Abstract
Objectives
Caspases, a family of cysteine proteases with unique substrate specificities, contribute to apoptosis, whereas autophagy‐related genes (ATGs) regulate cytoprotective autophagy or autophagic cell death in cancer. Accumulating evidence has recently revealed underlying mechanisms of apoptosis and autophagy; however, their intricate relationships still remain to be clarified. Identification of caspase/ATG switches between apoptosis and autophagy may address this problem.
Materials and methods
Identification of caspase/ATG switches was carried out using a series of elegant systems biology & bioinformatics approaches, such as network construction, hub protein identification, microarray analyses, targeted microRNA prediction and molecular docking.
Results
We computationally constructed the global human network from several online databases and further modified it into the basic caspase/ATG network. On the basis of apoptotic or autophagic gene differential expressions, we identified three molecular switches [including androgen receptor, serine/threonine‐protein kinase PAK‐1 (PAK‐1) and mitogen‐activated protein kinase‐3 (MAPK‐3)] between certain caspases and ATGs in human breast carcinoma MCF‐7 cells. Subsequently, we identified microRNAs (miRNAs) able to target androgen receptor, PAK‐1 and MAPK‐3, respectively. Ultimately, we screened a range of small molecule compounds from DrugBank, able to target the three above‐mentioned molecular switches in breast cancer cells.
Conclusions
We have systematically identified novel caspase/ATG switches involved in miRNA regulation, and predicted targeted anti‐cancer drugs. These findings may uncover intricate relationships between apoptosis and autophagy and thus provide further new clues towards possible cancer drug discovery.
Introduction
Cancer, a complex genetic disease resulting from mutations of oncogenes or tumour suppressors that can develop into alteration of signalling pathways, has been well known to have astonishingly numerous links to programmed cell death (PCD), including to apoptosis, autophagy and programmed necrosis 1, 2. Apoptosis, or type I PCD, leads to morphological characteristics of dying cells including cell shrinkage, membrane blebbing and formation of apoptotic bodies (with or without nuclear debris) that are phagocytosed by surrounding cells or by phagocytes 3. Autophagy, or type II PCD, is independent of phagocytes and differs from apoptosis by presence of autophagosomes, autolysosomes, and the cell nucleus remaining intact 4. Programmed necrosis, or type III PCD, involves cell swelling, organelle dysfunction and cell lysis 2. In contrast with apoptosis and programmed necrosis, autophagy is not only a molecular mechanism for cell suicide but also a survival response to either growth factor or nutrient deprivation 5. Importantly, co‐regulation of apoptosis and autophagy has been well characterized as participating in cancer cell death 6, 7.
Of note, the processes of PCD, particularly referring to apoptosis, are orchestrated by caspases, a family of cysteine proteases with unique substrate specificities 8. Apoptotic pathways are finely tuned by multiple signalling events, including direct phosphorylation of caspases, whereas kinases are often the substrates of active caspases. Caspase‐mediated cleavage of kinases can terminate pro‐survival signalling or generate pro‐death peptide fragments that may help execute the death program 9. Autophagy is highly regulated by a limited number of autophagy‐related genes (ATGs), playing key roles in autophagosome formation and autophagy regulation 10, 11, 12.
Apoptosis and autophagy bear their distinct morphological characteristics and physiological processes; however, there still exist some interrelationships between them. Under certain circumstances, apoptosis and autophagy can exert synergetic effects, whilst sometimes autophagy can be triggered only when apoptosis is suppressed 13. A comprehensive knowledge of apoptotic and/or autophagic signalling networks may provide a framework for better understanding relationships between them as an integrated system 2, 6. With increasing genome‐wide data on genetic, regulatory, functional and physical interactions, exploration of ‘molecular switches’ between apoptosis and autophagy is imperative for inferring their complex connections, in cancer 7, 13. Intriguingly, recent studies have reported that caspases are involved in some autophagic signalling pathways, indicating relationships between caspases and ATGs in cancer initiation and progression 14. Based on these aforementioned studies, identification of novel caspase/ATG switches may be a promising avenue for shedding light on exploration of the complicated connections between apoptosis and autophagy in cancer.
