Abstract
Abstract. Objectives: In this study, we have investigated whether secreted factors from embryonic stem cells (ESCs) could reprogramme keratinocytes and increase their potential to be directed into alternative cell lineages. Materials and methods: Contact and non‐contact co‐cultures of skin keratinocytes and murine ESCs were used initially to confirm any reprogramming ability of ESC‐conditioned medium (CM). Immunofluoresence was used to assess nuclear expression of octamer‐4 (Oct‐4), as well as to confirm neuronal protein expression in neuroectodermally directed keratinocytes. Transcript expression changes were evaluated using semiquantitative reverse transcription‐polymerase chain reaction. Western blotting, accompanied by densitometry analysis, was used to evaluate protein expression following morphology changes. Results: We found that keratinocytes treated with ESC‐CM changed their morphology and were stimulated to express the pluripotency regulator, Oct‐4, and its target transcripts, Sox‐2, Nanog, Utf1 and Rex‐1. We demonstrate that at least one of the reprogramming factors is bone morphogenetic factor‐4 (BMP4). Pre‐treated keratinocytes could be specifically directed to differentiate into cells of the neuronal lineage. The majority of responsive keratinocytes were the epidermal stem cell population, with a small percentage of transit‐amplifying cells also being affected. Conclusions: Our results suggest that ESC‐CM contains a number of factors, including BMP4, which are capable of reprogramming mouse skin keratinocytes to make them more developmentally potent, as evidenced by their ability to be re‐differentiated into cells of the neuronal lineage. Our findings also imply a continuum of differentiation within the basal keratinocyte population. An increase in developmental potential combined with directed differentiation could increase the therapeutic relevancy of somatic cells.
INTRODUCTION
Both the oocyte and the pre‐implantation embryo provide an environment uniquely rich in cytokines, growth factors and other morphogens. When a somatic cell nucleus is transferred into an enucleated egg, it undergoes a major reprogramming and a resultant increase in developmental potency (Wilmut et al. 1997; Byrne et al. 2003; Gurdon et al. 2003). Likewise, when a somatic cell is hybridized in vitro with an embryonic stem cell (ESC), the fused cell's genome is reset into a pluripotent state (Tada et al. 2001; Flasza et al. 2003; Pells & McWhir 2004; Cowan et al. 2005). When adult thymocytes were fused with ESCs, the inactivated X chromosome from the female thymocyte adopted some characteristics of an active X chromosome, and octamer‐4 (Oct‐4) expression was reactivated after 48 h. The embryonic stem (ES)‐thymocyte hybrid cells exhibited pluripotency, as evidenced by their contribution to all three embryonic germ layers, following blastocyst injection (Tada et al. 2001). Similar results were seen with human T‐lymphoma cells (Flasza et al. 2003), primary mouse brain cells (Pells & McWhir 2004) and human fibroblasts (Cowan et al. 2005). Even more recently, it has been found that cell‐free extracts from pluripotent cells can be used to reprogramme differentiated cells to increase their developmental potency (Collas et al. 2006).
Mouse ESC‐conditioned medium (CM) has been shown to contain several biologically active interleukins, colony‐stimulating factors, chemokines and other growth‐modulatory proteins, which enhanced survival of murine bone marrow myeloid progenitors (Guo et al. 2006). Goat embryonic stem‐like cells remained undifferentiated when cultured in ESC‐CM, indicating that the mouse ESCs secrete autocrine or paracrine factors capable of maintaining pluripotency (Tian et al. 2006). Wnt3a (Singla et al. 2006) and bone morphogenetic factor‐4 (BMP4) (Ying et al. 2003; Qi et al. 2004) have both been found to be independently capable of maintaining murine ESCs in their undifferentiated state. Wnt can function independently of leukaemia inhibitory factor (LIF) and STAT3 to maintain ESC pluripotency (Sato et al. 2004). More recently, CM prepared from cells expressing Wnt3a was shown to replace feeder layers and medium containing LIF in maintaining undifferentiated ESCs (Singla et al. 2006). BMPs are present in a wide variety of stem cell niches that maintain both embryonic and somatic stem cells (Li & Neaves 2006). BMP4 was found to act synergistically with LIF, to support ESC self‐renewal through inhibition of the mitogen‐activated protein kinase (MAPK) pathways, and to decrease upon ESC differentiation (Qi et al. 2004; Palmqvist et al. 2005).
Our laboratory has previously shown that when inter‐follicular epidermal stem cells (EpiSCs) are injected into a developing blastocyst, they are able to generate cell progeny derived from all three embryonic germ layers (Liang & Bickenbach 2002). Similar results have also been seen following blastocyst injection of neural and mesenchymal stem cells (Clarke et al. 2000; Jiang et al. 2002). Although such findings imply that these cells are multipotent, they do not consider their response to the factors secreted from the embryonic environment, which could affect their developmental potential and subsequent differentiation.
In this study, our results suggest that ESCs secrete factors that are capable of altering the gene expression profile of mouse inter‐follicular epidermal keratinocytes to more closely resemble that of ESCs. We found that ESC‐CM induced keratinocytes to express Oct‐4 and its target genes, Sox‐2, Nanog, Utf1 and Rex‐1, which have been shown to be essential for maintaining pluripotency (Ben‐Shushan et al. 1998; Fukushima et al. 1998; Okuda et al. 1998; Nishimoto et al. 1999; Pesce & Scholer 2000; Tomioka et al. 2002; Avilion et al. 2003; Constantinescu 2003; Mitsui et al. 2003; Wang et al. 2004; Nishimoto et al. 2005; Okumura‐Nakanishi et al. 2005; Rodda et al. 2005; Taranger et al. 2005). Our findings support that at least one of these secreted factors is BMP4. We demonstrate that the majority of the keratinocytes induced to express Oct‐4 represent the EpiSC population, with a small number of transient‐amplifying (TA) cells also responding, implying that a continuum of multipotent responsiveness exists within the inter‐follicular basal keratinocyte population. Additionally, we found that the reprogrammed cells could be specifically directed to differentiate into cells in the neuronal lineage, suggesting an increase in their developmental potential.
MATERIALS AND METHODS
Cells
Murine inter‐follicular epidermal basal keratinocytes were isolated from the dorsal and ventral skin of 2‐day‐old Hsd:ICR(CD‐1) mice (Harlan, Indianapolis, IN, USA; http://www.harlan.com) or 2‐day‐old transgenic C57BL/6 mice carrying the enhanced green fluorescent protein (EGFP) transgene (TgN(ACTbEGFP)1Osb) (originally obtained from Jackson Laboratories, Bar Harbor, ME, USA; http://www.jax.org; now bred and maintained at the University of Iowa's Animal Care Facility, Iowa City, IA, USA). All animal tissues were obtained in accordance with the University of Iowa Institutional Animal Care and Use Committee Guidelines, as previously described (Grinnell et al. 2006). Briefly, the skin was incubated in dispase II overnight at 4 °C, then the epidermis was gently separated from the underlying dermis. We have previously shown that with care, the separating plane occurs through the infundibulum, leaving the hair follicles in the dermis. The separated epidermal sheet was placed in 0.25% trypsin for 30 min at 37 °C, then medium was added to inactivate the trypsin; tubes were gently shaken to dissociate individual basal keratinocytes. Keratinocytes were plated on collagen type IV‐coated culture dishes in low‐calcium defined keratinocyte‐serum free medium (DKSFM) (Invitrogen, Grand Island, NY, USA).
Murine W4/129S6 ES cells were a gift from Dr Baoli Yang (the University of Iowa, Gene Targeting Core Facility). ESC were maintained in ESC medium [Knockout Dulbecco's modified Eagle's medium (DMEM) (Invitrogen), supplemented with 2 mm l‐glutamine (Invitrogen), 10 mm minimal essential medium non‐essential amino acids (Invitrogen), 0.1 mmβ‐mercaptoethanol (Sigma, St. Louis, MO, USA), 15% foetal bovine serum (HyClone, South Logan, UT, USA) and 1000 units/mL ESGRO® (LIF) (Chemicon, Temecula, CA, USA)].
Murine dermal fibroblasts were isolated from the body skin of 2‐day‐old ICR neonatal mice (Harlan). Incubation in dispase II overnight was followed by mechanical separation of the dermis from the epidermis. The dermis was then cut into small pieces and was placed in a collagenase solution [175 mg collagenase type I (Invitrogen) in 50 mL high‐glucose DMEM (Invitrogen)] at 37 °C for 1 h with frequent shaking. The solution was then filtered and the fibroblasts were pelleted by centrifugation and plated in DMEM.
Keratinocyte‐ESC co‐cultures
Contact co‐cultures between basal keratinocytes and ESCs were established as follows: 2.5 × 105 GFP+ C57BL/6 keratinocytes in DKSFM were centrally plated on a collagen IV‐coated 2‐well Permanox chamber slide (Nalge Nunc International, Rochester, NY, USA). Four drops containing 5.0 × 104 ESC in ESC medium were placed around the keratinocytes. The cells were left to plate in their respective media for 2 h. Wells were then rinsed and the cells were cultured in a 1 : 1 mixture of DKSFM and ESC medium for 24 or 48 h.
Non‐contact co‐cultures were established using 6‐well Transwell® filter plates (Corning Life Sciences, Acton, MA, USA). 400 000 ICR keratinocytes were seeded on to collagen IV‐coated Transwell filters in DKSFM above non‐collagen‐coated wells containing 3 × 105 ESC in ESC medium for 2 h. The filters and wells were then rinsed and cells were cultured in a 1 : 1 mixture of DKSFM and ESC medium for 24 or 48 h.
Production of CM and CM experiments
W4 ESC (3 × 106) were seeded on to 100‐mm tissues culture dishes in ESC medium. After 24 h, the medium was removed from the cultures and was passed through a 0.22‐µm filter, and was stored at –80 °C until used. It was designated CM.
