Abstract
Objectives
This study aimed to investigate effects of neurotrophin receptor‐mediated melanoma antigen‐encoding gene homology (NRAGE) on proliferation and odontoblastic differentiation of mouse dental pulp cells (mDPCs).
Materials and methods
Mouse dental pulp cells were infected with recombinant lentivirus to stably knockdown expression of NRAGE, and biological effects of NRAGE on the cells were detected. Proliferation and odontoblastic differentiation of mDPCs were observed. Simultaneously, mRNA and protein levels of NRAGE and nuclear factor‐κB (NF‐κB) protein expression were detected. Immunofluorescence assay was used to detect expression and location of NRAGE and NF‐κB.
Results
NRAGE mRNA and protein levels reduced significantly after mDPC odontoblastic induction. Knockdown of NRAGE inhibited the proliferation of mDPCs. However, knockdown of NRAGE enhanced their odontoblastic differentiation with up‐regulated ALPase activity. It also promoted mineral nodule formation as well as mRNA and protein expressions of ALP, DSPP and DMP1. Protein levels of NF‐κB/p50 significantly increased, whereas NF‐κB/p105 protein expression decreased in the mDPC/shNRG group. Immunofluorescence revealed that relocation of NF‐κB was similar to that of NRAGE during odontoblastic induction, in which NF‐κB translocated from the cytoplasm to the nucleus.
Conclusion
NRAGE is a potent regulator of proliferation and odontoblastic differentiation of mDPCs, which might be via the NF‐κB signalling pathway.
Introduction
Dental pulp is composed of ectomesenchymal components with neural crest‐derived cells, and contains mixed cell types including fibroblasts, odontoblasts and undifferentiated mesenchymal cells; it plays important roles in dentinogenesis 1, 2. Dental pulp cells (DPCs) can proliferate and differentiate into odontoblasts, thereby creating reparative dentin in response to appropriate stimuli 3, 4. This process involves migration, proliferation and differentiation of DPCs into odontoblast/odontoblast‐like cells 5. A variety of key growth factors and signalling molecules have been tested in this process. For example, nuclear factor I‐C regulates dentin sialophosphoprotein (DSPP) and E‐cadherin by control of Kruppel‐like factor 4 during dentinofenesis6. The TGF‐beta/BMP family of growth factors induce odontoblast differentiation and reparative dentin synthesis 7. HMGB1 promotes proliferation and differentiation of human DPCs. microRNA‐27 promotes differentiation of odontoblastic cells by activating Wnt/β‐catenin signalling 8. However, up to now, precise mechanisms underlying proliferation and differentiation of DPCs has remained unclear.
Neurotrophin receptor‐interacting melanoma antigen‐encoding gene homology (NRAGE) is also denominated as MAGE‐D1 or Dlxin‐1. It is a member of the Type II melanoma‐associated antigen (MAGE) family of proteins characterized by presence of a unique region of about 200 amino acids known as the MAGE homology domain. It has been identified as a molecule interacting with low affinity nerve growth factor receptor p75NTR 9, 10. Previous studies of NRAGE have mainly concerned apoptosis, the cell cycle and cell differentiation 11, 12. NRAGE plays an essential role in developmental apoptosis of sympathetic neurons in vivo 11, which may be involved in prion diseases 13. NRAGE suppresses metastasis of melanoma and pancreatic cancers in vivo and in in vitro studies 14; NRAGE also promotes proliferation of oesophageal carcinoma cells 15. Meanwhile, NRAGE provides endogenous regulation of neuronal proliferation and differentiation of PC12 cells 16. According to previous studies, NRAGE modulates function of Dlx/Msx to promote myogenic differentiation 17 and strong NRAGE mRNA signals have been found in cell layers surrounding cartilaginous elements in bone rudiments, during digit formation in embryogenesis. Further study has proven that NRAGE may act as a regulator of Dlx family members in bone formation 12. Dlx3 also plays a key role in odontoblast polarization and dentin formation 18. NRAGE participates in activating non‐canonical and alternative bone morphogenetic protein (BMP) signalling pathways 19, 20. Meanwhile, BMP signalling pathways play an important role in tooth development 21, 22, 23 and regulating proliferation and differentiation of DPCs 24, 25. Odontoblasts and osteoblasts share many similarities. Thus, we conjectured that NRAGE may exert effects on mouse DPCs (mDPCs) during tooth development.
In the present study, expression of NRAGE during mDPC differentiation was investigated using quantitative real‐time polymerase chain reaction (RT‐PCR) and western blot analysis. NRAGE was stably knocked down in mDPCs to determine its effects on their proliferation and odontoblastic differentiation.
Materials and methods
This study was performed according to the informed protocol approved by the Ethics Committee of Shanghai Tenth People's Hospital, Tongji University, Shanghai.
Isolation and culture of primary mDPCs
Primary cultured mDPCs were isolated from incisors of healthy 4‐week‐old mice. Pulp tissues were gently separated from teeth and cut into small pieces (~1 mm3) using ophthalmic scissors. Tissue pieces were cultured in high‐glucose Dulbecco's modified Eagle's medium (DMEM; Gibco‐BRL, Grand Island, NY, USA) supplemented with 10% foetal bovine serum (FBS; Gibco‐BRL Life Technologies, Paisley, UK) and antibiotics (100 U/ml penicillin and 100 U/ml streptomycin, Gibco‐BRL) in a humidified atmosphere of 5% CO2 at 37 °C. When the cells reached confluence, they were digested by trypsinization (0.2% trypsin and 0.02% ethylenediaminetetraacetic acid; Gibco) and cultured. Cultures between the third and sixth passages were used in the study, and culture medium was changed every 3 days.
