Abstract
Stress granules (SGs) and processing bodies (PBs) are non-membrane-enclosed RNA granules that dynamically sequester translationally inactive messenger ribonucleoprotein particles (mRNPs) into compartments that are distinct from the surrounding cytoplasm. mRNP remodeling, silencing, and/or storage involves the dynamic partitioning of closed-loop polyadenylated mRNPs into SGs, or the sequestration of deadenylated, linear mRNPs into PBs. SGs form when stress-activated pathways stall translation initiation but allow elongation and termination to occur normally, resulting in a sudden excess of mRNPs that are spatially condensed into discrete foci by protein:protein, protein:RNA, and RNA:RNA interactions. In contrast, PBs can exist in the absence of stress, when specific factors promote mRNA deadenylation, condensation, and sequestration from the translational machinery. The formation and dissolution of SGs and PBs reflect changes in messenger RNA (mRNA) metabolism and allow cells to modulate the proteome and/or mediate life or death decisions during changing environmental conditions.
Tight control of messenger RNA (mRNA) processing, trafficking, degradation, and translation are important in regulating gene expression. These processes are controlled by specific RNA-binding proteins (RBPs) that bind the mRNA within larger complexes called messenger ribonucleoprotein particles (mRNPs) (Mitchell and Parker 2014). In eukaryotes, such mRNPs are often localized to specific cellular compartments, both as a part of mRNA biogenesis under optimal conditions, and as a part of response to changing conditions. Recent data suggest that self-organization of mRNPs into various non-membrane-enclosed subcellular compartments, termed RNA granules, plays critical roles in mRNA metabolism (Shin and Brangwynne 2017). Two of the best-studied RNA granules are stress granules (SGs) and processing bodies (PBs), membraneless cytoplasmic foci formed by the condensation of translationally inactivated mRNPs. Although the composition of sequestered mRNAs and RBPs differs between SGs and PBs (Fig. 1), both RNA granules are linked to translational control events that modulate the proteome and/or influence cell survival. The accumulation and condensation of untranslating mRNPs into these discrete cytoplasmic granules are governed by similar events that are intimately connected to various aspects of translational control.
Figure 1.
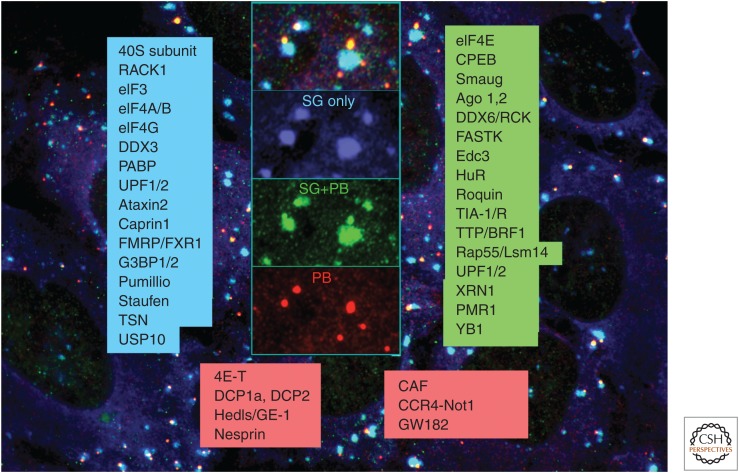
Selected stress granule (SG)- and processing body (PB)-associated proteins. Proteins (partial list) found exclusively in SGs (blue box), in both SGs and PB/GW-bodies (GWBs) (green box), or predominantly in PB/GWBs (red box). Image obtained using arsenite-treated U2OS cells stained for eukaryotic initiation factor 3b (eIF3b) (blue), DCP1a (red), and eIF4E (green).
The term “stress granules” was first used to describe phase-dense cytoplasmic particles that appeared in mammalian cells subjected to heat shock. These granules contained various heat-shock proteins (HSPs) (Collier and Schlesinger 1986; Collier et al. 1988), and similar particles were observed in heat-shocked tomato cells (Nover et al. 1983, 1989). Although initial compositional analysis revealed the presence of both HSPs and mRNAs in tomato “heat SGs” (Nover et al. 1983, 1989), later reports clarified that these SGs did not actually contain RNA and thus cannot be classified as RNA granules (Weber et al. 2008). However, before this revised report, the term “stress granules” was also used to describe cytoplasmic foci containing the translational repressor T-cell intracellular antigen 1 (TIA1), the translational enhancer poly(A)-binding protein (PABPC1), and polyadenylated mRNAs. Colocalization of these factors in discrete cytoplasmic granules was triggered by either heat-shock stress or sodium arsenite-induced oxidative stress (Kedersha et al. 1999). Unlike plant heat-shock granules, these mammalian mRNA-containing stress granules strictly required phosphorylation of eukaryotic translation initiation factor 2α (eIF2α) (Kedersha et al. 1999), thus linking SGs to translational control.
PBs were first described as “XRN1 foci” because of the granular cytoplasmic localization of the exoribonuclease XRN1 (Bashkirov et al. 1997). Subsequent observations revealed that other RNA decay-associated proteins were colocalized in these foci (Ingelfinger et al. 2002; van Dijk et al. 2002; Fenger-Gron et al. 2005; Wilczynska et al. 2005; Yu et al. 2005; Eulalio et al. 2007), leading to their designation as mRNA “processing bodies” (Sheth and Parker 2006). Proteins associated with mRNA silencing, such as the argonautes and glycine-tryptophan protein of 182 KDa (GW182)/trinucleotide repeat containing 6A, were also found in organized puncta described as “GW-bodies” (GWBs), which were often coincident with PBs (Eystathioy et al. 2003). For the purposes of this review, we will include GWBs under the umbrella term PBs, but note that they are not identical (reviewed in Stoecklin and Kedersha 2013).
STRESS GRANULES: COMPOSITION AND INITIATION
SGs consist of stalled preinitiation complexes that include small (40S), but not large (60S), ribosomal subunits, translation initiation factors eIF4F, eIF3, and PABP, and polyadenylated mRNAs (reviewed in Anderson and Kedersha 2009). Condensation of stalled preinitiation complexes (PICs) into SGs is mediated by specific RBPs, some of which show sequence-specific binding to mRNAs, and others that interact with the translational machinery. These two components, stalled PICs and SG-nucleating RBPs, together determine a threshold at which SGs form or disperse. Some SG-associated RBPs are shared with PBs, whereas other components are limited to SGs or PBs only. In terms of mRNA, SGs contain poly(A) mRNA, whereas PBs contain largely deadenylated mRNA. Figure 1 shows the SG/PB distribution of some of the best-characterized SG-specific proteins (blue), proteins common to both SGs and PBs (green), and proteins specific to PBs (red).
SGs and PBs are dynamic entities that are in equilibrium with polysomes (Kedersha et al. 2000, 2005). SG-associated mRNPs and RBPs dynamically shuttle between SGs and polysomes. Increasing the pool of translationally inactivated mRNA promotes SG assembly, whereas reducing the pool of untranslated mRNA causes SG disassembly. This balance is reflected by the antagonistic effects of the translation inhibitors emetine or cycloheximide and puromycin (Kedersha et al. 2000). Emetine and cycloheximide inhibit translation elongation, allowing slowly initiating/stalled PICs to complete initiation and be trapped in polysomes, reducing the free mRNP pool available for SGs. In the continued presence of stress, emetine treatment rapidly disassembles SGs, whereas PBs remain (see Movie 1). In the absence of stress, PBs are disassembled by 10–40 min of cycloheximide treatment (Andrei et al. 2005), suggesting that their steady-state integrity requires ongoing mRNP input. Conversely, puromycin-induced premature termination of translating mRNAs promotes SG formation (Kedersha et al. 2000). Mitotic cells do not form SGs or PBs, at least in part because of arrested elongation that prevents polysome disassembly (Sivan et al. 2007). The effects of these drugs highlight the link between SGs/PBs and translation, and distinguish them from other RNA granules (see Movie 1).
Movie 1.
U2OS cells stably expressing eIF4E-YFP (green) and mRFP-DDX6 (red), showing arsenite-induced SGs (green) are disassembled on the addition of the translational inhibitor emetine. Note that PBs (yellow) are not disassembled. Single confocal scans collected every 30 s, time elapsed 35 min.
SG disassembly occurs when cells adapt to stress, or when stress is removed and normal translational equilibrium is restored. Although SG assembly requires translationally stalled PICs, formation of SGs themselves is not required for translational arrest in the cells; specific stresses or knockdown of specific SG-associated proteins and SG nucleators uncouple SG formation from translational arrest (Ohn et al. 2008; Kedersha et al. 2016). Thus, a separate step beyond translational arrest is required for SG formation. For example, energy starvation induced by cold shock stalls the translational cycle, induces eIF2α phosphorylation, and triggers slow SG formation that takes hours rather than minutes, but these cold-shock SGs rapidly dissolve (5–10 min) when cells are warmed up, although dephosphorylation of eIF2α, polysome formation, and restoration of full translation take several hours (Hofmann et al. 2012). In this case, SG dissolution seems to result from a “decondensation” of mRNPs rather than restored translation that depletes the untranslated pool of mRNPs. Inhibiting the energy-sensing 5′AMP-activated protein kinase (AMPK) prevents cold-shock-induced SG formation, suggesting that as-yet-unidentified AMPK targets regulate the SG condensation event. In support of this idea, ubiquitin-specific peptidase 10 (USP10) is a protein that inhibits SG condensation but not translational arrest (Kedersha et al. 2016), and is both an activator and a substrate of AMPK (Deng et al. 2016).
