SUMMARY
In the postgenomic era, it is clear that the human genome encodes thousands of long noncoding RNAs (lncRNAs). Along the way, RNA imaging (e.g., RNA fluorescence in situ hybridization [RNA-FISH]) has been instrumental in identifying powerful roles for lncRNAs based on their subcellular localization patterns. Here, we explore how RNA imaging technologies have shed new light on how, when, and where lncRNAs may play functional roles. Specifically, we will synthesize the underlying principles of RNA imaging techniques by exploring several landmark lncRNA imaging studies that have illuminated key insights into lncRNA biology.
1. INTRODUCTION
It is becoming increasingly clear that RNA localization can influence numerous biological processes. Examples include specialized localization of mRNAs at opposing cellular poles resulting, on cellular division, in two different cell types, and the widespread rearrangement of RNA transcripts, on cellular stress, to subcellular compartments such as stress granules or P-bodies (Martin and Ephrussi 2009; Parton et al. 2014; Buxbaum et al. 2015; Van Treeck and Parker 2019). Yet, the information content present in subcellular RNA localization patterns is absent in most nonimaging-based expression measurements (e.g., RNA sequencing). Thus, the “when, where, and how” of RNA, which is critical to unraveling its biology, often remains a mystery.
The advent of RNA fluorescence in situ hybridization (RNA-FISH) provided an approach to address these questions. Moreover, such methods allow the measurement of how these patterns and properties vary between individual cells in a population. Together, this information provides powerful insights into the nature and mechanism of RNA-mediated cellular processes, including organization of nuclear subcompartments and the widespread interface between RNA and epigenetic regulation.
RNA-FISH is particularly relevant for noncoding RNA biology, in which the RNA itself is typically the final molecular effector. A classic example is the RNA-FISH visualization of the long noncoding RNA (lncRNA) Xist (Brown et al. 1992). Although it was known that Xist was sufficient to inactivate an entire female X chromosome during dosage compensation, its mechanism remained mysterious. Yet, with a single RNA-FISH experiment, it became clear that Xist remains in the nucleus and coats the inactive X chromosome in cis (Brown et al. 1992). This “seeing is believing” approach shows how visualization can provide insights critical for deciphering the mechanism of lncRNAs. Since Xist, many lncRNAs have been investigated using RNA imaging (Cabili et al. 2015; Dunagin et al. 2015; Orjalo and Johansson 2016). These studies have revealed many unexpected lncRNA localization patterns and in turn new biological insights (e.g., identification of new subcellular compartments).
Equally, as important as the spatiotemporal information provided by RNA imaging is the advantage of monitoring individual cells in a population. Specifically, RNA imaging can elucidate the heterogeneity of transcript abundance and localization within a population. The single-cell resolution of RNA-FISH can also provide key biological insights that would otherwise be missed in bulk population measurements (e.g., RNA sequencing). For example, a lncRNA could be highly correlated in expression with its neighboring gene according to RNA-seq, which could indicate a regulatory interaction. Alternatively, it could be that the lncRNA is expressed in half the cells and messenger RNA (mRNA) in the other half, in a mutually exclusive manner. Thus, on a population average, a positive interaction could be predicted but on a single-cell level, a repressive relationship would be revealed. Indeed, this is the case for at least two lncRNAs (linc-HOXA1 and lincRNA-Bxd) that have been implicated in promoter competition between the lncRNA and mRNA (Petruk et al. 2006; Maamar et al. 2013).
The single-cell nature of RNA imaging allows important insights into regulatory dynamics of RNA transcripts by looking at correlations at the single-cell level across subpopulations of cells. Yet, this is only the tip of the iceberg; one can—in parallel—glean information and correlate lncRNA expression patterns with many other cellular processes by simultaneously measuring, for example, at which stage in the cell cycle an RNA is expressed (e.g., cyclin mRNA), whether an RNA moves between the nucleus and cytoplasm (e.g., NRON is a lncRNA that sequesters the transcription factor NFAT in the cytoplasm and on release of RNA, NFAT localizes to the nucleus) or if an RNA is restricted to a subcellular compartment such as the nucleolus (pre-rRNA) (Willingham 2005; Cabili et al. 2015; Falahati et al. 2016). In other words, as the old aphorism goes: A picture is worth a thousand words.
Here, we will survey these examples and many more while highlighting different approaches and technologies for RNA-FISH and their application to lncRNA detection and visualization. We will explore lncRNA-FISH studies by introducing the basic experimental approaches, limitations, and controls. Then we will explore the application of lncRNA-FISH in the context of studies that have provided important insights into lncRNA biology. We will synthesize RNA-FISH, single-molecule RNA-FISH and live-cell RNA imaging by focusing on a few of these seminal examples.4
2. THE BASICS: COMPLEMENTARY PROBE PREPARATION FOR RNA-FISH
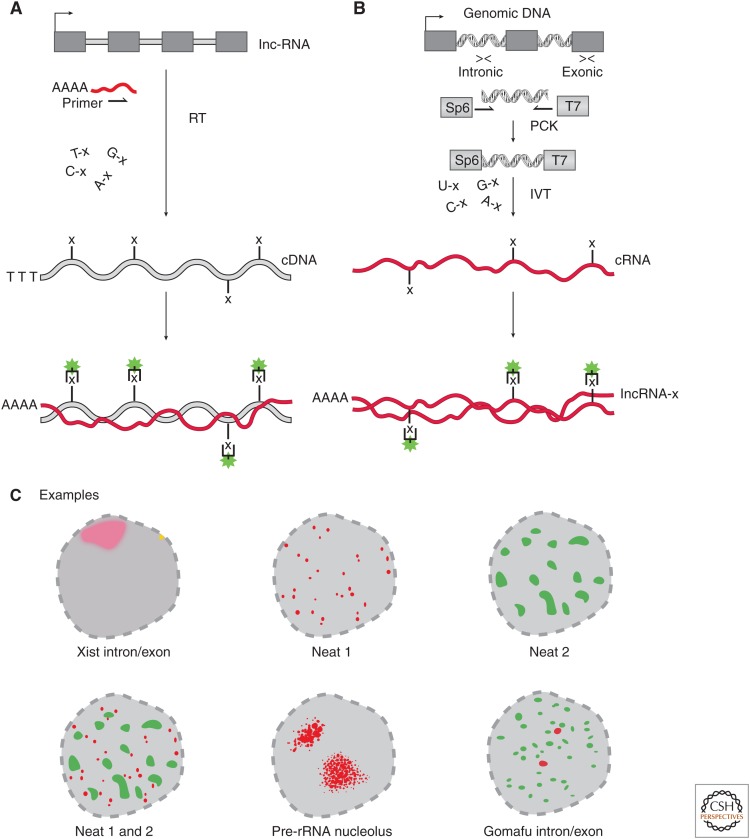
Before exploring the biological insights of lncRNA-FISH we will discuss the basic principles used to uncover lncRNA localization patterns. A common principle across most RNA-FISH approaches is the use of a DNA or RNA molecule(s) with sequence complementary to the endogenous lncRNA (Fig. 1). The first lncRNA-FISH studies used single probe designs that roughly distill into two principal methods used today.
cDNA probe: A complementary DNA (cDNA) can be made through reverse transcription of a target lncRNA with incorporation of “labeled” (e.g., digoxigenin [DIG] or biotin) DNA nucleotides (Fig. 1A). This complementary sequence or “probe” can hybridize in situ to the target lncRNA with tagged nucleotides; that in turn can be detected with secondary fluorescent binding molecules (e.g., antibodies for digoxigenin or streptavidin for biotin). These cDNA probes often span exon junctions, and thus are more likely to hybridize to the mature lncRNA and not the unspliced immature transcripts (producing large gaps in probe hybridization).
cRNA probe: Hybridization probes can also be generated by double-stranded DNA (dsDNA) templates with flanking in vitro transcription (IVT) promoters (Fig. 1B). This promoter is used in an IVT reaction that incorporates labeled RNA nucleotides that produces a complementary RNA (cRNA) probe (Fig. 1B). This cRNA probe is then targeted by secondary fluorescent molecules (e.g., antibody for digoxigenin or streptavidin for biotin). This approach also confers the advantage of generating a “sense” or noncomplementary control probe if an IVT promoter is placed on the opposite strand of the complementary probe. Thus, the same dsDNA template can generate a complementary probe for specific signal and a noncomplementary probe to determine nonspecific or background signal.