DMEM supplemented with 10% FBS, antibiotics, 50 μg/ml ascorbic acid, 10 mmol/l sodium β‐glycerophosphate and 10 nmol/l dexamethasone (Sigma, St Louis, MO, USA) was used as odontoblastic induction medium. mDPCs were plated in six‐well plates (Corning Inc., Corning, NY, USA) at 2 × 105 cells/well and then cultured in odontoblastic induction medium.
Plasmid construction and interfering RNA transfection
Lentiviral plasmids carrying small hairpin NRAGE interference (shNRG), targeting AAGATGAAAGTGCTGAGATTC sequence (hNRAGE, a human neurotrophin receptor interacting MAGE homologue) or non‐specific sequence (shCon), were constructed using the vector pLKO.1‐puro (Sigma), as reported in previous work 26. In brief, shCon‐plasmid and shRNA‐plasmid cotransfection were performed using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) in accordance with the manufacturer's protocol. shNRG or shCon were packaged in 293T and infected mDPCs using polybrene (Sigma) for 24 h, then 2 μg/ml puromycin (Sigma) was added to select positive transfected cells, for 4–7 days. A monoclonal population of stably infected cells was prepared by limiting dilution assay, and verified by RT‐PCR and western blotting.
Cell proliferation assay
Cell Counting Kit‐8 (CCK‐8; Dojindo Kagaku Co., Kumamoto, Japan) was used to analyse effects of NRAGE on mDPC proliferation, according to the manufacturer's protocols. Briefly, wild‐type, shCon‐ or shNRG‐infected mDPCs were seeded at 5 × 103 cells/well in four 96‐well plates (Corning Inc.), and then cultured overnight. Subsequently, fresh medium with 10% FBS was changed every day. After cells had been cultured for 1, 3, 5 and 7 days, numbers of cells were assessed using the cell counting kit. Absorbance was measured using a microplate reader at 450 nm to determine number of vitable cells in each well. A well containing medium and CCK‐8 solution, but without cells, was used as blank control. Cell proliferation was represented as mean ± SD of absorbance for five wells from each group.
Alizarin red and alkaline phosphatase (ALP) staining
Wild‐type, shCon‐ or shNRG‐infected mDPCs were incubated under odontoblastic induction for 14 days. Induction medium was replaced every 3 days. Mineralization was assessed by alizarin red staining. Briefly, 1% alizarin red (Sigma) was prepared in distilled water, adjusted to pH 4.3 and applied to cells in six‐well plates, for 30 min at 37 °C. For ALP staining, plates were harvested at 14 days. ALP colour development kit (Beyotime Institute of Biotechnology, Shanghai, China) was used, according to the manufacturer's protocols. All cells were washed three times in distilled water, then observed using phase contrast microscopy.
ALPase activity assay
ALPase activity was determined using cell lysates. Briefly, 220 μl 0.9 mm 2‐amino‐2‐methyl‐1‐propanol buffer was added to 5 μl samples or standards. Subsequently, 25 μl 15 mm p‐nitrophenyl phosphate was added and mixtures were incubated at 37 °C for 30 min. ALP activity was measured at 405 nm using an ELISA microplate reader (Bio‐Tek Instruments Inc., Winooski, VT, USA). ALPase activity (U/mg) was defined as release of 1 mol p‐nitrophenol per milligram of total cell protein.
RNA isolation and semi‐quantitative RT‐PCR analysis
Total RNA of wild‐type, shCon‐ and shNRG‐infected mDPCs cultured in odontoblastic induction medium for different times (0, 7 and 14 days) was isolated using TRIzol Reagent (Invitrogen) following the manufacturer's protocol. cDNA synthesized using PrimeScript RT reagent Kit with gDNA Eraser (Takara Bio, Shiga, Japan), was used as PCR template. Odontogenic differentiation of cells was monitored by analysing odontoblast‐related marker dental matrix protein 1 (dmp1), dspp and alp. Glyceraldehyde‐3‐phosphate dehydrogenase (gapdh) was used to normalize RNA expression. Sequences of specific primers used were as follows: dspp (forward: 5′‐TCGGTTACCGGTTGACATGG‐3′, reverse: 5′‐GCAGG CCCTGTTTCCTCAT‐3′); dmp1 (forward: 5′‐ACAGCCTGAACACATTCTCC‐3′, reverse: 5′‐ATGTTCTTGG GACGGATGTC‐3′); alp (forward: 5′‐TTCTCTCTTGGGCAGGCAAG‐3′, reverse: 5′‐ACCCCGCTATTCCAAACAGG‐3′); nrage (forward: 5′‐GGCATACT GGGAACGACCAA‐3′, reverse: 5′‐CCAGAGCATCCAAGGCTTCA‐3′); and gapdh (forward: 5′‐TGGGTGTGAACCATGAGAAGT‐3′, reverse: 5′‐TGAGTCCTTCCAC GATACCAA‐3′). Real‐time PCR reaction was amplified with SYBR Premix Ex Taq II (Takara Bio) in an ABI 7500 Real‐Time PCR System (Applied Biosystems, Foster City, CA, USA). Relative gene expression was calculated using the comparative 2−ΔΔCt method 27. Mean Ct value of the target gene was normalized to its averaged Ct values of gapdh, to obtain a ΔCt value, which was then normalized to control samples to obtain a ΔΔCt value. Each measurement was assessed in triplicate. Gene expression ratio is shown as mean ± SD from three independent experiments.
