Abstract
Objectives: Dental follicle cells (DFCs) provide the origin of periodontal tissues, and Runx2 is essential for bone formation and tooth development. In this study, pluripotency of DFCs was evaluated and effects of Runx2 on them were investigated.
Materials and methods: The DFCs were induced to differentiate towards osteoblasts, adipocytes or chondrocytes, and alizarin red staining, oil red O staining or alcian blue staining was performed to reveal the differentiated states. Bone marrow stromal cells (BMSCs) and primary mouse fibroblasts served as controls. DFCs were also infected with recombinant retroviruses encoding either full‐length Runx2 or mutant Runx2 without the VWRPY motif. Western blot analysis, real‐time real time RT‐PCR and in vitro mineralization assay were performed to evaluate the effects of full‐length Runx2 or mutant Runx2 on osteogenic/cementogenic differentiation of the cells.
Results: The above‐mentioned staining methods demonstrated that DFCs were successfully induced to differentiate towards osteoblasts, adipocytes or chondrocytes respectively, confirming the existence of pluripotent mesenchymal stem cells in dental follicle tissues. However, staining intensity in DFC cultures was weaker than in BMSC cultures. Real‐time PCR analysis indicated that mutant Runx2 induced a more pronounced increase in expression levels of OC, OPN, Col I and CP23 than full‐length Runx2. Mineralization assay also showed that mutant Runx2 increased mineralization nodule formation more prominently than full‐length Runx2.
Conclusions: Multipotent DFCs can be induced to differentiate towards osteoblasts, adipocytes or chondrocytes in vitro. Runx2 over‐expression up‐regulated expression levels of osteoblast/cementoblast‐related genes and in vitro enhanced osteogenic differentiation of DFCs. In addition, mutant Runx2‐induced changes in DFCs were more prominent than those induced by full‐length Runx2.
Introduction
Periodontal disease, the most common oral malady in adults, destroys tooth‐supporting alveolar bones and results in tooth loosening and eventual tooth loss. Much attention has been attracted to regeneration of periodontal tissues, but the molecular and cellular bases of periodontal tissue formation, repair and regeneration are still poorly understood, despite considerable research effort in this area (1). According to the classical theory of periodontal development, dental follicle cells (DFCs) are the origins of periodontal tissues, cementum, periodontal ligaments (PDL) and alveolar bone (2, 3, 4, 5, 6). It has been reported that in vitro, DFCs are plastic‐adherent colony‐forming cells that express putative mesenchymal stem cell markers (7). However, controversies still exist regarding the cellular mechanisms underlying periodontal development (8). Furthermore, attempts to regenerate periodontal tissues have not yet been successful, which may be in part due to a lesser degree of understanding of molecular events leading to initiation and development of root and periodontal tissues (1). Based on these facts, it is of great significance to isolate and characterize mesenchymal stem cells existing in dental follicle tissues.
Runx2 (also known as Cbfa1, OSF2, AML3, Pebp2αA) is a master transcription factor essential for skeletal development (9, 10). Analysis of Runx2‐deficient mice has revealed complete absence of mature osteoblasts and ossification; just few immature osteoblasts were observed in these mice, which weakly expressed alkaline phosphatase (ALP) but did not express osteopontin (OPN) or osteocalcin (OC) (11, 12). In addition, molar development in Runx2‐deficient mice was arrested at early cap stage, which indicates that Runx2 is also important in tooth formation (13). Furthermore, patients suffering from cleidocranial dysplasia (CCD) show multiple dental defects including supernumerary teeth, delayed tooth eruption and absence of cellular cementum formation (14, 15). Researchers also detected Runx2 expression in dental follicle tissues throughout development of periodontal tissues, suggesting a potential role of Runx2 in formation of periodontium (13, 16). In 1998, a structure‐function analysis revealed the existence of a five‐amino acid motif, VWRPY, at the C‐terminal end of Runx2, which is responsible for repressing transcriptional activation by Runx2. However, effects of this VWRPY motif on transcriptional activity of Runx2 in DFCs remained unclear.
In this study, pluripotency of DFCs was further investigated, and effects of full‐length Runx2 and mutant Runx2 without the VWPRY motif were evaluated.
Materials and methods
Cell culture
Retroviral packaging cell line PT67 (Clontech, Palo Alto, CA, USA) and murine fibroblast cell line NIH3T3 (American Type Culture Collection, Rockville, MD, USA) were maintained in α‐minimum essential medium (α‐MEM; HyClone, Logan, UT, USA) supplemented with 10% (v/v) foetal bovine serum (FBS; HyClone), 100 U/ml penicillin (Invitrogen, Carlsbad, CA, USA) and 100 μg/ml streptomycin (Invitrogen).
The DFCs were isolated from BALB/c mice (5–7 days old) as described previously (17). Briefly, bilateral first mandibular molar germs were removed using a dissecting microscope and treated with 1% trypsin at 4 °C for 1.5 h. After digestion, loose dental follicle tissues were isolated and cultured in α‐MEM supplemented with 20% (v/v) FBS, 100 U/ml penicillin and 100 μg/ml streptomycin at 37 °C in a 5% CO2 humidified incubator. Non‐adherent tissues were removed 24 h later and medium was changed every 3 days. Expressions of mesenchymal marker vimentin and epithelial marker cytokeratin in these cultured cells were determined by immunohistochemical analysis, and DFCs were identified as vimentin‐positive and cytokeratin‐negative cells as described previously (17, 18, 19, 20). DFCs at up to three passages were used for further experiments.
As described previously, bone marrow stromal cells (BMSCs) were obtained from BALB/c mice (6–8 weeks old) and were cultured under non‐differentiating conditions (α‐MEM with 20% FBS, 100 U/ml penicillin and 100 μg/ml streptomycin) (21). Oral fibroblasts were isolated from BALB/c mice (6–8 weeks old) and were cultured in DMEM with 10% FBS, 100 U/ml penicillin and 100 μg/ml streptomycin.
