Abstract
The translation of messenger RNA (mRNA) into protein and the folding of the resulting protein into an active form are prerequisites for virtually every cellular process and represent the single largest investment of energy by cells. Ribosome profiling-based approaches have revolutionized our ability to monitor every step of protein synthesis in vivo, allowing one to measure the rate of protein synthesis across the proteome, annotate the protein coding capacity of genomes, monitor localized protein synthesis, and explore cotranslational folding and targeting. The rich and quantitative nature of ribosome profiling data provides an unprecedented opportunity to explore and model complex cellular processes. New analytical techniques and improved experimental protocols will provide a deeper understanding of the factors controlling translation speed and its impact on protein function and cell physiology as well as the role of ribosomal RNA and mRNA modifications in regulating translation.
The translation of messenger RNA (mRNA) into protein and the folding of the resulting polypeptide into an active form connect genetic information to functional proteins—a prerequisite for virtually every cellular process. Translation is also a costly biosynthetic process that often comprises the single largest investment of energy by cells (Verduyn et al. 1991; Russell and Cook 1995). Translation is thus highly regulated to ensure that the right proteins are made in the right places within the cell. Ensuring that newly made proteins fold, assemble, and function properly is also a major challenge to the cell (Gloge et al. 2014). The biogenesis of functional proteins depends on cotranslational folding chaperones as well as the speed of translation itself, and quality control of aberrant translation suppresses the harmful effects of mutations and errors (Brandman and Hegde 2016).
The central role played by translation has motivated the development of experimental approaches to analyze both the proteins produced by the cell and the process of their synthesis. In particular, the advent of genomics and gene expression profiling has driven interest in extending such global analyses to the study of translation (Vogel and Marcotte 2012). Beyond taking a complete and quantitative inventory of proteins produced in the cell, there is also great interest in the dynamic, multistep process of synthesizing these proteins. The ribosome incorporates amino acids with varying chemical properties by translating mRNAs that show biased use of synonymous codons, show secondary structures, and are decorated with chemical modifications. It seems natural to ask how these features affect the speed of the ribosome and what consequences result from varying the speed of elongation (Plotkin and Kudla 2011). These variations, encoded within the mRNA sequence, impact mRNA stability (Presnyak et al. 2015; Bazzini et al. 2016; Chan et al. 2017) and the rate of protein synthesis (Gingold et al. 2014), as well as the identity (Kawakami et al. 1993), folding (Kimchi-Sarfaty et al. 2007; Zhang et al. 2009), and localization (Pechmann et al. 2014) of the resulting protein.
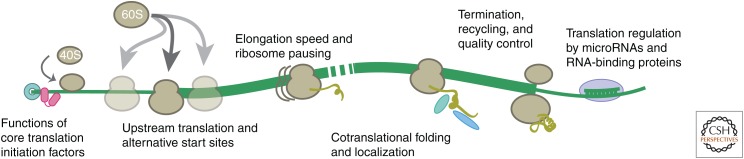
Ribosome profiling, in which next-generation sequencing is used to identify ribosome-protected mRNA fragments, thereby revealing the positions of the full set of ribosomes engaged in translation, has emerged as a transformative technique for enabling global analyses of in vivo translation and coupled, cotranslational events. Historically, it has been challenging to measure these even in vitro; now, ribosome profiling provides a comprehensive view in living cells. It has been applied to address a remarkably broad diversity of mechanistic and physiological questions (Fig. 1). In this review, we present the historical context for ribosome profiling and highlight studies that exemplify the insights it can provide. We cannot at this point hope to cover all uses of this technique. It has been reviewed extensively (Michel and Baranov 2013; Ingolia 2014, 2016; Brar and Weissman 2015), and so we place emphasis here on recent developments.
Figure 1.
Insights from ribosome profiling. Ribosome profiling experiments have addressed many aspects of the mechanisms of protein synthesis and its regulation in the cell as well as related, cotranslational processes.