Western blot analysis
Wild‐type, shCon‐ and shNRG‐infected mDPCs cultured in odontoblastic induction medium for 0, 7 and 14 days were lysed using a protein extraction kit (Piece, Rockford, IL, USA). Protein concentrations were determined employing a bicinchoninic acid protein assay kit (Piece). An equal amount of protein was separated, then transferred on to nitrocellulose membranes (Millipore Corporation, Billerica, MA, USA). After blocking, primary antibodies to mouse anti‐mouse NRAGE (Santa Cruz Biotechnology Inc., Dallas, TX, USA), rabbit anti‐mouse DMP1 (Bio Vision Research Products, Mountain View, CA, USA), rabbit anti‐mouse DSP (Santa Cruz Biotechnology Inc), rabbit anti‐mouse nuclear factor‐κB (NF‐κB) (Epitomics, Inc., Burlingame, CA, USA) and rabbit anti‐mouse β‐actin (Santa Cruz Biotechnology, Inc) were used. Then, secondary antibodies goat anti‐mouse immunoglobulin G (Licor Co., Lincoln, NE, USA) and goat anti‐rabbit immunoglobulin G (Licor Co.) were used. After the final wash, membranes were visualized using the Odyssey LI‐CDR system.
Immunofluorescence
Wild‐type mDPCs were seeded in chambers of 24‐well plates, then incubated overnight at 37 °C, in a humidified atmosphere of 5% CO2. Subsequently, medium was changed to odontoblastic induction for 0, 3, 7, 11 and 14 days. Cells in chambers were fixed in 4% paraformaldehyde at 4 °C for 25 min, permeabilized with 0.2% Triton‐X 100 (Sigma) for 15 min at room temperature and blocked in 5% bovine serum albumin in PBS. Subsequently, samples on slides were incubated with relevant primary antibodies in 2% bovine serum albumin overnight at 4 °C. They were washed and incubated with appropriate secondary antibodies for 1 h at 37 °C. 1:200 dilution mouse anti‐mouse NRAGE antibody (Santa Cruz Biotechnology, Inc), 1:300 rabbit anti‐mouse NF‐κB antibody (Epitomics, Inc.), goat anti‐mouse immunoglobulin G fragment (Alexa Fluor® 488 Conjugate) (Cell Signaling Technology, Inc., Boston, MA, USA) and goat anti‐rabbit immunoglobulin G fragment (Alexa Fluor® 555 Conjugate) (Cell Signaling Technology, Inc.) were used. After sample nuclei were stained for 10 min in Hoechst solution (Dojindo Laboratories, Kumamoto, Japan), they were visualized using confocal fluorescence microscopy (Carl Zeiss, German).
Statistical analysis
Experiments were performed in triplicate, and data are presented as mean ± SD. Results were evaluated by one‐way ANOVA using spss software (Version 10.0; SPSS, Chicago, IL, USA). P values less than 0.05 were considered statistically significant.
Results
Expression of NRAGE during odontoblastic differentiation of mDPCs
To detect expression of NRAGE in cell cultures, mRNA and protein levels of DMP‐1, DSPP, ALP and NRAGE were measured using real‐time PCR and western blot analysis during odontoblastic induction of mDPCs (Fig. 1). mRNA and protein levels of DMP‐1, DSPP and ALP increased during induction (Fig. 1a–c, f) and ALP activity exhibited the same trend as alp mRNA expression (Fig. 1d). In contrast, mRNA and protein levels of NRAGE were down‐regulated during odontoblastic differentiation (Fig. 1e, f).
Figure 1.
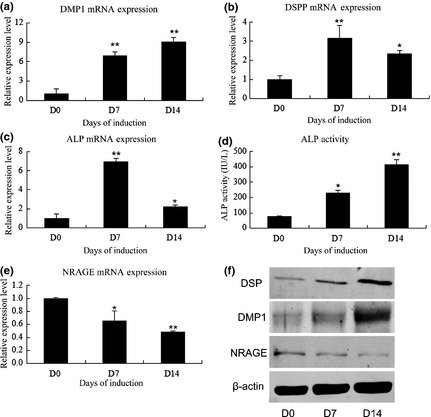
Expression of NRAGE during odontoblastic differentiation of mDPCs. mRNA expression of mineralization‐related markers and NRAGE on days 0, 7 and 14 after induction: (a) DMP1, (b) DSPP, (c) ALP, (d) ALP activity and (e) NRAGE. Glyceraldehyde‐3‐phosphate (GAPDH) was used as normalization control. (f) DSP, DMP1 and NRAGE protein levels on days 0, 7 and 14 after induction. *Significant difference (P < 0.05) versus day 0, **Significant difference (P < 0.01) versus day 0.
Stable knockdown of NRAGE in mDPCs
Stably transfected cell populations of mDPC/shCon and mDPC/shNRG were constructed. mRNA and protein levels of NRAGE were clearly lower in mDPC/shNRG than those in mDPC/wt and mDPC/shCon groups, after infection (Fig. 2). Further verification by RT‐PCR and western blotting demonstrated that expression of NRAGE remained down‐regulated until day 14 (Fig. 2). These results showed that NRAGE was stably knocked down in mDPC/shNRG.