For experiments, CM or ESC medium not conditioned by cells (uCM) was combined 1 : 1 with DKSFM and was placed on ICR keratinocyte cultures. Keratinocytes were exposed to the media (1 : 1 DKSFM : CM or 1 : 1 DKFSM : uCM) for 24 or 48 h. Respective media were renewed after 24 h.
Immunofluorescence
Immunocytochemistry was performed on cells plated on 2‐well Permanox chamber slides or Transwell filters, excised from their supports. For staining for Oct‐4 and neuronal nuclear protein (NeuN), cells were fixed in 4% paraformaldehyde for 30 min at room temperature, then were rinsed with 1× phosphate‐buffered saline (PBS) before being permeabilized with 0.1% Triton X‐100 for 20 min at room temperature. Following permeabilization, the cells were rinsed with 1× PBS and were blocked in 4% normal goat serum or 12% bovine serum albumin, respectively, for 1 h at room temperature. Cells were then incubated with a rabbit polyclonal antibody directed against Oct‐4 (1 : 750; catalogue no. H‐134, Santa Cruz Biotechnology, Santa Cruz, CA, USA) or a mouse monoclonal antibody directed against NeuN [1 : 600; catalogue no. MAB377; Millipore (Chemicon), Temecula, CA, USA] for 2 h at room temperature. After primary antibody incubation, cells were rinsed with 1× PBS and were incubated with a goat antirabbit or rabbit antimouse AlexaFluor488 antibody (1 : 500; Molecular Probes, Eugene, OR, USA) for 45 min at room temperature. For staining of keratin 14 (K14) and nestin, cells were fixed in ice‐cold 1 : 1 methanol : acetone for 20 min, were then rinsed with 1× PBS and were placed directly in a blocking solution (4% normal goat serum) for 1 h at room temperature, without permeabilization. They were then incubated with a rabbit polyclonal antibody against keratin 14 (1 : 500; catalogue no. PRB‐155P; Covance, Philadelphia, PA, USA) or a rabbit polyclonal directed against nestin (1 : 50; catalogue no. sc‐21249; Santa Cruz Biotechnology), followed by goat antirabbit AlexaFluor488 secondary antibody, as described above for Oct‐4. Co‐staining for bone morphogenetic protein receptor type 1A (BMPR1A) and Oct‐4 was performed by fixing the cells for 30 min in 4% paraformaldehyde at room temperature, permeabilizing in 0.1% Triton X‐100 for 20 min, and then incubating overnight at 4 °C in a combination of Oct‐4 antibody (1 : 750) and a goat polyclonal antibody directed against BMPR1A (1 : 200; catalogue no. sc‐5676S; Santa Cruz Biotechnology). For all staining, secondary antibody was removed through washing with 1× PBS and cells were mounted with VectaShield Mounting Medium with 4′,6‐diamidino‐2‐phenylindole (Vector Laboratories, Burlingame, CA, USA). Images were captured with a SPOT InSight digital camera (Diagnostic Instruments, Sterling Heights, MI, USA) on a Nikon Eclipse E600 epifluorescence microscope (Nikon, Melville, NY, USA).
RNA isolation and reverse transcription‐polymerase chain reaction
Total RNA was isolated from keratinocyte cultures using the RNAqueous kit (Ambion, Austin, TX, USA), according to the manufacturer's instructions. cDNA was generated through reverse transcription reactions consisting of the following: 1.5 µg of RNA; 50 µm Random Primer (Invitrogen); 5× RT buffer (Invitrogen); 0.1 m DTT (Invitrogen); 10 mm dNTP mix (Qiagen, Valencia, CA, USA); 40 IU RNasin (Promega, Madison, WI, USA); 200 IU RTase (Invitrogen) and nuclease‐free diethylpyrocarbonate water (Amresco, Solon, OH, USA). RNA, Random Primer and water were combined and heated at 70 °C for 10 min, then were chilled on ice. Following addition of other components, the RT reaction was incubated at 25 °C for 10 min, 42 °C for 1 h and then 70 °C for 15 min, using a GeneMate Genius thermocycler (ISC Bioexpress, Kaysville, UT, USA), and then samples were chilled on ice.
Two microlitres of cDNA template were then added to 23 µL of the following reaction mixture: 10× PCR Buffer (Qiagen); nuclease‐free diethylpyrocarbonate water; 2.5 mm dNTPs (Invitrogen); 5 IU Taq polymerase (Qiagen) and 10 µm each primer. Primers for Gapdh (GenBank M32599) were 5′‐GACTTCAACAGCAACTCCCAC‐3′ and 5′‐TCCACCACCCTGTTGCTGTA‐3′ (135 bp); primers for Oct‐4 (GenBank NM013633) were 5′‐TGGAGACTTTGCAGCCTGAG‐3′ and 5′‐TGAATGCATGGGAGAGCCCA‐3′ (800 bp); primers for Nanog (GenBank AY278951) were 5′‐AGGGTCTGCTACTGAGATGCTCTG‐3′ and 5′‐CAACCACTGGTTTTTCTGCCACCG‐3′ (360 bp); primers for Utf1 (GenBank NM009482) were 5′‐ACGTGGAGCATCTACGAGGT‐3′ and 5′‐TAGACTGGGGGTCGTTTCTG‐3′ (154 bp); primers for Rex‐1 (GenBank NM009556) were 5′‐GGCCAGTCCAGAATACCAGA‐3′ and 5′‐GAACTCGCTTCCAGAACCTG‐3′ (232 bp); primers for Sox‐2 (GenBank NM011443) were 5′‐TAGCACTTGTTGCCAGAACG‐3′ and 5′‐AAGCCGCTCTTCTCTTTTCC‐3′ (340 bp); primers for K14 (GenBank NM016958) were 5′‐CTAGCCGCATGTCCTCCATC‐3′ and 5′‐GCAGGAGGACATTGGCATTG‐3′ (513 bp). All PCR reactions were performed for 35 cycles.
Isolation of EpiSCs and TA cells
Basal keratinocytes were isolated from the backskin of 2‐day‐old ICR mice, as previously described (Grinnell et al. 2006). Following centrifugation, the cells were re‐suspended at 5 × 106 cells/mL in SMEM (Invitrogen) with 0.05 mm Ca2+, 7% chelexed foetal bovine serum, 1 mm 4‐(2‐hydroxyethyl)‐1‐piperazineethanesulfonic acid, 1% antibiotic/antimycotic (Invitrogen) and 5 µg/mL Hoechst 33342 (Sigma). The keratinocytes were then incubated in the medium/Hoechst dye mixture for 90 min at 37 °C, were centrifuged, re‐suspended in 1 µg/mL propidium iodide in medium, and then placed on ice until sorting.
Cell sorting was performed on a Becton Dickinson FACS DiVa, configured to set non‐rectilinear sort gates as shown in the bitmap histograms (Fig. S2). Forward and orthogonal scatter signals were generated using 100 mW of ultraviolet radiation on 488 nm Hoechst 33342 and propidium iodide at 351–364 nm (Liang & Bickenbach 2002). For these experiments, the keratinocytes were sorted into four populations (SC small, SC large, TA small and TA large). Hoechst fluorescence was measured through a 440/60 nm band‐pass filter and propidium iodide was measured through a 670/14 nm filter. All parameters were collected using linear amplification in listmode. The total EpiSC population was first sorted using a bitmap gate set to collect those cells with the lowest level of Hoechst fluorescence (4.6% of the basal population), and the TA cell population was collected using a gate set to collect those that retained the dye (30.9% of the basal cells) (Fig. S2A). From these gates, cells were then further sorted according to size. Two populations of stem cells were collected: SC small (1.4% of the basal cells; 34.0% of the SC Hoechst gate) and SC large (1.6% of the basal cells; 30.0% of the SC Hoechst gate) (Fig. S2B). Two populations of TA cells were also collected: TA small (18.4% of the basal cells; 59.8% of the TA Hoechst gate) and TA large (7.4% of the basal cells; 23.9% of the TA Hoechst gate) (Fig. S2C).
Neuronal differentiation studies
Medium conditions for the neuronal differentiation of keratinocytes were adopted from Jiang et al. (2003) and are detailed in Grinnell et al. (2006), and consisted of 60% DMEM‐low glucose (Invitrogen) and 40% MCDB‐201 (Sigma), supplemented with 1× insulin‐transferrin‐selenium (Sigma), 1× linoleic acid‐albumin (Sigma), 10−9 m dexamethasone (Sigma) and 104 ascorbic acid (Sigma). The basal medium was then supplemented with 100 ng/mL basic fibroblast growth factor (Sigma), 100 ng/mL sonic hedgehog (Cell Sciences), 10 ng/mL fibroblast growth factor‐8 (Cell Sciences) and 10 ng/mL brain‐derived neurotrophic factor (Cell Sciences). Following initial 24 h exposure to control conditions, P0 inter‐follicular keratinocytes were plated in collagen IV‐coated dishes, controls in 100% DKSFM or 1 : 1 DKSFM : uCM, and experiments in 1 : 1 DKSFM : CM or 1 : 1 DKSFM : uCM +100 ng/mL BMP4 for 24 h. Cells cultured in 100% DKSFM remained in this keratinocyte‐specific growth medium for 10 days. The other three keratinocyte groups received neuronal differentiation medium. Both the neuronal medium, with growth factors, as well as the control DKSFM, were renewed every 3 days.