Figure 1.
Classic RNA fluorescence in situ hybridization (RNA-FISH) probe design using complementary DNA (cDNA) and complementary RNA (cRNA) probes. (A) To generate cDNA probes for RNA-FISH, one performs reverse transcription (RT) with gene-specific primers to generate a cDNA to the targeted long noncoding RNA (lncRNA). During the RT reaction, nucleotides are randomly incorporated that have modifications (e.g., “x” = biotin or digoxigenin [DIG]). The labeled cDNA is hybridized to the endogenous lncRNA (red line), which can be imaged via addition of fluorescent secondary conjugates targeting the probe label. (B) Generation of cRNA RNA-FISH probes requires targeted polymerase chain reaction (PCR) of genomic DNA, targeting either the intronic or exonic regions of the lncRNA locus. PCR primers have 5′/3′ sequences appended encoding in vitro transcription (IVT) promoters that are incorporated into the final double-stranded DNA (dsDNA) product. The resulting dsDNA product is subjected to IVT either antisense (complementary) or antisense (noncomplementary) in the presence of modified nucleotides (same as in A) to produce a labeled cRNA probe (red line with “x”). The cRNA probe is then hybridized to the endogenous lncRNA target, which can be imaged via addition of fluorescent secondary conjugates targeting the probe label. (C) lncRNA-FISH examples of classic studies revealing new subnuclear structures (all images showing nucleus). (Top left) Xist showing intron and exon labeling of the Xist lncRNA showing a punctate site of transcription (yellow) and mature transcript as a “cloud” in red on the same chromosome as the site of transcription. (Top middle and right, bottom left) Cartoon rendition of the RNA-FISH nuclear localization pattern for Neat1, Neat2, and Neat1/2, respectively, with nonoverlapping “speckled” localization patterns. (Bottom middle) rRNA-FISH demarcates the nucleolus, the site of rRNA transcription. (Bottom right) Gomafu RNA-FISH with intronic (red) and exonic (green) RNA-FISH probes shows that the mature transcript localizes in large domains away from the site of transcription (intronic signal).
Often, it is useful to simultaneously visualize the site of transcription relative to the final location of mature lncRNA transcript. For example, this was useful in determining that Xist localizes to the same chromosome from which it is transcribed, thus working in cis. Probes to determine the site of transcription are generated by IVT using dsDNA templates comprising intronic sequences from genomic DNA (Fig. 1B) (Fremeau et al. 1986; Brown et al. 1992; Soler et al. 2017). Collectively, these approaches provide flexible and cost-efficient methods to generate RNA-FISH probes to visualize lncRNAs.
After probes are generated, there are many protocols and procedures for permeabilizing (or opening) cells followed by hybridization of probes and microscopy-based analyses. We will not discuss these aspects here other than to mention this often requires experimental optimization to account for many factors such as cell type and abundance of the lncRNA. The experimental details of these optimization protocols are nicely covered in the primary literature, textbooks, and reviews (see Lawrence and Singer 1986; Lawrence et al. 1988, 1989; Dirks et al. 1993; Tripathi et al. 2015; Querido et al. 2017). We do note, however, that permeability issues often affect longer cRNA probes more than oligonucleotide ssDNA probes, in particular for detection in the nucleus (discussed below).
3. NEAT RNA-FISH: CDNA AND CRNA PATTERN RECOGNITION OF LNCRNAS
The RNA-FISH approaches described above were seminal in understanding lncRNA biology, as they revealed fascinating and informative localization patterns. As early as 1992, it was clear from RNA-FISH studies that subnuclear compartments express lncRNAs (preribosomal RNA in the nucleolus and Xist in the Barr body; Fig. 1C), thus raising the question: could there be other subnuclear compartments demarcated by lncRNAs? It was not until the postgenomic era when two neat RNA-FISH experiments revealed that the answer was “yes.”
A seminal example of RNA-FISH leading to a new understanding of nuclear biology came from the discovery of nuclear-enriched abundant transcripts 1 and 2 (Neat1 and Neat2, the latter more recently referred to as Malat1) (Hutchinson et al. 2007). Fractionation RNA-sequencing of several primary human cell lines showed that these lncRNAs are exclusively nuclear. However, it was RNA-FISH of where in the nucleus these lncRNAs are expressed that led to an important insight. The cRNA probes targeting either Neat1 and Malat1 both showed “speckled” (Fig. 1C) localization patterns across a large expanse of the nucleus. Interestingly, parallel RNA-FISH of Neat1 and 2 revealed very little overlap between the two sets of speckles (Fig. 1C). Thus, Neat1 and Neat2/Malat1 lncRNAs demarcated two novel subnuclear domains: “nuclear speckles” and “paraspeckles,” respectively (Bond and Fox 2009; Mao et al. 2011; Spector and Lamond 2011). Thus, this single RNA-FISH experiment was able to reveal two different subnuclear compartments demarcated by a lncRNA.
RNA-FISH experiments led to our current understanding that many lncRNAs are fundamental constituents of subnuclear compartments (Quinodoz and Guttman 2014; Rinn and Guttman 2014; Vance and Ponting 2014; Cheng et al. 2016; Engreitz et al. 2016; Melé and Rinn 2016). Consistent with this notion, a similar and contemporary study of the lncRNA Gomafu (MIAT/Rncr2) showed a stunning nuclear localization pattern (Sone et al. 2007). Specifically, Gomafu shows widespread but solely nuclear localization that is associated with factors in the nuclear matrix. This study also used intronic probes that revealed two punctate foci representing the site of transcription for Gomafu (Fig. 1C). Together, these RNA-FISH images alone suggest a role for the mature Gomafu transcript in facilitating nuclear organization in trans, a property that would be very hard to identify by conventional population-based or even single-cell RNA sequencing.
Collectively, these and other seminal RNA-FISH studies suggested that lncRNAs are fundamental components of dynamic nuclear substructures that influence a plethora of biological processes.
4. SINGLE-MOLECULE RNA-FISH APPROACHES
Today, it is clear that the mammalian genome encodes thousands of lncRNAs. Along the way RNA-FISH has undergone many technical advances; including single-molecule detection sensitivity, accessible analysis software, and scalable probe generation techniques to target newly discovered lncRNAs. These single-molecule RNA-FISH (smRNA-FISH) studies provide quantification of individual RNA molecules, their subcellular position, and how those properties vary from cell to cell (Femino et al. 1998; Raj et al. 2008, 2010; Zenklusen et al. 2008; Larson et al. 2009; Raj and van Oudenaarden 2009; Batish et al. 2011; Trcek et al. 2011, 2012; Levesque et al. 2013; Shaffer et al. 2013; Dunagin et al. 2015; Querido et al. 2017; Tutucci et al. 2018a).
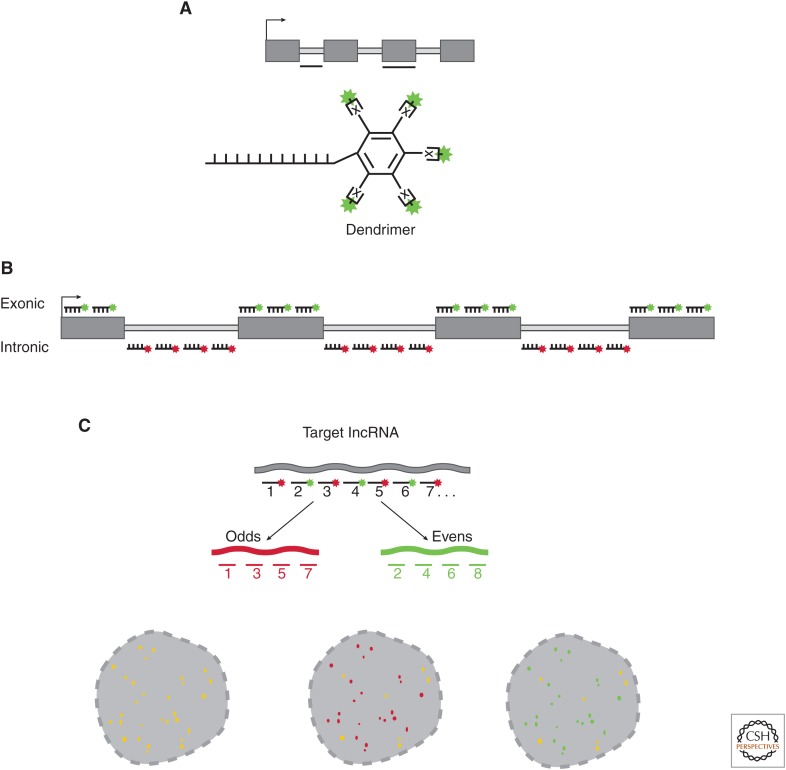
There are many variations of smRNA-FISH probe schemes, but most share a common principle of using DNA oligonucleotides (oligos) that are conjugated to fluorophores with defined stoichiometry. There are two basic approaches: (1) use of a small number of highly specific DNA oligos that provide a scaffold from which multiple sequences are hybridized in a “branched” manner termed a dendrimer (Wang et al. 2012; Sinnamon and Czaplinski 2014), in which the dendrimer is targeted by a complementary sequence conjugated to a fluorophore resulting in a signal that represents one molecule of the targeted lncRNA (Fig. 2A); and (2) use of pools of short (18–22-nt) DNA oligos—each with a 5′ or 3′-conjugated fluorophore—that tile individual lncRNA molecules (Fig. 2B) (Raj et al. 2008; Cabili et al. 2015).
