐Promoter. Bar indicates mean and standard deviation. Student’s t‐test: *P < 0.05 (n = 3). (b) Quantitative determination of miR‐494 expression level by Taqman real‐time RT‐PCR. Bar indicates mean and standard deviation. Student’s t‐test: *P < 0.05 (n = 3). (c) Establishment of miR‐494 stably expressing clones. A549G494 clones 1 to 4 were established by transfecting Green‐494 into A549 cells, and then selected by their resistance to G418 and GFP expression. Similarly, A549G was established by transfecting control pmR‐Zs‐Green1. These clones and parental A549 cells were subjected to luciferase assay using pGL3‐Promoter‐494. After 72 h transfection, relative luciferase activities were normalized by luciferase activities obtained from the pGL3‐Promoter set to 1. Bar indicates mean and standard deviation. Student’s t‐test: *P < 0.05 (n = 3, versus A549 and A549G respectively). (d) Colony formation ability of clones stably expressing miR‐494. A549G494 clones 1 to 4, control A549G and parental A549 cells were inoculated into 96‐well plates (1 cell per well), and after 7 days colony formation, the ratio was determined. Bar indicates mean and standard deviation. Student’s t‐test: *P < 0.05 (n = 3, versus A549 and A549G, respectively). (e) Cell proliferation rate of clones stably expressing miR‐494. A549G494 clones 1 to 4, control A549G and parental A549 cells were inoculated into 24‐well plates (500 cells per well), and then WST‐1 assay was performed at indicated time points. Bar indicates mean and standard deviation. Student’s t‐test: *P < 0.05 (n = 3, versus A549 and A549G, respectively). (f) Cell senescence of clones stably expressing miR‐494. β‐galactosidase activities of A549G494 clones 1 to 4, control A549G and parental A549 cells were detected with X‐gal staining. Bar indicates mean and standard deviation of blue stained cells. Student’s t‐test: *P < 0.05 (n = 3, versus A549 and A549G respectively).
Functional analysis of A549 cells stably expressing miR‐494
We next aimed to address the phenotypes of A549 cells stably expressing miR‐494. First, we analysed cell proliferation using colony formation ability as a criterion. For this, cells were inoculated into 96‐well plates (1 cell per well), and after 7 days, colony number was counted. In comparison to control cells, such as A549G and parental A549, all miR‐494 stably expressing clones A549G494‐1 to ‐4 had low colony forming ability (Fig. 1d). In addition, cell proliferation rate based on WST‐1 assay was suppressed in miR‐494 stably expressing clones A549G494‐1 to ‐4 compared to control cells such as A549G and parental A549 cells (Fig. 1e). In contrast, comparison between miR‐494 expressing clones and control clones resulted in no difference in cell cycle profiling as determined by flow cytometry (data not shown). Based on results obtained by WST‐1 assay, retardation of cell proliferation seen in miR‐494 stably transfected cells was significant, but mild in degree, and ranging approximately 50–75% as compared with that of control cells. This is the reason why we detected a slight difference between miR‐494 stably transfected cells and control cells. We further investigated any cell phenotypes related to suppression of cell proliferation. Based on β‐galactosidase activity, a marker of cell senescence, miR‐494 stably expressing clones A549G494‐1 to ‐4 showed a strong tendency towards senescence compared to A549G and parental A549 cells (Fig. 1f). In assays described in this section, no significant differences were observed between A549G and parental A549 cells.