BMP4/BMP2 studies
Primary ICR keratinocytes were cultured in DKSFM +100 ng/mL recombinant human BMP4 (R & D Systems Inc., Minneapolis, MN, USA) or a 1 : 1 mixture of DKSFM : uCM +100 ng/mL rBMP4. The medium, containing BMP4, was renewed every day. After the desired length of time, cells exposed to BMP4 were harvested and used for isolation of either RNA or protein. All experiments were repeated with 100 ng/mL human recombinant BMP2 (R & D Systems Inc.) and 100 ng/mL Wnt3a (R & D Systems Inc.), using the same method.
Immunoblotting
For Western blot analysis of nestin, NeuN, GluR1, NR1, K14 and actin, total protein extract was collected using radioimmunoprecipitation buffer (Pierce Biotechnology Inc., Rockford, IL, USA), supplemented with a protease inhibitor cocktail (Sigma, catalogue no. P8340) and two phosphatase inhibitor cocktails (Sigma, catalogue no. P2850, P5726). Analysis of soluble BMP4 and LIF was performed using protein purified from harvested ESC‐CM. Briefly, the conditioned culture medium was filtered through a 0.22‐µm filter, was frozen and was concentrated by lyophilization. Protein concentrations were then determined using a standard Bradford assay; 50 µg of each sample was loaded onto a 12% polyacrylamide gel and was run at 200 V for 1 h. The protein was then transferred to a nitrocellulose membrane at 100 mAmp at 4 °C for 12 h. After transfer, the membrane was blocked in a milk solution (4% non‐fat dry milk and 0.3% Tween‐20 in 1× PBS) for 2 h at room temperature. The membrane was then incubated with the appropriate primary antibody for 2 h at room temperature. Nestin (1 : 200), NeuN (1 : 500) and K14 (1 : 500) antibodies used were the same as listed above for immunocytochemistry. Actin was detected with a mouse monoclonal antibody used at a concentration of 1 : 1000 (catalogue no. AC‐40, Sigma). Tyrosine hydroxylase was detected with a rabbit polyclonal antibody used at a concentration of 1 : 1000 (catalogue no. PRB‐515P, Covance). The GluR1 and NR1 antibodies were a generous gift from Dr Johannes Hell (Department of Pharmacology, University of Iowa). The GluR1 antibody was made from a glutathione S‐transferase fusion protein of C‐terminal fragment of the GluR1 subunit of the α‐amino‐3‐hydroxy‐5‐methylisoxazole‐4‐propionic acid (AMPA) receptor. It is a serum antibody raised in rabbit and was used at a concentration of 1 : 1000. NR1 antibody is mouse monoclonal ascites‐derived and was used at a concentration of 1 : 1000. After primary antibody exposure, the membrane was washed and was incubated with an appropriate infrared secondary antibody (IRDye TM800 antirabbit IgG, 1 : 1000, 611‐132‐122; IRDye TM800 antimouse IgG, 1 : 1000, 610‐132‐121; Rockland Inc., Gilbertsville, PA, USA) for 45 min at room temperature. Following removal of secondary antibody, protein was detected with the use of an Odyssey infrared imaging system (LI‐COR Biotechnology, Lincoln, NE, USA). Densitometry analysis was performed using Odyssey infrared imaging software (LI‐COR Biotechnology).
RESULTS
Medium conditioned by mouse embryonic stem cells is sufficient to activate embryonic genes in inter‐follicular basal keratinocytes isolated from mouse epidermis
We had previously shown that when EpiSCs were injected into developing blastocysts, the resulting embryos exhibited differentiated cells derived from the injected keratinocytes (Liang & Bickenbach 2002). To determine whether direct contact with or close proximity to embryonic cells was required for keratinocytes to be reprogrammed, we examined mouse skin keratinocytes grown in contact and non‐contact co‐cultures with mouse ESCs for expression of Oct‐4.
For contact co‐cultures, clusters of mouse W4 ESCs (P21) were plated around a central colony of neonatal keratinocytes isolated from mice carrying the EGFP transgene (P0), in a 2‐well chamber slide, and were allowed to grow together. After 48 h in co‐culture, 1.6% of keratinocytes expressed Oct‐4 (Fig. S1). No staining of junctional proteins, E‐cadherin, desmoglein 1–3, zonula occludins‐1 (ZO‐1), or connexin 43 was seen between ESC and keratinocytes, although staining was seen between like cells (ESC–ESC, keratinocyte–keratinocyte) (data not shown). Thus continuous junctional attachment between the two cell types is not necessary to activate Oct‐4.
To determine if ESCs secreted factors that could activate Oct‐4 in keratinocytes, we plated mouse keratinocytes above mouse ESCs in Transwells®. By 24 h, ~4% of the keratinocytes cultured over the ESCs exhibited punctate nuclear staining of Oct‐4 (Fig. 1a–c). By 48 h, 4.5% of keratinocytes clearly expressed nuclear Oct‐4 (Fig. 1d–f), while none of the control keratinocytes cultured only in DKSFM (Fig. 1j–l) or in a 1 : 1 mixture of the keratinocyte medium, DKSFM and the ESC medium (not conditioned by cells) (1 : 1 DKFSM : uCM) (Fig. 1m–o) expressed Oct‐4. These findings suggest that ESCs secrete factor(s) that can stimulate Oct‐4 expression in mouse skin keratinocytes.
Figure 1.
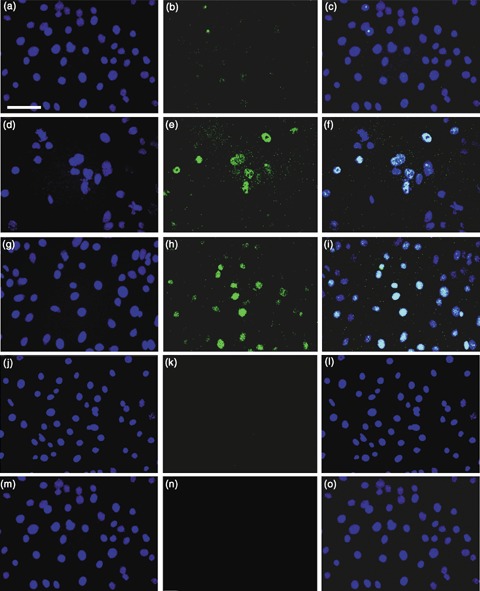
Expression of Oct‐4 in mouse keratinocytes in non‐contact co‐culture with mouse ESC or in cultures exposed to CM. (a–c) 24 h transwell cultures of P0 keratinocytes in 1 : 1 DKSFM : uCM above P22 W4 ESC. (a) DAPI (4′,6‐diamidino‐2‐phenylindole); (b) Anti‐Oct‐4; (c) Merged. Note, punctate nuclear Oct‐4 staining in ~4% of keratinocytes. (d–f) 48 h transwell cultures. (d) DAPI; (e) anti‐Oct‐4; (f) merged. Note, nuclear Oct‐4 in ~4.5% of keratinocytes. (g–i) Keratinocytes treated with CM for 48 h. (g) DAPI; (h) anti‐Oct‐4; (i) merged. Note, nuclear Oct‐4 in ~4.2% of keratinocytes. (j–l) Control P0 neonatal mouse keratinocytes grown in DKSFM without mouse ESC. (j) DAPI; (k) anti‐Oct‐4; (l) merged. (m–o) Control P0 keratinocytes grown in 1 : 1 DKSFM : uCM without mouse ESC. (m) DAPI; (n) anti‐Oct‐4; (o) merged. Note absence of Oct‐4 in control cultures. ×40 magnification. Scale bar = 50 µm.
In the Transwell experiments, ESCs were in close proximity to the keratinocytes and likely secreting Oct‐4‐inducing factor(s) continually into the medium. To determine if keratinocytes required close proximity to ESCs in order for the ESC‐secreted factor(s) to be effective and if the factor(s) were stable, we investigated whether ESC‐CM alone could activate Oct‐4 and its target genes in mouse keratinocytes. After a 48‐h exposure to CM, 4.2% of the keratinocytes expressed Oct‐4 (Fig. 1g–i), while none of the keratinocytes cultured in the control 1 : 1 DKSFM : uCM medium expressed Oct‐4. Reverse transcription‐polymerase chain reaction demonstrated that CM also stimulated the keratinocytes to express the Oct‐4 target genes, Nanog, Sox‐2, Utf1 and Rex‐1. By 24 h in the DKSFM : CM mixture, keratinocytes expressed Oct‐4 and all of its target transcripts (Fig. 2a). None of the transcripts were expressed when the keratinocytes were cultured in DKSFM : uCM (Fig. 2b). These results confirm that ESCs secrete factor(s) that are capable of reprogramming mouse keratinocytes to express Oct‐4, that the ESCs need not be within close proximity of keratinocytes, and that the factor(s) is/are stable in the medium.
Figure 2.
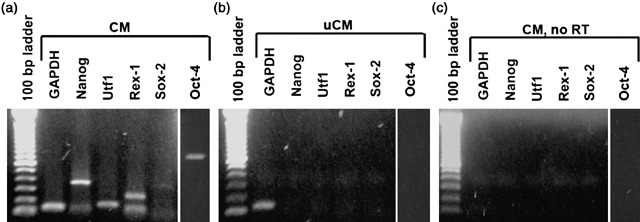
Embryonic gene expression in keratinocytes exposed to CM. (a) P0 neonatal keratinocytes expressed Oct‐4 (800 bp), Nanog (360 bp), Utf1 (154 bp), Rex‐1 (232 bp) and Sox‐2 (340 bp) after 24 h exposure to CM. (b) Keratinocytes, after 24 h exposure to uCM, did not express embryonic transcripts. (c) Control RT‐PCR without reverse transcriptase (RT).