In the study described here, we systematically identified several caspase/ATG switches and their pathways, involved in miRNA regulation and targeted drug prediction, which may not only uncover intricate relationships between apoptosis and autophagy, but provide more potential information towards novel drugs for cancer therapy.
Materials and methods
Retrieving functional genomic data
To build the global human PPI network, we collected diverse sets of biological evidence from five online databases. To predict pair‐wise PPIs, all data were pre‐processed into pair‐wise scores, reflecting similarity between protein pairs. We took advantage of several online databases including protein interaction data from Human Protein Reference Database (HPRD) 15, Biomolecular Object Network Databank (BOND) 16, IntAct 17, HomoMINT 18 and BioGRID 19.
Protein interaction network construction
In combination with caspases (Table S1) and ATGs (Table S2), we constructed the basic caspase/ATG network in which at least one non‐capsase or ATG member could interact with a caspase or ATG member. Subsequently, hub proteins could be based on the three following gold standards. First, degree of each protein in the function‐related network was calculated as number of links that one protein possessed to the other 20. Hub proteins, identified by their high degrees, were extracted based on the assumption that high‐degree proteins tended to play a key role in this network. Secondly, we indicated that hub proteins should connect caspases/ATGs, thereby making them worth particular attention when developing novel drug targets. This method was similar to previous studies that focussed on identifying novel cancer‐related genes 21. Thirdly, the network module could identify hub proteins at some level, as hubs often enrich within dense areas rather than in sparse areas 22.
Identifying molecular switches between apoptosis and autophagy
Proteins that interact with one another often possess similar gene expression patterns, thus genes that co‐express are more likely to interact than those genes that cannot co‐express. To identify genes that are co‐expressed, we used microarray data from human breast adenocarcinoma MCF‐7 cells, treated with 2.5 mM DTT to measure pair‐wise co‐expression of caspases in apoptosis (No. GSE22368) 23 and ATGs in autophagy (No. GSE26459) 24, treated with tamoxifen; co‐expression level was calculated as Pearson Correlation Coefficient ρ
where X and Y are expression level data vectors of length n for two genes, and are means, and σX and σY are the standard deviations.
Targeted microRNA prediction
As available prediction methods have strongly varying degrees of sensitivity and specificity, a combination of methods was assumed to profoundly mitigate the problem of presentation of false positives and negatives, and only accounted for interactions that were predicted by three algorithmically different methods. We assembled interactions between microRNAs and mRNAs, utilizing human‐specific data from TargetScan (stringent seed pairing, site number, site type, site context; option of ranking by likelihood of preferential conservation rather than site context) 25, MiRanda (moderately stringent seed pairing, site number, pairing to most of the miRNA) 26 and Diana‐MicroT (Hybridization energy threshold rules) 27.
Molecular docking
Initial three‐dimensional geometric co‐ordinates of the X‐ray crystal structures of androgen receptor (AR) (2am9), PAK‐1 (3fxz) and MAPK‐3 (2zoq) were downloaded from the Protein Data Bank (PDB) (http://www.pdb.org/pdb/home/home.do). We then constructed screening libraries for AR, PAK‐1 and MAPK‐3 containing all Food and Drug Administration (FDA)‐approved small molecule compounds, from the latest version of DrugBank (http://www.drugbank.ca/). To predict novel FDA‐approved small molecule drugs targeting AR, PAK‐1 and MAPK‐3, already established FDA‐approved drugs targeting the three proteins were removed from the original screening library. Then, the remaining drugs included in the screening library were subjected to OpenBabel toolbox to generate their 3D geometric co‐ordinates. Subsequently, 3D geometric co‐ordinates of the drugs were uploaded to the ZINC database to launch structure editing.