Figure 2.
Probe design for single-molecule RNA fluorescence in situ hybridization (smRNA-FISH). (A) The dendrimer or branched oligo approach. Oligo(s) are designed to specify an intronic or exonic region of the target long noncoding RNA (lncRNA). The dendrimer approach appends either multiple conjugation sites (x) to the 5′ or 3′ end of the oligo. Alternatively, “branched-oligos” are similar to dendrimers using multiple “branched” sequences that form a tree-like structure of conjugatable epitopes. (B) smRNA-FISH probe design using “oligo pools.” Each oligo ranges in length between 18 and 22 nucleotides (nt), and between 20 and 48 oligos are mixed into an oligo pool. Each oligo has modifications on the 5′ or 3′ end to allow conjugation of a fluorophore of choice. smRNA-FISH oligo pools are generated targeting either the intronic regions (site of lncRNA transcription) or exonic regions of the mature transcript. (C) smRNA-FISH probe validation. As a first test for oligo specificity, the pool can be split evenly and each half labeled with distinct fluorophores. If all oligos from the odd (red) and even (green) probe sets are specific to the targeted long noncoding RNA (lncRNA), the patterns will overlap producing yellow signal (left). If one or more of the odd (red) oligos hybridize to an off-target RNA, but most of the probes are specific, then yellow and red signal would be observed (middle). Similarly, if one of the even (green) probes had an off-target RNA, but some probes were specific, green and yellow signal would be observed (right).
Both dendrimer and oligo pool approaches can benefit greatly from computational design of optimal targeting oligos, as well as careful control experiments to determine the specificity of the smRNA-FISH probes. We will hereafter focus on smRNA-FISH oligo pools as a case study to discuss the basic principles in designing and validating smRNA-FISH probes.
4.1. Step 1: Oligo Design
Typically, each targeting oligo or probe is ∼18–22-nt long, with a 5′ or 3′ nucleotide amenable to conjugation. Approximately 20–48 oligos are pooled to target the lncRNA of interest (Fig. 2). Each probe is designed to target a unique region of the RNA of interest. Oligos can be designed using several online software programs, but their specificity requires empirical validation, as small subsequences within an oligo can cross-hybridize to highly abundant off-target transcripts, leading to spurious signals (see below).
This oligo-based scheme can easily accommodate the design and modification of probe sets for a specific question. For example, it is possible to visualize both the site of transcription and subcellular localization of a mature RNA transcript using individually labeled probe sets targeting the introns and exons, respectively. Furthermore, individual oligos in the pool can be tested for specificity (see below), and “rogue” oligonucleotides (that are often responsible for background signal) can easily be eliminated from the pool. It is critical to validate oligo specificity in new cellular contexts, as the expression of high abundance off-target RNAs may differ between cell lines or tissues.
4.2. Step 2: Oligo Conjugation
Following oligo design, the next step is to conjugate specific fluorophores to the modified 5′ or 3′ nucleotide (Raj et al. 2008; Orjalo and Johansson 2016) of each probe (or enzymatically labeled) (Gaspar et al. 2017). The unconjugated oligonucleotides must then be removed by high-performance liquid chromatography (HPLC)-based methods (unless the conjugation efficiency is reliably very high). The choice of fluorophores depends on the specific filter sets available in the microscope and the degree of spectral multiplexing required for the experiment. Photostable fluorophores should be used where possible, as high-powered lasers (such as those on a confocal microscope) can easily bleach most fluorophores. These parameters could also vary from cell type to cell type.
4.3. Step 3: Probe Set Validation
A nice feature of the pooled approach is that each individual oligo can be tested for specificity. For practical reasons, this specificity is usually tested by taking “odd” and “even” probes from the pool of 20–48 oligos and conjugating the odds and even with different fluorophores. These odd and even pools are then hybridized simultaneously. If the probes are specific to the intended target, the odd and even probes will overlap (Fig. 2C), and so the merged image of the odd and even fluorophores will show strong signal overlap (Fig. 2C) (Raj et al. 2008; Raj and van Oudenaarden 2009; Cabili et al. 2015; Dunagin et al. 2015).
However, if any nonoverlapping signals are present, this indicates that one or more oligos within the pool are hybridizing to off-target transcripts. If this is the case, then the rogue probe(s) can be identified by removing subsets of the pool until only overlapping signal is detected (Fig. 2C).
Controlling for off target hybridization is critical when investigating lncRNAs, which often have much lower abundance than mRNAs and unique localization patterns that can be easily misidentified because of background. In particular, highly abundant off-target transcripts can produce strong, false signals that can sometimes look like nuclear blobs (Cabili et al. 2015). This can even occur within small subsections of an individual oligo. For example, it was previously found that the putative expression pattern of a lowly expressed lncRNA could be attributed almost entirely to a single rogue oligo with a 15-nt region complementary to an off-target RNA with several 1000-fold higher abundance. (Cabili et al. 2015) Thus, even with the current best computational design tools, it is strongly recommended that one empirically validate the oligo pool as a whole using this “odds and evens” approach.
Often probe sets are designed using the above-described process to target intronic regions. Because introns degrade very rapidly after being spliced out of the nascent pre-mRNA, these probes often result in punctate foci surrounding the site of transcription. This provides a nice control, as signal at the site of transcription should be easily recognizable as a small “blob” of signal, whereas the mature transcripts may have more complex localization patterns with punctate foci. In summary, once a validated probe set is generated for a target lncRNA it allows for numerous single-cell and single-molecule analyses to quantitate the spatiotemporal properties of that target lncRNA.
5. LOCATION-TRACKING LNCRNAS: ONE MOLECULE AT A TIME
There are many advantages to single-molecule quantitation of lncRNAs using smRNA-FISH at the level of single cells. This includes the ability of smRNA-FISH to quantify the number of molecules of a lncRNA that are present, where they are present, and how they change across a population of cells. These properties can provide key insights into lncRNA biology that may otherwise be missed by other methods (e.g., single-cell RNA sequencing).
Imagine a scenario in which a lncRNA shows context-dependent expression in a small subset of a cell population, a phenomenon called “jackpotting” (Cabili et al. 2015; Shaffer et al. 2017). RNA-seq of the bulk population would reveal a very low average expression value, thus masking robust expression in a small number of cells. Fortunately, this scenario is rare, but it highlights the usage of transcript visualization by smRNA-FISH. It is conceivable that single-cell RNA-sequencing (SCS) methods would also identify “jackpotting” cells. However, SCS methods often do not sample lncRNAs, because typically only the most abundant mRNAs are measured in SCS, especially in the context of rare-cell analysis (Torre et al. 2018).
Overall, smRNA-FISH provides a unique three-dimensional perspective on lncRNA abundance and localization within single cells. Below, we discuss recent applications of smRNA-FISH that have uncovered fundamental and unexpected insights into lncRNA biology.