Animals used in this study were maintained and used in accordance with guidelines established by the Institutional Animal Care and Use Committee of Shandong University in Jinan, Shandong Province, P.R. China.
In vitro differentiation of DFCs
The DFCs were induced to differentiate towards osteoblasts, adipocytes and chondrocytes as described previously (22). Briefly for osteogenic differentiation, DFCs were cultured in α‐MEM supplemented with 10% FBS, 100 U/ml penicillin, 100 μg/ml streptomycin, 10−2 mβ‐glycerophosphate, 10−8 m dexamethasone and 10−4 m ascorbic acid. For chondrogenic differentiation, cells were cultured in the medium supplemented with 10−8 m dexamethasone and 50 μg/ml ascorbic acid. For adipogenic differentiation, cells were cultured in regular culture medium supplemented with 10−8 m dexamethasone and 6 ng/ml insulin. As described previously, alizarin red staining, alcian blue staining, or oil red O staining were performed to detect differentiation of these cells towards osteoblasts, chondroblasts or adipocytes respectively (22). In these experiments, BMSCs served as positive control, and primary murine oral fibroblasts served as negative control.
Quantification of alizarin red staining was performed as described previously (23, 24). Briefly, alizarin red‐S was extracted with 10% cetylpyridinium chloride and assessed at 562 nm. For quantification of alcian blue staining and oil red O staining, digital images of the stained cultures were captured using a Nikon Eclipse E600 microscope (Nikon Corporation, Tokyo, Japan). Percentage of positively stained areas was determined in six different fields of each well and in three independent experiments, using Spot Advanced software (Diagnostic Instruments, Sterling Heights, MI, USA).
Construction of Runx2 recombinant retroviral plasmids
Retroviral plasmid pLEGFP‐IRES‐Runx2, encoding full‐length mouse Runx2 cDNA with MASNSL as the N‐terminal sequence, was constructed as described previously (19). Mutant Runx2 without VWRPY motif was generated from plasmid pCMV5‐Cbfa1 (kindly provided by Dr Patricia Ducy, University of Texas M. D. Anderson Cancer Center). Primers were: 5′‐CTCGAGATGGTGGAGATCATCGCGGACCT‐3′ and 5′‐ACGCGTTTACAAAACAG TTGGGGAACTG‐3′. Construction of retroviral vector encoding mutant Runx2 was similar to that of pLEGFP‐IRES‐Runx2 (19), and the new construct was named pLEGFP‐IRES‐Runx2(M).
Production of viral stocks and viral infection
Retroviral stocks of pLEGFP‐IRES‐Runx2 or pLEGFP‐IRES‐Runx2(M) were prepared as described previously (19). Briefly, the retroviral construct was transfected into PT67 packaging cells using LipofectamineTM 2000 (Invitrogen), and the stable virus‐producing PT67 cells were selected with 400 μg/ml G418 (Takara) for 2 weeks. The supernatant of these stable virus‐producing PT67 cells was collected, filtered through a 0.45 μm filter and stored at −80 °C. The viral titre was determined indirectly using NIH3T3 cells as described previously (25). DFCs were then plated in six‐well plates (1 × 105 cells/well) and transduced with viral stocks of pLEGFP‐IRES‐Runx2, pLEGFP‐IRES‐Runx2(M) or empty vector in the presence of polybrene (6 μg/ml, Sigma, St Louis, MO, USA) for 8 h. Twenty‐four hours after infection, DFCs were subjected to G418 selection (300 μg/ml) for 2 weeks. The stably transduced DFCs were used for the following experiments, and stably transduced NIH3T3 cells served as controls.
Real‐time reverse‐transcriptase polymerase chain reaction
Real‐time RT‐PCR was performed as described previously (19). Briefly, total RNA was extracted and reverse transcribed. Real‐time PCR was then performed using LightCycler FastStart DNA Master SYBR Green I (Roche Applied Science, Mannheim, Germany). Sequences of the primers for amplification of mouse glyceraldehyde‐3‐phosphate dehydrogenase (GAPDH), β‐actin, ALP, α1(II) procollagen, lipoprotein lipase (LPL), Runx2, OPN, OC, bone sialoprotein (BSP), collagen I (Col I), cementum attachment protein (CAP) and cementum protein 23 (CP23) were exactly as reported previously (19, 22). Amount of mRNA was calculated for each sample, based on the standard curve using LightCycler Software 4.0 (Roche). β‐actin or GAPDH was used as internal control.
Western blot analysis
Cells were harvested using 0.25% trypsin and washed twice in PBS. Proteins were extracted using lysis buffer (25 mm Tris–HCl, pH 7.2, 1% Triton X‐100, 0.1% SDS, 1% sodium deoxycholate, 0.1 m NaCl and 1 mm EDTA) containing a protease inhibitor cocktail (Roche). Proteins were separated by SDS–polyacrylamide gel electrophoresis and transferred to a Protran nitrocellulose membrane, which was saturated overnight in Tris‐buffered saline with Tween (TBST) and 5% low fat milk. The membrane was incubated with anti‐Runx2 antibody (1:500, Santa Cruz, CA, USA) followed by incubation with rabbit horseradish peroxidase‐conjugated IgG (1:10,000, Sigma). Blots were visualized using diaminobenzidine, and β‐actin was used as internal control. Images of blots were analysed using JD801 IMAGE 4.11 program (Dahui Biotechnology, Guangzhou, China).