Figure 2.
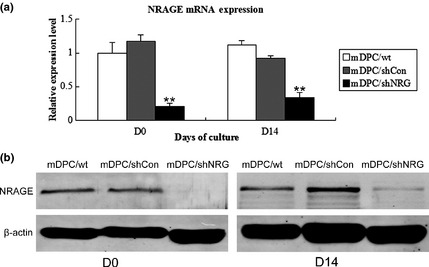
Stable knockdown of NRAGE in mDPCs. mRNA level (a) and protein level (b) on days 0 and 14 after mDPC infection. **Significant difference (P < 0.01) versus control.
Inhibition of mDPC proliferation after knockdown of NRAGE
We measured cell proliferation using CCK‐8 and results of the assay indicated that proliferation level of NRAGE‐depleted NRAGE mDPCs was much lower than that of mDPCs/wt and mDPC/shCon groups (P < 0.01) on days 1, 3, 5 and 7, whereas no significant difference was observed between mDPCs/wt and mDPCs/shCon groups (Fig. 3). All results demonstrated that cell population growth of NRAGE‐depleted mDPCs was greatly inhibited.
Figure 3.
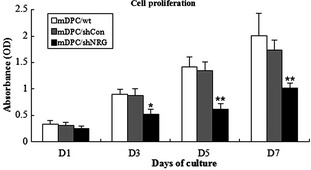
Effect of NRAGE knockdown on mDPC proliferation. *Significant differences (P < 0.05) versus control and **Significant difference (P < 0.01) versus control. OD, optical density.
Promotion of odontoblastic differentiation of mDPCs after knockdown of NRAGE
Mineralized nodule formation, ALP staining and ALP activity assay were performed to directly reveal effects of NRAGE on odontoblastic differentiation. After 14 days culture in odontoblastic induction medium, mineralized nodule formation was observed, using phase contrast microscopy. Amounts of nodules formed in the mDPC/shNRG group by 14 days were significantly higher than in mDPC/wt and mDPC/shCon groups (Fig. 4a). ALP staining had the same trend as mineralization in the three groups (Fig. 4b). ALP activity was also significantly higher in mDPC/shNRG group than in mDPC/wt and mDPC/shCon groups (Fig. 5d). To further document effects of endogenous NRAGE on regulating odontoblastic differentiation, mineralization‐related markers were selected. mRNA levels of dmp1, dspp and alp were significantly higher in mDPC/shNRG group than those in mDPC/wt and mDPC/shCon groups (P < 0.05; Fig. 5a–c). Western blot analysis indicated that DMP1 and DSP protein expression levels were much higher in mDPC/shNRG than those in mDPC/wt and mDPC/shCon groups (Fig. 5e). These results indicate that knockdown of NRAGE promoted odontoblastic differentiation of mDPCs.
Figure 4.
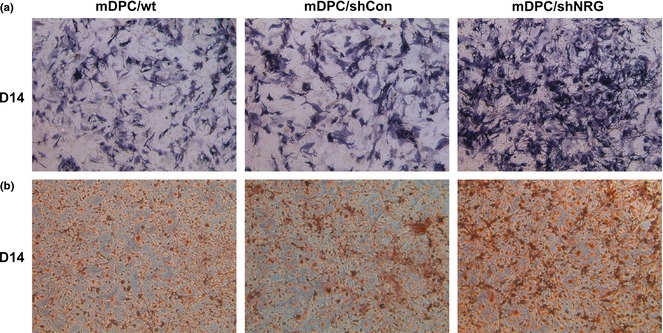
Mineralization and ALP staining of mDPC/wt, mDPC/shCon and mDPC/shNRG after odontoblastic induction for 14 days. (a) Result of ALP staining was higher in the mDPC/shNRG group than in mDPC/wt and mDPC/shCon groups (original magnification 50×). (b) Formation of mineralization nodules was much more in mDPC/shNRG group than in mDPC/wt and mDPC/shCon groups (original magnification 50×).
Figure 5.
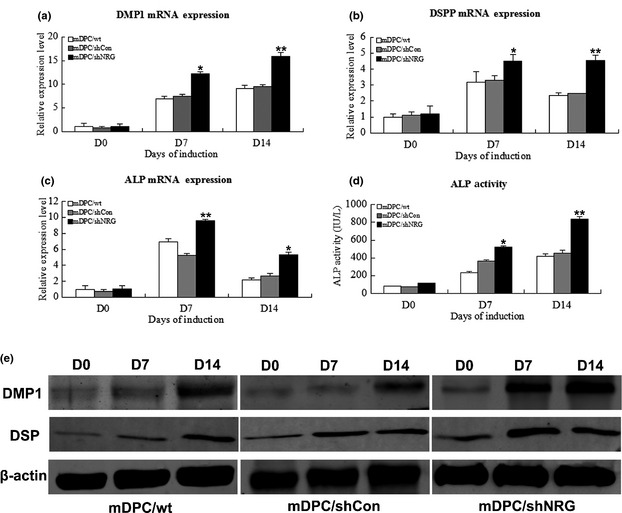
Effects of NRAGE knockdown on odontoblastic differentiation of mDPCs. Effect of NRAGE knockdown on mRNA level of odontoblastic marker genes in mDPCs on days 0, 7 and 14: (a) DMP1, (b) DSPP and (c) ALP. GAPDH was used as normalization control. (d) ALP activity in mDPC/shNRG group after 0, 7 and 14 days induction. Protein levels of DMP1 and DSP by western blot analysis on days 0, 7 and 14. *Significant difference (P < 0.05) versus control, **Significant difference (P < 0.01) versus control.
