Abstract
Objectives
Dental mesenchymal stem cells (MSCs) are easily obtained; however, mechanisms underlying directed differentiation of these cells remains unclear. Wnt/β‐catenin signalling is essential for mesenchymal cell commitment and differentiation, and Wnt inhibition is linked to stem cell maintenance and function. Secreted frizzled‐related protein 2 (SFRP2) competes with the Frizzled receptor for direct binding to Wnt and blocks activation of Wnt signalling. Here, we used stem cells derived from apical papillae (SCAPs) to study the functions of SFRP2.
Materials and methods
SCAPs were isolated from apical papillae of immature third molars. The cells were analysed using alkaline phosphatase activity assays, Alizarin red staining and quantitative calcium measurements. In addition, we evaluated expression profile of genes associated with osteogenesis and dentinogenesis (osteo‐/dentinogenesis), and conducted in vivo transplantation experiments to determine osteo‐/dentinogenic differentiation potential of SCAPs. ChIP assays were used to detect histone methylation at the SFRP2 promoter.
Results
We found that SFRP2 enhanced osteo‐/dentinogenic differentiation via Osterix, a key transcription factor in SCAPs. Furthermore, silencing SFRP2 induced SCAP cell death in osteogenic‐inducing medium, indicating that SFRP2 is a key factor in maintaining SCAP survival following osteo‐/dentinogenic commitment. Moreover, we found that silencing KDM2A, a histone demethylase and BCL6 co‐repressor, de‐repressed SFRP2 transcription by increasing histone H3K4 and H3K36 methylation at the SFRP2 promoter.
Conclusions
Our results have identified a new function of SFRP2 and shed new light on the molecular mechanism underlying directed differentiation of stem cells of dental origin.
Introduction
Mesenchymal stem cells (MSCs), originally isolated from bone marrow, are multipotent cells that can differentiate into different types of cells, including osteoblasts, chondrocytes, myocytes and adipocytes. Previous studies have demonstrated that MSCs are also present in non‐bone marrow tissues and that a plethora of adult MSCs are available for tissue engineering. MSCs derived from different tissues, such as bone marrow and periosteum, have similar epitope expression profiles. However, significant tissue‐specific differences have been observed in multiple MSC parameters, including differentiation, proliferation and migration potential 1, 2, 3, 4, 5. Recently, MSCs were isolated from various dental tissues, including stem cells from the periodontal ligament (PDLSCs), dental pulp stem cells (DPSCs) and stem cells from apical papilla (SCAPs) 6, 7, 8. These cells exhibit potent osteo‐/dentinogenic differentiation potential and are self‐renewable. When transplanted into animal models, dental tissue‐derived MSCs generate bone/dentin‐like mineralized tissues and can repair tooth defects 6, 7, 8, 9, 10. These cells are easily isolated and, in contrast to bone marrow stem cells, are more closely associated with dental tissues 6, 7, 8. However, their potential clinical applications are currently limited as the mechanism underlying their directed differentiation remains largely unknown.
MSC commitment and differentiation into osteocytes, chondrocytes and adipocytes requires Wnt/β‐catenin signalling 11, 12, 13, 14. Dkk1, a Wnt inhibitor, promotes MSC self‐renewal and osteogenic differentiation 15, 16. The proteins of the SFRP family, including SFRP1‐5, prevent the activation of Wnt signalling by directly binding Wnt 17, 18. A previous report demonstrated that SFRP1 enhanced MSCs surrounding neovessels, increasing vessel maturation and functionality 19. In a mouse model, a loss‐of‐function mutation in the Sfrp1 gene leads to a high bone mass phenotype, and Sfrp1 deletion enhances fracture repair by directing mesenchymal stem cells into the osteoblast lineage to promote early bone repair 20. SFRP2 is upregulated during MSC osteogenesis 21. It significantly enhances ALP activity in C3H10T1/2 cells 22. In addition, overexpression of SFRP2 increases cell survival in conditions of oxidative stress in MSCs derived from the bone marrow or umbilical cord 23. Animal studies demonstrated that intramyocardial implantation of MSCs overexpressing SFRP2 enhances cardiac wound repair 24, 25. SFRP3, another SFRP family member upregulated during MSC osteogenesis, promotes MSC osteoblastic differentiation and osteogenesis by inhibiting canonical Wnt signalling 21, 26. In an osteoblast apoptosis assay, SFRP3 was the only SFRP analysed that increased etoposide‐induced apoptosis in MC3T3‐E1 mouse osteoblasts 22. SFRP4 significantly increased ALP activity in C3H10T1/2 cells 22. However, another group demonstrated that the SFRP4 was downregulated during the induction of osteogenesis, thereby inhibiting osteoblastic differentiation in PDLSCs 26. In addition, SFRP4 is upregulated during adipogenic differentiation in human adipose tissue‐derived MSCs (ADSCs), and depletion of SFRP4 inhibits the differentiation of ADSCs into adipocytes 27, 28. SFRP5 expression is activated in fat tissue in obese humans and mice 29. SFRP5 expression gradually increases during adipocyte differentiation and is expressed at greater levels in mature adipocytes than in pre‐adipocytes 30, 31. Together, these data indicate that the SFRP family of genes may play an important role in the regulation of MSC differentiation; however, the precise mechanism by which SFRPs mediate this process remains unclear.
We previously demonstrated that BCOR inhibits MSC osteo‐/dentinogenic differentiation and proliferation and that a mutation in BCOR promotes MSC differentiation and growth. We compared the gene expression profile of SCAPs from healthy individuals with that of SCAPs from patients with oculo‐facio‐cardio‐dental (OFCD) syndrome using microarray analysis and demonstrated that SFRP2 was expressed at higher levels in SCAPs derived from patients with OFCD syndrome than in SCAPs derived from healthy individuals 32. These results suggest that SFRP2 may regulate differentiation or tissue regeneration in dental tissue‐derived MSCs. However, the precise function of SFRP2 in dental tissue‐derived MSCs remains unclear.
The present study revealed that SFRP2 enhanced osteo‐/dentinogenic differentiation in SCAPs. Knock‐down of BCOR or its co‐factor KDM2A directly de‐repressed the transcription of SFRP2 by increasing H3K4 and H3K36 methylation in the SFRP2 promoter, indicating that these molecules play a role in the regulation of MSC differentiation. These findings provide novel insight into the mechanism underlying the directed differentiation of dental tissue‐derived MSCs and have potentially valuable clinical applications.