Mineralization assay
Cells were incubated with mineralizing media (α‐MEM supplemented with 10% FBS, 100 U/ml penicillin, 100 μg/ml streptomycin, 10−2 mβ‐glycerophosphate, 10−8 m dexamethasone and 10−4 m ascorbic acid). In vitro mineralization assay was performed at weeks 2, 3 and 4 with von Kossa staining (26). Nodules were photographed using an Olympus camera and area covered by mineral nodules in each well was determined using image analysis system DT2000 software V1.0 (Tansi Technology Co. Ltd, Nanjing, China).
Statistical analysis
Data are shown as mean ± SE from at least three experiments and t‐test was used to test significance using SPSS (SPSS Inc., USA) 12.0 software. P < 0.05 was considered statistically significant.
Results
Pluripotency and differentiation of DFCs towards osteoblasts, chondrocytes and adipocytes in vitro
After chondrogenic and adipogenic induction, the corresponding special staining methods demonstrated existence of chondrocytes or adipocytes in DFC cultures and BMSC cultures. However, positive staining was barely detectable in oral fibroblast cultures. DFC cultures showed lower percentages of positively stained areas compared to BMSC cultures (Fig. 1a,b). After osteogenic induction, alizarin red staining showed deposition of densely stained extracellular matrix in both DFC cultures and BMSC cultures, with staining intensity being lower in DFC cultures than in BMSC cultures. In contrast, deposition of extracellular matrix was negligible in oral fibroblast cultures (Fig. 1c). Real‐time RT‐PCR analysis also showed that expression levels of osteoblast‐specific ALP, adipocyte‐specific LPL and chondrocyte‐specific α1(II) procollagen were significantly lower in DFCs than in BMSCs after differential induction (Fig. 1d).
Figure 1.
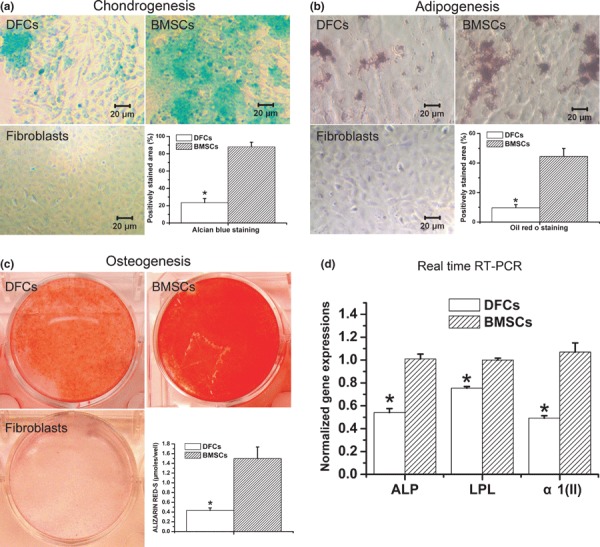
Multilineage differentiation of dental follicle cells. (a) Demonstrative photos and quantification of chondrogenesis in dental follicle cells (DFCs), bone marrow stromal cells (BMSCs), and primary oral fibroblasts (Fibroblasts). (b) Demonstrative photos and quantification of adipogenesis in DFCs, BMSCs and fibroblasts. (c) Demonstrative photos and quantification of osteogenesis in DFCs, BMSCs and fibroblasts. (d) Real time RT‐PCR analysis of expression levels of osteoblast‐specific ALP, adipocyte‐specific LPL, and chondrocyte‐specific α1(II) procollagen in DFC cultures and BMSC cultures after differential induction. *P < 0.05 vs. BMSC cultures.
Retroviral infection of DFCs
Recombinant retroviral vector pLEGFP‐IRES‐Runx2(M) was verified by digestion with restriction endonucleases. As shown in Fig. 2, size of Runx2(M) fragment was 1218 bp. Sequence of Runx2(M) was consistent with the sequence published in GenBank (Runx2: NM_009820).
Figure 2.
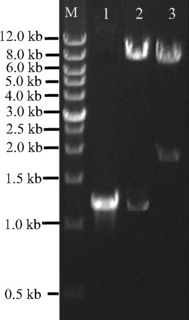
PCR products and restriction endonuclease digestion analysis of retroviral plasmids pLEGFP‐IRES‐Runx2(M). The recombinant retroviral plasmids pLEGFP‐IRES‐Runx2(M) was verified by digestion with restriction endonucleases. Lane M, wide range DNA marker (0.5–12.0 kb, Takara); lane 1, mutant Runx2 gene amplified from plasmid pCMV5‐Cbfa1; lane 2, plasmid pLEGFP‐IRESRunx2(M) digested with Xho I and Mlu I; lane 3, plasmid pLEGFP‐IRES‐Runx2(M) digested with Xho I and Sal I.
No significant morphological changes were observed in cells transduced with retroviral vectors, easily identified by expression of green fluorescence protein (GFP), using an inverted fluorescence microscope (Fig. 3a1,a2). Real‐time RT‐PCR and Western blot analysis also showed that expression level of Runx2 was higher in cells transduced with pLEGFP‐IRES‐Runx2 or pLEGFP‐IRES‐Runx2(M) compared to control cells transduced with empty vector (P < 0.01) (Fig. 3b1,b2,c1,c2,d1,d2).
Figure 3.