In addition, we used UCSF DOCK6.3 program with AMBER force field parameters to dock pre‐generated conformations of drugs into AR, PAK‐1 and MAPK‐3, for virtually screening their inhibitors 28. We performed flexible‐ligand docking to a rigid receptor with grid‐based scoring, in which the ligands (drugs) were allowed to be flexible and structurally rearranged in response to the receptor (AR, PAK‐1 and MAPK‐3). In docking processes, maximum number of orientations was set to 500. Subsequently, amber scoring function in DOCK6.3 was used to re‐rank the top 100 drugs (small molecule compounds) from the previous grid‐based scoring. During amber score calculation, the PDB2PQR server 29 was utilized to automate PDB file preparation and protonation state assignments of AR, PAK‐1 and MAPK‐3 with AMBER force‐field.
Results and discussion
Caspase/ATG network
In this study, we computationally constructed the global human PPI network covering almost all PPIs (about 200 000 protein pairs), based on five online databases including IntAct, HPRD, HomoMINT, BOND and BioGRID. To construct the set of true‐positive gene pairs, we achieved physical protein–protein interactions derived from manually curated PPI databases that include 58 976 protein pairs (8982 proteins) from BioGRID, 12 272 protein pairs (4073 proteins) from BOND, 25 256 protein pairs (7998 proteins) from HomoMINT, 39 240 protein pairs (9619 proteins) from HPRD, 49 235 protein pairs (8849 proteins) from IntAct and 12 975 protein pairs (4818 proteins) from DIP. Thus, we achieved a total number of 85 806 unique protein pairs (13 128 proteins) prepared as the global human PPI network (Fig. 1). It is well known that the PPI network is typically complex for its nature, with multiple connections amongst numerous signalling pathways; thus, it was necessary to represent the network by further integrating and analysing these high‐throughput data in a specific biological context, such as cancer. Subsequently, we extracted 12 caspase members (Table S1) and 23 ATG members (Table S2) and further modified the PPI network into the caspase/ATG network, composed of 42 proteins (76 protein pairs) (Fig. 1). Then, we analysed the three main aspects including differential gene expressions, their clustering and enrichment in a number of biological processes, and thus further identifying potential molecular switches between caspases and ATGs.
Figure 1.
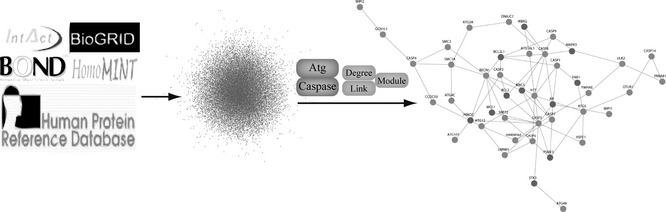
Construction of caspase/autophagy‐related gene network.
Microarray analyses of possible molecular switches
Significance analysis of microarray (SAM) analysis was performed on data of different expression microarrays, from tamoxifen‐treated human breast adenocarcinoma MCF‐7 cells, in apoptotic or autophagic stress, to identify divergent expression genes between normal and malignant cells (Fig. 2a). We indicate that proteins identified as divergent expression functional hub proteins, depend on gene co‐expression profiles, playing key roles as potential switches between apoptosis and autophagy in cancer (Fig. 2b). Due to these microarray data, we identified some potential molecular switches such as (AR), serine/threonine‐protein kinase PAK 1 (PAK‐1) and mitogen‐activated protein kinase 3 (MAPK‐3), between apoptosis and autophagy. Caspases are well known to possess many links to the Bcl‐2 family, which is also a key regulator of apoptosis and overexpressed in cancer 30.
Figure 2.
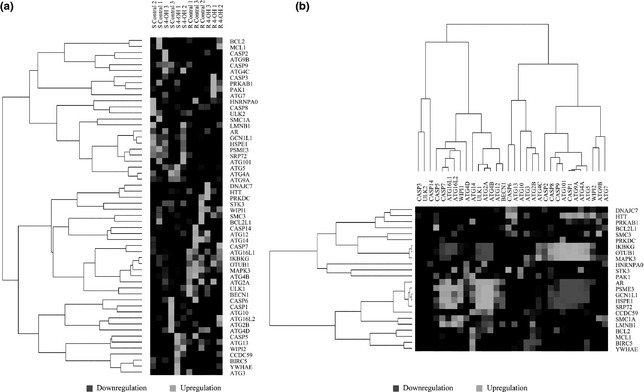
Microarray analyses of possible apoptosis/autophagy switches in MCF‐7 cells. (a) Microarray‐based differential expression analyses of potential switches in apoptosis and autophagy. (b) Microarray‐based correlation analyses of potential switches in apoptosis and autophagy.