BMP4 is present in CM and can stimulate embryonic gene expression and suppress K14 expression when added to keratinocyte cultures
In an attempt to identify what factor(s) was/were secreted by ESCs, we performed medium fractionation and two‐dimensional gel electrophoresis. Although these two approaches were inconclusive, they suggested that the factor(s) had a molecular weight of approximately 40–50 kDa. Using this information and a candidate approach, we identified two potential factors: Wnt3a (39 kDa) and BMP4 (25 kDa monomer; secreted as a 50 kDa dimer). Addition of 100 ng/mL of Wnt3a to P0 neonatal mouse keratinocytes resulted in no expression of Oct‐4 or its target genes (data not shown). When 100 ng/mL of BMP4 was added to keratinocytes (in 1 : 1 DKSFM : uCM), Oct‐4 and its target genes were expressed in 4.0% of the cells (Fig. 3c,d). No activation of Oct‐4 was found with 50 ng/mL BMP4 (Fig. 3b). Because a BMP‐like factor was reported present in bovine serum (Kodaira et al. 2006), we tested BMP4 activation of Oct‐4 and its targets with and without serum. Levels of Nanog, Utf1 and Rex‐1 did not differ under these conditions, and Sox‐2 and Oct‐4 expression levels were only slightly decreased (Fig. 3d). The absence of Oct‐4 and its downstream targets in cultures treated with uCM, which contains serum, confirmed that any BMP‐like factors in serum could not alone activate embryonic gene expression.
Figure 3.
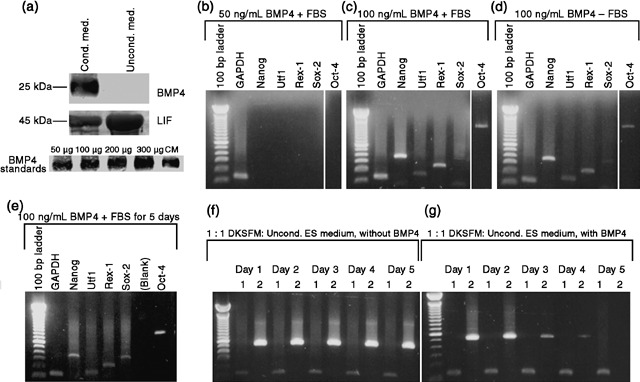
Bone morphogenetic factor‐4 (BMP4) in CM and effects of recombinant BMP4 on keratinocyte gene expression. (a) Immunoblot analysis of conditioned (CM) and unconditioned (uCM) media show BMP4 in the CM (24.6 ng/mL). LIF was 2.2‐fold higher in uCM than in CM. Various concentrations of recombinant BMP4 were used as standards by which to quantify the amount of BMP4 present in the CM. 3 mL lyophilized medium loaded per well. (b–g) RT‐PCR; (b) no activation of Oct‐4 or its target transcripts in keratinocytes exposed to 50 ng/mL BMP4 in the presence of FBS. (c) 100 ng/mL of BMP4 activated Oct‐4, Nanog, Utf1, Rex‐1 and Sox‐2 in the presence and (d) absence of FBS. (e) After 5 days exposure to 100 ng/mL BMP4, keratinocytes still expressed Oct‐4, Nanog, Utf1, Rex‐1 and Sox‐2. (f) Keratinocytes cultured in 1 : 1 DKSFM : uCM for 5 days without BMP4 expressed K14. (g) In keratinocytes cultured in 1 : 1 DKSFM : uCM, plus 100 ng/mL BMP4 (renewed daily), for 5 days, K14 was down‐regulated by Day 3, and extinguished by day 5. (f–g) Lane 1 = GAPDH (135 bp); Lane 2 = Keratin 14 (K14) (513 bp).
Bone morphogenetic factor‐4 was present in a concentration of 24.6 ng/mL in CM as determined by Western blot (Fig. 3a), with densitometry analysis. LIF was present in uCM and CM, but at a 2.2‐fold lower concentration in the CM. This is likely due to the fact that LIF is used by ESCs in culture; it must be replenished every 24 h for ESCs to remain undifferentiated. Its higher presence in the uCM confirms that LIF alone cannot activate Oct‐4 in mouse keratinocytes.
In order to examine the transcriptional effects of prolonged exposure to BMP4, mouse keratinocytes were cultured in the presence of the cytokine for 5 days. 100 ng/mL BMP4 was added to 1 : 1 DKSFM : uCM and placed on P0 keratinocytes, and was renewed every 24 h for 5 days. Oct‐4 and its target genes were expressed by 24 h (Fig. 3d), and remained expressed for 5 days of BMP4 exposure (Fig. 3e). Expression of the basal keratinocyte marker, K14, was found at roughly the same level in the control 1 : 1 DKSFM : uCM (Fig. 3f) and the experimental BMP4‐treated cultures (Fig. 3g) for the first 48 h. However, K14 expression was down‐regulated in BMP4‐treated cells by 72 h, and by day 5, it was undetectable (Fig. 3g). K14 was maintained in the control cultures (Fig. 3f). These results imply that continued exposure of mouse keratinocytes to BMP4 results in transcriptional changes, such as down‐regulation of the basal keratinocyte‐specific marker, K14, and the maintenance of Oct‐4 and its target embryonic transcripts.
In order to assess if keratinocytes undergo a permanent change in gene expression following exposure to BMP4 or if the response is dependent upon the continuous presence of BMP4, we exposed keratinocytes to 100 ng/mL of BMP4 for 24 h, then rinsed them with PBS, and cultured them in 1 : 1 DKSFM : uCM for 5 days. At 24 and 48 h after removal of BMP4, Nanog, Utf1 and Rex‐1 were still expressed; Sox‐2 was also expressed, but at a comparatively lower level (Fig. 4a,b). Oct‐4, however, only persisted for the first 24 h following BMP4 removal; it was gone at 48 h (Fig. 4a,b). Seventy‐two hours after BMP4 removal, Nanog and Sox‐2 were absent, while Utf1 and Rex‐1 continued to be expressed (Fig. 4c). Ninety‐six hours post‐BMP4, only Rex‐1 was barely detectable (Fig. 4d). At 120 h, no Oct‐4 target genes were detectable (Fig. 4e). Expression of K14 transcript is also re‐expressed at control levels by 120 h post‐BMP4 (Fig. 4f). Thus, continued exposure to BMP4 is necessary for keratinocytes to maintain expression of Oct‐4 and its target genes and to suppress expression of the basal keratinocyte marker, K14.
Figure 4.
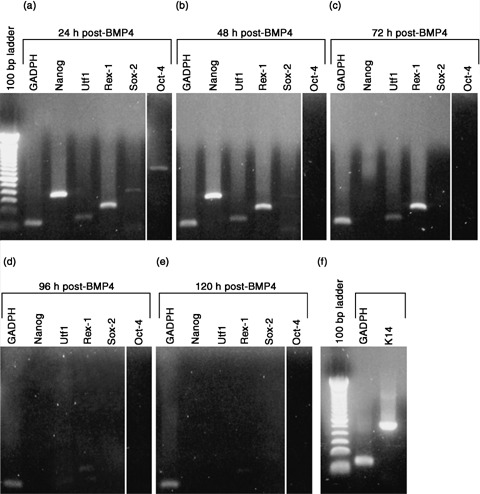
Embryonic gene expression in keratinocytes following BMP4 removal. P0 neonatal keratinocytes were exposed to 100 ng/mL BMP4 (in 1 : 1 DKSFM : uCM) for 24 h. After 24 h, BMP4 was removed and expression of Oct‐4 and its target genes examined over 5 days. (a) 24 h after removal of BMP4, Oct‐4, Nanog, Utf1, Rex‐1 and Sox‐2 were still expressed. (b) 48 h post‐BMP4, Oct‐4 was no longer expressed, but Nanog, Utf1, Rex‐1 and Sox‐2 persisted. (c) 72 h post‐BMP4, Oct‐4 remained off; Nanog and Sox‐2 were extinguished; Utf1 and Rex‐1 were still present. (d) 96 h post‐BMP4, Utf1 was extinguished, while Rex‐1 was barely detectable. (e) 120 h post‐BMP4, no embryonic transcripts were expressed. (f) K14 transcript level is restored to control by 120 h post‐BMP4.
CM‐ and BMP4‐responsive keratinocytes co‐express membrane BMPR1A and Oct‐4 in culture
We reasoned that if BMP4 activated Oct‐4 expression, then keratinocytes that were induced to express Oct‐4 should also express the BMP4 receptor, bone morphogenetic protein receptor type 1A (BMPR1A). We analysed the numbers of Oct‐4+ BMP1A+ cells in cultures of keratinocytes treated for 24 h with ESC‐CM or with BMP4‐treated medium. We found a similar number of cells induced to express nuclear Oct‐4, 6.3% of cells treated with CM and 5.0% of cells treated with BMP4 (Fig. 5c,h; Table 1). Unexpectedly, we found that fewer cells treated with BMP4 expressed BMPR1A than the cells treated with CM. 47% of Oct‐4+ cells in the BMP4‐treated cultures co‐expressed membranous BMPR1A, whereas 95% of CM‐treated cells co‐expressed Oct‐4 and BMPR1A (Fig. 5a–j; Table 1). Although 35.0% of BMP4‐treated cultures exhibited diffuse cytoplasmic BMPR1A staining, and 6.9% exhibited bright cytoplasmic staining, neither showed Oct‐4 expression (Fig. 5f–j; Table 1). Cells cultured under control conditions showed no surface BMPR1A or nuclear Oct‐4 expression (Fig. 5k–t; Table 1), although a small percentage of control cells exhibited some cytoplasmic BMPR1A staining (Fig. 5l,q; Table 1).
Figure 5.