Materials and methods
Cell cultures
Patient samples were obtained according to the approved guidelines established by Beijing Stomatological Hospital, Capital Medical University, and informed consent was obtained from all patients. Five human impacted third molars with immature roots (wisdom teeth) were collected from five healthy individuals (16–20 years old). The teeth were initially treated with 75% ethanol and washed with phosphate‐buffered saline. SCAPs were separated from the apical papilla of the root and digested in a solution of 3 mg/ml collagenase type I (Worthington Biochemical Corp., Lakewood, NJ, USA) and 4 mg/ml dispase (Roche Diagnostics Corp., Indianapolis, IN, USA) for 1 h at 37 °C. Single‐cell suspensions were acquired by passing the cells through a 70 μm strainer (Falcon; BD Biosciences, San Jose, CA, USA). SCAPs were cultured in DMEM (Invitrogen, Carlsbad, CA, USA) supplemented with 15% foetal bovine serum (Invitrogen) and 2 mmol/L glutamine, 100 U/ml penicillin and 100 μg/ml streptomycin (Invitrogen) in a humidified, 5% CO2 incubator at 37 °C. The culture medium was renewed every 3 days. The expression of phenotypic markers and lineage markers and the lineage differentiation potential of SCAPs were evaluated as previously described 33. The expression profiles of SCAPs were analysed using flow cytometry (Calibur; BD Biosciences) with phycoerythrin‐conjugated monoclonal antibodies against CD34, CD45 and CD105 (Biolegend, San Diego, CA, USA) (Fig. S1). Human embryonic kidney 293T cells were cultured in complete DMEM medium supplemented with 10% foetal bovine serum (Invitrogen), 100 μg/ml streptomycin and 100 U/ml penicillin (Invitrogen).
Plasmid construction and viral infection
Plasmids were constructed using routine methods and verified using appropriate restriction digestion and sequencing. Human full‐length SFRP2 cDNA was generated using a standard gene synthesis method and subcloned into a LV5 lentiviral vector (Genepharma Company, Suzhou, China). Specific short‐hairpin RNAs (shRNAs) were subcloned into a pLKO.1 lentiviral vector (Addgene, Cambridge, MA, USA). For the viral infection experiments, MSCs were infected with lentiviruses in the presence of 6 μg/ml polybrene (Sigma‐Aldrich, St. Louis, MO, USA) for 12 h. After 48 h, infected cells were selected with 2 μg/ml puromycin. The scramble shRNA control (Scramsh) was purchased from Addgene. The target sequences for the shRNAs are as follows: BCOR shRNA (BCORsh), 5′‐gatggcttcagtgctatat‐3′; KDM2A shRNA (KDM2Ash), 5′‐tttccaagccaatggtttc‐3′; SFRP2 shRNA (SFRP2sh), 5′‐ttgatgtaggttatctccttc‐3′; OSX shRNA (OSXsh), 5′‐cctcaggctatgctaatgatt‐3′; and LV3 shRNA (LV3sh), 5′‐ttctccgaacgtgtcacgtttc‐3′.
Reverse transcriptase polymerase chain reaction (RT‐PCR) and real‐time RT‐PCR
Total RNA was extracted from SCAPs using Trizol reagent (Invitrogen). We synthesized cDNA from 2 μg of RNA using random hexamers or oligo(dT)s and reverse transcriptase according to the manufacturer's protocol (Invitrogen). Real‐time PCR reactions were conducted using the QuantiTect SYBR Green PCR kit (Qiagen, Hilden, Germany) and an Icycler iQ Multi‐color Real‐time PCR Detection System. The primers used to amplify specific genes are provided in Table S1.
Alkaline phosphatase (ALP) and Alizarin red detection
SCAPs were grown in mineralization‐inducing media using the STEMPRO Osteogenesis Differentiation Kit (Invitrogen) for 2–3 weeks. ALP activity and Alizarin red staining assays were conducted as previously described 32. ALP activity was assayed using an ALP activity kit (Sigma‐Aldrich). Induced cells were fixed with 70% ethanol and stained with 2% Alizarin red (Sigma‐Aldrich).
Transplantation experiments
This study was approved by the Animal Care and Use Committee of Beijing Stomatological Hospital, Capital Medical University. Animal experiments were conducted in accordance with the National Institute of Health Guide for the Care and Use of Laboratory Animals (NIH Publications No.8023, revised 1978). The animals were purchased from the Institute of Animal Science of the Vital River Co., Ltd. The animals had not previously received drugs or other procedures. Approximately, 4.0 × 106 cells were combined with 40 mg of HA/tricalcium phosphate ceramic particles (Engineering Research Center for Biomaterials, Sichuan University, Chengdu, China) and subcutaneously transplanted into the dorsal skin of 10‐week‐old immunocompromised beige mice (nu/nu nude mice). The transplantation experiments were conducted in accordance with the approved animal protocol. Eight weeks after the procedure, the implants were harvested, fixed with 10% formalin, decalcified using 10% EDTA buffer (pH 8.0) and embedded in paraffin. Sections were stained with haematoxylin and eosin (H&E) and Goldner's trichrome. The bone/dentin‐like areas were measured in sections stained with H&E and qualitatively evaluated using the Image‐Pro Plus 6.0 program (Media Cybernetics, Rockville, MD, USA).
Goldner's trichrome staining
Goldner's trichrome staining (Catalog #26386; Electron Microscopy Sciences, Hatfield, PA, USA) assays were conducted according to the manufacturer's protocol. The sections were stained with Bouin's Fluid solution and washed in tap water. Then, they were incubated with Weigert's haematoxylin, stained with ponceau acid fuchsin solution and rinsed with 1% acetic acid. The sections were subsequently incubated in a phosphomolybdic acid‐orange G solution and washed with 1% acetic acid. Finally, the sections were stained using a Light Green stock solution. According to this protocol, red staining labelled mature bone, orange staining labelled red blood cells and blue staining labelled regenerated bone, osteoid or collagen fibres.
ChIP assays
A ChIP assay kit (Merck Millipore, Darmstadt, Germany) was used according to the manufacturer's protocol. Briefly, cells were incubated with 1% formaldehyde for 10 min at 37 °C. Approximately, 2 × 106 cells were used in each ChIP experiment. DNA was precipitated using 2 μg of a monoclonal antibody against HA (Clone No. C29F4, Cat No. 3724; Cell Signaling Technology, Beverly, MA, USA), a polyclonal antibody against trimethylated H3K4 (H3K4me3) (Cat No.07‐473; Merck Millipore) or a polyclonal antibody against dimethylated H3K36 (H3K36me2) (Cat No.07‐369; Merck Millipore). The immunoprecipitated DNA samples were analysed using real‐time PCR. The sequences of the primers used to amplify the KDM2A binding site in the SFRP2 promoter are as follows: forward, 5′‐cgtatgccatgtaaagttctgctcatacg‐3′; and reverse, 5′‐gttcagcagcctgtcggtgt‐3′.
Western blot analysis
Cells were harvested in RIPA buffer. Samples were separated on a 10% SDS polyacrylamide gel and transferred to polyvinylidene difluoride membranes using a semi‐dry transfer apparatus (Bio‐Rad Laboratories, Hercules, CA, USA). The membranes were blocked with a 5% milk solution for 2 h at room temperature and incubated with primary antibodies overnight. The blot was subsequently incubated with horseradish peroxidase‐conjugated anti‐rabbit or anti‐mouse IgG secondary antibodies (Promega, Madison, WI, USA), and the signal was detected using SuperSignal reagents (Pierce, Rockford, IL, USA). The following primary antibodies were used in the Western blot assays: anti‐HA (Clone No. C29F4, Cat No. 3724; Cell Signaling Technology), anti‐OSX (Cat No. ab22552; Abcam) and anti‐β‐actin (Cat No. C1313; Applygen, Beijing, China).