5.1. Promoter Tug-of-War
A good example of where smRNA-FISH can reveal surprising subpopulation dynamics came from the investigation of linc-HOXA1 in mouse embryonic stem cells (mESCs). Population-level RNA sequencing showed linc-HOXA1 (HAUNT) strongly correlated in expression with its neighboring gene, HOXA1, suggesting that the lncRNA could positively regulate HOXA1 (Maamar et al. 2013; Yin et al. 2015). However, smRNA-FISH revealed that the lncRNA and mRNA are expressed in a mutually exclusive manner: cells expressing HOXA1 did not express linc-HOXA1 and vice versa (Maamar et al. 2013).
Thus, on a single-cell level it was revealed that linc-HOXA1 may function as a repressor—rather than activator—of HOXA1. This repressive role was experimentally validated with antisense oligonucleotide (ASO) knockdown of linc-HOXA1, resulting in increased HOXA1 expression. Thus, linc-HOXA1 promoter activation inhibits HOXA1 transcription and vice versa (Maamar et al. 2013). A similar phenomenon was resolved in Drosophila between the Hox gene Ubx and an upstream lncRNA (lncRNA-bxd) that results in promoter competition (Petruk et al. 2006). Similarly, lncRNA-bxd is expressed in mutually exclusive subsets of cells from Ubx; again correlated in bulk, but repressive relationships are revealed at the single-cell level. Together, these examples show how smRNA-FISH can resolve complex regulatory relationships within subpopulations of cells.
5.2. lncRNAs Do Not Hit the Jackpot
Although some lncRNAs are highly abundant, the vast majority are estimated to be expressed at less than one molecule per cell (Cabili et al. 2011; Wang et al. 2011; Derrien et al. 2012; Hangauer et al. 2013). One possibility, described above, is “jackpotting” of lncRNA expression sub populations of cells. Thus, one hypothesis that lingered in the field was that lncRNAs are in fact highly abundant in a small subset of cells but are lowly expressed in most other cells. To address this question, one study examined the smRNA-FISH localization patterns of 61 lncRNAs and 49 mRNAs in three populations of different human cell lines (Cabili et al. 2015). It is important to note that more than 100 lncRNAs were targeted in this study, but only 61 had consistent signal in “odds and evens” probe validation. This further underscores the importance of probe optimization for lncRNAs (Fig. 3B). The 61 validated smRNA-FISH probe sets represented lncRNAs with a spectrum of cellular abundance (as measured by RNA-sequencing) and other properties (e.g., tissue specificity, which could be indicative of subpopulation expression dynamics). By quantifying lncRNA and mRNA abundance from cell to cell within a population, it was observed that overall, lncRNAs and mRNAs show similar cell-to-cell variability in expression and do not show any jackpot behavior. The scale and systematic approach of this smRNA-FISH study identified other fundamental lncRNA localization properties.
lncRNAs are more nuclear in their localization than mRNAs. For example, 55% of lncRNAs showed a significant bias for nuclear localization compared with only one mRNA that was found predominantly nuclear (Cabili et al. 2015). In contrast, some lncRNAs had exclusively cytoplasmic localization patterns, suggesting diverse subcellular functions for this class of RNA. Interestingly, these localization patterns were the same in three different human cell lines (85% of tested lncRNAs had the same nuclear to cytoplasmic ratios).
Nuclear chromatin localization of lncRNAs does not persist during mitosis. It remains a long-standing mystery how two chromosomes reestablish their transcriptional identity after DNA replication and mitosis. One hypothesis is that chromatin-bound lncRNAs influence this process. smRNA-FISH is uniquely suited to address this question by simultaneously determining subcellular abundance of a lncRNA. For example, how much of a lncRNA resides on chromatin and in which phase of the cell cycle (by simultaneous multicolor smRNA-FISH visualization of the lncRNA and cell cycle marker genes). Surprisingly, each of the six-known chromatin-associating lncRNAs show altered localization during mitosis: in almost all cases, the lncRNAs are released from chromatin and reside in the cytoplasm (Cabili et al. 2015). This included the well-known nuclear and chromatin associated lncRNAs (e.g., Xist), suggesting that they do not play a role in organizing chromatin after mitosis.
RNAs that are highly correlated are not necessarily involved in positive regulatory relationships. As in the case of linc-HOXA1 and lncRNA-Ubx above, correlations between lncRNAs and putative regulatory targets via smRNA-FISH can implicate a regulatory relationship. Indeed, certain highly correlated lncRNAs and mRNAs (including several that share the same promoter) show very different localization patterns (Cabili et al. 2015). This suggests diverse independent functions for these lncRNAs despite coregulation with neighboring mRNAs. Collectively, these findings and many emerging examples have provided unique insights about lncRNA expression and localization properties from smRNA-FISH.
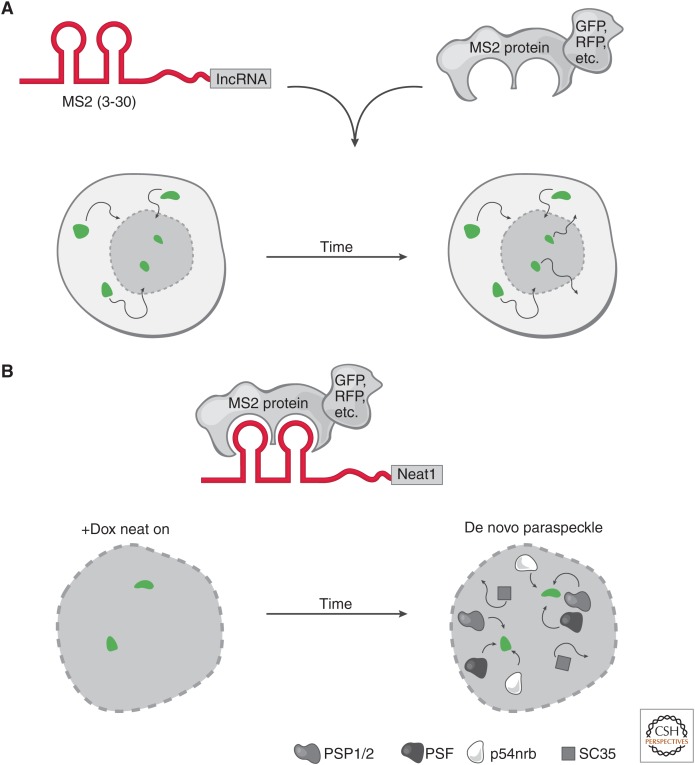
Figure 3.
Live-cell RNA imaging. (A) The targeted long noncoding RNA (lncRNA) has a specific RNA sequence/structure (aptamer) appended (e.g., a series of MS2 hairpins). In parallel, a protein that specifically and tightly binds to the RNA aptamer is fused to a fluorescent marker. These constructs can be integrated at the endogenous locus or ectopically expressed in cells. Conjugation of the RNA aptamer with the cognate fusion protein provides the ability to track the fluorescent signal in live cells through time. This highlights the movements (arrows) of a long noncoding RNA (lncRNA), such as its rate of nuclear and cytoplasmic transport (bottom nuclei). (B) Neat1 live-cell RNA imaging. Live-cell RNA imaging of Neat1 revealed key insights into the role of its transcription in the establishment and maintenance of nuclear paraspeckles. The Neat1 RNA was tracked by appending 24× MS2 hairpins and cognate MS2 protein fused to a fluorescent protein. Neat1 expression was induced (green foci) by doxycycline (+Dox) and over time, paraspeckle proteins (PSP1/2, PSF, p54nrb) were recruited to the de novo expression sites of Neat1 RNA. In contrast, nuclear speckle protein (SC35) was not recruited to the de novo paraspeckle. When Neat1 expression was turned off in the absence of Dox, the paraspeckle disassembles. Together, these observations show that expression of Neat1 precedes paraspeckle formation and is required to maintain paraspeckles in the nucleus.
6. ENTERING THE FOURTH DIMENSION: LIVE-CELL RNA IMAGING
Although much has been learned from the above lncRNA-FISH studies, the temporal dynamics of lncRNA localization in a living cell has been less well understood. For several decades, live-cell RNA imaging has been possible in many model organisms (e.g., Saccharomyces cerevisiae), cell lines and even living mice (Tyagi 2009; Larson et al. 2011; Park et al. 2014; Lee et al. 2016a; Nelles et al. 2016; Wu et al. 2016; Tutucci et al. 2018a). Although most of these studies have focused on mRNA there are ever-emerging examples of live-cell imaging of lncRNAs. For example, the telomeric lncRNA, telomeric repeat-containing RNA (TERRA), is activated in space and time specifically at chromosomes with short telomeres (Cusanelli et al. 2013). Another elegant live-cell RNA imaging study identified a lncRNA that over time switches between two transcriptional states to repress the neighboring GAL1 mRNA (Lenstra et al. 2015). These live-cell studies and others have begun to uncover the temporal properties of lncRNA synthesis, degradation, and localization (Tutucci et al. 2018a).