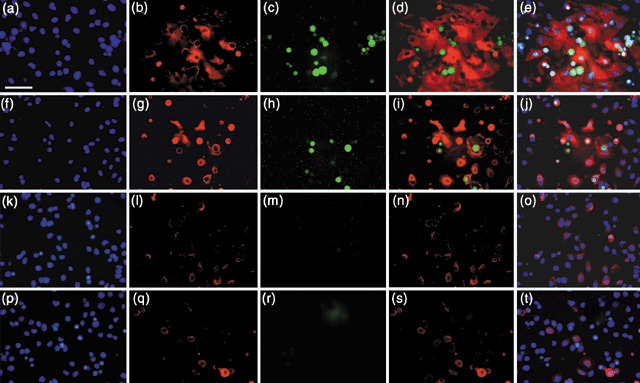
Expression of BMPR1A and Oct‐4 in keratinocytes following exposure to CM or recombinant BMP4. (a–e) Keratinocytes cultured in 1 : 1 DKSFM : CM for 24 h. (a) DAPI (4′,6‐diamidino‐2‐phenylindole); (b) anti‐BMPR1A; (c) anti‐Oct‐4; (d) merge BMPR1A and Oct‐4; (e) merge BMPR1A, Oct‐4 and DAPI. (f–j) Keratinocytes cultured in 1 : 1 DKSFM : uCM +100 ng/mL rBMP4, for 24 h. (f) DAPI; (g) anti‐BMPR1A; (h) anti‐Oct‐4; (i) merge BMPR1A and Oct‐4; (j) merged BMPR1A, Oct‐4 md DAPI. In a–j, note co‐staining of membrane BMPR1A and Oct‐4, and ESC‐like rounded morphology. Additionally, note small, rounded cells with bright cytoplasmic BMPR1A, but no Oct‐4, suggesting BMPR1A is expressed before Oct‐4. (k–o) Control 24 h DKSFM cultures. (k) DAPI; (l) anti‐BMPR1A; (m) anti‐Oct‐4; (n) merge BMPR1A and Oct‐4; (o) merge BMPR1A, Oct‐4 and DAPI. (p–t) Control 24 h 1 : 1 DKSFM : uCM cultures. (p) DAPI; (q) anti‐BMPR1A; (r) anti‐Oct‐4; (s) merge BMPR1A and Oct‐4; (t) merge BMPR1A, Oct‐4 and DAPI. Note, no Oct‐4 or membrane BMPR1A staining in controls. All images, ×40. Scale bar = 50 µm.
Table 1.
Expression of BMPR1A and Oct‐4 in cells treated with CM or recombinant BMP4
| 100% DKSFM | 1 : 1 DKSFM : uCM | 1 : 1 DKSFM : CM | 1 : 1 DKSFM : uCM + 100 ng/mL BMP4 | |
|---|---|---|---|---|
| % of Oct‐4‐positive cells co‐expressing membrane BMPR1A | 0.0 | 0.0 | 95.2 ± 8.3 | 47.2 ± 8.1 |
| % of cells expressing Oct‐4 | 0.0 | 0.0 | 6.3 ± 0.01 | 5.0 ± 0.80 |
| % of cells expressing diffuse cytoplasmic BMPR1A (no Oct‐4) | 5.2 ± 0.02 | 4.8 ± 0.12 | 75.3 ± 0.62 | 35.0 ± 0.65 |
| % of cells expressing bright cytoplasmic BMPR1A (no Oct‐4) | 0.8 ± 0.11 | 0.3 ± 0.20 | 1.2 ± 0.03 | 6.9 ± 2.8 |
means ± SD, n = 4 for each condition. BMPR1A, bone morphogenetic protein receptor type 1A; CM, conditioned medium; DKSFM defined keratinocyte‐serum free medium; Oct‐4, octamer‐4; uCM, unconditioned medium.
Additionally, we noted a morphological change in keratinocytes that co‐expressed surface BMPR1A and Oct‐4, and in keratinocytes with bright cytoplasmic BMPR1A. They seemed to lie on top of the monolayer and exhibited a morphology resembling that of murine ESCs, with reduced cytoplasmic : nuclear ratio and a smaller, rounded cell shape. These results suggest the existence of a subpopulation of mouse inter‐follicular basal keratinocytes that are capable of responding to ESC cues, one of which is BMP4. Such findings imply a relationship between signalling through the BMPR1A and expression of the embryonic transcription factor, Oct‐4.
Keratinocytes pre‐treated with CM or BMP4 can be directed towards a neuronal lineage in response to neuronal differentiation culture conditions
Because keratinocytes treated with CM or BMP4 responded by up‐regulation of Oct‐4 and its target embryonic transcripts, it suggested that these cells could be directed towards an alternative cell lineage. To investigate this, we first exposed keratinocytes to CM or to recombinant BMP4, then grew these treated cells in medium containing neuronal growth factors.
When the keratinocytes treated with CM were first exposed to neuronal growth medium, they exhibited the same cobblestone‐like morphology as the control populations (Fig. 6a,b,k,o). However, by day 7, the cells along the edges of the clusters became elongated, with multiple processes projecting out from the cell body (Fig. 6c). By day 10, these cells had greatly increased in number and populated ~90% of the dish (Fig. 6d). Higher magnification suggested that cells appeared to be in contact with one another through a network of processes (Fig. 6e), suggestive of dendrites seen in neuronal cultures. Keratinocytes exposed to 100 ng/mL BMP4 for 24 h, then placed in neuronal medium underwent a similar phenotypic transformation (Fig. 6f–j). By day 10, BMP4‐treated cultures were filled with cells possessing polygonal cell bodies and long processes (Fig. 6i). Higher magnification revealed areas of contact between projections of some cells and cell bodies of others (Fig. 6j), a property commonly seen in neuronal cultures, but never seen in keratinocyte cultures. Control keratinocytes grown in DKSFM exhibited a typical keratinocyte growth pattern of cobblestone‐like clusters, becoming completely confluent by day 10 (Fig. 6k–n). Control keratinocytes cultured in 1 : 1 DKSFM : uCM for 24 h, then placed in the neuronal differentiation medium exhibited a growth pattern similar to the DKSFM control cells at first (Fig. 6o–p). However, these keratinocytes underwent a rapid burst of growth, overcrowding the dish by day 7, lifting off the dish by day 10, leaving only large keratinocyte‐like cells behind (Fig. 6q–r).
Figure 6.
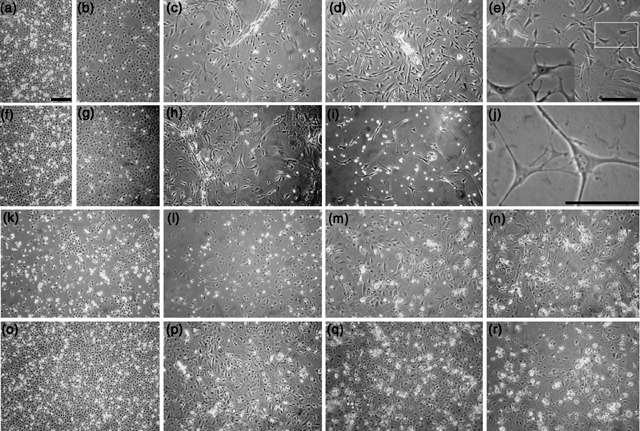
Neuronal morphology exhibited by keratinocytes exposed to CM or recombinant BMP4. Neonatal mouse skin keratinocytes were plated in 100% DKSFM for 24 h and cultured under the following conditions for 24 additional hours, then all cells exposed to neuronal differentiation medium for 10 days (note, days below refer to days in differentiation medium). (a–e) Keratinocytes treated with 1 : 1 DKSFM : CM. (a) Day 1; (b) day 3; (c) day 7; (d) day 10; (e) day 10, ×20 magnification. Insert represents enlargement of boxed area in e. (f–j) Keratinocytes treated with 1 : 1 DKSFM : uCM + BMP4 (f) Day 1; (g) day 3; (h) day 7; (i) day 10; (j) day 10, ×40 magnification. (k–n) Control keratinocytes cultured in 100% DKSFM for 10 days. (k) Day 1; (l) day 3; (m) day 7; (n) day 10. (o–r) Control, keratinocytes cultured in 1 : 1 DKSFM : uCM for 24 h, then cultured in neuronal differentiation medium for 10 days. (o) Day 1; (p) day 3; (q) day 7; (r) day 10. All images (except d and j) ×10 magnification. Scale bars = 200 µm.
Immunoblot analysis of these cultures at day 10 revealed that the cells’ phenotypic changes were accompanied by changes in protein expression to those of neuronal proteins, nestin and NeuN, the presynaptic AMPA receptor subunit, GluR1, and the postsynaptic NMDA receptor subunit, NR1, as well as dopaminergic enzyme, tyrosine hydroxylase (TH). Keratinocytes pre‐treated with CM or BMP4, not only changed their phenotype, but also expressed all of these neuronal proteins after 10 days of culture in neuronal differentiation medium (Fig. 7, lanes 2 and 3) with similar relative expression levels (Table S1). Control keratinocytes showed no neuronal protein expression (Fig. 7, lanes 1 & 4). The positive control, mouse ESCs cultured in the neuronal medium, also expressed the neuronal markers (Fig. 7, lane 5), whereas the fibroblast‐negative control was negative for nestin, NeuN, GluR1 and NR1, with a very faint band corresponding to TH (Fig. 7, lane 6). The negative results with fibroblasts demonstrate that elongated cells seen in experimental cultures are neuronal in lineage, rather than fibroblastic. Immunocytochemical analysis using antibodies to nestin, NeuN and K14 confirmed that the cells exhibiting a neuronal phenotype were the same cells expressing neuronal markers (Fig. 8). After pre‐treatment with CM or recombinant BMP4, followed by culture in neuronal differentiation medium for 7 days, cells with a neuronal‐like morphology, as well as cells emerging along the edge of clusters, expressed nestin (Fig. 8a–c,j). NeuN was seen primarily in the nuclei of the neuronal‐like cells (Fig. 8d–f). K14, present as filaments in the keratinocyte clusters, was absent in most neuronally differentiated cells, with a few cells showing perinuclear clumps (Fig. 8g–i,k).