Although SGs form over the course of minutes to hours, most SG protein components are in much more rapid dynamic equilibrium with the cytosol. Fluorescent recovery after photobleaching (FRAP) analysis indicates that the fluorescence recovery of SG proteins ranges from seconds to minutes. Rapidly shuttling SG RBPs include the translation silencers TIA1, Ras GTP-activating protein-binding protein 1 (G3BP1), tristetraprolin (TTP), and cytoplasmic polyadenylation element-binding protein (CPEB), which recover from bleaching within 10–30 sec. Fluorescence recovery of PABP is somewhat slower (30–60 sec), and recovery of Fas-activated serine-threonine kinase (FASTK) and the fragile-X proteins, fragile-X mental retardation protein (FMRP) and FMRP autosomal homolog 1 (FXR1) is very slow (minutes) (Kedersha et al. 2005; Bley et al. 2015). Thus, proteins that shuttle in and out of SGs show fast, medium, and slow recovery kinetics (Kedersha et al. 2000, 2005; Leung et al. 2006; Mollet et al. 2008). PB components also show a range of kinetics for eIF4E (fast), Lsm1 (medium), and eIF4E-T (slow) (Andrei et al. 2005), and support a model in which SG/PB structure arises from phase-transition-driven events (see below) rather than classic protein-based scaffolding or membrane-mediated partitioning. It must be noted that these FRAP studies used overexpressed, tagged proteins for measurements and used different cell lines; hence, some kinetic differences reported for specific situations may not be universal. As more SG proteins are knocked out and replaced by tagged versions using new technologies, better relative measurements should become available.
STRESS GRANULES AND TRANSLATIONAL MECHANISMS
Stress-induced phosphorylation of eIF2α (phospho-eIF2α) is necessary and sufficient for SG assembly (Fig. 2) (Kedersha et al. 1999, 2002). Early compositional analysis revealed that phospho-eIF2α-induced SGs contain most PIC factors but lack eIF5 and eIF2, proteins necessary for conversion of ribosomal preinitiation complexes into translationally competent ribosomes (Kedersha et al. 2002; Kimball et al. 2003). Mechanistically, stress-induced phospho-eIF2α inhibits translation initiation by depleting the eIF2•GTP-Met-tRNAi Met ternary complex that loads initiator Met-tRNAi Met onto the AUG start codon (reviewed in Jackson et al. 2010; Merrick and Pavitt 2018; Wek 2018). This results in assembly of a translationally stalled 48S initiation complex as shown in Figure 3 (yellow region), and allows elongating ribosomes to “run off” the mRNA, converting polysomes into the closed-loop mRNPs that are the core components of SGs (Kedersha et al. 2002; Kimball et al. 2003). During stress, transcripts bearing elements that evade phospho-eIF2α-mediated translational arrest (e.g., those possessing internal ribosome entry sites or upstream open reading frames) are selectively translated as they produce proteins that protect cells from stress-induced damage. For example, heat shock causes SG formation but HSP70 mRNAs (Kedersha and Anderson 2002) are selectively translated and excluded from SGs. Similarly, HSP90 mRNA is largely excluded from arsenite-induced SGs (Stohr et al. 2006). Thus, SGs are composed of translationally arrested mRNAs released from polysomes and remodeled into mRNPs, but mRNAs that remain actively translated are not remodeled.
Figure 2.
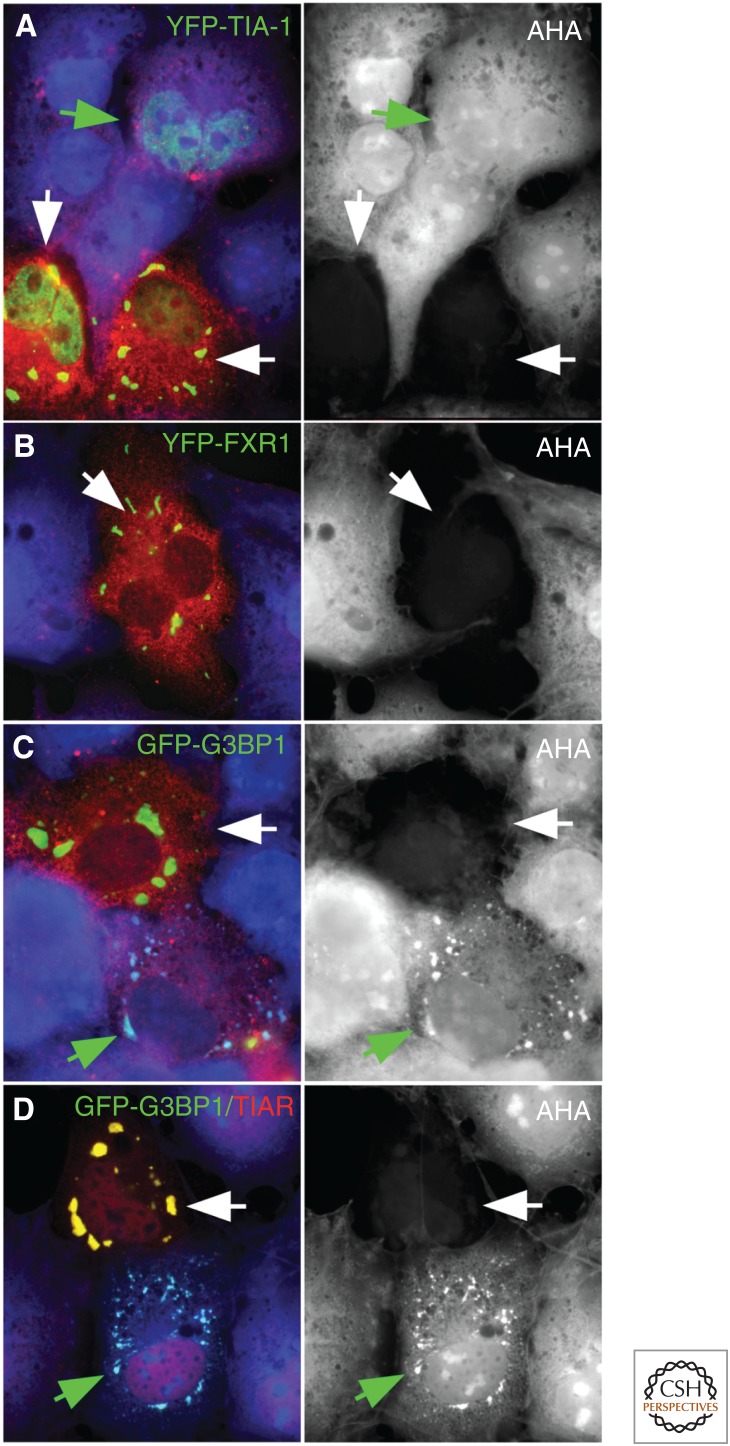
Stress granule (SG) nucleation, eukaryotic initiation factor 2α (eIF2α) phosphorylation, and global translation. Transient expression of (A) SG-nucleating proteins T-cell intracellular antigen 1 (TIA1), or (B) FMRP autosomal homolog 1 (FXR1) in COS7 cells triggers eIF2α phosphorylation (red) and prevents translation (blue in merged figure, shown separately in gray), as shown by labeling with the amino acid analog l-azidohomoalanine (AHA) in some transfectants (white arrows) but not those lacking SGs (green arrow). Overexpression of GTP-activating protein-binding protein 1 (G3BP1) (C,D) can nucleate SGs before/without triggering eIF2α phosphorylation or translational arrest (green arrows). Yellow indicates red/green colocalization.
Figure 3.
Regulatory stalling points in the translational cycle leading to stress granule (SG) formation. A closed-loop messenger RNA (mRNA) can stall at different points in the translation cycle, resulting in messenger ribonucleoproteins (mRNPs) of different composition eligible for condensation (mediated by GTP-activating protein-binding protein 1 [G3BP]) into SGs. Type I, or canonical SGs, result when eukaryotic initiation factor 2α (eIF2α) phosphorylation inhibits recharging of eIF2•GTP-Met-tRNAi Met, resulting in mRNPs that lack eIF2/5. Type II SGs form when eIF4A activities are inhibited and can be induced in cells lacking phospho-eIF2α, generating SGs that contain eIF2/5. Type III SGs result from xenobiotic stress, and lack eIF3. The mechanism shown here is hypothetical. Not shown are hyperosmotic/G3BP-independent SGs thought to arise from molecular crowding.
Stress and eIF2α phosphorylation are linked by a family of eIF2α kinases that comprise the integrated stress response (ISR), an intracellular surveillance system that monitors different parameters relevant to translation (see Wek 2018). Levels of charged transfer RNAs (tRNAs) are sensed by GCN2 (Wek et al. 1995), redox state and heme availability by HRI (McEwen et al. 2005), endoplasmic reticulum (ER) stress/unfolded protein levels by protein kinase R (PKR)-like ER kinase (PERK) (Harding et al. 2000), and double-stranded RNA (dsRNA) by PKR (Srivastava et al. 1998). Different stresses activate each kinase (reviewed in Donnelly et al. 2013), triggering eIF2α phosphorylation and initiating subsequent SG formation. Deletion or inactivation of any single eIF2α kinase renders cells unresponsive to the corresponding stress, for example, deletion of HRI prevents sodium arsenite-induced eIF2α phosphorylation and subsequent SG formation, but HRI knockout cells still respond to ER stress and PERK activation by both eIF2α phosphorylation and SG formation (Aulas et al. 2017). Knockout cells from mouse embryos homozygous for nonphosphorylatable mutant eIF2α (eIF2α[S51A], see below) evade stress-induced translational arrest and SG formation (Scheuner et al. 2001; McEwen et al. 2005).