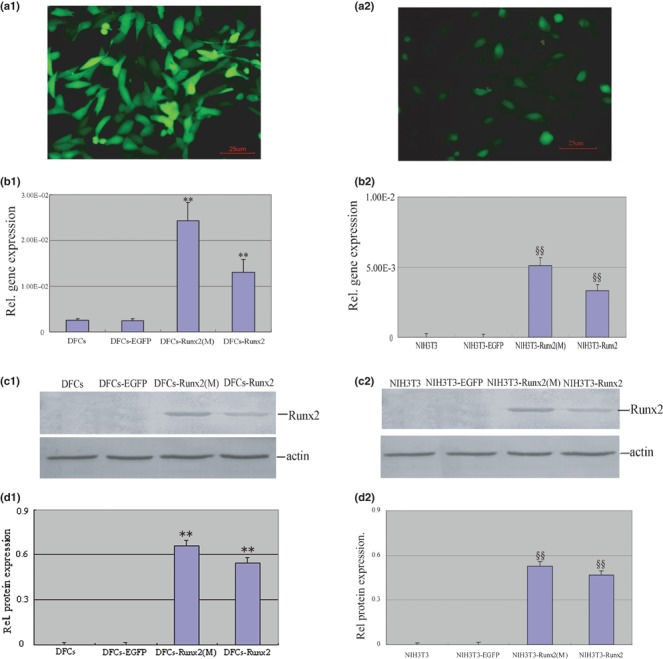
DFCs stably overexpressing Runx2. DFCs and NIH3T3 were infected by retrovirus and selected by G418. (a1, a2) Green fluorescence could be detected in the stably transduced cells but without significant morphological changes. (b1, b2) Real‐time RT‐PCR confirmed that the transduced cells overexpressed Runx2 gene. (c1, c2) Representative images demonstrated that the presence of Runx2 protein in cell extract could be detected by Western blot with Runx2 antibody. (d1, d2) Protein levels were normalized with that of β‐actin. Runx2 was up‐regulated in pLEGFPIRES‐Runx2 and pLEGFP‐IRES‐Runx2(M) infected cells, respectively. **P < 0.01 vs. DFCs and DFCs‐EGFP, §§P < 0.01 vs. NIH3T3 and NIH3T3‐EGFP. Statistical significance was determined using t‐test.
Effects of Runx2 over‐expression on osteoblast/cementoblast‐related gene expression
Expression levels of osteoblast/cementoblast‐related genes, including Col I, OPN, BSP, OC, CAP and CP23 were determined using real‐time RT‐PCR; all were detected in DFCs (Fig. 4a). Furthermore, in DFCs transduced with full‐length Runx2 or mutant Runx2, expression levels of these osteoblast/cementoblast‐related genes were higher than those in non‐transduced DFCs or DFCs transduced with empty vector (P < 0.01). Moreover, DFCs transduced with mutant Runx2 demonstrated a more prominent increase in expression levels of OC, OPN, Col I and CP23 compared to those transduced with full‐length Runx2 (P < 0.01). In contrast, CAP level in full‐length Runx2‐transduced DFCs was higher than that in mutant Runx2‐transduced DFCs (P < 0.01). There was no statistically significant difference in BSP expression level between full‐length Runx2‐transduced DFCs and mutant Runx2‐transduced DFCs (P > 0.05).
Figure 4.
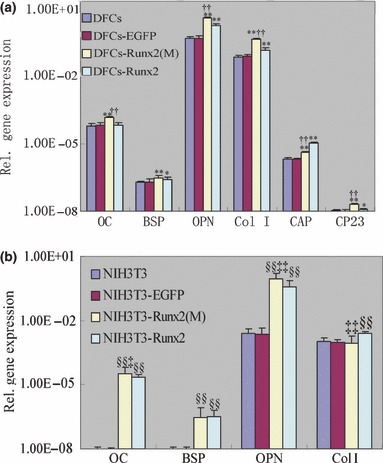
Effects of Runx2 overexpression on osteoblast/cementoblast‐related gene expressions. Total RNA was isolated from DFCs, NIH3T3 and the cells infected with pLEGFP‐N1, pLEGFP‐IRESRunx2 and pLEGFP‐IRES‐Runx2(M). Real‐time RT‐PCR was performed to investigate the effects of Runx2 overexpression on osteoblast/cementoblast‐related gene expressions, including Col I, OPN, BSP, OC, CAP and CP23. All values were normalized with β‐actin levels and the data were presented as the mean ± SE for three experiments. *P < 0.05 vs. DFCs and DFCs‐EGFP, **P < 0.01 vs. DFCs and DFCs‐EGFP, ††P < 0.01 vs. DFCs‐Runx2. §§P < 0.01 vs. NIH3T3 and NIH3T3‐EGFP, ‡‡P < 0.01 vs. NIH3T3‐Runx2, ‡P < 0.05 vs. NIH3T3‐Runx2. Statistical significance was determined using t‐test.
In NIH3T3 cells (Fig. 4b), over‐expression of full‐length Runx2 or mutant Runx2 also increased expressions of OPN, BSP, OC and Col I. In addition, expression levels of OC and OPN in NIH3T3 cells transduced with mutant Runx2 were higher than those in cells transduced with full‐length Runx2 (P < 0.05). However, expression level of Col I in mutant Runx2‐transduced cells was lower than in full‐length Runx2‐transduced cells (P < 0.01). There was no statistically significant difference in BSP expression level between full‐length Runx2‐transduced NIH3T3 cells and mutant Runx2‐transduced NIH3T3 cells (P > 0.05). Expression of neither CAP nor CP23 were detected in all four groups of NIH3T3 cells.
Effects of Runx2 over‐expression on in vitro cell mineralization
In vitro mineral nodule formation assay revealed that both full‐length Runx2‐transduced DFCs and mutant Runx2‐transduced DFCs exhibited a larger area covered by mineral nodules compared to non‐transduced DFCs or DFCs transduced with empty vector (Fig. 5a). In addition, the area covered by mineral nodules in DFCs transduced with mutant Runx2 was higher than in DFCs transduced with full‐length Runx2 (P < 0.05). Similar results were also observed in NIH3T3 cells (Fig. 5b).
Figure 5.