Figure 7.
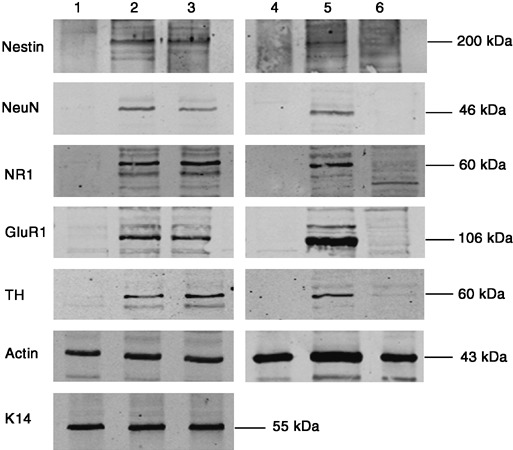
Expression of neuronal proteins and receptors in cells exposure of CM or BMP4, then cultured in neuronal differentiation medium. Western blot analysis of nestin, neuronal nuclear protein (NeuN), NMDA receptor 1 (NR1), glutamate receptor 1 (GluR1), tyrosine hydroxylase (TH) and actin. Lane 1 = Control, keratinocytes exposed to 1 : 1 DKSFM : uCM for 24 h, then neuronal differentiation medium for 10 days. Lane 2 = Keratinocytes exposed to 1 : 1 DKSFM : CM for 24 h, then neuronal differentiation medium for 10 days. Lane 3 = Keratinocytes exposed to 1 : 1 DKSFM : uCM +100 ng/mL BMP4, for 24 h, then neuronal differentiation medium for 10 days. Lane 4 = control, keratinocytes grown in 100% DKSFM for 10 days. Lane 5 = P22 W4 ESC grown in ESC medium for 24 h, then neuronal differentiation medium for 10 days. Lane 6 = Skin fibroblasts grown in DMEM for 10 days. 50 µg of protein loaded per lane.
Table 2.
Percentage of cells expressing Oct‐4 under various conditions
| Treatments | % cells expressing Oct‐4 protein |
|---|---|
| 100% DKSFM | 0.0 a |
| 1 : 1 DKSFM : uCM | 0.0 a |
| 1 : 1 DKSFM : ES‐CM | 4.2 ± 0.04 a |
| 50 ng/mL BMP4 | 0.0 a |
| 100 ng/mL BMP4 | 4.0 ± 0.30 a |
| 50 ng/mL BMP2 | 3.7 ± 2.11 b |
| 100 ng/mL BMP2 | 4.4 ± 2.08 b |
| 50 ng/mL BMP2 +50 ng/mL BMP4 | 7.9 ± 4.70 a |
| 100 ng/mL BMP2 +100 ng/mL BMP4 | 8.8 ± 5.77 a |
| Keratinocyte : ES cell contact co‐culture | 1.6 ± 0.02 a |
| Keratinocyte : ES cell non‐contact co‐culture | 4.5 ± 0.04 a |
means ± SD, a n = 4, b n = 2. BMP, bone morphogenetic protein; CM, conditioned medium; DKSFM defined keratinocyte‐serum free medium; ES, embryonic stem; Oct‐4, octamer‐4; uCM, unconditioned medium.
Figure 8.
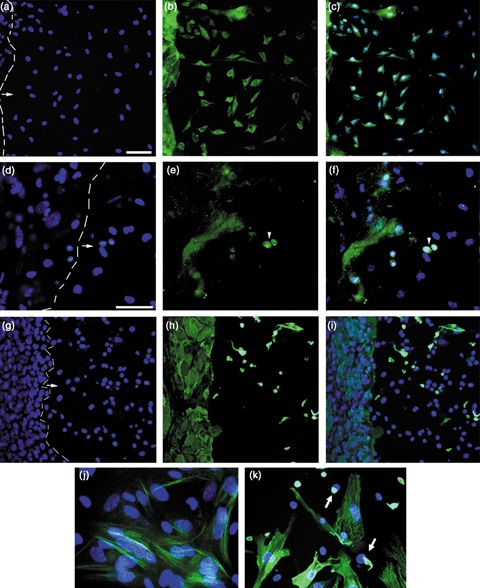
Expression of nestin, NeuN and K14 in cells grown for 7 days in neuronal differentiation medium. Keratinocytes were exposed to CM for 24 h then cultured in neuronal differentiation medium for 7 days. Dashed line marks border between keratinocyte colony and emerging neuronal‐type cells. Arrows indicate direction of neuron‐like cell migration. (a–c) Cultures stained with antibody to nestin. ×20 magnification. (a) DAPI (4′,6‐diamidino‐2‐phenylindole); (b) anti‐nestin; (c) merged. (d–f) Cultures stained with antibody to NeuN. ×40 magnification. (d) DAPI; (e) anti‐NeuN; (f) merged. Arrowheads mark NeuN+ nuclei. Note, multilayered colony edges showed non‐specific accumulation of NeuN antibody. (g–i) Cultures stained with antibody to keratin 14 (K14). ×20 magnification. (g) DAPI; (h) anti‐K14; (i) merged. Note, in the few K14+ neuronal‐type cells, the K14 filaments appeared collapsed around the nuclei. (j) ×60 magnification of antinestin/DAPI. (k) ×60 magnification of anti‐K14/DAPI. Arrows point to perinuclear clumps of keratin in cells. Scale bars = 50 µm.
Thus, the pre‐treatment with CM or recombinant BMP4 allowed the mouse skin keratinocytes to respond to neuronal differentiation factors in culture, by acquiring neuronal‐type morphology, with polygonal cell bodies and multiple projections (Fig. 6), and expressing two standard neuronal markers, nestin and NeuN, as well as the synaptic receptor proteins, GluR1 and NR1, and the dopamine‐associated enzyme, TH (Fig. 7). Such findings suggest that somatic cells can be directed towards alternative cell lineages.
Continuum of CM‐responsive inter‐follicular keratinocytes
Because only 4–8% of keratinocytes responded to the pre‐treatments (Table 2), we investigated whether the responsive cells were restricted to the EpiSC population. We modified our previously published methodology to sort EpiSC and TA cells (Liang & Bickenbach 2002). We fractionated the skin basal keratinocytes into four groups by performing a four‐way flow cytometric sort, based on exclusion of the DNA dye, Hoechst 33342 and cell size (Supplemental Fig. 2). Two populations of EpiSCs, EpiSC small and EpiSC large, and two populations of TA cells, TA small and TA large, were collected, cultured and either maintained in control media or pre‐treated with CM for 24 h. The CM pre‐treatment resulted in Oct‐4 expression in 96.0% of the EpiSC small and 30.5% of the EpiSC large cells, whereas only 7.2% of the TA small cells and none of the TA large cells expressed Oct‐4. This finding suggests that the responsive population is primarily of EpiSCs, as defined by Hoechst dye exclusion. It also suggests that a continuum of CM‐responsive cells may exist in the inter‐follicular keratinocyte population, which could be exploited for therapeutic use.
DISCUSSION
It was not clear whether somatic stem cells, injected into blastocysts, required junctional formation with embryonic cells in order to be reprogrammed (Krause et al. 2001; Liang & Bickenbach 2002). The data we present here provide evidence that cell–cell contact is not necessary. Rather, ESCs secrete relatively stable factor(s) into culture medium that are capable of transcriptionally reprogramming keratinocytes at a distance. One factor, BMP4, was present in CM at 24.6 ng/mL. It is unlikely that BMP4 is the sole reprogramming factor in CM, especially because we had to use 100 ng/mL recombinant BMP4 to reprogramme keratinocytes. Given the overlapping functions of BMP2 and BMP4 in vivo (Hager‐Theodorides et al. 2002), and the role of BMP2 in maintaining pluripotency (Wiese et al. 2006), we also assessed Oct‐4 activation in keratinocytes treated with BMP2. The results were similar to BMP4 (Table 2). Interestingly, combining BMP4 and BMP2 yielded twice the number of Oct‐4+ cells (8.8%), suggesting an additive effect. However, because BMP2 was only 2.9 ng/mL in CM, other factors likely participate in reprogramming. It is possibility that the high concentration of rBMP4 (100 ng/mL) compensates for the absence of other reprogramming factors. Alternatively, cytokines and serum factors in uCM may act with rBMP4 to induce expression of Oct‐4.
Recombinant BMP4 may reprogramme keratinocytes, but it was not permanent. Continued exposure was necessary to maintain expression of embryonic transcripts and to suppress K14. This led us to examine keratinocyte expression of BMPR1A (Alk3) (Gilboa et al. 2000; ten Dijke et al. 2003). Although transcript levels for all BMP receptors appeared to be the same in our control and BMP4‐exposed keratinocyte cultures (data not shown), our results support a link between cell surface BMPR1A expression and expression of Oct‐4. Oct‐4 mRNA expression can be activated through treatment with the DNA methylation inhibitor, 5‐aza‐2′‐deoxycytidine and can reverse ESC differentiation (Hattori et al. 2004; Tsuji‐Takayama et al. 2004). Expression of BMPR1A is also highly induced by 5‐aza‐2′‐deoxycytidine in progenitor cells (Oh et al. 2004), suggesting a possible link between Oct‐4 and BMPR1A at the level of stem cell activation, perhaps through the inhibition of DNA methyltransferase family members (Hattori et al. 2004). In addition, a recent study by Hartung et al. (2006) found that both the type I and type II BMPRs are continuously being endocytosed and that their localization to certain membrane regions is dependent upon the stimuli and signalling pathways involved. It is possible that the CM‐ and BMP4‐responsive cells, presumably made up largely of the EpiSC population, exhibit a decreased level of BMPR1A endocytosis compared to their more committed, unresponsive counterparts. The difference in co‐expression of BMPR1A and Oct‐4 in the rBMP4‐treated cultures compared to the CM‐treated cultures again implies that BMP4 is not the sole factor responsible for reprogramming the keratinocytes and that several ESC‐secreted factors are involved, not only with inducing expression of Oct‐4 and recruiting BMPR1A to the cell surface, but also with necessary demethylation events. BMP4 facilitates the self‐renewal of murine ESCs through inhibition of extracellular receptor kinase and p38 MAPK pathways (Qi et al. 2004). BMP4 (and BMP2) sustain murine ESC self‐renewal through activation the Smad pathway, which, then leads to activation of inhibitor of differentiation (Id) genes (Ying et al. 2003). It is possible that BMP4 functions through MAPK and Smad pathways, activating an alternative programme, as has been found for Smad7 in hair follicles (Han et al. 2006).