There are many approaches to live-cell RNA imaging but here, we will focus on one: appending an RNA aptamer that is recognized by a cognate protein (Fig. 3). One of the most frequently appended aptamers is the MS2 RNA sequence, which is specifically and tightly recognized by the MS2 protein (Beach et al. 1999; Eliscovich and Singer 2017; Lyon and Stasevich 2017; Tutucci et al. 2018a,b). By fusing MS2 protein to a fluorescent protein (e.g., GFP or RFP) the cognate RNA–protein interaction can be imaged in real time (Fig. 3). There are several other similar RNA–protein aptamers (e.g., PP7 RNA/protein); these can be multiplexed to allow for multiple RNAs to be uniquely tagged and tracked in living cells.
These tagged lncRNAs are introduced as exogenous copies in living cells followed by imaging of the targeted RNA’s spatiotemporal movements. One disadvantage of this approach is that endogenous (nontagged) copies of the lncRNA cannot be visualized. Alternatively, CRISPR–Cas9 can be used to engineer MS2 hairpins at the 3′ end of the endogenous lncRNA locus to overcome this limitation (see Engreitz et al. 2019).
As we discussed above, Neat1 and Neat2/Malat1, Xist, Gomafu, and pre-rRNA RNA-FISH each lead to the discovery of new nuclear sub compartments (Fig. 1). This raises several questions that can only be addressed using live-cell RNA imaging: (1) Do paraspeckles form first and then Neat1 localizes? (2) Does transcriptional activation of Neat1 result in paraspeckle formation? (3) Is the RNA a required component of maintaining the paraspeckle? Thus, only exploring these questions temporally in living cells will provided the needed insights into the functional role of Neat1 in paraspeckle formation.
To address these questions, a seminal study tagged Neat1 (both Menε/β isoforms) with 24 copies of the MS2 RNA aptamer on the 5′ end of Neat1 (Mao et al. 2010). This system was further engineered to induce Neat1 transcription via an inducible promoter (Tet-on: addition of Doxycycline [DOX] results in activation of Neat1). Importantly, the inducible-tagged Neat1 was placed in a dormant region of the genome (in mouse C2C12 cells). Together, these constructs allowed the visualization and tracking of Neat1 RNA at its site of transcription in parallel with protein constituents of the paraspeckle. This study found that indeed on induction of Neat1 transcription resulted in a concomitant formation of paraspeckles (Fig. 3B). In fact, the transcription of Neat1 recruited most paraspeckle proteins (e.g., PSP1, p54nrb, PSF, and PSP2), but not the protein components of the nuclear speckle associated with Malat1 (e.g., SC35). Moreover, it was shown that the newly formed paraspeckles were functional and stably formed over time. The reverse was true that shutdown of Neat1 transcription resulted, over time, in the disassembly of newly formed paraspeckles.
Together, these experiments brought our understanding of Neat1 into the fourth dimension and revealed the underlying dynamics of Neat1 induction and in turn the formation of paraspeckles. It was the aspect of time and living cells that provided these simple yet profound insights. Together, these results show that transcribing lncRNAs could facilitate changes in nuclear substructures and perhaps represents a more general role for nuclear lncRNAs.
However, many more questions remain about lncRNA biology that are suited to be tested by live-cell RNA imagining. For example, do lncRNAs that have both nuclear and cytoplasmic localization patterns shuttle between the nucleus and cytoplasm or is it the case that some molecules remain trapped in the nucleus whereas others shuttle to the cytoplasm (Fig. 3A)? If shuttling occurs, then one could dissect down the lncRNA to find the minimal RNA domain sufficient to shuttle RNA. These and many other questions are in the offing by applying live-cell imaging to lncRNA biology.
7. CRISPR-BASED VISUALIZATION OF LNCRNA LOCI
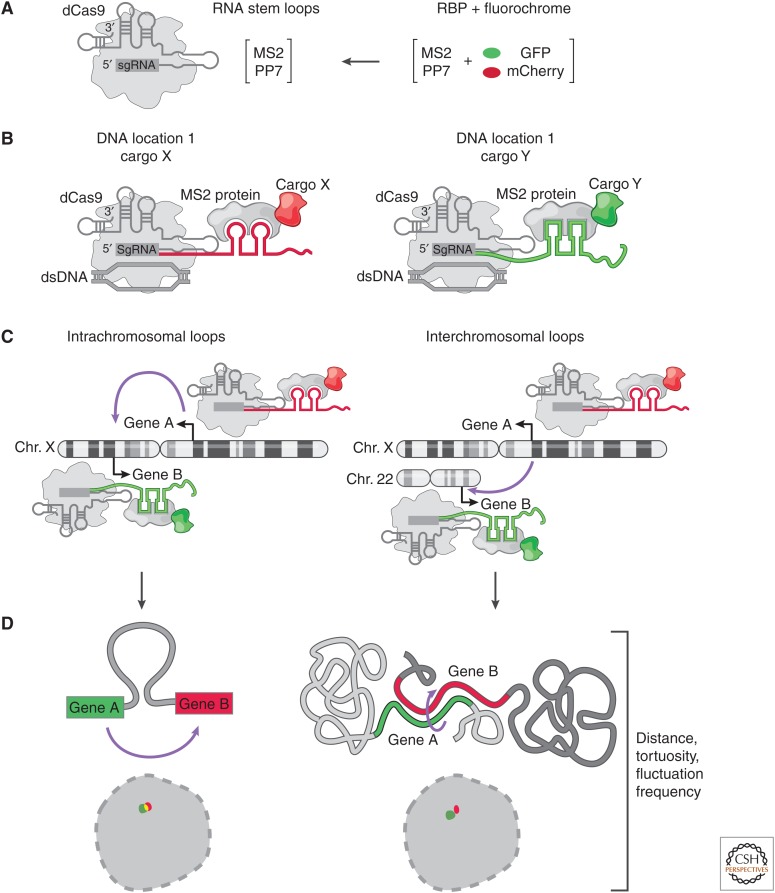
With the advent of CRISPR-based technologies, it has become possible to visualize the four-dimensional dynamics of lncRNA genomic loci (Chen et al. 2013, 2016; Ma et al. 2015; Shechner et al. 2015; Shao et al. 2016; Tu et al. 2016; Dreissig et al. 2017; Qin et al. 2017; Zhou et al. 2017; Maass et al. 2018a,b). These studies have made it possible to track DNA in living cells using a combination of CRISPR–Cas9 guide sequences (see Engreitz et al. 2019) modified to contain RNA aptamers (such as MS2) and catalytically inactive, or dead Cas9 (dCas9) (Fig. 4). More recently, these approaches have been used to explore the dynamic movements and interactions of noncoding DNA including lncRNA loci. For example, how often, stable and frequent are lncRNA-mediated intra- and interchromosomal interactions in the nucleus (Maass et al. 2018a,b) and the underlying dynamics of DNA enhancers finding their targets (Gu et al. 2018).
Figure 4.
Live-cell imaging of long noncoding RNA (lncRNA) loci. (A) Catalytically inactive CRISPR–Cas9 constructs with extended single-guide RNA (sgRNA) sequences that contain RNA aptamers (e.g., MS2 or PP7). The RNA binding protein (RBP) that recognizes sgRNA aptamers is fused to a fluorescent protein. (B) A CRISPR–dCas9/cargo complex, in which the modified sgRNA specifies the genomic location and acts as a scaffold for delivery of chosen “cargo” fused to the MS2 coat protein. (C) Application of guide sequence aptamer for CRISPR live-cell imaging (CLING). Each guide has a unique RNA aptamer that forms a defined complex with RBP:fluorescent protein fusion. This allows for real-time monitoring of lncRNA loci (or any genomic location) forming intra- or interchromosomal interactions in space and time. (D) Examples of using CLING to monitor the biophysical properties (tortuosity, distance, fluctuation rates) of DNA folding within and across chromosomes.