Regulation of the IGF2BP1‐IGF2 axis by miR‐494
To obtain any mechanistic insight related to cell population growth retardation and senescence promotion seen in miR‐494 stably expressing clones, we searched for downstream targets of miR‐494. The Target Scan 5.1 database (http://www.targetscan.org/) predicted several cell cycle‐related mRNAs including IGF2BP1, JUND and PTEN. First, we focused on IGF2BP1 as IGF2BP1 has been shown to be down‐regulated by let‐7 in A549 cells, and this down‐regulation could account for suppression of cell proliferation (9). IGF2BP1 mRNA (GenBank Accession No. NM_006546) is 8769 base pairs (bp) long and its coding region is 335–2068 bp. The Target Scan 5.1 database predicted four miR‐494 target sites in the 3′‐UTR of IGF2BP1 mRNA, namely 1814–1835, 2392–2413, 6615–6636 and 6644–6665 bp (the relative location as the first nucleotide, 2069 bp, of 3′‐UTR is set as 1 bp) (Fig. 2a). These four sites showed complete complementarity to the seed region (2–8 bp) of miR‐494 and are well conserved between man, mouse and rat. Two regions, 6615–6636 and 6644–6665 bp, were predicted to be very close to each other. Thus, we constructed a reporter plasmid where the 6615–6665 bp region of IGF2BP1 3′‐UTR was inserted into the 3′‐UTR of the luciferase gene (hereafter referred to as pGL3‐Promoter‐IGF2BP1). Over‐expression of miR‐494, but not of control RNA, with pGL3‐Promoter‐IGF2BP1 resulted in decrease in luciferase activity (Fig. 2b), suggesting that the 6615–6665 bp region of IGF2BP1 3′‐UTR is recognized by miR‐494. As 6615–6665 bp region of IGF2BP1 3′‐UTR was sufficient to be targeted by miR‐494, we did not test the possibility of whether 1814–1835 and 2392–2413 bp regions of IGF2BP1 3′‐UTR could be a target of miR‐494. On the other hand, JUND mRNA (GenBank Accession No. NM_005354) is 1890 base pairs (bp) long and its coding region is 139–1182 bp. The Target Scan 5.1 database predicted one miR‐494 target site in the 3′‐UTR of JUND mRNA, namely 416–438 bp (relative location as the first nucleotide, 1183 bp, of 3′‐UTR is set as 1 bp). On this basis, we constructed pGL3‐Promoter‐JUND. In addition, the result obtained from pGL3‐Promoter‐IGF2BP1, miR‐494 suppressed activity of pGL3‐Promoter‐JUND (Fig. 2b). We also constructed pGL3‐Promoter‐PTEN (details are available upon request). The miR‐494 suppressed activity of pGL3‐Promoter‐PTEN (data not shown). The result matched well that of a previous report, in that PTEN was a target of miR‐494 (10).
Figure 2.
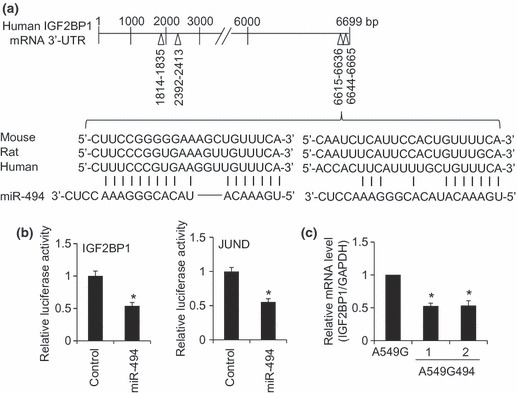
Downregulation of IGF2BP1 in A549 cells expressing miR‐494. (a) Prediction for miR‐494 binding sites in the 3′‐UTR of human IGF2BP1 mRNA with Target Scan database. Among four miR‐494 binding sites (1814–1835, 2392–2413, 6615–6636 and 6644–6665 bp), two adjacent target sites near the 3′ end are aligned in man, mouse and rat. (b) Verification of IGF2BP1 and JUND mRNA as miR‐494 targets. Two adjacent target sites (6615–6636 and 6644–6665 bp) were inserted into the luciferase gene 3′‐UTR to construct the pGL3‐Promoter‐IGF2BP1 reporter. Similarly, pGL3‐Promoter‐JUND reporter was constructed. The pGL3‐Promoter‐IGF2BP1 or pGL3‐Promoter‐JUND was co‐transfected with control RNA or miR‐494 into A549 cells, and after 24 h transfection, luciferase activity was measured. Relative luciferase activities between control and miR‐494 were normalized by luciferase activities obtained from the pGL3‐Promoter set to 1. Bar indicates mean and standard deviation. Student’s t‐test: *P < 0.05 (n = 3). (c) Measurement of IGF2BP1 mRNA in clones stably expressing miR‐494. Taqman real‐time RT‐PCR analysis was performed for A549G494 clones 1 and 2, and control A549G. Relative IGF2BP1 mRNA levels among the clones were normalized by the GAPDH mRNA level set to 1. Bar indicates mean and standard deviation. Student’s t‐test: *P < 0.05 (n = 3).
We then set out to investigate IGF2BP1 mRNA level in A549 cells stably expressing miR‐494. A recent report has concluded that regulation of gene expression through miRNA is primarily achieved by mRNA degradation rather than translational suppression, in mammalian cells (11). Taqman real‐time RT‐PCR revealed that IGF2BP1 mRNA levels were significantly reduced in A549G494‐1 and ‐2 clones compared to the control A549G clone (Fig. 2c). In human MCF7 breast cancer cells, IGF2BP1 knockdown has been shown to result in increase in IGF2 mRNA levels and inhibition of cell proliferation (12). To confirm whether suppressed IGF2BP1 mRNA levels seen in miR‐494 stably expressing cells accompanied stabilization of IGF2 mRNA, we performed RT‐PCR. This revealed that IGF2 mRNA was more abundant in A549G494‐1 and ‐2 clones than in control A549G clone (Fig. 3a). To show that IGF2 mRNA stabilization was an miR‐494 expression‐specific event, we used an miR494 inhibitor. Based on pGL3‐Promoter‐494 reporter activity, suppression of reporter activity mediated by miR‐494 was clearly rescued by addition of miR494 inhibitor (Fig. 3b). The miR‐494 inhibitor itself had no effect on IGF2 mRNA in control A549G cells, but when miR‐494 inhibitor was introduced into miR‐494 stably expressing cells (A549G494‐1 and ‐2), increased levels of IGF2 mRNA were significantly reduced (Fig. 3c). Taken together, our present experiments suggest that the IGF2BP1‐IGF2 axis could be a pivotal downstream target for miR‐494, and this regulation could partly account for observed phenotypes of miR‐494 stably expressing, A549 cells.