Our findings suggest that pre‐treating somatic cells with CM and rBMP4 can change their developmental potential and allow them to be directed towards an alternative cell lineage. Keratinocytes that up‐regulated Oct‐4 and its targets responded to neuronal differentiation factors by altering their morphology and protein expression (Lendahl et al. 1990; Mullen et al. 1992; Wolf et al. 1996; Jiang et al. 2003). The presence of both the presynaptic AMPA receptor subunit, GluR1, and the postsynaptic NMDA receptor subunit, NR1, as well as the dopamine‐associated enzyme, tyrosine hydroxylase (TH) implies that the cells changed functionally and may be capable of transmitting electrical impulses (Nishikawa 2005; Wang et al. 2005; Zeiss 2005). Although we assume that these Oct‐4+ were not completely de‐differentiated into ESCs, our data indicate that the cells underwent at least a partial, de‐differentiation, marked by up‐regulation of pluripotency‐associated markers and down‐regulation of the basal keratinocyte‐specific K14. We did not see expression of HoxB4, or lineage markers, including CD45 and Sca‐1. Such de‐differentiation events were not permanent as continuous exposure to rBMP4 was required to maintain Oct‐4 expression. It appears that the Oct‐4+ cells become a ‘transitioning cell’, with a potential between that of a somatic cell and an ESC. At this stage, they can be re‐differentiated along a different lineage pathway, such as the neuronal pathway. The question we have not answered is what happens to these cells when they are place in vivo. It may be that the pre‐treated cells differentiate in response to the new environment or remain undifferentiated. Alternatively, they may return to the keratinocyte lineage no matter what the environment. Of course, the only way to determine this is to inject them into an animal model, which must be the next step.
Although we were unable to assess if the ability to change lineage was restricted to the Oct‐4+ cells, we observed that the percentage of keratinocytes induced to express Oct‐4 seemed to correlate with the number of cells that initially responded to the neuronal factors. In our neuronal cultures, the majority of keratinocytes grew in clusters, from which the responding cells sprouted. Our data suggest a clonal activation of Oct‐4, with the Oct‐4+ cells responding to neuronal growth conditions. The cells that failed to activate Oct‐4 remained as keratinocytes, assessed by continued expression of K14 and maintenance of keratinocyte morphology. The number of keratinocytes that activate Oct‐4 and the clonal expansion of these cells suggested that the responsive cells might be restricted to the EpiSC compartment. Modifying our previously published method of sorting EpiSC (Liang & Bickenbach 2002), we determined that both EpiSC and a small number of TA small cells could express Oct‐4 in response to treatment. Unpublished observations in our laboratory indicate that the neural crest‐derived melanocytes and merkel cells present in relatively lower numbers in the epidermis were only found within the TA large gate (Fig. S2). Thus, there may exist a continuum of differentiation within the basal keratinocyte population, and that some of the cells defined as TA cells are perhaps not fully committed to terminal keratinocyte differentiation.
Our results are the first to demonstrate that a somatic cell can be reprogrammed by exposure to ESC‐secreted factors in culture, and that the reprogrammed cell can be directed towards an alternative cell lineage. Such findings may help to elucidate how somatic cells are reprogrammed by embryonic cells in vivo and shed light on the mechanisms by which certain somatic populations may be made more developmentally potent.
Supporting information
Supporting info item
Supporting info item
Supporting info item
ACKNOWLEDGEMENTS
We thank the other members of the Bickenbach laboratory for their generous insights and the staff of the University of Iowa Flow Cytometry Facility, Jason Fishbaugh, Gene Hess and George Rasmussen, for their expert assistance in sorting the EpiSCs. We also gratefully acknowledge Dr Baoli Yang, for his helpful advice and donation of ESC, and Dr Johannes Hell and Dr Michelle Merrill, for their valuable suggestions and gift of their GluR1 and NR1 antibodies. This work was supported by the National Institutes of Health (R01‐AG20913).
REFERENCES
- Avilion AA, Nicolis SK, Pevny LH, Perez L, Vivian N, Lovell‐Badge R (2003) Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev. 17, 126–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben‐Shushan E, Thompson JR, Gudas LJ, Bergman Y (1998) Rex‐1, a gene encoding a transcription factor expressed in the early embryo, is regulated via Oct‐3/4 and Oct‐6 binding to an octamer site and a novel protein, Rox‐1, binding to an adjacent site. Mol. Cell. Biol. 18, 1866–1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne JA, Simonsson S, Western PS, Gurdon JB (2003) Nuclei of adult mammalian somatic cells are directly reprogrammed to oct‐4 stem cell gene expression by amphibian oocytes. Curr. Biol. 13, 1206–1213. [DOI] [PubMed] [Google Scholar]
- Clarke DL, Johansson CB, Wilbertz J, Veress B, Nilsson E, Karlstrom H, Lendahl U, Frisen J (2000) Generalized potential of adult neural stem cells. Science 288, 1660–1663. [DOI] [PubMed] [Google Scholar]
- Collas P, Taranger CK, Boquest AC, Noer A, Dahl JA (2006) On the way to reprogramming cells to pluripotency using cell‐free extracts. Reprod. Biomed. Online 12, 762–770. [DOI] [PubMed] [Google Scholar]
- Constantinescu S (2003) Stemness, fusion and renewal of hematopoietic and embryonic stem cells. J. Cell. Mol. Med. 7, 103–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowan CA, Atienza J, Melton DA, Eggan K (2005) Nuclear reprogramming of somatic cells after fusion with human embryonic stem cells. Science 309, 1369–1373. [DOI] [PubMed] [Google Scholar]
- Ten Dijke P, Korchynskyi O, Valdimarsdottir G, Goumans MJ (2003) Controlling cell fate by bone morphogenetic protein receptors. Mol. Cell. Endocrinol. 211, 105–113. [DOI] [PubMed] [Google Scholar]
- Flasza M, Shering AF, Smith K, Andrews PW, Talley P, Johnson PA (2003) Reprogramming in inter‐species embryonal carcinoma‐somatic cell hybrids induces expression of pluripotency and differentiation markers. Cloning Stem Cells 5, 339–354. [DOI] [PubMed] [Google Scholar]
- Fukushima A, Okuda A, Nishimoto M, Seki N, Hori TA, Muramatsu M (1998) Characterization of functional domains of an embryonic stem cell coactivator UTF1 which are conserved and essential for potentiation of ATF‐2 activity. J. Biol. Chem. 273, 25840–25849. [DOI] [PubMed] [Google Scholar]
- Gilboa L, Nohe A, Geissendorfer T, Sebald W, Henis YI, Knaus P (2000) Bone morphogenetic protein receptor complexes on the surface of live cells: a new oligomerization mode for serine/threonine kinase receptors. Mol. Biol. Cell 11, 1023–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grinnell KL, Yang B, Eckert RL, Bickenbach JR (2007) De‐differentiation of mouse interfollicular keratinocytes by the embryonic transcription factor Oct‐4. J. Invest. Dermatol. 127, 372–380. [DOI] [PubMed] [Google Scholar]
- Guo Y, Graham‐Evans B, Broxmeyer HE (2006) Murine embryonic stem cells secrete cytokines/growth modulators that enhance cell survival/anti‐apoptosis and stimulate colony formation of murine hematopoietic progenitor cells. Stem Cells 24, 850–856. [DOI] [PubMed] [Google Scholar]
- Gurdon JB, Byrne JA, Simonsson S (2003) Nuclear reprogramming and stem cell creation. Proc. Natl Acad. Sci. USA 100 (Suppl. 1), 11819–11822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hager‐Theodorides AL, Outram SV, Shah DK, Sacedon R, Shrimpton RE, Vicente A, Varas A, Crompton T (2002) Bone morphogenetic protein 2/4 signaling regulates early thymocyte differentiation. J. Immunol. 169, 5496–5504. [DOI] [PubMed] [Google Scholar]
- Han G, Li AG, Liang Y‐Y, Owens P, He W, Lu S, Yoshimatsu Y, Wang D, Ten Dijke P, Lin X, Wang X‐J (2006) Smad7‐induced b‐catenin degradation alters epidermal appendage development. Dev. Cell 11, 301–312. [DOI] [PubMed] [Google Scholar]