Interestingly, a new emerging caspase substrate is Beclin‐1, a component of PI3KCIII complex required in autophagy. Beclin‐1 has lately been demonstrated to be a substrate of caspases −3, −7 and −8, suggesting a key role in cross‐talk between apoptotic and autophagic pathways 13, 14. Thus, our results demonstrate that some Bcl‐2 family members (for example, Bcl‐2, Mcl‐1 and BCL‐2L1) may link caspases to ATGs in apoptosis and autophagy and seems to be more highly dependable as a basic framework for apoptosis and autophagy in cancer. Protein kinases can orchestrate activation of signalling cascades in response to extracellular and intracellular stimuli to control cell proliferation, survival and apoptosis.
Targeted microRNA prediction of molecular switches
In this study, we took advantage of TargetScan, MiRanda and Diana‐MicroT to predict targeted miRNAs, respectively (Fig. 3a). Then, we integrated these predicted miRNAs into a combinatory result that some miRNAs were shown to target the above‐mentioned switches, respectively, in the context of PCD (Fig. 3b). Subsequently, ULK2‐MAPK‐3‐caspase‐8/‐9 pathway was predicted to be regulated by miR‐132, miR‐212 and miR‐613 (targeting MAPK‐3); and ATG5‐PAK‐1‐caspase‐1 pathway was predicted to be modulated by the let‐7 family, miR‐26a/b, miR‐96 and miR‐1271 (targeting PAK‐1). Additionally, WIPI1‐AR‐caspase‐1/‐3/‐7/‐8 pathway was predicted to be modulated by miR‐124, miR‐454 and miR‐506 (targeting AR) (Fig. 3b).
Figure 3.
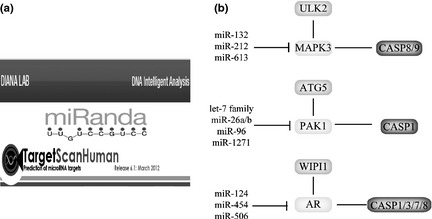
Molecular switches between apoptosis and autophagy involved in microRNA regulation. (a) Prediction of some microRNAs targeting autophagic hub proteins by TargetScan, MiRanda and Diana‐MicroT. (b) Switches and relevant microRNAs: (i) MAPK‐3‐mediated pathways and relevant miRNAs, (ii) PAK‐1‐mediated pathways and relevant miRNAs, (iii) AR‐mediated pathways and relevant miRNAs.
Of note, miRNAs, highly conserved, non‐coding endogenous RNAs ∼22 nucleotides (nt) in length, have been well known to regulate apoptotic and autophagic pathways in cancer, indicating miRNAs' function as oncogenes or tumour suppressors 31, 32, 33. In our study, we found that some predicted miRNAs could target caspase/ATG switches such as AR, PAK‐1 and MAPK‐3 in breast cancer cells, suggesting that miRNAs play important roles in cancer cell apoptosis and/or autophagy. Our previous studies have reported that a naïve Bayesian model‐based apoptotic network could identify some microRNAs that may target apoptotic hub proteins such as TP53, SRC, M3K3/5/8, cyclin‐dependent kinase2/6, TNFR16/19 and TGF‐β receptor 1/2 31. Additionally, our recent studies have demonstrated that the autophagic PPI network could confirm 9 autophagic hub proteins and 13 relevant oncogenic and tumour suppressive miRNAs in human breast carcinoma MCF‐7 cells 32, 33. In comparison to these previous studies in cancer cell apoptosis or autophagy, we report first that some predicted miRNAs can target three possible apoptosis/autophagy switches, AR, MAPK‐3 and PAK‐1 in breast cancer cells.