Figure 3.
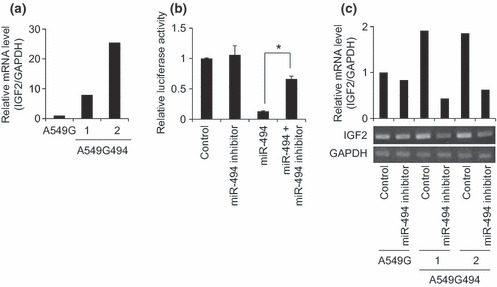
Accumulation of IGF2 mRNA in clones stably expressing miR‐494. (a) Semi‐quantitative detection of IGF2 mRNA. RT‐PCR analysis was performed for A549G494 clones 1 and 2, and control A549G. Relative IGF2 mRNA levels among the clones were normalized by GAPDH mRNA. Number of PCR cycles for IGF2 was 37. Gel images were quantified with Quantity One software, and shown by normalization with GAPDH set to 1. (b) Verification of the miR‐494 inhibitor towards inhibition of the miR‐494 effect. The pGL3‐Promoter‐494 reporter was co‐transfected with control RNA, the miR‐494 inhibitor, miR‐494 and miR‐494 plus the miR‐494 inhibitor, and after 24 h transfection, luciferase activity was measured. Relative luciferase activities were normalized by luciferase activities obtained from the pGL3‐Promoter set to 1. Bar indicates mean and standard deviation. Student’s t‐test: *P < 0.05 (n = 3). (c) Effect of miR‐494 inhibitor on IGF2 mRNA levels in clones stably expressing miR‐494. Control RNA or the miR‐494 inhibitor was transfected into A549G494 clones 1 and 2, and control A549G. After 72 h transfection, RT‐PCR analysis was performed. Relative IGF2 mRNA levels among the clones were normalized by GAPDH mRNA. Number of PCR cycles for IGF2 was 36. Gel images were quantified using Quantity One software, and shown by normalization with GAPDH set to 1.
Discussion
In our previous investigations, miR‐494 has been identified as one of the transcriptional targets regulated by cell proliferation signalling (5). In the present study, we have revealed that constitutive expression of miR‐494 in A549 cells resulted in suppression of cell proliferation and induction of senescence with stabilizing IGF2 mRNA, probably through IGF2BP1 mRNA degradation by miR‐494.
miR‐494 was initially identified as up‐regulated miRNA in retinoblastoma (6) and has been shown to regulate the tumour suppressor gene PTEN (10). Ectopic expression of miR‐494 has been shown to inhibit caspase‐3 activity and lead to suppression of apoptosis in adult heart muscle cells (13). These reports support a notion that miR‐494 promotes cell proliferation, whereas in miR‐494 stably expressing A549 cells established in this study, cell proliferation was suppressed and senescence was induced. This discrepancy can be explained by the following: one miRNA can regulate approximately 200 kinds of mRNA due to allowance of mismatch base pairing with mRNAs (14). Considering a further fact, that mRNA repertory varies according to individual cell and tissue type, it is likely that one miRNA can exert an opposite effect in different cells. Indeed, it has been indicated that miR‐494 potentially acts as a suppressor for both pro‐apoptotic and anti‐apoptotic factors (13). In this study, caspase‐3 and ‐7 activities remained unchanged between miR‐494 stably expressing cells and control cells (data not shown).
As an RNA binding protein, IGF2BP1 negatively regulates IGF2 mRNA (7, 8). Consistent with these reports, a decrease in IGF2BP1 mRNA levels in miR‐494 stably expressing cells accompanied increased levels of IGF2 mRNA. In melanoma, IGF2BP1 knockdown has been shown to inhibit cell proliferation and induce apoptosis via suppression of NF‐κB activity (15). On the other hand, a further study has reported that IGF2 promoted cell proliferation and inhibited apoptosis (16). Taken together, the suppressive effect of miR‐494 on cell proliferation could be partly explained by decreased levels of IGF2BP1 and increased levels of IGF2 in miR‐494 expressing cells.