Mechanistically, eIF2α phosphorylation does not directly impact eIF2 function in the preinitiation complex (recognition of an AUG initiation codon on an mRNA). Instead, it inhibits eIF2B, the guanine nucleotide exchange factor that reloads eIF2 with GTP. As eIF2B is limiting in cells, even a small amount of phospho-eIF2α efficiently inhibits eIF2B activity, causing a consequent depletion of the ternary complex that inhibits translation and initiates SG assembly (Jackson et al. 2010; Merrick and Pavitt 2018; Wek 2018). Consequently, pharmacological manipulations targeting eIF2α phosphorylation and eIF2B also modulate SG assembly/disassembly. Integrated stress response inhibitor (ISRIB), a chemical inhibitor of PERK signaling, reverses the inhibitory effects of phospho-eIF2α (Sidrauski et al. 2013, 2015a) by activating eIF2B and antagonizing phospho-eIF2α effects. Treatment with ISRIB reverses the ISR by rapidly restoring translation and causing SG disassembly, despite continuously elevated phospho-eIF2α (Sidrauski et al. 2015a,b). As in the case of emetine-enforced SG disassembly (see Movie 1), ISRIB treatment disassembles SGs but not PBs (Sidrauski et al. 2015a).
Although stress-induced translational repression triggers synchronous SG assembly in cultured cells, it is less recognized that isolated cells in otherwise healthy cultures can transiently form SGs. Movie 2 shows individual cells displaying transient SG formation and resolution while neighboring cells divide and grow normally, suggesting that SG formation occurs during metabolic changes associated with normal growth but is often overlooked. In support of this idea, SG-positive cells have been found in a subset of hypoxia-sensitive neurons in intact brain (DeGracia et al. 2007), in cochlear hair cells exposed to ototoxins (Mangiardi et al. 2004), in individual cells in hypoxic tumors (Moeller et al. 2004), and in virally infected cell cultures (reviewed by Lloyd 2013). In hepatitis C virus (HCV)-infected cultures, individual cells show oscillating cycles of SG formation/disassembly that parallel eIF2α phosphorylation and dephosphorylation, and global protein translation. These cycles result from the opposing effects on translation of the eIF2α kinase PKR and the eIF2α phosphatase protein phosphatase 1 (PP1), respectively (see Movie 2) (Ruggieri et al. 2012).
U2OS stably expressing GFP-G3BP1, adapting to a CO2-independent media over 12 h, imaged using confocal microscopy (volume-rendered Z-stacks) at 5-min intervals.
In situ metabolic labeling using the methionine analog l-azidohomoserine (AHA) reveals the tight correlation between SG formation, eIF2α phosphorylation, and translational arrest (Fig. 2). However, other noncanonical modes of SG formation are triggered by phospho-eIF2α–independent mechanisms. A model summarizing the major stalling points in the translational cycle linked to SG formation depicts a closed-loop mRNA that can be paused at different checkpoints, initiating polysome disassembly at these points and resulting in SGs of different composition (Fig. 3). Whereas phospho-eIF2α depletes ternary complex to promote the assembly of canonical “type I” SGs lacking eIF2, other agents (pateamine A [Low et al. 2005], hippuristanol [Mazroui et al. 2006)], rocaglates [Sadlish et al. 2013]) or lipid mediators (15-deoxy-Δ12,14-prostaglandin J2, or 15-d-PGJ2 [Kim et al. 2007]) inactivate the eIF4A helicase that helps unwind the 5′ untranslated region (UTR) of mRNAs to promote translation initiation. Failure of translation initiation via this mechanism triggers the formation of phospho-eIF2α-independent “type II” SGs that contain eIF2 and eIF5 (Fig. 3, blue), and can be triggered in the absence of phospho-eIF2α. Whereas type I SGs are generally cytoprotective, type II SGs may be cytotoxic (Fujimura et al. 2012; Anderson et al. 2015), although the mechanisms by which these compositionally distinct granules mediate opposing effects on cell survival are not known. Another subclass of SGs lacks eIF3, and is induced by chemotherapeutic drugs such as selenite, UV, or other xenobiotic agents (Fujimura et al. 2012; Anderson et al. 2015). UV-induced SGs do not require phospho-eIF2α but are disassembled by cycloheximide (Moutaoufik et al. 2014). We suggest that these eIF3-negative “type III” SGs may result from stalling before the recycling of terminating 40S subunits into new 43S preinitiation complexes at the 5′ end of the mRNA, a process that requires eIF3 (Pisarev et al. 2007, 2010) and is the commitment step to another round of translation (Fig. 3, top gray-shaded area). Finally, hypertonic conditions trigger assembly of another noncanonical, phospho-eIF2α-independent SG (Bevilacqua et al. 2010; Bounedjah et al. 2012); the mechanism for their formation is related to molecular crowding, but the details are unknown.
Many stresses that trigger the ISR, such as UV irradiation, heat shock or oxidative stress, also activate endogenous RNases such as angiogenin (Lyons et al. 2017), which cleaves tRNAs in their anticodon loops to produce tRNA halves (Fu et al. 2009; Yamasaki et al. 2009) known as tiRNAs (tRNA-derived, stress-induced small RNAs) (Yamasaki et al. 2009). A subset of tiRNAs (5′ tiRNAs derived from tRNAAla and tRNACys) inhibit translation (Emara et al. 2010) by displacing the eIF4F complex from the m7GTP cap, a step that is required for cap-dependent translation (Ivanov et al. 2011a, 2014). Transfected 5′ tiRNAAla and 5′ tiRNACys induce SG assembly that does not require phospho-eIF2α (Emara et al. 2010; Ivanov et al. 2011b), but does require the SG protein YB-1 (Lyons et al. 2016), a cold-shock, domain-containing protein also found in PBs (Yang and Bloch 2007). The mechanisms whereby tiRNAs collaborate with YB1 to induce SGs are unknown, but are under active investigation.
SG/PB CONDENSATION
In addition to stalled PIC mRNPs, SG formation also requires SG-nucleating proteins (Panas et al. 2016). Overexpression of any of these SG nucleators drives “spontaneous” SG assembly (Kedersha and Anderson 2007; Reineke et al. 2015), and knockdown of SG nucleators can impair SG formation. SG nucleating proteins include CAPRIN1, G3BP1, and G3BP2 (collectively referred to as G3BP), TIA1 and TIAR, FMRP, FXR1, and FXR2, TTP, BRF1, FASTK, and CPEB (reviewed in Anderson and Kedersha 2008). Many of these proteins act as translational silencers and display high-affinity, sequence-specific RNA-binding activity, and most require concurrent activation of PKR to trigger eIF2α phosphorylation to induce SGs (Kedersha and Anderson 2007). Figure 2 shows that overexpressed TIA1 (A) or FXR1 (B) nucleates SGs in cells that display both elevated phospho-eIF2α and globally repressed translation (white arrows). In the case of G3BP, overexpressed protein localizes to granule-like regions before activating the eIF2α kinase PKR (Reineke et al. 2012) in a feedforward manner (Fig. 2C,D). Cells lacking G3BP (G3BP-null cells) are unable to assemble mRNPs into SGs, despite overexpression of other SG nucleating proteins (Kedersha et al. 2016); hence, G3BP appears to play a direct role in SG condensation (see below) in addition to activating PKR.
Whereas stalling of translation initiates SG formation by promoting polysome disassembly into PIC mRNPs, the physical condensation of these mRNPs into granules is a separate event that requires G3BP. Two proteins, Caprin1 and USP10, competitively bind G3BP to promote (Solomon et al. 2007) or inhibit SG condensation, respectively (Kedersha et al. 2016). G3BP-null cells are unable to assemble either phospho-eIF2α-dependent type I or eIF4A-dependent type II SGs (Kedersha et al. 2016). SG formation is rescued in these cells by expression of either wild-type G3BP, or G3BP containing a point mutation that renders it unable to bind USP10 or Caprin1. Thus, G3BP is uniquely required for the formation of both type I and II SGs, and its activity is regulated by Caprin1 and USP10, but does not require these proteins to cause SG condensation. In addition, several viruses (Semliki Forest, Chikungunya, and herpes simplex) encode SG-inhibiting proteins that share the same binding motif (FGDF motif) within USP10 that binds to G3BP (Panas et al. 2015). The FGDF motif is sufficient to bind (and in some cases relocalize) G3BP, blocking its ability to mediate SG condensation. The importance of G3BP to SG condensation is further illustrated by the number of other viruses (including poliovirus, encephalomyocarditis virus [EMCV], and Coxsackievirus) that encode specific proteases that cleave G3BP to inhibit SG formation and disable the host antiviral response (White et al. 2007; Fung et al. 2013; Ng et al. 2013).
PROCESSING BODIES AND TRANSLATIONAL CONTROL
PBs share some components (Fig. 1) and properties with SGs, but differ from SGs in composition, behavior, and proposed functions (reviewed in Decker and Parker 2012; Stoecklin and Kedersha 2013). Shared components include eIF4E, mRNA, and selected RBPs, but PBs lack SG-associated eIF3, PABP, small ribosomal subunits, and many signaling proteins (Fig. 1). Generally, SGs house proteins involved in mRNA translation (discussed above), whereas PBs house proteins associated with mRNA decay (DCP1, DCP2, XRN1, EDC3, hedls/EDC4) or mRNA silencing (GW182, argonautes). This led to the proposal that PBs are sites of active mRNA decay (reviewed in Decker and Parker 2012). However, subsequent studies showed that repressed mRNAs can move from PBs into the cytosol where they reenter translation (Brengues et al. 2005), and disruption of PBs does not prevent global or specialized pathways of mRNA decay (Eulalio et al. 2007). Recent studies confirm that mRNAs segregated into PBs are translationally repressed but not degraded (Hubstenberger et al. 2017) and represent one-fifth of the mRNA transcriptome. The PB-associated subset of mRNAs is enriched for transcripts encoding regulatory proteins, especially regulatory subunits of multiprotein complexes. The sequestration of these mRNAs in PBs allows for their rapid mobilization into the translationally active pool, bypassing transcription, mRNA processing, and nuclear export, and thus enabling local control of PB-localized mRNAs (Hubstenberger et al. 2017). As PBs (and related GWBs) are also distinguished by the inclusion of RNA-induced silencing complex components (argonautes, GW182/TNRC6), at least part of their regulatory control is likely to be miRNA dependent (Eulalio et al. 2008). Importantly, various RBPs cosegregate into PBs with their mRNA targets, providing an alternative mechanism for the transport of specific mRNAs into and out of the PB storage depots to modulate protein production in response to cellular needs (Hubstenberger et al. 2017). A schematic illustrating the relationship between polysomes, SGs, and PBs is shown in Figure 4.