Counting viable cells
SCAPs were grown in mineralization‐inducing media using the STEMPRO Osteogenesis Differentiation Kit (Invitrogen) for 48 h. Cells were digested with 0.25% trypsin (Invitrogen) and resuspended in 1 ml of PBS. The cells were quantified using an automated cell counter (TC10TM, Bio‐Rad Laboratories). Trypan blue was added to the cell suspension to label and eliminate non‐viable cells.
Statistics
All statistical calculations were performed using SPSS 13 statistical software. Student's t‐test or one‐way ANOVA was used to determine statistical significance. P ≤ 0.05 was considered statistically significant.
Results
Overexpression of SFRP2 enhanced the osteo‐/dentinogenic differentiation potential of MSCs
We first examined the expression of SFRP2 in MSCs at different stages of osteogenic differentiation. During the induction of SCAP differentiation into osteo‐/dentinogenic cells, SFRP2 mRNA levels were upregulated in a time‐dependent manner (Fig. 1). To determine the function of SFRP2 in MSCs, we ectopically overexpressed SFRP2 by infecting SCAPs with a lentiviral construct expressing SFRP2. Transduced cells were selected after 7 days of treatment using 2 μg/ml puromycin, and ectopic SFRP2 expression was confirmed using real‐time RT‐PCR analysis (Fig. 2a).
Figure 1.
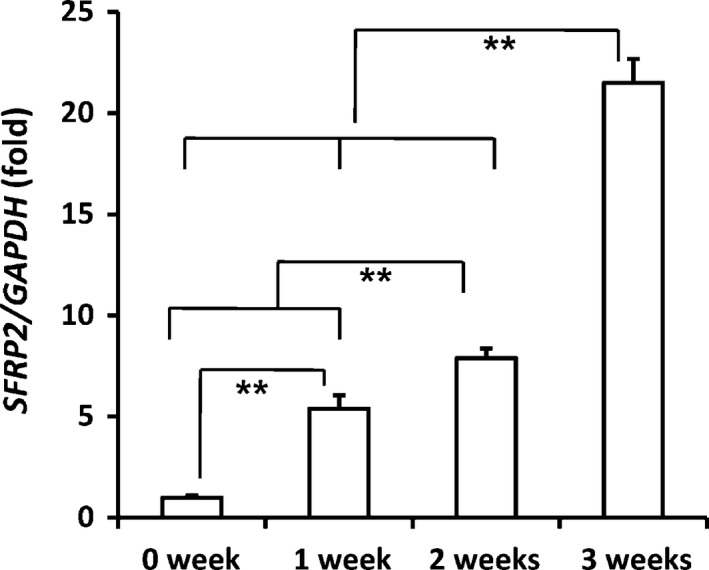
SFRP 2 expression increased in differentiated SCAP s. Real‐time RT‐PCR analysis of SFRP2 mRNA levels in differentiated SCAPs. GAPDH was used as an internal control. One‐way ANOVA was used to calculate statistical significance. Error bars represent the SD (n = 3). **P ≤ 0.01.
Figure 2.
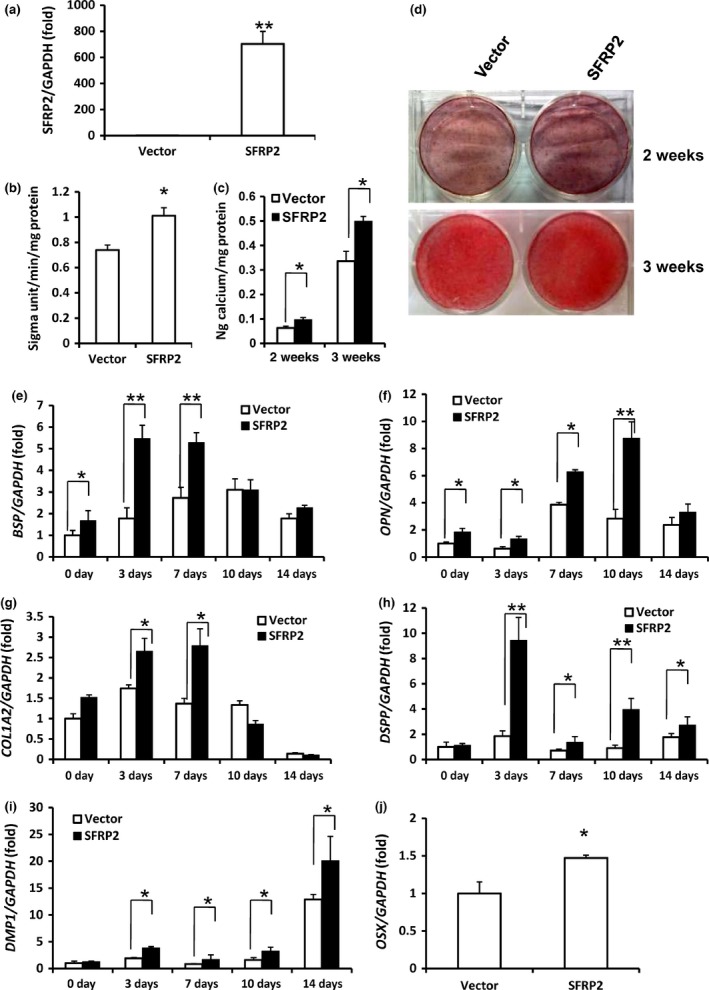
SFRP 2 overexpression enhanced SCAP osteo‐/dentinogenic differentiation. (a) Real‐time RT‐PCR verified that SCAPs infected with the lentivirus construct overexpressed SFRP2. (b) ALP activity. (c) Quantitative calcium analysis. (d) Alizarin red staining. (e–j) Real‐time RT‐PCR analysis of the expression of BSP (e), OPN (f), COL1A2 (g), DSPP (h), DMP1 (i) and OSX (j). GAPDH was used as an internal control. Student's t‐test was used to determine statistical significance. Error bars represent the SD (n = 3). *P ≤ 0.05; **P ≤ 0.01.
The transduced SCAP cells were cultured in osteogenic‐inducing medium for 5 days. Overexpression of wild‐type SFRP2 increased ALP activity, an early marker of osteo‐/dentinogenic differentiation in SCAPs (Fig. 2b). Accordingly, 2 and 3 weeks after induction, mineralization was markedly enhanced in SCAPs overexpressing SFRP2 compared with cells infected with the empty vector, as demonstrated by Alizarin red staining and calcium levels (Fig. 2c,d). Consistent with these findings, real‐time RT‐PCR analysis revealed that the osteogenic marker gene bone sialoprotein (BSP), which encodes bone extracellular matrix proteins, was expressed at greater levels in SCAP‐SFRP2 cells than in control SCAP cells on 0, 3 and 7 days after induction (Fig. 2e). The expression of another osteogenic marker, osteopontin (OPN), also increased on 0, 3, 7 and 10 days after induction in SCAP‐SFRP2 cells compared with SCAP control cells (Fig. 2f). In addition, expression of the osteogenic marker collagen type I alpha 2 (COL1A2) was strongly enhanced on 3 and 7 days (Fig. 2g) after induction in SCAP‐SFRP2 cells compared with SCAP control cells. The dentinogenic markers, dentin sialophosphoprotein (DSPP) and dentin matrix protein 1 (DMP1), both of which encode extracellular matrix proteins of dentin, were expressed at higher levels in SCAP‐SFRP2 cells compared with SCAP control cells on 3, 7, 10 and 14 days after induction (Fig. 2h,i).