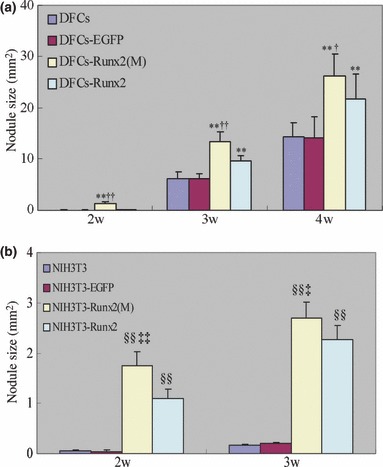
Effects of Runx2 overexpression on mineral nodule formation. Cells were cultured in mineralizing media and von Kossa staining was performed to evaluate the mineral nodule formation on week 2, 3, 4. The area covered by mineral nodules in each well was determined. The data were presented as the mean ± SE for three experiments. **P < 0.01 vs. DFCs and DFCs‐EGFP, †P < 0.05 vs. DFCs‐Runx2, ††P < 0.01 vs. DFCs‐Runx2, §§P < 0.01 vs. NIH3T3 and NIH3T3‐EGFP, ‡‡P < 0.01 vs. NIH3T3‐Runx2, ‡P < 0.05 vs. NIH3T3‐Runx2. Statistical significance was determined using t‐test.
Discussion
Although exciting advancements have been made in research of early tooth development, attempts to regenerate periodontal tissues have not been successful (1). This may be, at least in part, due to an incomplete degree of understanding of molecular and cellular events leading to initiation and development of periodontal tissues (8). The dental follicle is of loose connective tissue derived from ectomesenchyme, surrounding the enamel organ and dental papilla of the developing tooth germ prior to eruption (27). Although controversies still exist regarding formation of periodontal tissues, it has been widely accepted that dental follicle tissues are the origin of periodontal tissues or at least play an essential role during development of periodontal tissues (5, 6, 28, 29, 30). Like BMSCs, in vitro DFCs are plastic‐adherent and colony‐forming cells, and can differentiate into osteoblast‐like cells (31). DFCs can also differentiate into connective tissue cells and produce mineral nodules (32). In addition, DFCs express typical markers of PDL and cementum, such as collagen type XII (Col 12) and CAP (33). In a recently published study, DFCs were subjected to flow cytometric surface marker expression analysis (7). It was reported that they positively expressed the putative stem cell marker STRO‐1, adhesion molecules CD29, CD105 and CD106, receptor molecule CD44 and extracellular matrix protein CD90 (7), which confirmed existence of mesenchymal stem cells in dental follicle tissues. In this study, DFCs were successfully induced to differentiate towards osteoblasts, adipocytes and chondrocytes in vitro, which further confirmed them pluripotent mesenchymal stem cells in dental follicle tissues.
The BMSCs, also known as non‐haematopoietic bone marrow‐derived mesenchymal stem cells (BMMSCs), have long been used in adult stem cell investigation and tissue engineering. BMSCs are also pluripotent and can be induced to differentiate towards osteoblasts, chondrocytes and adipocytes in vitro (34). In this study, pluripotent BMSCs were used as positive control to evaluate ability of DFCs to differentiate towards multiple cell types. Our results demonstrated that after differential induction, staining intensities or percentages of positively stained areas, in DFC cultures were lower than those in BMSC cultures. Moreover, expression levels of osteoblast‐specific ALP, adipocyte‐specific LPL and chondrocyte‐specific α1(II) procollagen were significantly lower in DFCs than in BMSCs after differential induction. These results indicated that pluripotency of DFCs was weaker than BMSCs, or else stem cell percentage in the DFC cultures was lower than BMSC cultures. Extracellular matrix of differentiated cells has been reported to induce differentiation of mesenchymal stem cells. For example, the extracellular matrix of osteoblasts was shown to enhance expression of osteoblast‐specific ALP and deposition of calcium (35). During development of periodontal tissues, the microenvironment of DFCs is filled with molecular signals from odontoblasts, epithelial root sheath cells and other differentiated cells, which may induce DFCs to differentiate. In contrast, low oxygen tension is thought to be characteristic of the native bone marrow microenvironment, which is important in maintenance of proliferation and pluripotency of stem cells (36, 37). Thus, the microenvironment of BMSCs is more appropriate to maintain stemness of these cells, which may be the reason that here the percentage of undifferentiated stem cells in DFCs was lower than that of BMSCs.
Forced expression of Runx2 can induce expression of osteoblast‐specific genes in non‐osteoblastic cells (9). Furthermore, the core binding factor site of Runx2 protein is found in promoter regions of all major extracellular matrix proteins of mineralizing tissues such as OC, Col I, BSP, OPN and ALP (38). Our data also showed that Runx2 over‐expression in DFCs increased expression levels of OPN, Col I, OC, BSP, CP23 and CAP. Also, mineralization assay demonstrated that Runx2 over‐expression enhanced in vitro mineralization of DFCs. These results clearly indicated that Runx2 over‐expression enhanced differentiation of DFCs towards osteoblasts/cementoblasts.
In this study, transcriptional activity of a mutant Runx2, without terminal five amino acids (the VWRPY motif) was also investigated. VWRPY motif has been reported to bind to co‐repressor proteins of TLE or Grg family, which is considered to be a potential mechanism for Runx2 repression during cell differentiation (39, 40). For instance, researchers found that transactivation function of Runx2 was inhibited by interaction of Runx2 at its VWRPY motif with TLE2, which is a mammalian homologue of Drosophila Groucho (41). Consistent with these findings, we found that mutant Runx2 resulted in higher levels of OPN, Col I and CP23 in dental follicle cells compared to full‐length Runx2.