Figure 4.
Relationship between polysomes, stress granules (SGs), and processing bodies (PBs). Polysomes are maintained when translation initiation and termination occur at balanced rates on a single messenger RNA (mRNA). When termination occurs more frequently than initiation, polysome disassembly results, resulting in stalled, circularized messenger ribonucleoproteins (mRNPs). Agents that prevent elongation (such as emetine and cycloheximide) inhibit polysome/mRNP conversion, and hence reduce the pool of mRNPs, whereas puromycin promotes premature termination and accelerates the polysome-mRNP conversion. A pool of free mRNPs is necessary, but not sufficient, for SG assembly, which requires the dynamic condensation of mRNPs out of the surrounding cytoplasm. Condensation (and possibly decondensation) requires the mRNA-binding protein G3BP, and is regulated by G3BP-binding proteins Caprin1 (which promotes) and USP10 (which inhibits) the G3BP-mediated condensation of mRNPs into SGs. PBs contain deadenylated mRNPs and/or mRNAs undergoing deadenylation, and PB condensation requires multiple proteins, including EDC4, LSM14, 4-ET, and DDX6 (reviewed in Luo et al. 2018).
SG/PB FORMATION AND THE “LIQUID/LIQUID PHASE TRANSITION” CONCEPT
As shown in Movie 2, SGs fuse and show liquid-like behavior, similar to that of other RNA-containing structures (reviewed in Shin and Brangwynne 2017). Although SGs can persist for hours, the residence time of SG proteins (Kedersha et al. 2000, 2005), mRNAs (Mollet et al. 2008), and PB proteins (Andrei et al. 2005) is on the order of seconds to minutes. This dynamic behavior in seemingly stable structures distinguishes SGs and PBs from more static RNA granules and from membrane-bound organelles. SG/PB proteins are highly enriched in intrinsically disordered/low complexity (ID/LC) regions (reviewed in Uversky 2017), which do not assume classic structured domains. These regions are highly flexible, able to assume multiple conformations influenced by posttranslational modifications or templating interactions with other molecules (reviewed in Kedersha et al. 2013; Shin and Brangwynne 2017; Uversky 2017). The high proportion of ID/LC regions in SG-nucleating proteins supports the hypothesis that SG/PB formation is mediated by a liquid/liquid phase separation event (Han et al. 2012; Kato et al. 2012; Weber and Brangwynne 2012) that drives SG and PB components to condense out of the surrounding cytosol. This model suggests that the rapid shutting of ID/LC proteins into and out of SGs may parallel rapid conformational changes in their ID/LC regions.
Most SG-nucleating proteins are ID/LC-rich (reviewed in Kedersha et al. 2013; Panas et al. 2016), consistent with a phase transition model. One such protein is the mRNA decay-promoting protein TTP, which nucleates both SGs and PBs. Phosphorylation of TTP by MAPKAP2 (mitogen-activated protein kinase-activated protein kinase 2) promotes TTP binding to 14–3–3 proteins, and this interaction regulates TTP trafficking by eliminating the ability of TTP to condense into SGs (Stoecklin et al. 2004). As seen in Movie 3, arsenite-triggered phosphorylation of TTP results in its rapid export from SGs, but not PBs (Lykke-Andersen and Wagner 2005; Franks and Lykke-Andersen 2007). As 14–3–3 binding to TTP does not inhibit its binding to target mRNAs, regulated binding to 14–3–3 proteins may allow TTP to escort its target mRNAs out of SGs and into PBs (Stoecklin et al. 2004). An attractive model to explain this behavior is that the ID/LC regions of TTP are templated into a fixed conformation on binding to the highly ordered 14–3–3 proteins, “freezing” TTP into a state incompatible with the dynamics required for shuttling into SGs (see Movie 3).
Movie 3.
TTP leaving SGs but not PBs, in response to sodium arsenite, mRFP-TTP (red) was transiently expressed in U2OS cells stably expressing GFP-G3BP1, and TTP expression nucleates both SGs (yellow) and PBs (red). Sodium arsenite (200 µm) treatment causes p38/MPKAP2 kinase-driven phosphorylation of TTP and subsequent 14–3–3 binding, causing TTP to leave SGs but not PBs (red, do not colocalize with green G3BP). At the same time, arsenite activates eIF2α phosphorylation resulting in SG assembly in the neighboring cells. Movie 3 is assembled from volume-rendered Z-stacks collected at 1 min intervals.
Most SG-nucleating proteins (TIA-1/TIAR, Caprin1, CPEB1) bind mRNAs in a sequence-specific manner, and likely concentrate and selectively condense their target mRNAs when their translation is stalled. In addition to binding a subset of mRNAs, other SG nucleators such as G3BP1/2 and FMR1/FXR1/2 interact with ribosomal subunits. G3BP binds 40S ribosomal subunits (Simsek et al. 2017) through its RGG region (Kedersha et al. 2016; Simsek et al. 2017). Truncated G3BP lacking the RGG region loses both its ribosome-binding ability and its ability to restore SG assembly to the G3BP-null cells. The G3BP-binding protein Caprin1 is unable to nucleate SG formation in the absence of G3BP, thus it seems likely that G3BP:Caprin1 complexes cooperate to recruit Caprin1-specific transcripts into SGs (Solomon et al. 2007). In contrast to G3BP, the FMR/FXR1/FXR2 proteins bind large ribosomal subunits (Chen et al. 2014; Simsek et al. 2017), and paradoxically their overexpression strongly nucleates SGs despite the fact that SGs lack 60S subunits. Higher-order complexes containing the FMRP/FXR1 proteins and G3BP/Caprin1 proteins have been reported (El Fatimy et al. 2012; Baumgartner et al. 2013) that hint at cooperative effects between these two SG nucleating families, but details are not yet clear. A recent paper notes that these two families show mRNA binding that is regulated by adenosine methylation at position N6 (m6A). This study finds that m6A-modified RNA preferentially binds to FMRP/FXR1 and is translationally repressed, whereas G3BP preferentially binds and stabilizes mRNA that is not m6A modified (Edupuganti et al. 2017).
PROPOSED ROLES OF SGs IN CELL SIGNALING
Cellular stress requires rapid reprogramming of RNA metabolism as part of the ISR. SG assembly occurs downstream of stress-induced translational arrest, and the resulting pool of translationally arrested mRNAs constitutes a scaffold for RNA-binding proteins that promote SG condensation. Subsequently, specific mRNA-RBP complexes recruit a variety of signaling molecules that can promote cell survival and adaptation/recovery or induce apoptosis, suggesting that granule condensation may assemble signaling centers that allow cross talk between the ISR and other signaling pathways. The interplay between signaling pathways and SGs occurs at many levels (reviewed in Kedersha et al. 2013). First, signaling events directed by eIF2α kinases regulate cellular translation arrest that initiates SG assembly. Second, stress-modulated posttranslational modifications of SG proteins such as phosphorylation, O-GlcNAc addition, poly-ADP-ribosylation, and arginine methylation directly impact SG assembly and disassembly (reviewed in Ohn and Anderson 2010). Third, SGs modulate signaling cascades during stress by forming transient signaling hubs that coordinate signaling events in both the cytoplasm and the nucleus. Such modulation is achieved by selective sequestration of specific signal transduction-related molecules (such as RACK1, TRAF2, RSK2, etc.) into, or their exclusion from, SGs. As signaling centers, SG formation communicates a “state of emergency,” and their transient existence rewires the network of signaling events to control cell fate (Kedersha et al. 2013).
SGs, PBs, AND DISEASE
Given the varied effects that RNA granules have on metabolism and cell signaling, it is not surprising that they are also implicated in many diseases. Defects in SG dynamics are found in cancer, neurodegenerative disease, viral infection, and autoimmune disease. Several recent reviews discuss in detail the roles that RNA granules play in disease pathogenesis (Wolozin 2014; Anderson et al. 2015; Shukla and Parker 2016; Taylor et al. 2016; McCormick and Khaperskyy 2017).
Viruses depend on host translational machinery for their propagation, and several aspects of interactions between cellular translation and viruses directly or indirectly impact dynamics of RNA granules (Stern-Ginossar et al. 2018). In response to different viruses, SG and PB formation can be triggered or repressed (Lloyd 2013). Some viruses physically interact with SGs/PBs or hijack specific SG/PB components to favor viral replication. In some cases, SGs sequester key translation components, thus reducing the availability of ribosomal subunits and associated protein synthesis factors needed for translation of viral transcripts. Some viruses (e.g., EMCV, poliovirus) encode proteases that cleave the key SG nucleator G3BP to enforce SG disassembly and enhance viral replication (White et al. 2007; Fung et al. 2013; Ng et al. 2013). Other viruses (such as influenza A virus) block SG formation via expression of viral proteins (NS1) that directly bind and antagonize the eIF2α kinase PKR, a critical sensor of viral dsRNA (Khaperskyy et al. 2012). Flaviviruses (such as West Nile virus, Dengue virus) hijack multiple SG components (e.g., TIA1, TIAR, G3BP) via their sequestration by viral RNA 3′ stem-loop structures, thus suppressing SG formation (Li et al. 2002; Emara et al. 2008). Semliki Forest virus co-opts the SG nucleator G3BP (Panas et al. 2012), whereas HCV recruits both G3BP and multiple PB components and diverts them into lipid droplet-associated foci containing HCV replication/assembly complexes (Ariumi et al. 2011; Pager et al. 2013).