If only one color and one position are being monitored, then simply fusing a fluorescent protein to the Cas9 protein with a guide to a specific DNA location is suitable. Yet, as soon as two different locations need to be monitored simultaneously, the guide sequence needs to be “coded” with an RNA aptamer (such as MS2 RNA described above) (Fig. 4). Otherwise, combining two different Cas9-fluorescent fusions with two different guides would result in a random mixing of guides and fluorescent Cas9. In other words, the “color” would not be specified by the guide.
By way of analogy, each Cas9 is like a drone with cargo. The location of the drone is determined by the guide sequence. The RNA sequences (e.g., MS2 RNA sequence) appended to the guide specify the cargo that is to be delivered to that location (e.g., MS2 protein fused to a fluorescent protein or other fusions) (Fig. 4). Thus, each Cas9 drone brings a specific cargo to specific genomic location. Finally, several drones can be deployed in a single-cell, each with a specific genomic location and cargo (Fig. 4). Interestingly, guide RNA sequences are amenable to adding very long RNA cargos, even entire lncRNAs.
Overall, the CRISPR–Cas systems are universally applicable to deliver varieties of RNA cargo to and/or demarcate specific genomic locations. In principle, this simplifies to CRISPR–Cas9-based live-cell DNA imaging, for coding and noncoding regions alike. Several recent studies have used the MS2 and PP7 RNA aptamers to monitor DNA dynamics in three and four dimensions. This has shed new light on diverse aspects of nuclear biology: from how telomeres are organized in the living nucleus, to how specific chromosomal regions move around the nucleus, to DNA packaging dynamics. More recently, these approaches have been applied to monitor the dynamics of intra- and interchromosomal interactions by lncRNA loci (Chen et al. 2013, 2016; Ma et al. 2015; Shechner et al. 2015; Shao et al. 2016; Tu et al. 2016; Dreissig et al. 2017; Qin et al. 2017; Zhou et al. 2017; Maass et al. 2018a,b). Collectively, live-cell RNA and DNA imaging are synergistic approaches to understanding the dynamic roles of RNA, facilitating nuclear subcompartments in parallel with the resulting reorganization DNA by lncRNAs.
8. CONCLUDING REMARKS
Herein, we have covered several RNA imaging approaches and in turn how they have illuminated fundamental properties of lncRNA biology. As we hope to have highlighted, RNA imaging has a unique advantage of “seeing” where, when, and how a lncRNA is expressed in each cell. Thus, RNA imaging is often the best place to start before moving forward with mechanistic studies. For example, if a lncRNA has an exclusively nuclear localization versus a predominantly cytoplasmic pattern, which would lead to very different mechanistic hypotheses. Indeed, a wide variety of localization patterns and lncRNA functional insights have been gleaned from RNA imaging: from NORAD, with its exquisite cytoplasmic localization led to an understanding of its role in translation regulation (Lee et al. 2016b; Tichon et al. 2016); to CONCR, which is specifically activated in the nucleus during S-phase and regulates sister chromatid pairing (Marchese et al. 2016); to SLERT, which localizes to the nucleolus and forms ring-shaped structures that influence RNA polymerase I transcription (Xing et al. 2017); to 7SK that localizes to promoters to regulate transcription (Prasanth et al. 2010). With each emerging lncRNA-FISH study, fascinating new localization patterns, dynamics, and biological functions are being revealed for lncRNAs.
Collectively, the key findings from these and other RNA imaging studies raises a fundamental question about lncRNA biology. Could certain lncRNA localization patterns be indicative of specific functional lncRNA “classes” or “categories”? A general bifurcation is cytoplasmic versus nuclear localized lncRNAs that appear to a first approximation to indicate common functionalities within these subcellular regions. The screening of a small panel of lncRNAs revealed all possible lncRNA localization patterns, ranging from exclusively nuclear, nucleoplasmic, nuclear, and cytoplasmic or exclusively cytoplasmic (Cabili et al. 2015). Yet, in each category there were at least a few lncRNAs sharing similar expression patterns that were consistent across several cell lines. Thus, it is tantalizing to speculate that lncRNAs sharing specific spatial-temporal localization patterns could also have common mechanistic roles. Consistent with this notion, many lncRNAs show two nuclear foci in which intronic and exonic probes overlap. This localization pattern could suggest that such lncRNAs are acting in cis (see Engreitz et al. 2019). Indeed, this localization feature is shared by many lncRNAs that either function as enhancer RNAs (eRNAs) that “loop” enhancers to gene targets or facilitate global nuclear architecture through interactions with nuclear lamina (Ørom and Shiekhattar 2011; Lam et al. 2014; Melé and Rinn 2016; Nozawa et al. 2017). As more lncRNA mechanisms are elucidated, these association between localization and function may become more evident.
Teasing apart associations between different lncRNAs will further require RNA imaging advancements to simultaneously image numerous RNA species in situ. These limitations are starting to be overcome, with several multiplexing methods now available for the detection of hundreds or even thousands of RNAs in individual cells (Levsky et al. 2002; Lubeck and Cai 2012; Levesque and Raj 2013; Lubeck et al. 2014; Chen et al. 2015). These methods will greatly expand and enable a more discovery-oriented approach to lncRNA biology. Yet, perhaps more importantly, these methods will allow direct detection of common localization patterns and perhaps functional classes across hundreds of lncRNAs at once.
Moving forward, it is important to note that lncRNAs have a long-standing track record of nuclear regulatory roles. This may perhaps be caused by observational bias, but from the limited examples covered here we have seen that lncRNAs play roles in establishing nuclear domains such as the nucleolus (pre-rRNA), paraspeckles (Neat1), nuclear speckles (Malat1), nuclear matrix (Gomafu), and the Barr body (Xist). Moreover, many lncRNAs have been found to have focal localization patterns that could be indicative of nuclear organization on a smaller scale. Thus, understanding how these exquisite lncRNA localization patterns relate to function is a key goal of future studies.
ACKNOWLEDGMENTS
We thank all the students and postdocs that have provided numerous insights into lncRNA biology using RNA imaging techniques. We also thank Tom Cech for editorial advice and editing, Alexandra and Mark McCorkindale for manuscript edits, and Sigrid Knemeyer for illustrations.
Authorship was listed alphabetically.
We apologize in advance for not being able to cover all the existing and emerging RNA-FISH studies targeting lncRNA. Here we aim to focus only on a few examples to cover basic principles of RNA-FISH in the context of insights into lncRNA biology.
Editors: Thomas R. Cech, Joan A. Steitz, and John F. Atkins
Additional Perspectives on RNA Worlds available at www.cshperspectives.org
REFERENCES
*Reference is also in this collection.