Interestingly, we also found that in NIH3T3 cells, mutant Runx2 resulted in higher level of OPN but lower level of Col I when compared with full‐length Runx2. In addition, the level of another putative molecular marker for cementoblasts, CAP, was found to be lower in DFCs transduced with mutant Runx2 when compared to those transduced with full‐length Runx2. C terminus of Runx2 protein has been reported to support either activation or repression of cell type‐specific gene expression, and the overall effect of Runx2 on target genes depends on cell type, regulatory organization of target promoter, existence of certain co‐activators or co‐repressors, and subnuclear localization of Runx2 protein (39). It was reported that ALY, a context‐dependent co‐activator of Runx family proteins, binds to C terminus of Runx2 and activates its transcriptional activity (42). Transcriptional co‐activator with PDZ‐binding motif (TAZ), CCAAT/enhancer binding protein β (C/EBPβ) and p300 also act as multifunctional transcriptional co‐activators, binding to C terminus of Runx2 and serving as adapters for this potent osteogenic transcription factor (43, 44, 45). Consistent with these previous findings, our results suggested that transcriptional activity of Runx2 is subtly regulated, and the final effect of Runx2 may not only be cell‐specific, but also gene‐specific.
CAP (46) and CP23 (47) are both putative molecular markers synthesized by cementoblasts, which are expressed in cementum and cementoblast subpopulations and progenitor cells of human periodontal ligaments. CAP is a 56‐kDa collagenous protein, which has been shown to promote several biological activities, such as cell attachment, chemotaxis and differentiation (48, 49). This protein preferentially promotes adhesion of osteoblastic cells, and periodontal cells strongly binding to CAP have been reported to form cementum‐like mineralized tissue in vitro (50). CAP was also shown to promote proliferation of monkey BMSCs, while having little effects on mineralization (51). In contrast, CP23 plays an important role during the early stage of biomineralization processes by promoting octacalcium phosphate crystal nucleation (52). This protein has been implicated in regulating deposition rate, composition and morphology of hydroxyapatite crystals formed by putative human cementoblasts (53). For the first time, our results have indicated that expression of these cementum‐derived matrix proteins is stringently regulated during cementogenesis and root formation, and expression of CAP is more prominently enhanced by Runx2 when compared to that of CP23. As an important transcription factor for both osteogenesis and cementogenesis, Runx2 protein is present in early proliferative osteoblasts/cementoblasts, a developmental stage at which cell proliferation is still required to obtain enough committed cells for matrix formation. A higher level of CAP at this early stage of cementogenesis is consistent with its function in promoting cell proliferation. In contrast, CP23 has a more important role in mineralization, and moderate expression level of CP23 at this early stage may better control the process of mineralization.
In conclusion, our study further has confirmed pluripotency of DFCs. We also showed that Runx2 over‐expression in DFCs enhanced expression of osteoblast/cementoblast‐related genes, and mutant Runx2 without the VWRPY motif showed different transcriptional activities compared to full‐length Runx2, indicating an important regulatory effect of this motif. Although the in vitro findings may not necessarily represent actual events in vivo, these results strongly indicate that Runx2 is important in differentiation of DFCs towards osteoblasts/cementoblasts.
Acknowledgements
This study was supported by grants from the national Natural Science Foundation of China (No. 30772425), Beijing, China, and Shandong Provincial Natural Science Foundation (No. Y 2007C 091), Jinan, China.
References
- 1. Polimeni G, Xiropaidis AV, Wikesjo UM (2006) Biology and principles of periodontal wound healing/regeneration. Periodontol. 2000 41, 30–47. [DOI] [PubMed] [Google Scholar]
- 2. Bosshardt DD, Schroeder HE (1996) Cementogenesis reviewed: a comparison between human premolars and rodent molars. Anat. Rec. 245, 267–292. [DOI] [PubMed] [Google Scholar]
- 3. Chai Y, Jiang X, Ito Y, Bringas P Jr, Han J, Rowitch DH et al. (2000) Fate of the mammalian cranial neural crest during tooth and mandibular morphogenesis. Development 127, 1671–1679. [DOI] [PubMed] [Google Scholar]
- 4. Cho MI, Garant PR (1988) Ultrastructural evidence of directed cell migration during initial cementoblast differentiation in root formation. J. Periodontal. Res. 23, 268–276. [DOI] [PubMed] [Google Scholar]
- 5. Cho MI, Garant PR (2000) Development and general structure of the periodontium. Periodontol. 2000 24, 9–27. [DOI] [PubMed] [Google Scholar]
- 6. Paynter KJ, Pudy G (1958) A study of the structure, chemical nature, and development of cementum in the rat. Anat. Rec. 131, 233–251. [DOI] [PubMed] [Google Scholar]
- 7. Lindroos B, Maenpaa K, Ylikomi T, Oja H, Suuronen R, Miettinen S (2008) Characterisation of human dental stem cells and buccal mucosa fibroblasts. Biochem. Biophys. Res. Commun. 368, 329–335. [DOI] [PubMed] [Google Scholar]