Interactions between viruses, the translational machinery, and RNA granules directly impact immunity. Virus-induced SGs regulate production of interferons that counteract virus propagation, thus identifying SGs as part of the innate immune system (reviewed in McCormick and Khaperskyy 2017). By preventing SG assembly or dissolving existing SGs, viruses evade the host defense. Several innate immune system-related proteins reside in SGs, including both interferon-inducers (such as PKR, RIG-I, MDA5) and interferon-effectors (RNase L and OASes) (Onomoto et al. 2012). The consequences and importance of colocalization of these factors with SGs for innate immunity are complex and not well understood. SGs may constitute platforms linking immune responses with other cell signaling pathways, or regulate selective production of specific immune factors such as cytokines. However, it is also possible that some SG-associated proteins are directly involved in innate immune responses irrespective of their localization.
SGs are implicated in the pathogenesis of neurodegenerative diseases such as amyotrophic lateral sclerosis, frontotemporal dementia and Alzheimer’s disease (reviewed in Wolozin 2012, 2014). The connection between SGs and these diseases comes from histological observations linking SG proteins to intracellular and extracellular protein aggregates that are pathological hallmarks of neurodegeneration (Wolozin 2012, 2014). The finding that mutations in several SG proteins are found in patients with neurodegenerative disease has led to speculation that disease-causing forms of SG proteins (e.g., TDP43, FUS/TLS, hnRNPA1, SMN) modulate SG dynamics in ways that impact neuronal survival (Shukla and Parker 2016; Taylor et al. 2016; Maziuk et al. 2017). Whereas evidence from multiple in vitro and cell culture models using disease-associated mutants of SG-associated proteins supports this hypothesis and shows that SG-related mechanisms can impair neuronal functions leading to neuron loss (Shukla and Parker 2016; Taylor et al. 2016; Maziuk et al. 2017), these studies require further evaluation in humans.
SGs are also associated with cancer (Anderson et al. 2015) and are found in tumors of different histological origins including colorectal cancer, carcinomas, and glioblastomas (Adjibade et al. 2015; El-Naggar et al. 2015; Vilas-Boas Fde et al. 2016). Responding and adapting to stress is important for cancer progression as cancer cells encounter hostile tumor microenvironments characterized by hypoxia, nutrient starvation, high levels of reactive oxygen species and hyperosmolarity, all of which trigger SG formation. Importantly, SG formation is often found as a part of the response to both radio- and chemotherapies (Moeller et al. 2004; Szaflarski et al. 2016). As SGs are generally prosurvival, their induction by chemotherapy can inadvertently compromise cancer treatment. Therefore, preventing SG assembly may prove to be a valuable therapeutic option in promoting tumor cell death. SGs are also linked to metastasis, where they contribute to cancer cell survival during tumor invasion and dissemination. Mouse xenograft sarcoma models reveal that increased levels of the SG nucleator G3BP1 promote SG formation in sarcoma cells to enhance their survival, resistance to chemotherapy, and metastasis to lungs (Somasekharan et al. 2015). By reducing G3BP1 expression and SG formation, metastasis and tumor invasion are significantly diminished (Somasekharan et al. 2015). Similarly, cancer cells can up-regulate SG assembly and enhance resistance to chemotherapy by producing and secreting the lipid mediator 15-deoxy-Δ12,14-prostaglandin J2 (Grabocka and Bar-Sagi 2016); this stimulates SG formation in an autocrine and paracrine manner to increase adaptation to tumor cell environments (Grabocka and Bar-Sagi 2016).
CONCLUDING REMARKS
Adjusting protein synthesis to adapt to changing conditions is crucial for growth and survival, and requires highly coordinated, rapid, specific, and localized reprogramming of cellular translation. Such regulation requires modulating global translation control as well as selective translation of mRNA subpopulations. Although global translation regulation pathways such as the integrated stress response and the mechanistic target of rapamycin (mTOR)/AMPK nutrient and energy regulatory cascades are understood in some detail, the mechanisms underlying selective translation remain largely unexplored, and RNA granules appear important in these processes. We anticipate that regulatory sequences, modified nucleotides (methylations, editing) (Peer et al. 2018), microRNAs (Duchaine and Fabian 2018), and structures within mRNAs mediate their selective recruitment to or exclusion from the translation machinery and/or RNA granules. Such regulatory mRNA elements may be recognized by specific RBPs that ferry them into RNA granules, and/or dictate their availability for translation. Posttranslational modifications of RBPs and their interactions with other proteins influence their localization, aggregation, and functions; we are only beginning to catalog these so we can ask larger questions.
Very recent studies using new technologies of proximity labeling are finally cataloging the range of proteins and mRNAs that shuttle through SGs and PBs. Using the BirA/BioID (proximity-dependent biotin identification) enzymatic system that biotin-labels interacting proteins over time (several hours), multiple BirA-tagged SG/PB baits were used to identify complex networks of cumulative protein interactors, but the long labeling period was unable to distinguish between stress-specific and normal interactions (Youn et al. 2018). Another study (Markmiller et al. 2018) used a faster labeling system based on the engineered peroxidase APEX2, revealing a 1-min snapshot labeling of proteins in close proximity to APEX2-tagged G3BP. This study detected stress-specific interactions between G3BP and the translational machinery (eIF3, eIF4G), but also revealed a complex network of interactions specific to certain types of stress, interactions that are stress-independent, and many which are cell-type-specific. We eagerly anticipate future studies that marry proximity labeling with techniques such as photoactivatable ribonucleoside-enhanced cross-linking and immunoprecipitation (PAR-CLIP) to obtain a census of SG/PB-associated RNAs under different conditions. Such studies are needed to address global questions such as: How are distinct subsets of mRNAs coordinately regulated in translation? Which signaling pathways regulate cross talk between global and selective translation control mechanisms? How are these integrated with biological outcomes of cell proliferation, motility, and death?
Both SGs and PBs are dynamic, and their contents are in flux. RNA helicases, which are required to facilitate the protein:RNA and RNA:RNA transitions, may play a direct role in remodeling stalled polysomes into mRNPs eligible for condensation into RNA granules. Specific helicases eIF4A, DDX3, and DDX6 contribute to SG and PB formation and resolution (Bordeleau et al. 2006; Dang et al. 2006; Ohn et al. 2008; Shih et al. 2008; Cencic et al. 2009; Ayache et al. 2015; Iwasaki et al. 2016). These play important functions in translational control as well as signaling (reviewed in Heerma van Voss et al. 2017). Future studies will elucidate their specific roles in SG/PB dynamics.
Physiological and environmental changes often trigger pathophysiological events underlying human disorders, ranging from infectious diseases to cancer and neurodegeneration. Translation control maintains homeostasis in response to environment change to ensure survival. RNA granule formation is both a biomarker of stress-induced translational arrest, and a mechanism for regulating selective translation. SGs influence mRNA sorting and sequestration, potentiate eIF2α phosphorylation, and constitute signaling centers that integrate RNA metabolism with other aspects of cellular physiology. PBs contain a subset of mRNAs, but how these are removed from PBs and translationally activated is unknown. We have only begun to elucidate the mechanisms whereby RNA granule-associated proteins and their client mRNAs fine-tune translation in response to environmental and physiological cues.
ACKNOWLEDGMENTS
We thank members of the Ivanov and Anderson laboratories for helpful discussions. This work is supported by the National Institutes of Health (NS094918 to P.I., GM121410, and GM111700 to P.A.). We apologize to our colleagues whose work could not be mentioned owing to space constraints.
Footnotes
Editors: Michael B. Mathews, Nahum Sonenberg, and John W.B. Hershey
Additional Perspectives on Translation Mechanisms and Control available at www.cshperspectives.org
REFERENCES
*Reference is also in this collection.