Possible role of the NF‐κB pathway in regulating odontoblastic differentiation of mDPCs
Western blot analysis reveaed two bands for NF‐κB protein. NF‐κB/p105 was down‐regulated in mDPC/shNRG group, while NF‐κB/p50 was up‐regulated (Fig. 6). Immunofluorescence staining showed that NRAGE was located homogeneously in the nucleus and cytoplasm of mDPCs without any induction, while NF‐κB was mainly located in the cytoplasm (Fig. 7). Under odontoblastic induction, NRAGE gradually translocated to nuclei of mDPCs, but was still found in both cytoplasm and nucleus (Fig. 7). Meanwhile, NF‐κB translocated from the cytoplasm into nuclei of mDPCs during the same process (Fig. 7). Moreover, locations of NRAGE and NF‐κB were almost the same after induction for 14 days (Fig. 7). These results suggest that NRAGE regulated odontoblastic differentiation of mDPCs via the NF‐κB pathway.
Figure 6.
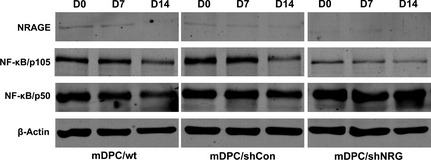
Protein levels of NRAGE and NF‐κB after odontoblastic induction for 0, 7 and 14 days in mDPC/wt, mDPC/shCon and mDPC/shNRG groups. NRAGE was stably knocked down during induction in the mDPC/shNRG group. Protein levels of NF‐κB/p50 and NF‐κB/p105 by western blot analysis.
Figure 7.
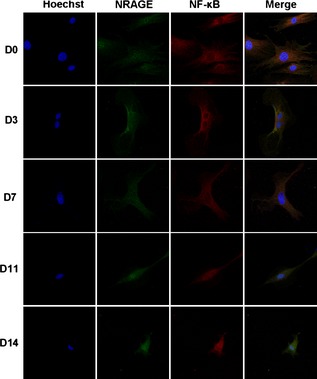
Immunofluorescence staining of NRAGE and NF‐κB relocation in mDPCs during odontoblastic induction for days 0, 3, 7, 11 and 14 (400× magnification). Immunofluorescence staining with anti‐NRAGE antibody (green), anti‐NF‐κB antibody (red) and Hoechst (blue) was performed. Negative control staining was performed with immunoglobulin G control antibody.
Discussion
Development, morphology and repair of damaged dental pulp tissue are dependent on proliferation and differentiation of DPCs. Proliferation and odontoblastic differentiation of dental pulp involves various growth factors, signalling molecules and transcription factors, such as BMP‐2 and BMP‐4 28, 29, MSX1 30, Dlx1/Dlx2 31, HMGB1 32 and more. This present study is the first to analyse function of NRAGE during proliferation and odontoblastic differentiation of mDPCs.
DMP1, ALP and DSPP are commonly used as mineralization markers in odontoblast/odontoblast‐like differentiation of DPCs 33. In particular, DSP protein and DSPP mRNA have been reported to be odontoblast‐specific markers 34, 35. In the current study, ALP expression and ALP activity increased during odontoblastic induction. DMP1 and DSPP were up‐regulated during the same procedure. These results demonstrate that mDPCs differentiated into odontoblast‐like cells; meanwhile, mRNA and protein levels of NRAGE were significantly down‐regulated in the process. Thus, we concluded that NRAGE possibly regulated odontoblastic differentiation of mDPCs.
NRAGE has been identified as a binding partner of the intracellular domain of p75 neurotrophin receptor, thereby facilitating apoptosis in neural progenitors 36. NRAGE has been investigated for participating in neurone apoptosis 36, cell cycle regulation 37, renal branching morphogenesis 38 and bone formation 20. It is expressed in most developing and adult tissues 39 and plays important roles in activating the BMP MAPK and p38MAPK pathways 39 and in tooth development, proliferation and differentiation of DPCs involves numerous overlapping tracts, including BMP MAPK 21 and p38MAPK pathways 40. Thus, we presumed that NRAGE possibly participated in tooth development and reparative dentin formation.
Lentiviral vectors provide efficient gene knockout in vitro and infect non‐dividing cells. Results of CCK‐8 demonstrate that cell proliferation of the mDPC/shNRG group was significantly inhibited. No significant differences between mDPC/wt and mDPC/shCon groups were observed in terms of either proliferation or differentiation. These findings indicate that recombinant lentivirus infection did not disturb the properties of mDPCs. NRAGE knockdown inhibited proliferation of mDPCs, identical to the previously published effect of NRAGE on oesophageal squamous cell carcinomas 26.
In this study, real‐time PCR and western blotting showed that odontoblastic differentiation markers, including DMP1, DSP and ALP, were up‐regulated in the mDPC/shNRG group, and to confirm this result, ALP activity, ALP staining and mineralization assays were performed. ALP activity and ALP staining increased with ALP mRNA level in the mDPC/shNRG group. The mineralization assay showed that NRAGE knockdown promoted mineralization potential of mDPCs. These results suggest that NRAGE knockout forced odontoblastic differentiation of mDPCs, which shows a potential role for NRAGE in tooth development and reparative dentin formation.