- 8. Foster BL, Popowics TE, Fong HK, Somerman MJ (2007) Advances in defining regulators of cementum development and periodontal regeneration. Curr. Top. Dev. Biol. 78, 47–126. [DOI] [PubMed] [Google Scholar]
- 9. Ducy P, Zhang R, Geoffroy V, Ridall AL, Karsenty G (1997) Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell 89, 747–754. [DOI] [PubMed] [Google Scholar]
- 10. Stein GS, Lian JB, Van Wijnen AJ, Stein JL, Montecino M, Javed A et al. (2004) Runx2 control of organization, assembly and activity of the regulatory machinery for skeletal gene expression. Oncogene 23, 4315–4329. [DOI] [PubMed] [Google Scholar]
- 11. Komori T, Yagi H, Nomura S, Yamaguchi A, Sasaki K, Deguchi K et al. (1997) Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell 89, 755–764. [DOI] [PubMed] [Google Scholar]
- 12. Otto F, Thornell AP, Crompton T, Denzel A, Gilmour KC, Rosewell IR et al. (1997) Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell 89, 765–771. [DOI] [PubMed] [Google Scholar]
- 13. D’Souza RN, Aberg T, Gaikwad J, Cavender A, Owen M, Karsenty G et al. (1999) Cbfa1 is required for epithelial–mesenchymal interactions regulating tooth development in mice. Development 126, 2911–2920. [DOI] [PubMed] [Google Scholar]
- 14. Hitchin AD (1975) Cementum and other root abnormalities of permanent teeth in cleidocranial dysostosis. Br. Dent. J. 139, 313–318. [DOI] [PubMed] [Google Scholar]
- 15. Jensen BL, Kreiborg S (1990) Development of the dentition in cleidocranial dysplasia. J. Oral Pathol. Med. 19, 89–93. [DOI] [PubMed] [Google Scholar]
- 16. Bronckers AL, Engelse MA, Cavender A, Gaikwad J, D’Souza RN (2001) Cell‐specific patterns of Cbfa1 mRNA and protein expression in postnatal murine dental tissues. Mech. Dev. 101, 255–258. [DOI] [PubMed] [Google Scholar]
- 17. Wise GE, Lin F, Fan W (1992) Culture and characterization of dental follicle cells from rat molars. Cell Tissue Res. 267, 483–492. [DOI] [PubMed] [Google Scholar]
- 18. Hou LT, Liu CM, Chen YJ, Wong MY, Chen KC, Chen J et al. (1999) Characterization of dental follicle cells in developing mouse molar. Arch. Oral Biol. 44, 759–770. [DOI] [PubMed] [Google Scholar]
- 19. Pan KQ, Yan SG, Ge SH, Li S, Zhao YH, Yang PS (2009) Effects of core binding factor alpha1 or bone morphogenic protein‐2 overexpression on osteoblast/cementoblast‐related gene expressions in NIH3T3 mouse cells and dental follicle cells. Cell Prolif. 42, 364–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Webb PP, Moxham BJ, Benjamin M, Ralphs JR (1996) Changing expression of intermediate filaments in fibroblasts and cementoblasts of the developing periodontal ligament of the rat molar tooth. J. Anat. 188, 529–539. [PMC free article] [PubMed] [Google Scholar]
- 21. Li S, Tu Q, Zhang J, Stein G, Lian J, Yang PS et al. (2008) Systemically transplanted bone marrow stromal cells contributing to bone tissue regeneration. J. Cell. Physiol. 215, 204–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tu Q, Valverde P, Chen J (2006) Osterix enhances proliferation and osteogenic potential of bone marrow stromal cells. Biochem. Biophys. Res. Commun. 341, 1257–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Stanford CM, Jacobson PA, Eanes ED, Lembke LA, Midura RJ (1995) Rapidly forming apatitic mineral in an osteoblastic cell line (UMR 106‐01 BSP). J. Biol. Chem. 270, 9420–9428. [DOI] [PubMed] [Google Scholar]
- 24. Tu Q, Zhang J, Paz J, Wade K, Yang P, Chen J (2008) Haploinsufficiency of Runx2 results in bone formation decrease and different BSP expression pattern changes in two transgenic mouse models. J. Cell. Physiol. 217, 40–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kwon YJ, Huang G, Anderson WF, Peng CA, Yu H (2003) Determination of infectious retrovirus concentration from colony‐forming assay with quantitative analysis. J. Virol. 77, 5712–5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Puchtler H, Meloan SN (1978) Demonstration of phosphates in calcium deposits: a modification of von Kossa’s reaction. Histochemistry 56, 177–185. [DOI] [PubMed] [Google Scholar]
- 27. Ten Cate AR (1997) The development of the periodontium – a largely ectomesenchymally derived unit. Periodontol. 2000 13, 9–19. [DOI] [PubMed] [Google Scholar]
- 28. Diekwisch TG (2001) The developmental biology of cementum. Int. J. Dev. Biol. 45, 695–706. [PubMed] [Google Scholar]
- 29. Lezot F, Davideau JL, Thomas B, Sharpe P, Forest N, Berdal A (2000) Epithelial Dlx‐2 homeogene expression and cementogenesis. J. Histochem. Cytochem. 48, 277–284. [DOI] [PubMed] [Google Scholar]
- 30. MacNeil RL, Thomas HF (1993) Development of the murine periodontium II. Role of the epithelial root sheath in formation of the periodontal attachment. J. Periodontol. 64, 285–291. [DOI] [PubMed] [Google Scholar]
- 31. Morsczeck C, Schmalz G, Reichert TE, Vollner F, Galler K, Driemel O (2008) Somatic stem cells for regenerative dentistry. Clin. Oral Investig. 12, 113–118. [DOI] [PubMed] [Google Scholar]
- 32. Morsczeck C (2006) Gene expression of runx2, Osterix, c‐fos, DLX‐3, DLX‐5, and MSX‐2 in dental follicle cells during osteogenic differentiation in vitro . Calcif. Tissue Int. 78, 98–102. [DOI] [PubMed] [Google Scholar]