We also examined the expression of key transcription factors involved in MSC osteo‐/dentinogenic differentiation, including runt‐related transcription factor 2 (RUNX2), Osterix (OSX) and distal‐less homeobox factors (DLX) 2, 3 and 5. The mRNA levels of OSX were significantly greater in SCAP‐SFRP2 cells than in SCAP control cells on the day that the cells were infected (Fig. 2j), but the expression levels of other genes did not significantly change (data not shown).
Finally, we investigated if SFRP2 overexpression affects SCAP osteo‐/dentinogenesis in vivo. To this end, we subcutaneously transplanted control or SFRP2‐overexpressing SCAPs into nude mice. The transplanted tissues were harvested 8 weeks after cell transplantation. H&E and Goldner's trichrome staining revealed an increase in bone/dentin‐like tissues in tissues derived from transplanted SFRP2‐overexpressing SCAPs (35.02 ± 6.92%) compared with tissues derived from transplanted control SCAPs (14.98 ± 3.07%) (Fig. 3a–c). Taken together, these data demonstrated that SFRP2 expression strongly enhanced osteo‐/dentinogenic differentiation in SCAPs in vivo.
Figure 3.
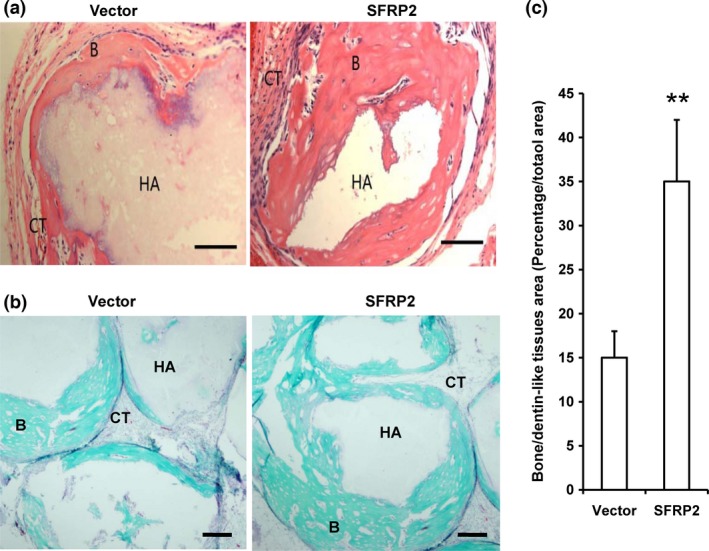
SFRP 2 overexpression enhanced bone/dentin‐like tissue formation in SCAP s. (a) H&E‐stained micrographs of SFRP2‐overexpressing SCAPs transplanted into nude mice for 8 weeks exhibited bone/dentin‐like tissues. (b) The transplants were examined using Goldner's trichrome staining. (c) Qualitative measurement of bone/dentin‐like tissues in tissue samples. Student's t‐test was used to determine statistical significance. Error bars represent the SD (n = 5). **P ≤ 0.01. Bar: 100 μm. B, bone/dentin‐like tissues; HA, hydroxyapatite tricalcium carrier; CT, connective tissue.
Depletion of SFRP2 inhibited the osteo‐/dentinogenic differentiation potential of MSCs
To investigate the effect of SFRP2 depletion on MSC osteo‐/dentinogenic differentiation, we designed a shRNA lentiviral construct targeting SFRP2 and introduced it into SCAPs (SCAP‐SFRP2sh cells). The knock‐down efficiency of the shRNA (90%) was determined using real‐time RT‐PCR (Fig. 4a). The mRNA level of OSX significantly decreased in SCAP‐SFRP2sh cells compared with SCAP cells infected with Scramsh control shRNA (SCAP‐Scramsh) (Fig. 4b). Interestingly, shRNA‐mediated SFRP2 silencing induced cell death in SCAPs cultured in osteogenic‐inducing medium for 48 h. In contrast, shRNA‐mediated OSX silencing did not affect cell survival in SCAPs cultured in osteogenic‐inducing medium for 48 h. In addition, mineralization was successfully induced by osteogenic medium in OSX‐depleted SCAPs in vitro (Fig. S2).
Figure 4.
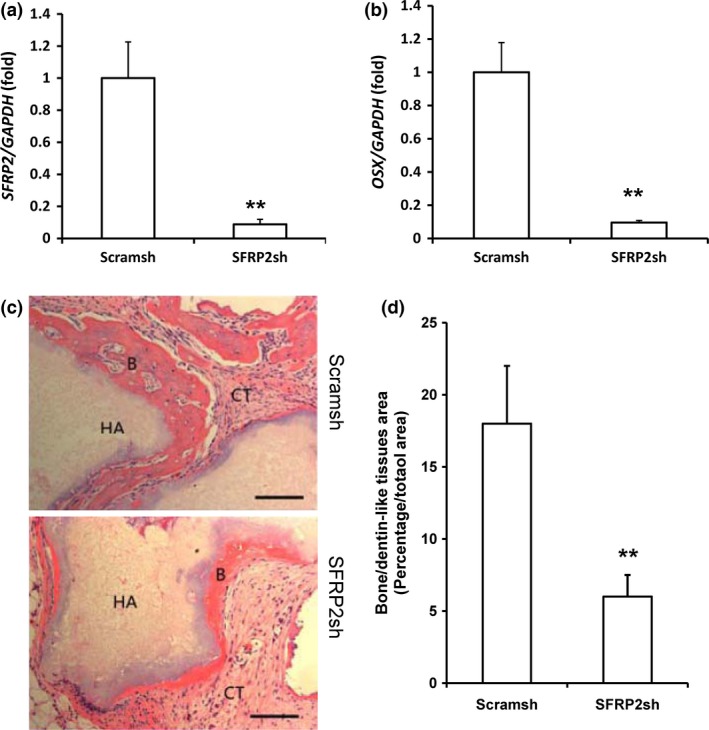
SFRP 2 knock‐down inhibited osteo‐/dentinogenic potential in SCAP s. (a) Real‐time RT‐PCR analysis of SFRP2 expression after SFRP2 depletion in SCAPs. (b) Real‐time RT‐PCR analysis of OSX expression in SFRP2‐depleted SCAPs. GAPDH was used as an internal control. Error bars represent the SD (n = 3). (c) H&E‐stained micrographs of bone/dentin‐like tissue formation derived from SFRP2‐shRNA cells transplanted into nude mice for 8 weeks. (d) Qualitative measurement of bone/dentin‐like tissues in transplant tissue samples. Student's t‐test was used to determine statistical significance. Error bars represent the SD (n = 5). **P ≤ 0.01. Bar: 100 μm. B, bone/dentin‐like tissues; HA, hydroxyapatite tricalcium carrier; CT, connective tissue.