- Adjibade P, St-Sauveur VG, Huberdeau MQ, Fournier MJ, Savard A, Coudert L, Khandjian EW, Mazroui R. 2015. Sorafenib, a multikinase inhibitor, induces formation of stress granules in hepatocarcinoma cells. Oncotarget 6:43927–43943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson P, Kedersha N. 2008. Stress granules: The Tao of RNA triage. Trends Biochem Sci 33: 141–150. [DOI] [PubMed] [Google Scholar]
- Anderson P, Kedersha N. 2009. Stress granules. Curr Biol 19: R397–R398. [DOI] [PubMed] [Google Scholar]
- Anderson P, Kedersha N, Ivanov P. 2015. Stress granules, P-bodies and cancer. Biochim Biophys Acta 1849: 861–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrei MA, Ingelfinger D, Heintzmann R, Achsel T, Rivera-Pomar R, Luhrmann R. 2005. A role for eIF4E and eIF4E-transporter in targeting mRNPs to mammalian processing bodies. RNA 11: 717–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariumi Y, Kuroki M, Kushima Y, Osugi K, Hijikata M, Maki M, Ikeda M, Kato N. 2011. Hepatitis C virus hijacks P-body and stress granule components around lipid droplets. J Virol 85: 6882–6892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aulas A, Fay MM, Lyons SM, Achorn CA, Kedersha N, Anderson P, Ivanov P. 2017. Stress-specific differences in assembly and composition of stress granules and related foci. J Cell Sci 130: 927–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayache J, Benard M, Ernoult-Lange M, Minshall N, Standart N, Kress M, Weil D. 2015. P-body assembly requires DDX6 repression complexes rather than decay or Ataxin2/2L complexes. Mol Biol Cell 26: 2579–2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashkirov VI, Scherthan H, Solinger JA, Buerstedde JM, Heyer WD. 1997. A mouse cytoplasmic exoribonuclease (mXRN1p) with preference for G4 tetraplex substrates. J Cell Biol 136: 761–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgartner R, Stocker H, Hafen E. 2013. The RNA-binding proteins FMR1, rasputin and caprin act together with the UBA protein lingerer to restrict tissue growth in Drosophila melanogaster. PLoS Genet 9: e1003598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevilacqua E, Wang X, Majumder M, Gaccioli F, Yuan CL, Wang C, Zhu X, Jordan LE, Scheuner D, Kaufman RJ, et al. 2010. eIF2α phosphorylation tips the balance to apoptosis during osmotic stress. J Biol Chem 285: 17098–17111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bley N, Lederer M, Pfalz B, Reinke C, Fuchs T, Glass M, Moller B, Huttelmaier S. 2015. Stress granules are dispensable for mRNA stabilization during cellular stress. Nucleic Acids Res 43: e26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordeleau ME, Cencic R, Lindqvist L, Oberer M, Northcote P, Wagner G, Pelletier J. 2006. RNA-mediated sequestration of the RNA helicase eIF4A by Pateamine A inhibits translation initiation. Chem Biol 13: 1287–1295. [DOI] [PubMed] [Google Scholar]
- Bounedjah O, Hamon L, Savarin P, Desforges B, Curmi PA, Pastre D. 2012. Macromolecular crowding regulates assembly of mRNA stress granules after osmotic stress: New role for compatible osmolytes. J Biol Chem 287: 2446–2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brengues M, Teixeira D, Parker R. 2005. Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science 310: 486–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cencic R, Carrier M, Galicia-Vazquez G, Bordeleau ME, Sukarieh R, Bourdeau A, Brem B, Teodoro JG, Greger H, Tremblay ML, et al. 2009. Antitumor activity and mechanism of action of the cyclopenta[b]benzofuran, silvestrol. PLoS ONE 4: e5223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen E, Sharma MR, Shi X, Agrawal RK, Joseph S. 2014. Fragile X mental retardation protein regulates translation by binding directly to the ribosome. Mol Cell 54: 407–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier NC, Schlesinger MJ. 1986. The dynamic state of heat shock proteins in chicken embryo fibroblasts. J Cell Biol 103: 1495–1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier NC, Heuser J, Levy MA, Schlesinger MJ. 1988. Ultrastructural and biochemical analysis of the stress granule in chicken embryo fibroblasts. J Cell Biol 106: 1131–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang Y, Kedersha N, Low WK, Romo D, Gorospe M, Kaufman R, Anderson P, Liu JO. 2006. Eukaryotic initiation factor 2α-independent pathway of stress granule induction by the natural product pateamine A. J Biol Chem 281: 32870–32878. [DOI] [PubMed] [Google Scholar]
- Decker CJ, Parker R. 2012. P-bodies and stress granules: Possible roles in the control of translation and mRNA degradation. Cold Spring Harb Perspect Biol 4: a012286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeGracia DJ, Rudolph J, Roberts GG, Rafols JA, Wang J. 2007. Convergence of stress granules and protein aggregates in hippocampal Cornu Ammonis 1 at later reperfusion following global brain ischemia. Neuroscience 146: 562–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng M, Yang X, Qin B, Liu T, Zhang H, Guo W, Lee SB, Kim JJ, Yuan J, Pei H, et al. 2016. Deubiquitination and activation of AMPK by USP10. Mol Cell 61: 614–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly N, Gorman AM, Gupta S, Samali A. 2013. The eIF2α kinases: Their structures and functions. Cell Mol Life Sci 70: 3493–3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Duchaine TF, Fabian MR. 2018. Mechanistic insights into microRNA-mediated gene silencing. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a032771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edupuganti RR, Geiger S, Lindeboom RGH, Shi H, Hsu PJ, Lu Z, Wang SY, Baltissen MPA, Jansen P, Rossa M, et al. 2017. N6-methyladenosine (m6A) recruits and repels proteins to regulate mRNA homeostasis. Nat Struct Mol Biol 24: 870–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Fatimy R, Tremblay S, Dury AY, Solomon S, De Koninck P, Schrader JW, Khandjian EW. 2012. Fragile X mental retardation protein interacts with the RNA-binding protein Caprin1 in neuronal RiboNucleoProtein complexes [corrected]. PLoS ONE 7: e39338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Naggar AM, Veinotte CJ, Cheng H, Grunewald TG, Negri GL, Somasekharan SP, Corkery DP, Tirode F, Mathers J, Khan D, et al. 2015. Translational activation of HIF1α by YB-1 promotes sarcoma metastasis. Cancer Cell 27: 682–697. [DOI] [PubMed] [Google Scholar]
- Emara MM, Liu H, Davis WG, Brinton MA. 2008. Mutation of mapped TIA-1/TIAR binding sites in the 3′ terminal stem-loop of West Nile virus minus-strand RNA in an infectious clone negatively affects genomic RNA amplification. J Virol 82: 10657–10670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emara MM, Ivanov P, Hickman T, Dawra N, Tisdale S, Kedersha N, Hu GF, Anderson P. 2010. Angiogenin-induced tRNA-derived stress-induced RNAs promote stress-induced stress granule assembly. J Biol Chem 285: 10959–10968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A, Behm-Ansmant I, Schweizer D, Izaurralde E. 2007. P-body formation is a consequence, not the cause, of RNA-mediated gene silencing. Mol Cell Biol 27: 3970–3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A, Huntzinger E, Izaurralde E. 2008. GW182 interaction with Argonaute is essential for miRNA-mediated translational repression and mRNA decay. Nat Struct Mol Biol 15: 346–353. [DOI] [PubMed] [Google Scholar]
- Eystathioy T, Jakymiw A, Chan EK, Seraphin B, Cougot N, Fritzler MJ. 2003. The GW182 protein colocalizes with mRNA degradation associated proteins hDcp1 and hLSm4 in cytoplasmic GW bodies. RNA 9: 1171–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenger-Gron M, Fillman C, Norrild B, Lykke-Andersen J. 2005. Multiple processing body factors and the ARE binding protein TTP activate mRNA decapping. Mol Cell 20: 905–915. [DOI] [PubMed] [Google Scholar]
- Franks TM, Lykke-Andersen J. 2007. TTP and BRF proteins nucleate processing body formation to silence mRNAs with AU-rich elements. Genes Dev 21: 719–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H, Feng J, Liu Q, Sun F, Tie Y, Zhu J, Xing R, Sun Z, Zheng X. 2009. Stress induces tRNA cleavage by angiogenin in mammalian cells. FEBS Lett 583: 437–442. [DOI] [PubMed] [Google Scholar]
- Fujimura K, Sasaki AT, Anderson P. 2012. Selenite targets eIF4E-binding protein-1 to inhibit translation initiation and induce the assembly of non-canonical stress granules. Nucleic Acids Res 40: 8099–8110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung G, Ng CS, Zhang J, Shi J, Wong J, Piesik P, Han L, Chu F, Jagdeo J, Jan E, et al. 2013. Production of a dominant-negative fragment due to G3BP1 cleavage contributes to the disruption of mitochondria-associated protective stress granules during CVB3 infection. PLoS ONE 8: e79546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabocka E, Bar-Sagi D. 2016. Mutant KRAS enhances tumor cell fitness by upregulating stress granules. Cell 167: 1803–1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han TW, Kato M, Xie S, Wu LC, Mirzaei H, Pei J, Chen M, Xie Y, Allen J, Xiao G, et al. 2012. Cell-free formation of RNA granules: Bound RNAs identify features and components of cellular assemblies. Cell 149: 768–779. [DOI] [PubMed] [Google Scholar]
- Harding HP, Novoa I, Zhang Y, Zeng H, Wek R, Schapira M, Ron D. 2000. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol Cell 6: 1099–1108. [DOI] [PubMed] [Google Scholar]
- Heerma van Voss MR, Vesuna F, Bol GM, Afzal J, Tantravedi S, Bergman Y, Kammers K, Lehar M, Malek R, Ballew M, et al. 2017. Targeting mitochondrial translation by inhibiting DDX3: A novel radiosensitization strategy for cancer treatment. Oncogene 37: 63–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann S, Cherkasova V, Bankhead P, Bukau B, Stoecklin G. 2012. Translation suppression promotes stress granule formation and cell survival in response to cold shock. Mol Biol Cell 23: 3786–3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubstenberger A, Courel M, Benard M, Souquere S, Ernoult-Lange M, Chouaib R, Yi Z, Morlot JB, Munier A, Fradet M, et al. 2017. P-body purification reveals the condensation of repressed mRNA regulons. Mol Cell 68: 144–157. [DOI] [PubMed] [Google Scholar]