- 33. Morsczeck C, Gotz W, Schierholz J, Zeilhofer F, Kuhn U, Mohl C et al. (2005) Isolation of precursor cells (PCs) from human dental follicle of wisdom teeth. Matrix Biol. 24, 155–165. [DOI] [PubMed] [Google Scholar]
- 34. Pittenger MF, Mackay AM, Beck SC, Jaiswal RK, Douglas R, Mosca JD et al. (1999) Multilineage potential of adult human mesenchymal stem cells. Science 284, 143–147. [DOI] [PubMed] [Google Scholar]
- 35. Datta N, Holtorf HL, Sikavitsas VI, Jansen JA, Mikos AG (2005) Effect of bone extracellular matrix synthesized in vitro on the osteoblastic differentiation of marrow stromal cells. Biomaterials 26, 971–977. [DOI] [PubMed] [Google Scholar]
- 36. Covello KL, Kehler J, Yu H, Gordan JD, Arsham AM, Hu CJ et al. (2006) HIF‐2alpha regulates Oct‐4: effects of hypoxia on stem cell function, embryonic development, and tumor growth. Genes Dev. 20, 557–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Grayson WL, Zhao F, Izadpanah R, Bunnell B, Ma T (2006) Effects of hypoxia on human mesenchymal stem cell expansion and plasticity in 3D constructs. J. Cell. Physiol. 207, 331–339. [DOI] [PubMed] [Google Scholar]
- 38. Ducy P (2000) Cbfa1: a molecular switch in osteoblast biology. Dev. Dyn. 219, 461–471. [DOI] [PubMed] [Google Scholar]
- 39. Javed A, Guo B, Hiebert S, Choi JY, Green J, Zhao SC et al. (2000) Groucho/TLE/R‐esp proteins associate with the nuclear matrix and repress RUNX (CBF (alpha)/AML/PEBP2 (alpha) dependent activation of tissue‐specific gene transcription. J. Cell Sci. 113, 2221–2231. [DOI] [PubMed] [Google Scholar]
- 40. McLarren KW, Theriault FM, Stifani S (2001) Association with the nuclear matrix and interaction with Groucho and RUNX proteins regulate the transcription repression activity of the basic helix loop helix factor Hes1. J. Biol. Chem. 276, 1578–1584. [DOI] [PubMed] [Google Scholar]
- 41. Thirunavukkarasu K, Mahajan M, McLarren KW, Stifani S, Karsenty G (1998) Two domains unique to osteoblast‐specific transcription factor Osf2/Cbfa1 contribute to its transcriptional function and its inability to heterodimerize with Cbfβ. Mol. Cell. Biol. 18, 4197–4208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bruhn L, Munnerlyn A, Grosschedl R (1997) ALY, a context‐dependent coactivator of LEF‐1 and AML‐1, is required for TCRalpha enhancer function. Genes Dev. 11, 640–653. [DOI] [PubMed] [Google Scholar]
- 43. Hata K, Nishimura R, Ueda M, Ikeda F, Matsubara T, Ichida F et al. (2005) A CCAAT/enhancer binding protein beta isoform, liver‐enriched inhibitory protein, regulates commitment of osteoblasts and adipocytes. Mol. Cell. Biol. 25, 1971–1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hong JH, Hwang ES, McManus MT, Amsterdam A, Tian Y, Kalmukova R et al. (2005) TAZ, a transcriptional modulator of mesenchymal stem cell differentiation. Science 309, 1074–1078. [DOI] [PubMed] [Google Scholar]
- 45. Sierra J, Villagra A, Paredes R, Cruzat F, Gutierrez S, Javed A et al. (2003) Regulation of the bone‐specific osteocalcin gene by p300 requires Runx2/Cbfa1 and the vitamin D3 receptor but not p300 intrinsic histone acetyltransferase activity. Mol. Cell. Biol. 23, 3339–3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Saito M, Iwase M, Maslan S, Nozaki N, Yamauchi M, Handa K et al. (2001) Expression of cementum‐derived attachment protein in bovine tooth germ during cementogenesis. Bone 29, 242–248. [DOI] [PubMed] [Google Scholar]
- 47. Arzate H, Jiménez‐García LF, Alvarez‐Pérez MA, Landa A, Bar‐Kana I, Pitaru S (2002) Immunolocalization of a human cementoblastoma conditioned medium‐derived protein. J. Dent. Res. 81, 541–546. [DOI] [PubMed] [Google Scholar]
- 48. Pitaru S, Savion N, Hekmati H, Olsen S, Narayanan AS (1992) Binding of a cementum attachment protein to extracellular matrix components and to dental surfaces. J. Periodontal. Res. 27, 640–646. [DOI] [PubMed] [Google Scholar]
- 49. Pitaru S, Narayanan AS, Olson S, Savion N, Hekmati H, Alt I et al. (1995) Specific cementum attachment protein enhances selectively the attachment and migration of periodontal cells to root surfaces. J. Periodontal. Res. 30, 360–368. [DOI] [PubMed] [Google Scholar]
- 50. Van Den Bos T, Handoko G, Niehof A, Ryan LM, Coburn SP, Whyte MP et al. (2005) Cementum and dentin in hypophosphatasia. J. Dent. Res. 84, 1021–1025. [DOI] [PubMed] [Google Scholar]
- 51. Xie Y, Song Z, Shu R, Sun Y, Luo M, Zhang X (2008) Effects of cementum attachment proteins on the proliferation and mineralization of monkey bone marrow stromal cells in vitro . Chin. J. Stomatol. Res. (Electronic Version) 2, 14–18. [Google Scholar]
- 52. Villarreal‐Ramírez E, Moreno A, Mas‐Oliva J, Chávez‐Pacheco JL, Narayanan AS, Gil‐Chavarría I et al. (2009) Characterization of recombinant human cementum protein 1 (hrCEMP1): primary role in biomineralization. Biochem. Biophys. Res. Commun. 384, 49–54. [DOI] [PubMed] [Google Scholar]
- 53. Alvarez Pérez MA, Pitaru S, Alvarez Fregoso O, Reyes Gasga J, Arzate H (2003) Anti‐cementoblastoma‐derived protein antibody partially inhibits mineralization on a cementoblastic cell line. J. Struct. Biol. 143, 1–13. [DOI] [PubMed] [Google Scholar]