We then subcutaneously transplanted SCAP‐Scramsh or SCAP‐SFRP2sh cells into nude mice and harvested the transplanted tissues 8 weeks later. H&E staining revealed that less bone/dentin‐like tissues were present in tissues derived from SCAP‐SFRP2sh cells (6.04 ± 1.58%) than in tissues derived from SCAP‐Scramsh cells (17.93 ± 4.12%) (Fig. 4c,d). These experiments demonstrated that SCAP‐SFRP2sh inhibited bone/dentin‐like tissue generation, indicating that SFRP2 knock‐down significantly inhibited osteo‐/dentinogenic differentiation in SCAPs in vivo.
Depletion of KDM2A or BCOR promoted SFRP2 transcription by increasing histone H3K4 and H3K36 methylation of the SFRP2 promoter
BCOR shRNA (BCORsh) was used to silence BCOR in SCAPs, and real‐time RT‐PCR analysis revealed that the knock‐down efficiency of BCORsh was 60% relative to the Scramsh control in SCAPs (Fig. 5a). Real‐time RT‐PCR analysis revealed that the expression of SFRP2 increased in BCORsh‐infected SCAPs compared with Scramsh‐infected SCAPs (Fig. 5b). In addition, we silenced KDM2A using a KDM2A shRNA (KDM2Ash) lentiviral construct in SCAPs, and real‐time RT‐PCR analysis demonstrated that the knock‐down efficiency of KDM2Ash was 70% relative to Scramsh (Fig. 5c). Real‐time RT‐PCR results indicated that the expression of SFRP2 increased in KDM2A‐depleted SCAPs compared with Scramsh‐infected SCAPs (Fig. 5d). Next, we investigated if the KDM2A–BCOR complex can demethylate histones H3K36 or H3K4 in the SFRP2 promoter. Using ChIP assays, we found that KDM2A knock‐down increased H3K36me2 and H3K4me3 levels in the SFRP2 promoter (Fig. 5e–g), and similar results were observed with BCOR knock‐down (Fig. 5h,i). To further investigate the activity of the KDM2A–BCOR complex at the SFRP2 promoter, we infected SCAPs with retroviruses overexpressing HA‐KDM2A or HA‐BCOR or with an empty control HA‐vector (Fig. 5j,l) and analysed the cells using ChIP assays with anti‐HA antibodies. Compared with the HA control, significantly greater levels of HA‐KDM2A and HA‐BCOR precipitated with the SFRP2 promoter (Fig. 5k,m).
Figure 5.
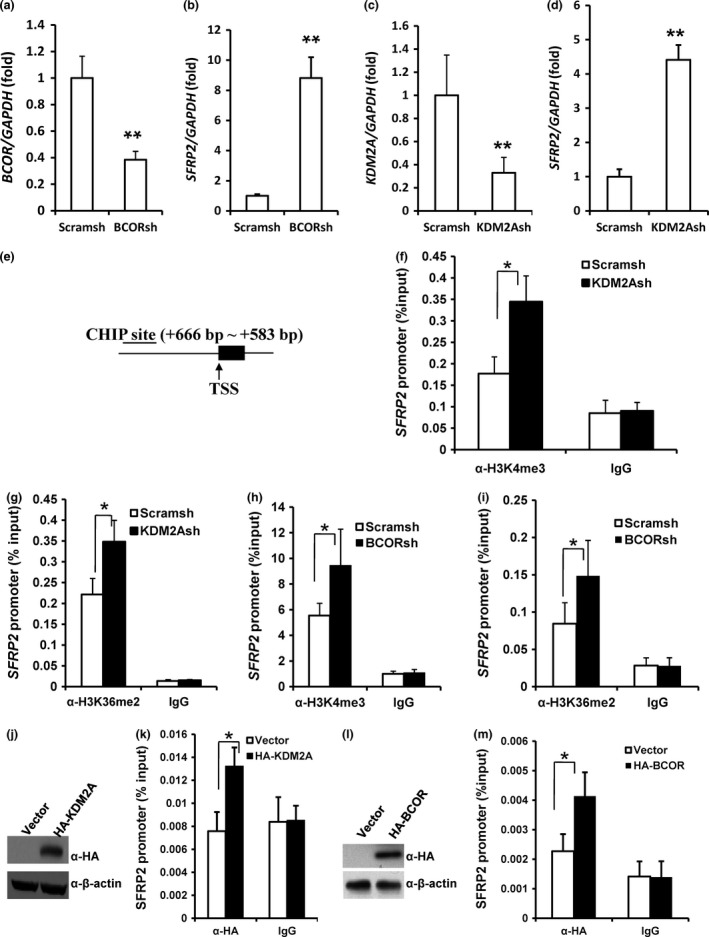
Depletion of KDM 2A or BCOR increased SFRP 2 expression in SCAP s by increasing histone K4/36 methylation at the SFRP 2 promoter. Real‐time RT‐PCR analysis of BCOR (a), SFRP2 (b, d) and KDM2A (c) in cells infected with BCOR shRNA (a, b) or KDM2A shRNA (c, d). GAPDH was used as an internal control. (e) Location of ChIP assay primers that targeted the SFRP2 promoter 666 to 583 bp upstream of the transcription start site (TSS). (f–i) ChIP analysis of changes in histone methylation in the SFRP2 promoter caused by infection with shRNA to silence either KDM2A (f, g) or BCOR (h, i) expression. (j) Western blot assays demonstrated that HA‐KDM2A‐infected SCAPs overexpressed KDM2A. (k) HA‐KDM2A overexpression enhanced the levels of KDM2A observed at the SFRP2 promoter. (l) Western blot assays demonstrated that HA‐BCOR‐infected SCAPs overexpressed BCOR. (m) HA‐BCOR overexpression enhanced the level of BCOR observed at the SFRP2 promoter. H3K36me2, Histone K36 dimethylation; H3K4me3, histone K4 trimethylation. Student's t‐test was used to determine statistical significance. Error bars represent the SD (n = 3). *P ≤ 0.05; **P ≤ 0.01.
Discussion
Recent years have witnessed significant progress in the field of MSC‐mediated tissue regeneration 3, 6, 8, 9. However, the mechanism underlying the directed differentiation of MSCs remains unclear.
MSC commitment and differentiation into osteocytes, chondrocytes and adipocytes requires Wnt/β‐catenin signalling 11, 12, 13, 14. In an animal model of accelerated ageing, the secreted protein Klotho directly binds to Wnt and prevents the activation of Wnt signalling, and the early‐onset ageing phenotype of Klotho‐deficient mice results from chronic activation of Wnt signalling 34. In addition, muscle stem cells derived from aged mice preferentially differentiate into fibrogenic rather than myogenic cell types, and this effect is associated with the activation of Wnt signalling 35. These findings indicate that Wnt signalling plays an important role in stem cell maintenance and function 13, 14, 34, 35.