The BMP pathways regulate tooth development through two representative methods, namely, canonical and non‐canonical pathways. In the non‐canonical BMP pathway, NRAGE facilitates TAK1–TAB 1–XIAP complex, and drives BMP receptor and p38 activation 41. Previous researchers have shown that NF‐κB pathway links to non‐canonical BMP pathway via formation of the TAK1–TAB 1–XIAP complex 39, 42, 43, 44. NRAGE has been reported to regulate neuronal progenitor cell differentiation by up‐regulating NF‐κB activity and activating the BMP pathway 20. NF‐κB pathway is activated at sites of injury, and regulates apoptosis and the cell cycle 19. Activation of NF‐κB promotes odontoblastic differentiation against oxidative stress in DPCs 45 and furthermore, β‐oestradiol enhances odonto/osteogenic potency of DPSCs via activation of NF‐κB 46. Most importantly, Lee et al. have shown that NF‐κB is activated on promotion of proliferation, migration and differentiation in recombinant human DSP‐treated DPCs 47. Thus, the NF‐κB pathway may also regulate reparative dentin formation. However, activation of NF‐κB inhibits odonto/osteogenic differentiation of DPSCs in oestrogen‐deficient rats 48. These data suggest that activation of NF‐κB in different cell types may result in distinctive differentiation potency through different upstream and downstream signalling pathways.
In unstimulated cells, NF‐κB is sequestered in the cytoplasm, and translocates to nuclei when activated 39. Generally, NF‐κB/p105 is retained in the cytoplasm to inhibit activation of transcription. In response to stimuli, NF‐κB/p50 is released rapidly and translocates to the nucleus, whereas NF‐κB/p105 is degraded in proteasomes. These observations are in accordance with our results. Immunofluorescence revealed that NF‐κB gradually translocated from cytoplasm to nuclei during odontoblastic induction, and locations of NRAGE and NF‐κB were similar on day 14 after induction. Western blotting demonstrated that protein level of NF‐κB/p105 was down‐regulated compared with that of controls, whereas protein level of NF‐κB/p50 was up‐regulated. Thus it seems that, NRAGE possibly regulated odontoblastic differentiation of mDPCs by activation of the NF‐κB pathway, in our study. More intensive investigations are necessary to elucidate relationships between NRAGE and the NF‐κB pathway in odontoblastic differentiation.
In summary, NRAGE regulated odontoblastic differentiation of mDPCs, which seemed to depend on the NF‐κB pathway. Our studies showed that NRAGE could be a new regulation marker for use in regenerative dentistry. We will further elucidate the signalling pathway of NRAGE on proliferation and odontoblastic differentiation of DPCs; primary human DPCs will be used in our future studies.
Acknowledgements
This work was supported by the grants from the National Natural Science Foundation of China (81371913 and 81171778). All the authors denied any conflicts of interest.
The first three authors contributed equally to this study.
References
- 1. Sinanan AC, Hunt NP, Lewis MP (2004) Human adult craniofacial muscle‐derived cells: neural‐cell adhesion‐molecule (NCAM; CD56)‐expressing cells appear to contain multipotential stem cells. Biotechnol. Appl. Biochem. 40, 25–34. [DOI] [PubMed] [Google Scholar]
- 2. d'Aquino R, De Rosa A, Laino G, Caruso F, Guida L, Rullo R et al (2009) Human dental pulp stem cells: from biology to clinical applications. J. Exp. Zool. B Mol. Dev. Evol. 312B, 408–415. [DOI] [PubMed] [Google Scholar]
- 3. Huang GT, Gronthos S, Shi S (2009) Mesenchymal stem cells derived from dental tissues vs. those from other sources: their biology and role in regenerative medicine. J. Dent. Res. 88, 792–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wu Y, Sun H, Song F, Fu D, Wang J (2014) DDIT3 overexpression increases odontoblastic potential of human dental pulp cells. Cell Prolif. 47, 249–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Magloire H, Joffre A, Bleicher F (1996) An in vitro model of human dental pulp repair. J. Dent. Res. 75, 1971–1978. [DOI] [PubMed] [Google Scholar]
- 6. Lee HK, Lee DS, Park SJ, Cho KH, Bae HS, Park JC. (2014) Nuclear factor I‐C (NFIC) regulates dentin sialophosphoprotein (DSPP) and E‐cadherin via control of Kruppel‐like factor 4 (KLF4) during dentinogenesis. J. Biol. Chem. 289, 28225–28236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Durand SH, Romeas A, Couble ML, Langlois D, Li JY, Magloire H et al (2007) Expression of the TGF‐beta/BMP inhibitor EVI1 in human dental pulp cells. Arch. Oral Biol. 52, 712–719. [DOI] [PubMed] [Google Scholar]