Candidate compounds targeting apoptosis/autophagy switches
In our study, we screened structure‐based candidate drugs that could target AR, MAPK‐3 and PAK‐1, respectively, based on FDA‐approved and ongoing experimental drugs from DrugBank. Also, we found some drugs that could target AR, MAPK‐3 and PAK‐1 very well (see in Table 1). Subsequently, we found some candidate small molecule compounds that can target MAPK‐3, such as Lansoprazole, Flubendazole, Miconazole, Bicalutamide, Carvedilol, Baclofen and Rizatriptan (Fig. 4a).
Table 1.
Small molecule compounds targeting novel apoptosis/autophagy switches

Figure 4.
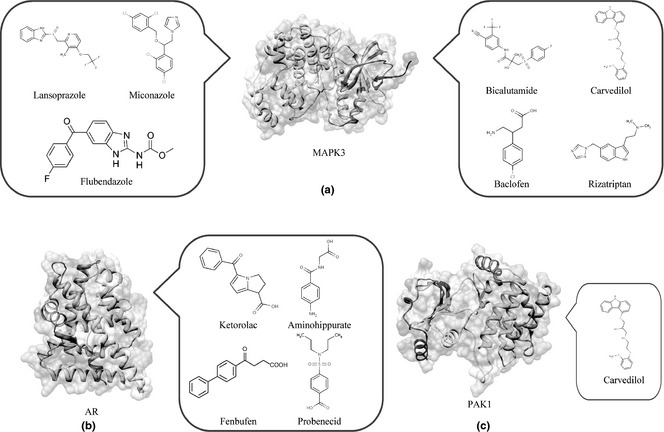
Small molecule compounds targeting MAPK‐3, PAK‐1 and AR in breast cancer cells. (a) lansoprazole, flubendazole, miconazole, bicalutamide, carvedilol, baclofen and rizatriptan target MAPK‐3. (b) ketorolac, aminohippuric, fenbufen and probenecid target AR; (c) carvedilol targets PAK‐1.
We report for the first time that, to the best of our knowledge, MAPK‐3 can be regarded as a new drug target for cancer treatment. Moreover, we found that further selective candidate small molecule compounds such as Ketorolac, Aminohippuric, Fenbufen and Probenecid could target AR, which is well known as a key point for anti‐cancer therapy (Fig. 4b). In addition, we found that Carvedilol could target PAK‐1 in malignant cell apoptosis and autophagy (Fig. 4c). Recent anti‐cancer drug discovery has significantly benefited from rapid progress in understanding how to target protein kinases with small molecules 34. Thus, we identified other protein kinases such as PAK‐1 and MAPK‐3 that may be more promising for utilization as potential therapeutic targets in network‐driven anti‐cancer drug discovery 35, 36.
Conclusion
In summary, we computationally constructed the global human protein–protein interaction (PPI) network based on several online databases, and identified the caspase/ATG network. In combination with gene ontology (GO) annotation and microarray analyses, we identified three possible caspase/ATG switches including AR, PAK‐1 and MAPK‐3, and their relative signalling pathways, in apoptosis and autophagy. In addition, we found targeted miRNAs on the three switches (that is, miR‐132, miR‐212 and miR‐613 targeting MAPK‐3, let‐7 family, miR‐26a/b, miR‐96 and miR‐1271 targeting PAK‐1, and miR‐124, miR‐454 and miR‐506 targeting AR) and further screened a variety of candidate small molecule compounds targeting them in breast cancer (Lansoprazole, Flubendazole, Miconazole, Bicalutamide, Carvedilol, Baclofen and Rizatriptan targeting MAPK‐3, Ketorolac, Aminohippuric, Fenbufen and Probenecid targeting AR, and Carvedilol targeting PAK‐1). Therefore, these findings demonstrate that caspase/ATG switches may lead to distinctive modes of PCD: apoptosis, autophagy or both, in breast cancer cells, which shed new light on elucidating the complicated mechanisms of apoptosis and autophagy in cancer pathogenesis, thereby exploring more new candidate drugs for cancer therapeutics.
Supporting information
Table S1. Caspase family members.