SFRP2 directly binds to Wnt, thereby preventing activation of Wnt signalling 17, 18, 36. Using microarray analysis, we previously found that SFRP2 expression was 15 times greater in SCAPs derived from patients with OFCD than in SCAPs derived from healthy individuals 32. In the present study, we found that SFRP2 was upregulated during osteogenic differentiation in a time‐dependent manner in MSCs derived from dental tissue. Moreover, in gain‐of‐function studies, SFRP2 enhanced osteo‐/dentinogenic differentiation in SCAPs and increased the expression of BSP, COL1A2, OPN, DSPP and DMP1 genes that encode various extracellular matrix proteins found in bone and dentin. RUNX2 and OSX are the two key transcription factors required for osteogenic differentiation 37. RUNX2 mediates the activation of osteo‐/dentinogenic differentiation, and OSX is a downstream target gene of RUNX2 38. Members of the DLX gene family, specifically DLX2, DLX3 and DLX5, play important roles in osteoblast differentiation 39, 40, 41, 42, 43, and DLX transcription factors can induce the expression of RUNX2 and OSX 39, 40, 41, 42, 43, 44, 45. We demonstrated that SFRP2 was able to activate the expression of the key transcription factor OSX but did not affect RUNX2, DLX2, DLX3 and DLX5 expression. These findings indicated that SFRP2 activation of OSX expression is independent of the transcription factor genes RUNX2, DLX2, DLX3 and DLX5. Then, shRNA‐mediated silencing of SFRP2 induced cell death in SCAPs after 48 h in osteogenic‐inducing culture medium, indicating that SFRP2 is critical to maintaining SCAP survival following osteo‐/dentinogenic commitment. An in vitro study demonstrated that SFRP2 protected mammalian cell lines from UV‐ and tumour necrosis factor‐induced apoptosis 46. SFRP2 has also been reported to protect cardiomyocytes from hypoxia‐induced apoptosis 47. In addition, SFRP2 increases MSC survival in oxidative stress 23. Based on these findings, we hypothesized that SFRP2 depletion induced cell death in SCAPs cultured in osteogenic‐inducing medium by depleting OSX levels. However, we found that OSX depletion did not affect cell survival in SCAPs cultured in osteogenic‐inducing medium. These data indicated that SFRP2 depletion‐mediated cell death was independent of OSX. Interestingly, the expression of OSX significantly decreased in SFRP2‐depleted SCAPs, suggesting that SFRP2 knock‐down reduces the osteo‐/dentinogenic differentiation potential of SCAPs and that SFRP2 promotes osteo‐/dentinogenic differentiation.
The therapeutic potential of MSCs can be assessed using in vitro assays that measure cell growth, proliferation and viability 48. MSCs that exhibit robust growth, proliferation and viability have greater potential to generate vascularized granulation tissue and facilitate long‐term MSC engraftment 48, 49. These discoveries strongly suggest that increasing growth, proliferation and viability of MSCs may enhance vascularization and tissue regeneration potential. Together, our findings that SFRP2 promotes MSC differentiation in vitro support the hypothesis that SFRP2 enhances tissue regeneration potential. Indeed, SFRP2 considerably enhanced osteo‐/dentinogenesis in transplantation experiments in vivo.
Previous studies using microarray analysis demonstrated that mutant BCOR increased the expression of SFRP2 compared with WT BCOR 32. In the present study, we confirmed that SFRP2 is a downstream target of BCOR. BCOR forms a protein complex with KDM2A that regulates downstream targets and MSC functions 50. Therefore, we hypothesized that KDM2A and BCOR targeted SFRP2. Indeed, we found that silencing KDM2A or BCOR increased the expression of SFRP2 and that inhibiting KDM2A or BCOR function increased the expression of osteo‐/dentinogenic markers and enhanced osteo‐/dentinogenesis in vivo 32, 50. Together these findings indicate that the enhanced osteo‐/dentinogenesis resulting from KDM2A or BCOR inhibition may be mediated by SFRP2; however, further investigation is required to elucidate the precise mechanism by which this occurs.
In addition, we investigated if KDM2A or BCOR can demethylate H3K36me2 or H3K4me3 at the SFRP2 promoter and thus regulate SFRP2 transcription. We found that silencing either KDM2A or BCOR increased H3K36me2 and H3K4me3 levels at the SFRP2 promoter and upregulated SFRP2 expression. The presence of KDM2A and BCOR at the SFRP2 promoter was enhanced in KDM2A‐ or BCOR‐overexpressing SCAPs compared with control cells, indicating that KDM2A or BCOR can bind to the SFRP2 promoter. These results also indicated that KDM2A demethylated histones at the SFRP2 promoter by directly binding BCOR to form a protein complex that inhibited SFRP2 transcription in SCAPs.
In conclusion, depletion of KDM2A and BCOR increased H3K4 and H3K36 methylation at the SFRP2 promoter and resulted in the depression of SFRP2 transcription. SFRP2 enhanced the osteo‐/dentinogenic differentiation potential of SCAPs by activating the key transcription factor OSX. Our work identified novel SFRP2 functions and provided insight into the molecular mechanisms underlying the directed differentiation of MSCs. These findings may contribute to the identification of potential therapeutic gene targets that may enhance the efficacy of tissue regeneration approaches.
Conflicts of interest
The authors declare no potential conflicts of interest.
Supporting information
Fig. S1 Flow cytometry analysis of cell surface makers. The results indicated that SCAPs were positive for CD105 and negative for CD34 and CD45. The peak dividing the positive and negative sides depicts the cell population stained with the appropriate antibody.
Fig. S2 Cell survival was unaffected in SCAPs depleted of OSX. (a) Western blot analysis of OSX knock‐down in SCAPs. Beta‐actin was used as an internal control. (b) Alizarin red staining. (c) SCAPs were cultured with osteogenic medium for 48 h. No significant difference in the number of viable cells was observed between the OSX knock‐down group and the control group. Student's t‐test was used to determine statistical significance. Error bars represent the SD (n = 3). NS, no significant difference.
Table S1 Primers sequences used in the real‐time RT‐PCR.
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (81271100 and 81570936 to R.D.), the Beijing Natural Science Foundation (5132011 to Z.P.F.), the Program for New Century Excellent Talents in University (NCET‐12‐0611 to Z.P.F.), the Program for “Hundred‐Thousand‐Ten thousand” Talents in Beijing (2014006 to Z.P.F.), High‐level Talents of the Beijing Health System (2013‐3‐034 to R.D., 2013‐3‐035 to Z.P.F.) and the Beijing Municipal Government (TJSHG201310025005 to S.L.W.).