- 8. Park MG, Kim JS, Park SY, Lee SA, Kim HJ, Kim CS et al (2014) MicroRNA‐27 promotes the differentiation of odontoblastic cell by targeting APC and activating Wnt/beta‐catenin signaling. Gene 538, 266–272. [DOI] [PubMed] [Google Scholar]
- 9. Frade JM (2000) NRAGE and the cycling side of the neurotrophin receptor p75. Trends Neurosci. 23, 591–592. [DOI] [PubMed] [Google Scholar]
- 10. Sang M, Wang L, Ding C, Zhou X, Wang B, Wang L et al (2011) Melanoma‐associated antigen genes – an update. Cancer Lett. 302, 85–90. [DOI] [PubMed] [Google Scholar]
- 11. Bertrand MJ, Kenchappa RS, Andrieu D, Leclercq‐Smekens M, Nguyen HN, Carter BD et al (2008) NRAGE, a p75NTR adaptor protein, is required for developmental apoptosis in vivo. Cell Death Differ. 15, 1921–1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Masuda Y, Sasaki A, Shibuya H, Ueno N, Ikeda K, Watanabe K (2001) Dlxin‐1, a novel protein that binds Dlx5 and regulates its transcriptional function. J. Biol. Chem. 276, 5331–5338. [DOI] [PubMed] [Google Scholar]
- 13. Bragason BT, Palsdottir A (2005) Interaction of PrP with NRAGE, a protein involved in neuronal apoptosis. Mol. Cell Neurosci. 29, 232–244. [DOI] [PubMed] [Google Scholar]
- 14. Chu CS, Xue B, Tu C, Feng ZH, Shi YH, Miao Y et al (2007) NRAGE suppresses metastasis of melanoma and pancreatic cancer in vitro and in vivo. Cancer Lett. 250, 268–275. [DOI] [PubMed] [Google Scholar]
- 15. Yang Q, Ou C, Liu M, Xiao W, Wen C, Sun F (2014) NRAGE promotes cell proliferation by stabilizing PCNA in a ubiquitin‐proteasome pathway in esophageal carcinomas. Carcinogenesis 35, 1643–1651. [DOI] [PubMed] [Google Scholar]
- 16. Feng Z, Li K, Liu M, Wen C (2010) NRAGE is a negative regulator of nerve growth factor‐stimulated neurite outgrowth in PC12 cells mediated through TrkA‐ERK signaling. J. Neurosci. Res. 88, 1822–1828. [DOI] [PubMed] [Google Scholar]
- 17. Kuwajima T, Taniura H, Nishimura I, Yoshikawa K (2004) Necdin interacts with the Msx2 homeodomain protein via MAGE‐D1 to promote myogenic differentiation of C2C12 cells. J. Biol. Chem. 279, 40484–40493. [DOI] [PubMed] [Google Scholar]
- 18. Choi SJ, Song IS, Feng JQ, Gao T, Haruyama N, Gautam P et al (2010) Mutant DLX 3 disrupts odontoblast polarization and dentin formation. Dev. Biol. 344, 682–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Matluk N, Rochira JA, Karaczyn A, Adams T, Verdi JM (2010) A role for NRAGE in NF‐kappaB activation through the non‐canonical BMP pathway. BMC Biol. 8, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kang J, Chung KC (2014). The F‐box protein FBXO7 positively regulates bone morphogenetic protein‐mediated signaling through Lys‐63‐specific ubiquitination of neurotrophin receptor‐interacting MAGE (NRAGE). Cell. Mol. Life Sci. 72, 181–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dong X, Shen B, Ruan N, Guan Z, Zhang Y, Chen Y et al (2014) Expression patterns of genes critical for BMP signaling pathway in developing human primary tooth germs. Histochem. Cell Biol. 142, 657–665. [DOI] [PubMed] [Google Scholar]
- 22. Cao H, Jheon A, Li X, Sun Z, Wang J, Florez S et al (2013) The Pitx2:miR‐200c/141:noggin pathway regulates Bmp signaling and ameloblast differentiation. Development 140, 3348–3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Schurr T, Grover NB (1990) Analysis of a model for minichromosome segregation in Escherichia coli. J. Theor. Biol. 146, 395–406. [DOI] [PubMed] [Google Scholar]
- 24. Qin W, Lin ZM, Deng R, Li DD, Song Z, Tian YG et al (2012) p38a MAPK is involved in BMP‐2‐induced odontoblastic differentiation of human dental pulp cells. Int. Endod. J. 45, 224–233. [DOI] [PubMed] [Google Scholar]
- 25. Kim DS, Jue SS, Lee SY, Kim YS, Shin SY, Kim EC (2014) Effects of glutamine on proliferation, migration, and differentiation of human dental pulp cells. J. Endod. 40, 1087–1094. [DOI] [PubMed] [Google Scholar]
- 26. Yang Q, Ou C, Liu M, Xiao W, Wen C, Sun F (2014) NRAGE promotes cell proliferation by stabilizing PCNA in a ubiquitin‐proteasome pathway in esophageal carcinomas. Carcinogenesis 35, 1643–1651. [DOI] [PubMed] [Google Scholar]
- 27. Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real‐time quantitative PCR and the 2(‐Delta Delta C(T)) Method. Methods 25, 402–408. [DOI] [PubMed] [Google Scholar]
- 28. Yang W, Harris MA, Cui Y, Mishina Y, Harris SE, Gluhak‐Heinrich J (2012) Bmp2 is required for odontoblast differentiation and pulp vasculogenesis. J. Dent. Res. 91, 58–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Vainio S, Karavanova I, Jowett A, Thesleff I (1993) Identification of BMP‐4 as a signal mediating secondary induction between epithelial and mesenchymal tissues during early tooth development. Cell 75, 45–58. [PubMed] [Google Scholar]