- Batish M, Raj A, Tyagi S. 2011. Single molecule imaging of RNA in situ. Methods Mol Biol 714: 3–13. [DOI] [PubMed] [Google Scholar]
- Beach DL, Salmon ED, Bloom K. 1999. Localization and anchoring of mRNA in budding yeast. Curr Biol 9: 569–578. [DOI] [PubMed] [Google Scholar]
- Bond CS, Fox AH. 2009. Paraspeckles: Nuclear bodies built on long noncoding RNA. J Cell Biol 186: 637–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CJ, Hendrich BD, Rupert JL, Lafrenière RG, Xing Y, Lawrence J, Willard HF. 1992. The human XIST gene: Analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell 71: 527–542. [DOI] [PubMed] [Google Scholar]
- Buxbaum AR, Haimovich G, Singer RH. 2015. In the right place at the right time: Visualizing and understanding mRNA localization. Nat Rev Mol Cell Biol 16: 95–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, Rinn JL. 2011. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev 25: 1915–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabili MN, Dunagin MC, McClanahan PD, Biaesch A, Padovan-Merhar O, Regev A, Rinn JL, Raj A. 2015. Localization and abundance analysis of human lncRNAs at single-cell and single-molecule resolution. Genome Biol 16: 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Gilbert LA, Cimini BA, Schnitzbauer J, Zhang W, Li G-W, Park J, Blackburn EH, Weissman JS, Qi LS, et al. 2013. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell 155: 1479–1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen KH, Boettiger AN, Moffitt JR, Wang S, Zhuang X. 2015. RNA imaging. Spatially resolved, highly multiplexed RNA profiling in single cells. Science 348: aaa6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Guan J, Huang B. 2016. Imaging specific genomic DNA in living cells. Annu Rev Biophys 45: 1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L, Ming H, Zhu M, Wen B. 2016. Long noncoding RNAs as organizers of nuclear architecture. Sci China Life Sci 59: 236–244. [DOI] [PubMed] [Google Scholar]
- Cusanelli E, Romero CAP, Chartrand P. 2013. Telomeric noncoding RNA TERRA is induced by telomere shortening to nucleate telomerase molecules at short telomeres. Mol Cell 51: 780–791. [DOI] [PubMed] [Google Scholar]
- Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, et al. 2012. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res 22: 1775–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirks RW, van de Rijke FM, Fujishita S, van der Ploeg M, Raap AK. 1993. Methodologies for specific intron and exon RNA localization in cultured cells by haptenized and fluorochromized probes. J Cell Sci 104: 1187–1197. [DOI] [PubMed] [Google Scholar]
- Dreissig S, Schiml S, Schindele P, Weiss O, Rutten T, Schubert V, Gladilin E, Mette MF, Puchta H, Houben A. 2017. Live-cell CRISPR imaging in plants reveals dynamic telomere movements. Plant J 91: 565–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunagin M, Cabili MN, Rinn J, Raj A. 2015. Visualization of lncRNA by single-molecule fluorescence in situ hybridization. Methods Mol Biol 1262: 3–19. [DOI] [PubMed] [Google Scholar]
- Eliscovich C, Singer RH. 2017. RNP transport in cell biology: The long and winding road. Curr Opin Cell Biol 45: 38–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engreitz JM, Ollikainen N, Guttman M. 2016. Long non-coding RNAs: Spatial amplifiers that control nuclear structure and gene expression. Nat Rev Mol Cell Biol 17: 756–770. [DOI] [PubMed] [Google Scholar]
- *.Engreitz J, Abudayyeh O, Gootenberg J, Zhang F. 2019. CRISPR tools for systematic studies of RNA regulation. Cold Spring Harb Perspect Biol 11: a035386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falahati H, Pelham-Webb B, Blythe S, Wieschaus E. 2016. Nucleation by rRNA dictates the precision of nucleolus assembly. Curr Biol 26: 277–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Femino AM, Fay FS, Fogarty K, Singer RH. 1998. Visualization of single RNA transcripts in situ. Science 280: 585–590. [DOI] [PubMed] [Google Scholar]
- Fremeau RT, Lundblad JR, Pritchett DB, Wilcox JN, Roberts JL. 1986. Regulation of pro-opiomelanocortin gene transcription in individual cell nuclei. Science 234: 1265–1269. [DOI] [PubMed] [Google Scholar]
- Gaspar I, Wippich F, Ephrussi A. 2017. Enzymatic production of single-molecule FISH and RNA capture probes. RNA 23: 1582–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu B, Swigut T, Spencley A, Bauer MR, Chung M, Meyer T, Wysocka J. 2018. Transcription-coupled changes in nuclear mobility of mammalian cis-regulatory elements. Science 359: 1050–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hangauer MJ, Vaughn IW, McManus MT. 2013. Pervasive transcription of the human genome produces thousands of previously unidentified long intergenic noncoding RNAs. PLoS Genet 9: e1003569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson JN, Ensminger AW, Clemson CM, Lynch CR, Lawrence JB, Chess A. 2007. A screen for nuclear transcripts identifies two noncoding RNAs associated with SC35 splicing domains. BMC Genomics 8: 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam MTY, Li W, Rosenfeld MG, Glass CK. 2014. Enhancer RNAs and regulated transcriptional programs. Trends Biochem Sci 39: 170–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson DR, Singer RH, Zenklusen D. 2009. A single molecule view of gene expression. Trends Cell Biol 19: 630–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson DR, Zenklusen D, Wu B, Chao JA, Singer RH. 2011. Real-time observation of transcription initiation and elongation on an endogenous yeast gene. Science 332: 475–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence JB, Singer RH. 1986. Intracellular localization of messenger RNAs for cytoskeletal proteins. Cell 45: 407–415. [DOI] [PubMed] [Google Scholar]
- Lawrence JB, Singer RH, Villnave CA, Stein JL, Stein GS. 1988. Intracellular distribution of histone mRNAs in human fibroblasts studied by in situ hybridization. Proc Natl Acad Sci 85: 463–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence JB, Singer RH, Marselle LM. 1989. Highly localized tracks of specific transcripts within interphase nuclei visualized by in situ hybridization. Cell 57: 493–502. [DOI] [PubMed] [Google Scholar]
- Lee BH, Bae S-W, Shim JJ, Park SY, Park HY. 2016a. Imaging single-mRNA localization and translation in live neurons. Mol Cells 39: 841–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Kopp F, Chang T-C, Sataluri A, Chen B, Sivakumar S, Yu H, Xie Y, Mendell JT. 2016b. Noncoding RNA NORAD regulates genomic stability by sequestering PUMILIO proteins. Cell 164: 69–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenstra TL, Coulon A, Chow CC, Larson DR. 2015. Single-molecule imaging reveals a switch between spurious and functional ncRNA transcription. Mol Cell 60: 597–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levesque MJ, Raj A. 2013. Single-chromosome transcriptional profiling reveals chromosomal gene expression regulation. Nat Methods 10: 246–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levesque MJ, Ginart P, Wei Y, Raj A. 2013. Visualizing SNVs to quantify allele-specific expression in single cells. Nat Meth 10: 865–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levsky JM, Shenoy SM, Pezo RC, Singer RH. 2002. Single-cell gene expression profiling. Science 297: 836–840. [DOI] [PubMed] [Google Scholar]
- Lubeck E, Cai L. 2012. Single-cell systems biology by super-resolution imaging and combinatorial labeling. Nat Methods 9: 743–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubeck E, Coskun AF, Zhiyentayev T, Ahmad M, Cai L. 2014. Single-cell in situ RNA profiling by sequential hybridization. Nat Methods 11: 360–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyon K, Stasevich TJ. 2017. Imaging translational and post-translational gene regulatory dynamics in living cells with antibody-based probes. Trends Genet 33: 322–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H, Naseri A, Reyes-Gutierrez P, Wolfe SA, Zhang S, Pederson T. 2015. Multicolor CRISPR labeling of chromosomal loci in human cells. Proc Natl Acad Sci 112: 3002–3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maamar H, Cabili MN, Rinn J, Raj A. 2013. linc-HOXA1 is a noncoding RNA that represses Hoxa1 transcription in cis. Genes Dev 27: 1260–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maass PG, Barutcu AR, Shechner DM, Weiner CL, Melé M, Rinn JL. 2018a. Spatiotemporal allele organization by allele-specific CRISPR live-cell imaging (SNP-CLING). Nat Struct Mol Biol 25: 176–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maass PG, Barutcu AR, Weiner CL, Rinn JL. 2018b. Inter-chromosomal contact properties in live-cell imaging and in Hi-C. Mol Cell 70: 188–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao YS, Sunwoo H, Bin Zhang, Spector DL. 2010. Direct visualization of the co-transcriptional assembly of a nuclear body by noncoding RNAs. Nat Cell Biol 13: 95–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao YS, Zhang B, Spector DL. 2011. Biogenesis and function of nuclear bodies. Trends Genet 27: 295–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchese FP, Grossi E, Marín-Béjar O, Bharti SK, Raimondi I, González J, Martínez-Herrera DJ, Athie A, Amadoz A, Brosh RM, et al. 2016. A long noncoding RNA regulates sister chromatid cohesion. Mol Cell 63: 397–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin KC, Ephrussi A. 2009. mRNA localization: Gene expression in the spatial dimension. Cell 136: 719–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melé M, Rinn JL. 2016. “Cat’s cradling” the 3D genome by the act of lncRNA transcription. Mol Cell 62: 657–664. [DOI] [PubMed] [Google Scholar]