References
- 1. Jahagirdar BN, Verfaillie CM (2005) Multipotent adult progenitor cell and stem cell plasticity. Stem Cell Rev. 1, 53–59. [DOI] [PubMed] [Google Scholar]
- 2. Phinney DG, Prockop DJ (2007) Concise review: mesenchymal stem/multipotent stromal cells: the state of transdifferentiation and modes of tissue repair–current views. Stem Cells 25, 2896–2902. [DOI] [PubMed] [Google Scholar]
- 3. Dulak J, Szade K, Szade A, Nowak W, Józkowicz A (2015) Adult stem cells: hopes and hypes of regenerative medicine. Acta Biochim. Pol. 62, 329–337. [DOI] [PubMed] [Google Scholar]
- 4. Caplan AI (2015) Adult mesenchymal stem cells: when, where, and how. Stem Cells Int. 2015, 628767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Elahi KC, Klein G, Avci‐Adali M, Sievert KD, MacNeil S, Aicher WK (2016) Human mesenchymal stromal cells from different sources diverge in their expression of cell surface proteins and display distinct differentiation patterns. Stem Cells Int. 2016, 5646384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Huang GT, Gronthos S, Shi S (2009) Mesenchymal stem cells derived from dental tissues vs. those from other sources: their biology and role in regenerative medicine. J. Dent. Res. 88, 792–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Saito MT, Silvério KG, Casati MZ, Sallum EA, Nociti FH Jr (2015) Tooth‐derived stem cells: update and perspectives. World J. Stem Cells 7, 399–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Park YJ, Cha S, Park YS (2016) Regenerative applications using tooth derived stem cells in other than tooth regeneration: a literature review. Stem Cells Int. 2016, 9305986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wei F, Song T, Ding G, Liu Y, Liu D, Fan Z et al (2013) Functional tooth restoration by allogeneic mesenchymal stem cell‐based bio‐root regeneration in swine. Stem Cells Dev. 22, 1752–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Liu O, Xu J, Ding G, Liu D, Fan Z, Zhang C et al (2013) Periodontal ligament stem cells regulate B lymphocyte function via programmed cell death protein 1. Stem Cells 31, 1371–1382. [DOI] [PubMed] [Google Scholar]
- 11. Gaur T, Lengner CJ, Hovhannisyan H, Bhat RA, Bodine PV, Komm BS et al (2005) Canonical WNT signaling promotes osteogenesis by directly stimulating Runx2 gene expression. J. Biol. Chem. 280, 33132–33140. [DOI] [PubMed] [Google Scholar]
- 12. Yano F, Kugimiya F, Ohba S, Ikeda T, Chikuda H, Ogasawara T et al (2005) The canonical Wnt signaling pathway promotes chondrocyte differentiation in a Sox9‐dependent manner. Biochem. Biophys. Res. Commun. 333, 1300–1308. [DOI] [PubMed] [Google Scholar]
- 13. D'Alimonte I, Lannutti A, Pipino C, Di Tomo P, Pierdomenico L, Cianci E et al (2013) Wnt signaling behaves as a”master regulator” in the osteogenic and adipogenic commitment of human amniotic fluid mesenchymal stem cells. Stem Cell Rev. 9, 642–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Visweswaran M, Pohl S, Arfuso F, Newsholme P, Dilley R, Pervaiz S et al (2015) Multi‐lineage differentiation of mesenchymal stem cells ‐ To Wnt, or not Wnt. Int. J. Biochem. Cell Biol. 68, 139–147. [DOI] [PubMed] [Google Scholar]
- 15. Gregory CA, Perry AS, Reyes E, Conley A, Gunn WG, Prockop DJ (2005) Dkk‐1‐derived synthetic peptides and lithium chloride for the control and recovery of adult stem cells from bone marrow. J. Biol. Chem. 280, 2309–2323. [DOI] [PubMed] [Google Scholar]
- 16. Liu Q, Hu CH, Zhou CH, Cui XX, Yang K, Deng C et al (2015) DKK1 rescues osteogenic differentiation of mesenchymal stem cells isolated from periodontal ligaments of patients with diabetes mellitus induced periodontitis. Sci. Rep. 5, 13142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kawano Y, Kypta R (2003) Secreted antagonists of the Wnt signalling pathway. J. Cell Sci. 116, 2627–2634. [DOI] [PubMed] [Google Scholar]
- 18. Mii Y, Taira M (2011) Secreted Wnt”inhibitors” are not just inhibitors: regulation of extracellular Wnt by secreted Frizzled‐related proteins. Dev. Growth Differ. 53, 911–923. [DOI] [PubMed] [Google Scholar]
- 19. Dufourcq P, Descamps B, Tojais NF, Leroux L, Oses P, Daret D et al (2008) Secreted frizzled‐related protein‐1 enhances mesenchymal stem cell function in angiogenesis and contributes to neovessel maturation. Stem Cells 26, 2991–3001. [DOI] [PubMed] [Google Scholar]
- 20. Gaur T, Wixted JJ, Hussain S, O'Connell SL, Morgan EF, Ayers DC et al (2009) Secreted frizzled related protein 1 is a target to improve fracture healing. J. Cell. Physiol. 220, 174–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Boland GM, Perkins G, Hall DJ, Tuan RS (2004) Wnt 3a promotes proliferation and suppresses osteogenic differentiation of adult human mesenchymal stem cells. J. Cell. Biochem. 93, 1210–1230. [DOI] [PubMed] [Google Scholar]
- 22. Cho SW, Her SJ, Sun HJ, Choi OK, Yang JY, Kim SW et al (2008) Differential effects of secreted frizzled‐related proteins (sFRPs) on osteoblastic differentiation of mouse mesenchymal cells and apoptosis of osteoblasts. Biochem. Biophys. Res. Commun. 367, 399–405. [DOI] [PubMed] [Google Scholar]
- 23. Pomduk K, Kheolamai P, U‐Pratya Y, Wattanapanitch M, Klincumhom N, Issaragrisil S (2015) Enhanced human mesenchymal stem cell survival under oxidative stress by overexpression of secreted frizzled‐related protein 2 gene. Ann. Hematol. 94, 319–327. [DOI] [PubMed] [Google Scholar]
- 24. Alfaro MP, Pagni M, Vincent A, Atkinson J, Hill MF, Cates J et al (2008) The Wnt modulator sFRP2 enhances mesenchymal stem cell engraftment, granulation tissue formation and myocardial repair. Proc. Natl Acad. Sci. USA 105, 18366–18371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Alfaro MP, Vincent A, Saraswati S, Thorne CA, Hong CC, Lee E et al (2010) sFRP2 suppression of bone morphogenic protein (BMP) and Wnt signaling mediates mesenchymal stem cell (MSC) self‐renewal promoting engraftment and myocardial repair. J. Biol. Chem. 285, 35645–35653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yamada A, Iwata T, Yamato M, Okano T, Izumi Y (2013) Diverse functions of secreted frizzled‐related proteins in the osteoblastogenesis of human multipotent mesenchymal stromal cells. Biomaterials 34, 3270–3278. [DOI] [PubMed] [Google Scholar]