In conclusion, we have characterized miR‐494 as a miRNA with a suppressive effect on cell proliferation. In addition, we present evidence that IGF2BP1 could be a target of miR‐494. Notably, decreased IGF2BP1 mRNA levels in miR‐494 stably expressing A549 cells accompanied stabilization of IGF2 mRNA, a downstream target of IGF2BP1. In the near future, knowing the precise regulatory mechanisms on miR‐494 function in various cell types, it is highly possible that miR‐494 could be applied in development of molecular markers and RNA‐based medicine, intended to protect against proliferative disease.
Acknowledgements
This study is partly supported by a grant from Organization for the Strategic Coordination of Research and intellectual Property, Meiji University.
References
- 1. Kawaji H, Hayashizaki Y (2008) Exploration of small RNAs. PLoS Genet. 4, e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zhang C (2009) Novel functions for small RNA molecules. Curr. Opin. Mol. Ther. 11, 641–651. [PMC free article] [PubMed] [Google Scholar]
- 3. Lai EC (2002) Micro RNAs are complementary to 3′ UTR sequence motifs that mediate negative post‐transcriptional regulation. Nat. Genet. 30, 363–364. [DOI] [PubMed] [Google Scholar]
- 4. Zhang B, Pan X, Cobb GP, Anderson TA (2007) microRNAs as oncogenes and tumor suppressors. Dev. Biol. 302, 1–12. [DOI] [PubMed] [Google Scholar]
- 5. Ohdaira H, Sasaki T, Yoshida K (2010) A subset of microRNAs potentially acts as a convergent hub for upstream transcription factors in cancer cells. Oncol. Rep. 24, 1371–1381. [DOI] [PubMed] [Google Scholar]
- 6. Zhao JJ, Yang J, Lin J, Yao N, Zhu Y, Zheng J et al. (2009) Identification of miRNAs associated with tumorigenesis of retinoblastoma by miRNA microarray analysis. Childs Nerv. Syst. 1, 13–20. [DOI] [PubMed] [Google Scholar]
- 7. Nielsen J, Christiansen J, Lykke‐Andersen J, Johnsen AH, Wewer UM, Nielsen FC (1999) A family of insulin‐like growth factor II mRNA‐binding proteins represses translation in late development. Mol. Cell. Biol. 19, 1262–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ioannidis P, Mahaira L, Papadopoulou A, Teixeira MR, Heim S, Andersen JA et al. (2003) CRD‐BP: a c‐Myc mRNA stabilizing protein with an oncofetal pattern of expression. Anticancer Res. 23, 2179–2183. [PubMed] [Google Scholar]
- 9. Boyerinas B, Park SM, Shomron N, Hedegaard MM, Vinther J, Andersen JS et al. (2008) Identification of let‐7‐regulated oncofetal genes. Cancer Res. 68, 2587–2591. [DOI] [PubMed] [Google Scholar]
- 10. Liu L, Jiang Y, Zhang H, Greenlee AR, Han Z (2010) Overexpressed miR‐494 down‐regulates PTEN gene expression in cells transformed by anti‐benzo(a)pyrene‐trans‐7,8‐dihydrodiol‐9,10‐epoxide. Life Sci. 86, 192–198. [DOI] [PubMed] [Google Scholar]
- 11. Guo H, Ingolia NT, Weissman JS, Bartel DP (2010) Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 466, 835–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ioannidis P, Mahaira LG, Perez SA, Gritzapis AD, Sotiropoulou PA, Kavalakis GJ et al. (2005) CRD‐BP/IMP1 expression characterizes cord blood CD34+ stem cells and affects c‐myc and IGF‐II expression in MCF‐7 cancer cells. J. Biol. Chem. 280, 20086–20093. [DOI] [PubMed] [Google Scholar]
- 13. Wang X, Zhang X, Ren XP, Chen J, Liu H, Yang J et al. (2010) MicroRNA‐494 targeting both proapoptotic and antiapoptotic proteins protects against ischemia/reperfusion‐induced cardiac injury. Circulation 122, 1308–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Krek A, Grün D, Poy MN, Wolf R, Rosenberg L, Epstein EJ et al. (2005) Combinatorial microRNA target predictions. Nat. Genet. 37, 495–500. [DOI] [PubMed] [Google Scholar]
- 15. Elcheva I, Tarapore RS, Bhatia N, Spiegelman VS (2008) Overexpression of mRNA‐binding protein CRD‐BP in malignant melanomas. Oncogene 27, 5069–5074. [DOI] [PubMed] [Google Scholar]
- 16. Polychronakos C, Giannoukakis N, Deal CL (1995) Imprinting of IGF2, insulin‐dependent diabetes, immune function, and apoptosis: a hypothesis. Dev. Genet. 17, 253–262. [DOI] [PubMed] [Google Scholar]