- Nelles DA, Fang MY, O’Connell MR, Xu JL, Markmiller SJ, Doudna JA, Yeo GW. 2016. Programmable RNA tracking in live cells with CRISPR/Cas9. Cell 165: 488–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozawa R-S, Boteva L, Soares DC, Naughton C, Dun AR, Buckle A, Ramsahoye B, Bruton PC, Saleeb RS, Arnedo M, et al. 2017. SAF-A regulates interphase chromosome structure through oligomerization with chromatin-associated RNAs. Cell 169: 1214–1227.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orjalo AV, Johansson HE. 2016. Stellaris® RNA fluorescence in situ hybridization for the simultaneous detection of immature and mature long noncoding RNAs in adherent cells. Methods Mol Biol 1402: 119–134. [DOI] [PubMed] [Google Scholar]
- Ørom UA, Shiekhattar R. 2011. Noncoding RNAs and enhancers: Complications of a long-distance relationship. Trends Genet 27: 433–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HY, Lim H, Yoon YJ, Follenzi A, Nwokafor C, Lopez-Jones M, Meng X, Singer RH. 2014. Visualization of dynamics of single endogenous mRNA labeled in live mouse. Science 343: 422–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parton RM, Davidson A, Davis I, Weil TT. 2014. Subcellular mRNA localisation at a glance. J Cell Sci 127: 2127–2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petruk S, Sedkov Y, Riley KM, Hodgson J, Schweisguth F, Hirose S, Jaynes JB, Brock HW, Mazo A. 2006. Transcription of bxd noncoding RNAs promoted by trithorax represses Ubx in cis by transcriptional interference. Cell 127: 1209–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasanth KV, Camiolo M, Chan G, Tripathi V, Denis L, Nakamura T, Hübner MR, Spector DL. 2010. Nuclear organization and dynamics of 7SK RNA in regulating gene expression. Mol Biol Cell 21: 4184–4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin P, Parlak M, Kuscu C, Bandaria J, Mir M, Szlachta K, Singh R, Darzacq X, Yildiz A, Adli M. 2017. Live cell imaging of low- and non-repetitive chromosome loci using CRISPR-Cas9. Nat Commun 8: 14725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Querido E, Dekakra-Bellili L, Chartrand P. 2017. RNA fluorescence in situ hybridization for high-content screening. Methods 126: 149–155. [DOI] [PubMed] [Google Scholar]
- Quinodoz S, Guttman M. 2014. Long noncoding RNAs: An emerging link between gene regulation and nuclear organization. Trends Cell Biol 24: 651–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj A, van Oudenaarden A. 2009. Single-molecule approaches to stochastic gene expression. Annu Rev Biophys 38: 255–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj A, van den Bogaard P, Rifkin SA, van Oudenaarden A, Tyagi S. 2008. Imaging individual mRNA molecules using multiple singly labeled probes. Nat Methods 5: 877–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj A, Rifkin SA, Andersen E, van Oudenaarden A. 2010. Variability in gene expression underlies incomplete penetrance. Nature 463: 913–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn J, Guttman M. 2014. RNA Function. RNA and dynamic nuclear organization. Science 345: 1240–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer SM, Wu M-T, Levesque MJ, Raj A. 2013. Turbo FISH: A method for rapid single molecule RNA FISH. PLoS ONE 8: e75120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer SM, Dunagin MC, Torborg SR, Torre EA, Emert B, Krepler C, Beqiri M, Sproesser K, Brafford PA, Xiao M, et al. 2017. Rare cell variability and drug-induced reprogramming as a mode of cancer drug resistance. Nature 546: 431–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao S, Zhang W, Hu H, Xue B, Qin J, Sun C, Sun Y, Wei W, Sun Y. 2016. Long-term dual-color tracking of genomic loci by modified sgRNAs of the CRISPR/Cas9 system. Nucleic Acids Res 19: e86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shechner DS, Hacisuleyman E, Younger ST, Rinn JL. 2015. Multiplexable, locus-specific targeting of long RNAs with CRISPR-display. Nat Methods 12: 664–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinnamon JR, Czaplinski K. 2014. RNA detection in situ with FISH-STICs. RNA 20: 260–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soler M, Boque-Sastre R, Guil S. 2017. RNA-FISH to study regulatory RNA at the site of transcription. Methods Mol Biol 1543: 221–229. [DOI] [PubMed] [Google Scholar]
- Sone M, Hayashi T, Tarui H, Agata K, Takeichi M, Nakagawa S. 2007. The mRNA-like noncoding RNA Gomafu constitutes a novel nuclear domain in a subset of neurons. J Cell Sci 120: 2498–2506. [DOI] [PubMed] [Google Scholar]
- Spector DL, Lamond AI. 2011. Nuclear speckles. Cold Spring Harb Perspect Biol 3: a000646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tichon A, Gil N, Lubelsky Y, Havkin Solomon T, Lemze D, Itzkovitz S, Stern-Ginossar N, Ulitsky I. 2016. A conserved abundant cytoplasmic long noncoding RNA modulates repression by Pumilio proteins in human cells. Nat Commun 7: 12209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torre E, Dueck H, Shaffer S, Gospocic J, Gupte R, Bonasio R, Kim J, Murray J, Raj A. 2018. Rare cell detection by single-cell RNA sequencing as guided by single-molecule RNA FISH. Cell Syst 6: 171–179.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trcek T, Larson DR, Moldón A, Query CC, Singer RH. 2011. Single-molecule mRNA decay measurements reveal promoter-regulated mRNA stability in yeast. Cell 147: 1484–1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trcek T, Chao JA, Larson DR, Park HY, Zenklusen D, Shenoy SM, Singer RH. 2012. Single-mRNA counting using fluorescent in situ hybridization in budding yeast. Nature Protoc 7: 408–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi V, Fei J, Ha T, Prasanth KV. 2015. RNA fluorescence in situ hybridization in cultured mammalian cells. Methods Mol Biol 1206: 123–136. [DOI] [PubMed] [Google Scholar]
- Tu L-C, Naseri A, Huisman M, Zhang S, Grunwald D, Pederson T, Ma H. 2016. Multiplexed labeling of genomic loci with dCas9 and engineered sgRNAs using CRISPRainbow. Nat Biotechnol 34: 1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tutucci E, Livingston NM, Singer RH, Wu B. 2018a. Imaging mRNA in vivo, from birth to death. Annu Rev Biophys 47: 85–106. [DOI] [PubMed] [Google Scholar]
- Tutucci E, Vera M, Biswas J, Garcia J, Parker R, Singer RH. 2018b. An improved MS2 system for accurate reporting of the mRNA life cycle. Nat Methods 15: 81–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyagi S. 2009. Imaging intracellular RNA distribution and dynamics in living cells. Nat Methods 6: 331–338. [DOI] [PubMed] [Google Scholar]
- Vance KW, Ponting CP. 2014. Transcriptional regulatory functions of nuclear long noncoding RNAs. Trends Genet 30: 348–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Van Treeck B, Parker R. 2019. Principles of stress granules revealed by imaging approaches. Cold Spring Harb Perspect Biol 11: a033068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, Lajoie BR, Protacio A, Flynn RA, Gupta RA, et al. 2011. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature 472: 120–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang DO, Matsuno H, Ikeda S, Nakamura A, Yanagisawa H, Hayashi Y, Okamoto A. 2012. A quick and simple FISH protocol with hybridization-sensitive fluorescent linear oligodeoxynucleotide probes. RNA 18: 166–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willingham AT. 2005. A strategy for probing the function of noncoding RNAs finds a repressor of NFAT. Science 309: 1570–1573. [DOI] [PubMed] [Google Scholar]
- Wu B, Eliscovich C, Yoon YJ, Singer RH. 2016. Translation dynamics of single mRNAs in live cells and neurons. Science 352: 1430–1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y-H, Yao R-W, Zhang Y, Guo C-J, Jiang S, Xu G, Dong R, Yang L, Chen L-L. 2017. SLERT regulates DDX21 rings associated with Pol I transcription. Cell 169: 664–678.e16. [DOI] [PubMed] [Google Scholar]
- Yin Y, Yan P, Lu J, Song G, Zhu Y, Li Z, Zhao Y, Shen B, Huang X, Zhu H, et al. 2015. Opposing roles for the lncRNA Haunt and its genomic locus in regulating HOXA gene activation during embryonic stem cell differentiation. Cell Stem Cell 16: 504–516. [DOI] [PubMed] [Google Scholar]
- Zenklusen D, Larson DR, Singer RH. 2008. Single-RNA counting reveals alternative modes of gene expression in yeast. Nat Struct Mol Biol 15: 1263–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Wang P, Tian F, Gao G, Huang L, Wei W, Xie XS. 2017. Painting a specific chromosome with CRISPR/Cas9 for live-cell imaging. Cell Res 27: 1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]