Table S2. Selective autophagy‐related genes.
Acknowledgements
We are grateful to He‐jiao Bian (Boston University), Qian Liu (National University of Singapore) and Ming‐wei Min (University of Cambridge) for their critical reviews on this manuscript. We also thank Qi‐jia Yu, Shu‐ya Wang, Xu Zhao, Cheng‐cheng Zhou and Na An (Sichuan University) for their good suggestions on this work. This work was supported in part by the grants from Young teacher's fund of Sichuan University (No. 2010SCU11066), the Science Foundation for Post Doctorate Research of China (No. 20110491725) and the Major State Basic Research Development Program of China (973 Program, No. 2010cb529900).
L.‐L. Fu, Y. Yang and H.‐L. Xu contributed equally to this work.
References
- 1. Hanahan D, Weinberg RA (2011) Hallmarks of cancer: the next generation. Cell 144, 646–674. [DOI] [PubMed] [Google Scholar]
- 2. Ouyang L, Shi Z, Zhao S, Wang FT, Zhou TT, Liu B et al (2012) Programmed cell death pathways in cancer: a review of apoptosis, autophagy and programmed necrosis. Cell Prolif. 45, 487–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cotter TG (2009) Apoptosis and cancer: the genesis of a research field. Nat. Rev. Cancer 9, 501–507. [DOI] [PubMed] [Google Scholar]
- 4. Liu B, Bao JK, Yang JM, Cheng Y (2012) Targeting autophagic pathways for cancer drug discovery. Chin. J. Cancer. doi: 10.5732/cjc.012.10010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wang SY, Yu QJ, Zhang RD, Liu B (2011) Core signaling pathways of survival/death in autophagy‐related cancer networks. Int. J. Biochem. Cell Biol. 43, 1263–1266. [DOI] [PubMed] [Google Scholar]
- 6. Wen X, Lin ZQ, Liu B, Wei YQ (2012) Targeting caspase‐mediated programmed cell death pathways for cancer therapy. Cell Prolif. 45, 217–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Giansanti V, Torriglia A, Scovassi AI (2011) Conversation between apoptosis and autophagy: is it your turn or mine? Apoptosis 16, 321–333. [DOI] [PubMed] [Google Scholar]
- 8. Hengartner MO (2000) The biochemistry of apoptosis. Nature 407, 770–776. [DOI] [PubMed] [Google Scholar]
- 9. Kurokawa M, Kornbluth S (2009) Caspases and kinases in a death grip. Cell 138, 838–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Liu B, Cheng Y, Liu Q, Bao JK, Yang JM (2010) Autophagic pathways as new targets for cancer drug development. Acta Pharmacol. Sin. 31, 1154–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Moreau K, Luo S, Rubinsztein DC (2010) Cytoprotective roles for autophagy. Curr. Opin. Cell Biol. 22, 206–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Liang C, Jung JU (2010) Autophagy genes as tumor suppressors. Curr. Opin. Cell Biol. 22, 226–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Liu JJ, Lin M, Yu JY, Liu B, Bao JK (2011) Targeting apoptotic and autophagic pathways for cancer therapeutics. Cancer Lett. 300, 105–114. [DOI] [PubMed] [Google Scholar]
- 14. Wirawan E, Vande Walle L, Kersse K, Cornelis S, Claerhout S, Vanoverberghe I et al (2010) Caspase‐mediated cleavage of Beclin‐1 inactivates Beclin‐1‐induced autophagy and enhances apoptosis by promoting the release of proapoptotic factors from mitochondria. Cell Death Dis. 1, e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mishra GR, Suresh M, Kumaran K, Kannabiran N, Suresh S, Bala P et al (2006) Human protein reference database–2006 update. Nucleic Acids Res. 34, D411–D414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Alfarano C, Andrade CE, Anthony K, Bahroos N, Bajec M, Bantoft K et al (2005) The Biomolecular Interaction Network Database and related tools 2005 update. Nucleic Acids Res. 33, D418–D424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kerrien S, Alam‐Faruque Y, Aranda B, Bancarz I, Bridge A, Derow C et al (2007) IntAct–open source resource for molecular interaction data. Nucleic Acids Res. 35, D561–D565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Persico M, Ceol A, Gavrila C, Hoffmann R, Florio A, Cesareni G (2005) HomoMINT: an inferred human network based on orthology mapping of protein interactions discovered in model organisms. BMC Bioinformatics 6 (Suppl. 