- Hartung A, Bitton‐Worms K, Rechtman MM, Wenzel V, Boergermann JH, Hassel S, Henis YI, Knaus P (2006) Different routes of bone morphogenic protein (BMP) receptor endocytosis influence BMP signaling. Mol. Cell. Biol. 26, 7791–7805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori N, Nishino K, Ko YG, Hattori N, Ohgane J, Tanaka S, Shiota K (2004) Epigenetic control of mouse Oct‐4 gene expression in embryonic stem cells and trophoblast stem cells. J. Biol. Chem. 279, 17063–17069. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Henderson D, Blackstad M, Chen A, Miller RF, Verfaillie CM (2003) Neuroectodermal differentiation from mouse multipotent adult progenitor cells. Proc. Natl Acad. Sci. USA 100 (Suppl. 1), 11854–11860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Jahagirdar BN, Reinhardt RL, Schwartz RE, Keene CD, Ortiz‐Gonzalez XR, Reyes M, Lenvik T, Lund T, Du Blackstad MJ, Aldrich S, Lisberg A, Low WC, Largaespada DA, Verfaillie CM (2002) Pluripotency of mesenchymal stem cells derived from adult marrow. Nature 418, 41–49. [DOI] [PubMed] [Google Scholar]
- Kodaira K, Imada M, Goto M, Tomoyasu A, Fukuda T, Kamijo R, Suda T, Higashio K, Katagiri T (2006) Purification and identification of a BMP‐like factor from bovine serum. Biochem. Biophys. Res. Commun. 345, 1224–1231. [DOI] [PubMed] [Google Scholar]
- Krause DS, Theise ND, Collector MI, Henegariu O, Hwang S, Gardner R, Neutzel S, Shariks SJ (2001) Multi‐organ, multi‐lineage engraftment by a single bone marrow‐derived stem cell. Cell 105, 369–377. [DOI] [PubMed] [Google Scholar]
- Lendahl U, Zimmerman LB, McKay RD (1990) CNS stem cells express a new class of intermediate filament protein. Cell 60, 585–595. [DOI] [PubMed] [Google Scholar]
- Li L, Neaves WB (2006) Normal stem cells and cancer stem cells: the niche matters. Cancer Res. 66, 4553–4557. [DOI] [PubMed] [Google Scholar]
- Liang L, Bickenbach JR (2002) Somatic epidermal stem cells can produce multiple cell lineages during development. Stem Cells 20, 21–31. [DOI] [PubMed] [Google Scholar]
- Mitsui K, Tokuzawa Y, Itoh H, Segawa K, Murakami M, Takahashi K, Maruyama M, Maeda M, Yamanaka S (2003) The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell 113, 631–642. [DOI] [PubMed] [Google Scholar]
- Mullen RJ, Buck CR, Smith AM (1992) NeuN, a neuronal specific nuclear protein in vertebrates. Development 116, 201–211. [DOI] [PubMed] [Google Scholar]
- Nishikawa T (2005) Metabolism and functional roles of endogenous d‐serine in mammalian brains. Biol. Pharm. Bull. 28, 1561–1565. [DOI] [PubMed] [Google Scholar]
- Nishimoto M, Fukushima A, Okuda A, Muramatsu M (1999) The gene for the embryonic stem cell coactivator UTF1 carries a regulatory element which selectively interacts with a complex composed of Oct‐3/4 and Sox‐2. Mol. Cell. Biol. 19, 5453–5465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimoto M, Miyagi S, Yamagishi T, Sakaguchi T, Niwa H, Muramatsu M, Okuda A (2005) Oct‐3/4 maintains the proliferative embryonic stem cell state via specific binding to a variant octamer sequence in the regulatory region of the UTF1 locus. Mol. Cell. Biol. 25, 5084–5094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh H, Chi X, Bradfute SB, Mishina Y, Pocius J, Michael LH, Behringer RR, Schwartz RJ, Entman ML, Schneider MD (2004) Cardiac muscle plasticity in adult and embryo by heart‐derived progenitor cells. Ann. N. Y. Acad. Sci. 1015, 182–189. [DOI] [PubMed] [Google Scholar]
- Okuda A, Fukushima A, Nishimoto M, Orimo A, Yamagishi T, Nabeshima Y, Kuro OM, Nabeshima Y, Boon K, Keaveney M, Stunnenberg HG, Muramatsu M (1998) UTF1, a novel transcriptional coactivator expressed in pluripotent embryonic stem cells and extra‐embryonic cells. EMBO J. 17, 2019–2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okumura‐Nakanishi S, Saito M, Niwa H, Ishikawa F (2005) Oct‐3/4 and Sox2 regulate Oct‐3/4 gene in embryonic stem cells. J. Biol. Chem. 280, 5307–5317. [DOI] [PubMed] [Google Scholar]
- Palmqvist L, Glover CH, Hsu L, Lu M, Bossen B, Piret JM, Humphries RK, Helgason CD (2005) Correlation of murine embryonic stem cell gene expression profiles with functional measures of pluripotency. Stem Cells 23, 663–680. [DOI] [PubMed] [Google Scholar]
- Pells S, McWhir J (2004) Studying nuclear reprogramming with cell hybrids. Methods Mol. Biol. 254, 301–312. [DOI] [PubMed] [Google Scholar]
- Pesce M, Scholer HR (2000) Oct‐4: control of totipotency and germline determination. Mol. Reprod. Dev. 55, 452–457. [DOI] [PubMed] [Google Scholar]
- Qi X, Li TG, Hao J, Hu J, Wang J, Simmons H, Miura S, Mishina Y, Zhao GQ (2004) BMP4 supports self‐renewal of embryonic stem cells by inhibiting mitogen‐activated protein kinase pathways. Proc. Natl Acad. Sci. USA 101, 6027–6032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodda DJ, Chew JL, Lim LH, Loh YH, Wang B, Ng HH, Robson P (2005) Transcriptional regulation of nanog by OCT4 and SOX2. J. Biol. Chem. 280, 24731–24737. [DOI] [PubMed] [Google Scholar]
- Sato N, Meijer L, Skaltsounis L, Greengard P, Brivanlou AH (2004) Maintenance of pluripotency in human and mouse embryonic stem cells through activation of Wnt signaling by a pharmacological GSK‐3‐specific inhibitor. Nat. Med. 10, 55–63. [DOI] [PubMed] [Google Scholar]
- Singla DK, Schneider DJ, Lewinter MM, Sobel BE (2006) wnt3a but not wnt11 supports self‐renewal of embryonic stem cells. Biochem. Biophys. Res. Commun. 345, 789–795. [DOI] [PubMed] [Google Scholar]
- Tada M, Takahama Y, Abe K, Nakatsuji N, Tada T (2001) Nuclear reprogramming of somatic cells by in vitro hybridization with ES cells. Curr. Biol. 11, 1553–1558. [DOI] [PubMed] [Google Scholar]
- Taranger CK, Noer A, Sorensen AL, Hakelien AM, Boquest AC, Collas P (2005) Induction of dedifferentiation, genomewide transcriptional programming, and epigenetic reprogramming by extracts of carcinoma and embryonic stem cells. Mol. Biol. Cell 16, 5719–5735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian HB, Wang H, Sha HY, Xu XJ, Zhu M, Wu YB, Cheng SH, Chen JQ, Shi YX, Bai ZL, Cheng GX (2006) Factors derived from mouse embryonic stem cells promote self‐renewal of goat embryonic stem‐like cells. Cell Biol. Int. 30, 452–458. [DOI] [PubMed] [Google Scholar]
- Tomioka M, Nishimoto M, Miyagi S, Katayanagi T, Fukui N, Niwa H, Muramatsu M, Okuda A (2002) Identification of Sox‐2 regulatory region which is under the control of Oct‐3/4‐Sox‐2 complex. Nucleic. Acids Res. 30, 3202–3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji‐Takayama K, Inoue T, Ijiri Y, Otani T, Motoda R, Nakamura S, Orita K (2004) Demethylating agent, 5‐azacytidine, reverses differentiation of embryonic stem cells. Biochem. Biophys. Res. Commun. 323, 86–90. [DOI] [PubMed] [Google Scholar]
- Wang JQ, Arora A, Yang L, Parelkar NK, Zhang G, Liu X, Choe ES, Mao L (2005) Phosphorylation of AMPA receptors: mechanisms and synaptic plasticity. Mol. Neurobiol. 32, 237–249. [DOI] [PubMed] [Google Scholar]
- Wang QT, Piotrowska K, Ciemerych MA, Milenkovic L, Scott MP, Davis RW, Zernicka‐Goetz M (2004) A genome‐wide study of gene activity reveals developmental signaling pathways in the preimplantation mouse embryo. Dev. Cell 6, 133–144. [DOI] [PubMed] [Google Scholar]
- Wiese C, Rolletschek A, Kania G, Navarrete‐Santos A, Anisimov SV, Steinfarz B, Tarasov KV, Brugh SA, Zahanich I, Ruschenschmidt C, Beck H, Blyszczuk P, Czyz J, Heubach JF, Ravens U, Horstmann O, St‐Onge L, Braun T, Brustle O, Boheler KR, Wobus AM (2006) Signals from embryonic fibroblasts induce adult intestinal epithelial cells to form nestin‐positive cells with proliferation and multilineage differentiation capacity in vitro . Stem Cells 24, 2085–2097. [DOI] [PubMed] [Google Scholar]
- Wilmut I, Schnieke AE, McWhir J, Kind AJ, Campbell KH (1997) Viable offspring derived from fetal and adult mammalian cells. Nature 385, 810–813. [DOI] [PubMed] [Google Scholar]
- Wolf HK, Buslei R, Schmidt‐Kastner R, Schmidt‐Kastner PK, Pietsch T, Wiestler OD, Blumcke I (1996) NeuN: a useful neuronal marker for diagnostic histopathology. J. Histochem. Cytochem. 44, 1167–1171. [DOI] [PubMed] [Google Scholar]
- Ying QL, Nichols J, Chambers I, Smith A (2003) BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self‐renewal in collaboration with STAT3. Cell 115, 281–292. [DOI] [PubMed] [Google Scholar]
- Zeiss CJ (2005) Neuroanatomical phenotyping in the mouse: the dopaminergic system. Vet. Pathol. 42, 753–773. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting info item
Supporting info item
Supporting info item