- 27. Park JR, Jung JW, Lee YS, Kang KS (2008) The roles of Wnt antagonists Dkk1 and sFRP4 during adipogenesis of human adipose tissue‐derived mesenchymal stem cells. Cell Prolif. 41, 859–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Visweswaran M, Schiefer L, Arfuso F, Dilley RJ, Newsholme P, Dharmarajan A (2015) Wnt antagonist secreted frizzled‐related protein 4 upregulates adipogenic differentiation in human adipose tissue‐derived mesenchymal stem cells. PLoS ONE 10, e0118005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mori H, Prestwich TC, Reid MA, Longo KA, Gerin I, Cawthorn WP et al (2012) Secreted frizzled‐related protein 5 suppresses adipocyte mitochondrial metabolism through WNT inhibition. J. Clin. Invest. 122, 2405–2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lv C, Jiang Y, Wang H, Chen B (2012) Sfrp5 expression and secretion in adipocytes are up‐regulated during differentiation and are negatively correlated with insulin resistance. Cell Biol. Int. 36, 851–855. [DOI] [PubMed] [Google Scholar]
- 31. Wang R, Hong J, Liu R, Chen M, Xu M, Gu W et al (2014) SFRP5 acts as a mature adipocyte marker but not as a regulator in adipogenesis. J. Mol. Endocrinol. 53, 405–415. [DOI] [PubMed] [Google Scholar]
- 32. Fan Z, Yamaza T, Lee JS, Yu J, Wang S, Fan G et al (2009) BCOR regulates mesenchymal stem cell function by epigenetic mechanisms. Nat. Cell Biol. 11, 1002–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cao Y, Xia DS, Qi SR, Du J, Ma P, Wang SL et al (2013) Epiregulin can promote proliferation of stem cells from the dental apical papilla via MEK/Erk and JNK signalling pathways. Cell Prolif. 46, 447–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Liu H, Fergusson MM, Castilho RM, Liu J, Cao L, Chen J et al (2007) Augmented Wnt signaling in a mammalian model of accelerated aging. Science 317, 803–806. [DOI] [PubMed] [Google Scholar]
- 35. Brack AS, Conboy MJ, Roy S, Lee M, Kuo CJ, Keller C et al (2007) Increased Wnt signaling during aging alters muscle stem cell fate and increases fibrosis. Science 317, 807–810. [DOI] [PubMed] [Google Scholar]
- 36. Suzuki H, Watkins DN, Jair KW, Schuebel KE, Markowitz SD, Chen WD et al (2004) Epigenetic inactivation of SFRP genes allows constitutive WNT signaling in colorectal cancer. Nat. Genet. 36, 417–422. [DOI] [PubMed] [Google Scholar]
- 37. Komori T (2006) Regulation of osteoblast differentiation by transcription factors. J. Cell. Biochem. 99, 1233–1239. [DOI] [PubMed] [Google Scholar]
- 38. Chen S, Gluhak‐Heinrich J, Wang YH, Wu YM, Chuang HH, Chen L et al (2009) Runx2, osx, and dspp in tooth development. J. Dent. Res. 88, 904–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hassan MQ, Javed A, Morasso MI, Karlin J, Montecino M, van Wijnen AJ et al (2004) Dlx3 transcriptional regulation of osteoblast differentiation: temporal recruitment of Msx2, Dlx3, and Dlx5 homeodomain proteins to chromatin of the osteocalcin gene. Mol. Cell. Biol. 24, 9248–9261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Viale‐Bouroncle S, Felthaus O, Schmalz G, Brockhoff G, Reichert TE, Morsczeck C (2012) The transcription factor DLX3 regulates the osteogenic differentiation of human dental follicle precursor cells. Stem Cells Dev. 21, 1936–1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Holleville N, Matéos S, Bontoux M, Bollerot K, Monsoro‐Burq AH (2007) Dlx5 drives Runx2 expression and osteogenic differentiation in developing cranial suture mesenchyme. Dev. Biol. 304, 860–874. [DOI] [PubMed] [Google Scholar]
- 42. Li H, Marijanovic I, Kronenberg MS, Erceg I, Stover ML, Velonis D et al (2008) Expression and function of Dlx genes in the osteoblast lineage. Dev. Biol. 316, 458–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Qu B, Liu O, Fang X, Zhang H, Wang Y, Quan H et al (2014) Distal‐less homeobox 2 promotes the osteogenic differentiation potential of stem cells from apical papilla. Cell Tissue Res. 357, 133–143. [DOI] [PubMed] [Google Scholar]
- 44. Hassan MQ, Tare RS, Lee SH, Mandeville M, Morasso MI, Javed A et al (2006) BMP2 commitment to the osteogenic lineage involves activation of Runx2 by DLX3 and a homeodomain transcriptional network. J. Biol. Chem. 281, 40515–40526. [DOI] [PubMed] [Google Scholar]
- 45. Lee MH, Kim YJ, Kim HJ, Park HD, Kang AR, Kyung HM et al (2003) BMP‐2‐induced Runx2 expression is mediated by Dlx5, and TGF‐beta 1 opposes the BMP‐2‐induced osteoblast differentiation by suppression of Dlx5 expression. J. Biol. Chem. 278, 34387–34394. [DOI] [PubMed] [Google Scholar]
- 46. Lee JL, Chang CJ, Chueh LL, Lin CT (2006) Secreted frizzled related protein 2 (sFRP2) decreases susceptibility to UV‐induced apoptosis in primary culture of canine mammary gland tumors by NF‐kappaB activation or JNK suppression. Breast Cancer Res. Treat. 100, 49–58. [DOI] [PubMed] [Google Scholar]
- 47. Zhang Z, Deb A, Zhang Z, Pachori A, He W, Guo J et al (2009) Secreted frizzled related protein 2 protects cells from apoptosis by blocking the effect of canonical Wnt3a. J. Mol. Cell. Cardiol. 46, 370–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Deskins DL, Bastakoty D, Saraswati S, Shinar A, Holt GE, Young PP (2013) Human mesenchymal stromal cells: identifying assays to predict potency for therapeutic selection. Stem Cells Transl. Med. 2, 151–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Feng J, Mantesso A, De Bari C, Nishiyama A, Sharpe PT (2011) Dual origin of mesenchymal stem cells contributing to organ growth and repair. Proc. Natl Acad. Sci. USA 108, 6503–6508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Du J, Ma Y, Ma P, Wang S, Fan Z (2013) Demethylation of epiregulin gene by histone demethylase FBXL11 and BCL6 corepressor inhibits osteo/dentinogenic differentiation. Stem Cells 31, 126–136. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Flow cytometry analysis of cell surface makers. The results indicated that SCAPs were positive for CD105 and negative for CD34 and CD45. The peak dividing the positive and negative sides depicts the cell population stained with the appropriate antibody.
Fig. S2 Cell survival was unaffected in SCAPs depleted of OSX. (a) Western blot analysis of OSX knock‐down in SCAPs. Beta‐actin was used as an internal control. (b) Alizarin red staining. (c) SCAPs were cultured with osteogenic medium for 48 h. No significant difference in the number of viable cells was observed between the OSX knock‐down group and the control group. Student's t‐test was used to determine statistical significance. Error bars represent the SD (n = 3). NS, no significant difference.
Table S1 Primers sequences used in the real‐time RT‐PCR.