4), S21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Winter AG, Wildenhain J, Tyers M (2011) BioGRID REST Service, BiogridPlugin2 and BioGRID WebGraph: new tools for access to interaction data at BioGRID. Bioinformatics 27, 1043–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Agarwal S, Deane CM, Porter MA, Jones NS (2010) Revisiting date and party hubs: novel approaches to role assignment in protein interaction networks. PLoS Comput. Biol. 6, e1000817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ostlund G, Lindskog M, Sonnhammer EL (2010) Network‐based Identification of novel cancer genes. Mol. Cell. Proteomics 9, 648–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Qi Y, Ge H (2006) Modularity and dynamics of cellular networks. PLoS Comput. Biol. 2, e174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Taylor KJ, Sims AH, Liang L, Faratian D, Muir M, Walker G et al (2010) Dynamic changes in gene expression in vivo predict prognosis of tamoxifen‐treated patients with breast cancer. Breast Cancer Res. 12, R39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gonzalez‐Malerva L, Park J, Zou L, Hu Y, Moradpour Z, Pearlberg J et al (2011) High‐throughput ectopic expression screen for tamoxifen resistance identifies an atypical kinase that blocks autophagy. Proc. Natl. Acad. Sci. USA. 108, 2058–2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Friedman RC, Farh KK, Burge CB, Bartel DP (2009) Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 19, 92–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Betel D, Wilson M, Gabow A, Marks DS, Sander C (2008) The microRNA.org resource: targets and expression. Nucleic Acids Res. 36, D149–D153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Maragkakis M, Alexiou P, Papadopoulos GL, Reczko M, Dalamagas T, Giannopoulos G et al (2009) Accurate microRNA target prediction correlates with protein repression levels. BMC Bioinformatics 10, 295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lang PT, Brozell SR, Mukherjee S, Pettersen EF, Meng EC, Thomas V et al (2009) DOCK 6: combining techniques to model RNA‐small molecule complexes. RNA 15, 1219–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dolinsky TJ, Czodrowski P, Li H, Nielsen JE, Jensen JH, Klebe G et al (2007) PDB2PQR: expanding and upgrading automated preparation of biomolecular structures for molecular simulations. Nucleic Acids Res. 35, W522–W525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chipuk JE, Moldoveanu T, Llambi F, Parsons MJ, Green DR (2010) The BCL‐2 family reunion. Mol. Cell 37, 299–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fu LL, Wen X, Bao JK, Liu B (2012) MicroRNA‐modulated autophagic signaling networks in cancer. Int. J. Biochem. Cell Biol. 44, 733–736. [DOI] [PubMed] [Google Scholar]
- 32. Fu LL, Zhao X, Xu HL, Wen X, Wang SY, Liu B et al (2012) Identification of microRNA‐regulated autophagic pathways in plant lectin‐induced cancer cells. Cell Prolif. 45, 477–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wang N, Xu HL, Zhao X, Wen X, Wang FT, Wang SY et al (2012) Network‐based identification of novel connections among apoptotic signaling pathways in cancer. Appl. Biochem. Biotech. 167, 621–631. [DOI] [PubMed] [Google Scholar]
- 34. Brognard J, Hunter T (2011) Protein kinase signaling networks in cancer. Curr. Opin. Genet. Dev. 21, 4–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Schadt EE, Friend SH, Shaywitz DA (2009) A network view of disease and compound screening. Nat. Rev. Drug. Discov. 8, 286–295. [DOI] [PubMed] [Google Scholar]
- 36. Knight ZA, Lin H, Shokat KM (2010) Targeting the cancer kinome through polypharmacology. Nat. Rev. Cancer 10, 130–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Caspase family members.
Table S2. Selective autophagy‐related genes.