- Ingelfinger D, Arndt-Jovin DJ, Luhrmann R, Achsel T. 2002. The human LSm1–7 proteins colocalize with the mRNA-degrading enzymes Dcp1/2 and Xrnl in distinct cytoplasmic foci. RNA 8: 1489–1501. [PMC free article] [PubMed] [Google Scholar]
- Ivanov P, Emara MM, Villen J, Gygi SP, Anderson P. 2011a. Angiogenin-induced tRNA fragments inhibit translation initiation. Mol Cell 43: 613–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov P, Kedersha N, Anderson P. 2011b. Stress puts TIA on TOP. Genes Dev 25: 2119–2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov P, O'Day E, Emara MM, Wagner G, Lieberman J, Anderson P. 2014. G-quadruplex structures contribute to the neuroprotective effects of angiogenin-induced tRNA fragments. Proc Natl Acad Sci 111: 18201–18206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki S, Floor SN, Ingolia NT. 2016. Rocaglates convert DEAD-box protein eIF4A into a sequence-selective translational repressor. Nature 534: 558–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson RJ, Hellen CU, Pestova TV. 2010. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol 11: 113–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M, Han TW, Xie S, Shi K, Du X, Wu LC, Mirzaei H, Goldsmith EJ, Longgood J, Pei J, et al. 2012. Cell-free formation of RNA granules: Low complexity sequence domains form dynamic fibers within hydrogels. Cell 149: 753–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N, Anderson P. 2002. Stress granules: Sites of mRNA triage that regulate mRNA stability and translatability. Biochem Soc Trans 30: 963–969. [DOI] [PubMed] [Google Scholar]
- Kedersha N, Anderson P. 2007. Mammalian stress granules and processing bodies. Methods Enzymol 431: 61–81. [DOI] [PubMed] [Google Scholar]
- Kedersha NL, Gupta M, Li W, Miller I, Anderson P. 1999. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 α to the assembly of mammalian stress granules. J Cell Biol 147: 1431–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N, Cho MR, Li W, Yacono PW, Chen S, Gilks N, Golan DE, Anderson P. 2000. Dynamic shuttling of TIA-1 accompanies the recruitment of mRNA to mammalian stress granules. J Cell Biol 151: 1257–1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N, Chen S, Gilks N, Li W, Miller IJ, Stahl J, Anderson P. 2002. Evidence that ternary complex (eIF2-GTP-tRNA(i)(Met))-deficient preinitiation complexes are core constituents of mammalian stress granules. Mol Biol Cell 13: 195–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N, Stoecklin G, Ayodele M, Yacono P, Lykke-Andersen J, Fritzler MJ, Scheuner D, Kaufman RJ, Golan DE, Anderson P. 2005. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J Cell Biol 169: 871–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N, Ivanov P, Anderson P. 2013. Stress granules and cell signaling: More than just a passing phase? Trends Biochem Sci 38: 494–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N, Panas MD, Achorn CA, Lyons S, Tisdale S, Hickman T, Thomas M, Lieberman J, McInerney GM, Ivanov P, et al. 2016. G3BP-Caprin1-USP10 complexes mediate stress granule condensation and associate with 40S subunits. J Cell Biol 212: 845–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaperskyy DA, Hatchette TF, McCormick C. 2012. Influenza A virus inhibits cytoplasmic stress granule formation. FASEB J 26: 1629–1639. [DOI] [PubMed] [Google Scholar]
- Kim WJ, Kim JH, Jang SK. 2007. Anti-inflammatory lipid mediator 15d-PGJ2 inhibits translation through inactivation of eIF4A. EMBO J 26: 5020–5032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimball SR, Horetsky RL, Ron D, Jefferson LS, Harding HP. 2003. Mammalian stress granules represent sites of accumulation of stalled translation initiation complexes. Am J Physiol Cell Physiol 284: C273–C284. [DOI] [PubMed] [Google Scholar]
- Leung AK, Calabrese JM, Sharp PA. 2006. Quantitative analysis of Argonaute protein reveals microRNA-dependent localization to stress granules. Proc Natl Acad Sci 103: 18125–18130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Li Y, Kedersha N, Anderson P, Emara M, Swiderek KM, Moreno GT, Brinton MA. 2002. Cell proteins TIA-1 and TIAR interact with the 3′ stem-loop of the West Nile virus complementary minus-strand RNA and facilitate virus replication. J Virol 76: 11989–12000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd RE. 2013. Regulation of stress granules and P-bodies during RNA virus infection. Wiley Interdiscip Rev RNA 4: 317–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low WK, Dang Y, Schneider-Poetsch T, Shi Z, Choi NS, Merrick WC, Romo D, Liu JO. 2005. Inhibition of eukaryotic translation initiation by the marine natural product pateamine A. Mol Cell 20: 709–722. [DOI] [PubMed] [Google Scholar]
- Luo Y, Na Z, Slavoff SA. 2018. P-bodies: Composition, properties, and functions. Biochemistry 10.1021/acs.biochem.7b01162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen J, Wagner E. 2005. Recruitment and activation of mRNA decay enzymes by two ARE-mediated decay activation domains in the proteins TTP and BRF-1. Genes Dev 19: 351–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons SM, Achorn C, Kedersha NL, Anderson PJ, Ivanov P. 2016. YB-1 regulates tiRNA-induced stress granule formation but not translational repression. Nucleic Acids Res 44: 6949–6960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons SM, Fay MM, Akiyama Y, Anderson PJ, Ivanov P. 2017. RNA biology of angiogenin: Current state and perspectives. RNA Biol 14: 171–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangiardi DA, McLaughlin-Williamson K, May KE, Messana EP, Mountain DC, Cotanche DA. 2004. Progression of hair cell ejection and molecular markers of apoptosis in the avian cochlea following gentamicin treatment. J Comp Neurol 475: 1–18. [DOI] [PubMed] [Google Scholar]
- Markmiller S, Soltanieh S, Server KL, Mak R, Jin W, Fang MY, Luo EC, Krach F, Yang D, Sen A, et al. 2018. Context-dependent and disease-specific diversity in protein interactions within stress granules. Cell 172: 590–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maziuk B, Ballance HI, Wolozin B. 2017. Dysregulation of RNA binding protein aggregation in neurodegenerative disorders. Front Mol Neurosci 10: 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazroui R, Sukarieh R, Bordeleau ME, Kaufman RJ, Northcote P, Tanaka J, Gallouzi I, Pelletier J. 2006. Inhibition of ribosome recruitment induces stress granule formation independently of eukaryotic initiation factor 2α phosphorylation. Mol Biol Cell 17: 4212–4219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick C, Khaperskyy DA. 2017. Translation inhibition and stress granules in the antiviral immune response. Nat Rev Immunol 17: 647–660. [DOI] [PubMed] [Google Scholar]
- McEwen E, Kedersha N, Song B, Scheuner D, Gilks N, Han A, Chen JJ, Anderson P, Kaufman RJ. 2005. Heme-regulated inhibitor kinase-mediated phosphorylation of eukaryotic translation initiation factor 2 inhibits translation, induces stress granule formation, and mediates survival upon arsenite exposure. J Biol Chem 280: 16925–16933. [DOI] [PubMed] [Google Scholar]
- *.Merrick WC, Pavitt GD. 2018. Protein synthesis initiation in eukaryotic cells. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a033092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell SF, Parker R. 2014. Principles and properties of eukaryotic mRNPs. Mol Cell 54: 547–558. [DOI] [PubMed] [Google Scholar]
- Moeller BJ, Cao Y, Li CY, Dewhirst MW. 2004. Radiation activates HIF-1 to regulate vascular radiosensitivity in tumors: Role of reoxygenation, free radicals, and stress granules. Cancer Cell 5: 429–441. [DOI] [PubMed] [Google Scholar]
- Mollet S, Cougot N, Wilczynska A, Dautry F, Kress M, Bertrand E, Weil D. 2008. Translationally repressed mRNA transiently cycles through stress granules during stress. Mol Biol Cell 19: 4469–4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moutaoufik MT, El Fatimy R, Nassour H, Gareau C, Lang J, Tanguay RM, Mazroui R, Khandjian EW. 2014. UVC-induced stress granules in mammalian cells. PLoS ONE 9: e112742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng CS, Jogi M, Yoo JS, Onomoto K, Koike S, Iwasaki T, Yoneyama M, Kato H, Fujita T. 2013. Encephalomyocarditis virus disrupts stress granules, the critical platform for triggering antiviral innate immune responses. J Virol 87: 9511–9522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nover L, Scharf KD, Neumann D. 1983. Formation of cytoplasmic heat shock granules in tomato cell cultures and leaves. Mol Cell Biol 3: 1648–1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nover L, Scharf KD, Neumann D. 1989. Cytoplasmic heat shock granules are formed from precursor particles and are associated with a specific set of mRNAs. Mol Cell Biol 9: 1298–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohn T, Anderson P. 2010. The role of posttranslational modifications in the assembly of stress granules. Wiley Interdiscip Rev RNA 1: 486–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohn T, Kedersha N, Hickman T, Tisdale S, Anderson P. 2008. A functional RNAi screen links O-GlcNAc modification of ribosomal proteins to stress granule and processing body assembly. Nat Cell Biol 10: 1224–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onomoto K, Jogi M, Yoo JS, Narita R, Morimoto S, Takemura A, Sambhara S, Kawaguchi A, Osari S, Nagata K, et al. 2012. Critical role of an antiviral stress granule containing RIG-I and PKR in viral detection and innate immunity. PLoS ONE 7: e43031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pager CT, Schutz S, Abraham TM, Luo G, Sarnow P. 2013. Modulation of hepatitis C virus RNA abundance and virus release by dispersion of processing bodies and enrichment of stress granules. Virology 435: 472–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panas MD, Varjak M, Lulla A, Eng KE, Merits A, Karlsson Hedestam GB, McInerney GM. 2012. Sequestration of G3BP coupled with efficient translation inhibits stress granules in Semliki Forest virus infection. Mol Biol Cell 23: 4701–4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panas MD, Schulte T, Thaa B, Sandalova T, Kedersha N, Achour A, McInerney GM. 2015. Viral and cellular proteins containing FGDF motifs bind G3BP to block stress granule formation. PLoS Pathog 11: e1004659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panas MD, Ivanov P, Anderson P. 2016. Mechanistic insights into mammalian stress granule dynamics. J Cell Biol 215: 313–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Peer E, Moshitch-Moshkovitz S, Rechavi G, Dominissini D. 2018. The epitranscriptome in translation regulation. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a032623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisarev AV, Hellen CU, Pestova TV. 2007. Recycling of eukaryotic posttermination ribosomal complexes. Cell 131: 286–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisarev AV, Skabkin MA, Pisareva VP, Skabkina OV, Rakotondrafara AM, Hentze MW, Hellen CU, Pestova TV. 2010. The role of ABCE1 in eukaryotic posttermination ribosomal recycling. Mol Cell 37: 196–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reineke LC, Dougherty JD, Pierre P, Lloyd RE. 2012. Large G3BP-induced granules trigger eIF2α phosphorylation. Mol Biol Cell 23: 3499–3510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reineke LC, Kedersha N, Langereis MA, van Kuppeveld FJ, Lloyd RE. 2015. Stress granules regulate double-stranded RNA-dependent protein kinase activation through a complex containing G3BP1 and Caprin1. mBio 6: e02486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggieri A, Dazert E, Metz P, Hofmann S, Bergeest JP, Mazur J, Bankhead P, Hiet MS, Kallis S, Alvisi G, et al. 2012. Dynamic oscillation of translation and stress granule formation mark the cellular response to virus infection. Cell Host Microbe 12: 71–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadlish H, Galicia-Vazquez G, Paris CG, Aust T, Bhullar B, Chang L, Helliwell SB, Hoepfner D, Knapp B, Riedl R, et al. 2013. Evidence for a functionally relevant rocaglamide binding site on the eIF4A-RNA complex. ACS Chem Biol 8: 1519–1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheuner D, Song B, McEwen E, Liu C, Laybutt R, Gillespie P, Saunders T, Bonner-Weir S, Kaufman RJ. 2001. Translational control is required for the unfolded protein response and in vivo glucose homeostasis. Mol Cell 7: 1165–1176. [DOI] [PubMed] [Google Scholar]
- Sheth U, Parker R. 2006. Targeting of aberrant mRNAs to cytoplasmic processing bodies. Cell 125: 1095–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih JW, Tsai TY, Chao CH, Wu Lee YH. 2008. Candidate tumor suppressor DDX3 RNA helicase specifically represses cap-dependent translation by acting as an eIF4E inhibitory protein. Oncogene 27: 700–714. [DOI] [PubMed] [Google Scholar]
- Shin Y, Brangwynne CP. 2017. Liquid phase condensation in cell physiology and disease. Science 357: eaaf4382. [DOI] [PubMed] [Google Scholar]
- Shukla S, Parker R. 2016. Hypo- and hyper-assembly diseases of RNA-protein complexes. Trends Mol Med 22: 615–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidrauski C, Acosta-Alvear D, Khoutorsky A, Vedantham P, Hearn BR, Li H, Gamache K, Gallagher CM, Ang KK, Wilson C, et al. 2013. Pharmacological brake-release of mRNA translation enhances cognitive memory. eLife 2: e00498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidrauski C, McGeachy AM, Ingolia NT, Walter P. 2015a. The small molecule ISRIB reverses the effects of eIF2α phosphorylation on translation and stress granule assembly. eLife 4: e05033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidrauski C, Tsai JC, Kampmann M, Hearn BR, Vedantham P, Jaishankar P, Sokabe M, Mendez AS, Newton BW, Tang EL, et al. 2015b. Pharmacological dimerization and activation of the exchange factor eIF2B antagonizes the integrated stress response. eLife 4: e07314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simsek D, Tiu GC, Flynn RA, Byeon GW, Leppek K, Xu AF, Chang HY, Barna M. 2017. The mammalian ribo-interactome reveals ribosome functional diversity and heterogeneity. Cell 169: 1051–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivan G, Kedersha N, Elroy-Stein O. 2007. Ribosomal slowdown mediates translational arrest during cellular division. Mol Cell Biol 27: 6639–6646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon S, Xu Y, Wang B, David MD, Schubert P, Kennedy D, Schrader JW. 2007. Distinct structural features of caprin-1 mediate its interaction with G3BP-1 and its induction of phosphorylation of eukaryotic translation initiation factor 2α, entry to cytoplasmic stress granules, and selective interaction with a subset of mRNAs. Mol Cell Biol 27: 2324–2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somasekharan SP, El-Naggar A, Leprivier G, Cheng H, Hajee S, Grunewald TG, Zhang F, Ng T, Delattre O, Evdokimova V, et al. 2015. YB-1 regulates stress granule formation and tumor progression by translationally activating G3BP1. J Cell Biol 208: 913–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava SP, Kumar KU, Kaufman RJ. 1998. Phosphorylation of eukaryotic translation initiation factor 2 mediates apoptosis in response to activation of the double-stranded RNA-dependent protein kinase. J Biol Chem 273: 2416–2423. [DOI] [PubMed] [Google Scholar]
- *.Stern-Ginossar N, Thompson SR, Mathews MB, Mohr I. 2018. Translational control in virus-infected cells. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a033001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoecklin G, Kedersha N. 2013. Relationship of GW/P-bodies with stress granules. Adv Exp Med Biol 768: 197–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoecklin G, Stubbs T, Kedersha N, Wax S, Rigby WF, Blackwell TK, Anderson P. 2004. MK2-induced tristetraprolin: 14–3–3 complexes prevent stress granule association and ARE-mRNA decay. EMBO J 23: 1313–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stohr N, Lederer M, Reinke C, Meyer S, Hatzfeld M, Singer RH, Huttelmaier S. 2006. ZBP1 regulates mRNA stability during cellular stress. J Cell Biol 175: 527–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szaflarski W, Fay MM, Kedersha N, Zabel M, Anderson P, Ivanov P. 2016. Vinca alkaloid drugs promote stress-induced translational repression and stress granule formation. Oncotarget 7: 30307–30322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JP, Brown RH Jr, Cleveland DW. 2016. Decoding ALS: From genes to mechanism. Nature 539: 197–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uversky VN. 2017. Intrinsically disordered proteins in overcrowded milieu: Membrane-less organelles, phase separation, and intrinsic disorder. Curr Opin Struct Biol 44: 18–30. [DOI] [PubMed] [Google Scholar]
- van Dijk E, Cougot N, Meyer S, Babajko S, Wahle E, Seraphin B. 2002. Human Dcp2: A catalytically active mRNA decapping enzyme located in specific cytoplasmic structures. EMBO J 21: 6915–6924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilas-Boas Fde A, da Silva AM, de Sousa LP, Lima KM, Vago JP, Bittencourt LF, Dantas AE, Gomes DA, Vilela MC, Teixeira MM, et al. 2016. Impairment of stress granule assembly via inhibition of the eIF2α phosphorylation sensitizes glioma cells to chemotherapeutic agents. J Neurooncol 127: 253–260. [DOI] [PubMed] [Google Scholar]
- Weber SC, Brangwynne CP. 2012. Getting RNA and protein in phase. Cell 149: 1188–1191. [DOI] [PubMed] [Google Scholar]
- Weber C, Nover L, Fauth M. 2008. Plant stress granules and mRNA processing bodies are distinct from heat stress granules. Plant J 56: 517–530. [DOI] [PubMed] [Google Scholar]
- *.Wek RC. 2018. Role of eIF2α kinases in translational control and adaptation to cellular stresses. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a032870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wek SA, Zhu S, Wek RC. 1995. The histidyl-tRNA synthetase-related sequence in the eIF-2 α protein kinase GCN2 interacts with tRNA and is required for activation in response to starvation for different amino acids. Mol Cell Biol 15: 4497–4506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JP, Cardenas AM, Marissen WE, Lloyd RE. 2007. Inhibition of cytoplasmic mRNA stress granule formation by a viral proteinase. Cell Host Microbe 2: 295–305. [DOI] [PubMed] [Google Scholar]
- Wilczynska A, Aigueperse C, Kress M, Dautry F, Weil D. 2005. The translational regulator CPEB1 provides a link between dcp1 bodies and stress granules. J Cell Sci 118: 981–992. [DOI] [PubMed] [Google Scholar]
- Wolozin B. 2012. Regulated protein aggregation: Stress granules and neurodegeneration. Mol Neurodegener 7: 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolozin B. 2014. Physiological protein aggregation run amuck: Stress granules and the genesis of neurodegenerative disease. Discov Med 17: 47–52. [PMC free article] [PubMed] [Google Scholar]
- Yamasaki S, Ivanov P, Hu GF, Anderson P. 2009. Angiogenin cleaves tRNA and promotes stress-induced translational repression. J Cell Biol 185: 35–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang WH, Bloch DB. 2007. Probing the mRNA processing body using protein macroarrays and “autoantigenomics.” RNA 13: 704–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youn JY, Dunham WH, Hong SJ, Knight JDR, Bashkurov M, Chen GI, Bagci H, Rathod B, MacLeod G, Eng SWM, et al. 2018. High-density proximity mapping reveals the subcellular organization of mRNA-associated granules and bodies. Mol Cell 69: 517–532. [DOI] [PubMed] [Google Scholar]
- Yu JH, Yang WH, Gulick T, Bloch KD, Bloch DB. 2005. Ge-1 is a central component of the mammalian cytoplasmic mRNA processing body. RNA 11: 1795–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]