- 30. Feng XY, Zhao YM, Wang WJ, Ge LH (2013) Msx1 regulates proliferation and differentiation of mouse dental mesenchymal cells in culture. Eur. J. Oral Sci. 121, 412–420. [DOI] [PubMed] [Google Scholar]
- 31. Lezot F, Thomas B, Greene SR, Hotton D, Yuan ZA, Castaneda B et al (2008) Physiological implications of DLX homeoproteins in enamel formation. J. Cell. Physiol. 216, 688–697. [DOI] [PubMed] [Google Scholar]
- 32. Qi SC, Cui C, Yan YH, Sun GH, Zhu SR (2013) Effects of high‐mobility group box 1 on the proliferation and odontoblastic differentiation of human dental pulp cells. Int. Endod. J. 46, 1153–1163. [DOI] [PubMed] [Google Scholar]
- 33. Gronthos S, Mankani M, Brahim J, Robey PG, Shi S (2000) Postnatal human dental pulp stem cells (DPSCs) in vitro and in vivo. Proc. Natl. Acad. Sci. USA 97, 13625–13630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chen S, Gluhak‐Heinrich J, Wang YH, Wu YM, Chuang HH, Chen L et al (2009) Runx2, osx, and dspp in tooth development. J. Dent. Res. 88, 904–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Iejima D, Sumita Y, Kagami H, Ando Y, Ueda M (2007) Odontoblast marker gene expression is enhanced by a CC‐chemokine family protein MIP‐3alpha in human mesenchymal stem cells. Arch. Oral Biol. 52, 924–931. [DOI] [PubMed] [Google Scholar]
- 36. Salehi AH, Roux PP, Kubu CJ, Zeindler C, Bhakar A, Tannis LL et al (2000) NRAGE, a novel MAGE protein, interacts with the p75 neurotrophin receptor and facilitates nerve growth factor‐dependent apoptosis. Neuron 27, 279–288. [DOI] [PubMed] [Google Scholar]
- 37. Kendall SE, Goldhawk DE, Kubu C, Barker PA, Verdi JM (2002) Expression analysis of a novel p75(NTR) signaling protein, which regulates cell cycle progression and apoptosis. Mech. Dev. 117, 187–200. [DOI] [PubMed] [Google Scholar]
- 38. Nikopoulos GN, Martins JF, Adams TL, Karaczyn A, Adams D, Vary C et al (2009) NRAGE: a potential rheostat during branching morphogenesis. Mech. Dev. 126, 337–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rochira JA, Matluk NN, Adams TL, Karaczyn AA, Oxburgh L, Hess ST et al (2011) A small peptide modeled after the NRAGE repeat domain inhibits XIAP‐TAB 1‐TAK1 signaling for NF‐kappaB activation and apoptosis in P19 cells. PLoS One 6, e20659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chou MY, Kao CT, Hung CJ, Huang TH, Huang SC, Shie MY et al (2014) Role of the P38 pathway in calcium silicate cement‐induced cell viability and angiogenesis‐related proteins of human dental pulp cell in vitro. J. Endod. 40, 818–824. [DOI] [PubMed] [Google Scholar]
- 41. Kendall SE, Battelli C, Irwin S, Mitchell JG, Glackin CA, Verdi JM (2005) NRAGE mediates p38 activation and neural progenitor apoptosis via the bone morphogenetic protein signaling cascade. Mol. Cell. Biol. 25, 7711–7724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lu M, Lin SC, Huang Y, Kang YJ, Rich R, Lo YC et al (2007) XIAP induces NF‐kappaB activation via the BIR1/TAB 1 interaction and BIR1 dimerization. Mol. Cell 26, 689–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hofer‐Warbinek R, Schmid JA, Stehlik C, Binder BR, Lipp J, de Martin R (2000) Activation of NF‐kappa B by XIAP, the X chromosome‐linked inhibitor of apoptosis, in endothelial cells involves TAK1. J. Biol. Chem. 275, 22064–22068. [DOI] [PubMed] [Google Scholar]
- 44. Shibuya H, Iwata H, Masuyama N, Gotoh Y, Yamaguchi K, Irie K et al (1998) Role of TAK1 and TAB 1 in BMP signaling in early Xenopus development. EMBO J. 17, 1019–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lee YH, Kang YM, Heo MJ, Kim GE, Bhattarai G, Lee NH et al (2013) The survival role of peroxisome proliferator‐activated receptor gamma induces odontoblast differentiation against oxidative stress in human dental pulp cells. J. Endod. 39, 236–241. [DOI] [PubMed] [Google Scholar]
- 46. Wang Y, Zheng Y, Wang Z, Li J, Wang Z, Zhang G et al (2013) 10(‐7) m 17beta‐oestradiol enhances odonto/osteogenic potency of human dental pulp stem cells by activation of the NF‐kappaB pathway. Cell Prolif. 46, 677–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lee SY, Kim SY, Park SH, Kim JJ, Jang JH, Kim EC (2012) Effects of recombinant dentin sialoprotein in dental pulp cells. J. Dent. Res. 91, 407–412. [DOI] [PubMed] [Google Scholar]
- 48. Wang Y, Yan M, Yu Y, Wu J, Yu J, Fan Z (2013) Estrogen deficiency inhibits the odonto/osteogenic differentiation of dental pulp stem cells via activation of the NF‐kappaB pathway. Cell Tissue Res. 352, 551–559. [DOI] [PubMed] [Google Scholar]