RIBOSOME FOOTPRINT PROFILING OF IN VIVO TRANSLATION
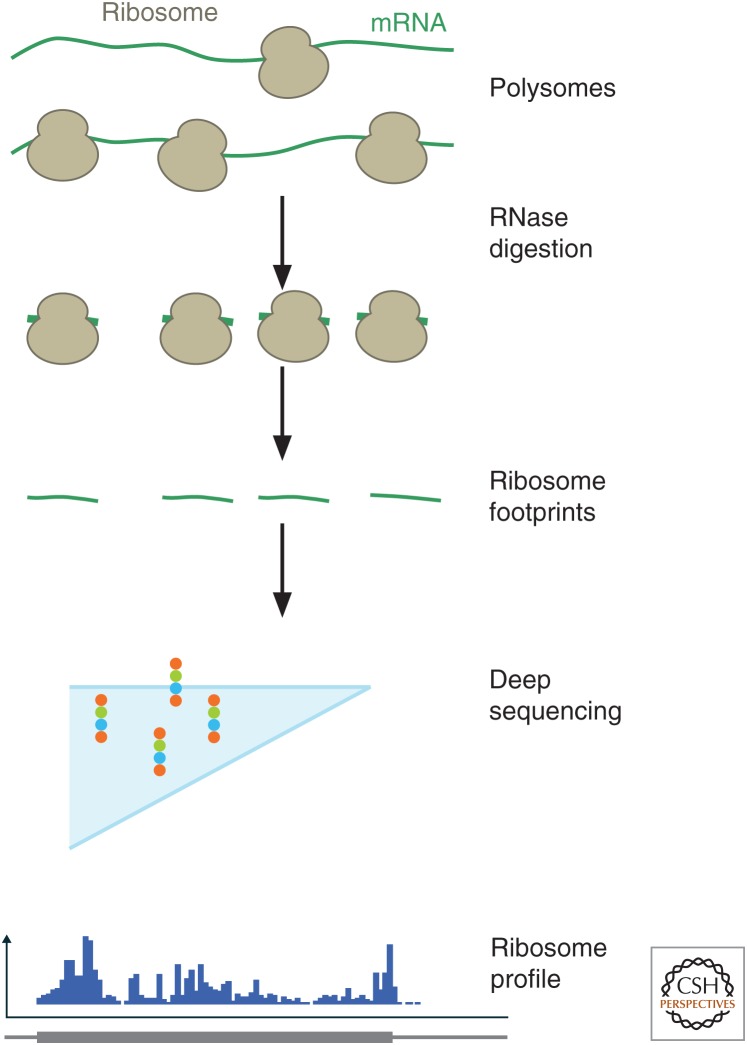
Ribosome profiling relies on deep sequencing of ribosome footprints—the short (typically, ∼30 nucleotide [nt]) fragments of mRNA that are physically enclosed by the ribosome and shielded from nuclease digestion (Fig. 2). These footprints are converted into a library of DNA fragments and analyzed by next-generation sequencing (Ingolia et al. 2012; McGlincy and Ingolia 2017). Each sequenced footprint reports on the position of one ribosome, revealing what transcript that ribosome was translating and where along the coding sequence it was captured during cell lysis, often with single-nucleotide resolution. Current deep-sequencing technologies analyze hundreds of millions of individual short reads in one experiment. When applied to libraries of ribosome footprints, this sequencing yields a comprehensive view of the translational landscape that can address many fundamental questions about translation (Fig. 1). Whereas there remain important experimental and analytical challenges to fully exploiting these data, especially in regard to the analysis of ribosome pause sites as discussed below, the presence of ribosome footprints indicates which sequences are being translated in the cell, and thus what protein is being produced. The overall density of ribosome footprints reflects the rate of translation occurring on different transcripts, allowing a direct and quantitative measure of how rapidly a cell is producing each of its proteins. These densities must be corrected for differences in the average elongation rate of each mRNA, which, when explored, have been found to be relatively modest (Li et al. 2014). The detailed pattern of ribosome footprints within a coding sequence varies substantially, however, and reveals the relative speed of the ribosome along the transcript. As described below, these rich data can be augmented further, using drugs that modulate translation or targeted purification of interesting ribosomal subpopulations to address a wide array of biological questions.
Figure 2.
Ribosome footprint profiling. Steps in a typical ribosome profiling experiment are shown. Polysomes reflecting in vivo translation are isolated from cells and subjected to RNase digestion, which degrades unprotected messenger RNA (mRNA). The ribosome-protected footprints are analyzed by deep sequencing, schematized by the flowcell (light blue) with clusters of fluorescently labeled DNA attached to it (colored dots). Aligning these footprint sequences back to the transcriptome produces a quantitative profile of ribosome occupancy.
QUANTIFYING GENE-SPECIFIC TRANSLATION
An important antecedent to the ribosome profiling approach was a study from Joan Steitz (1969), who mapped the sites of translation initiation in bacteriophage RNA by analyzing the RNase-resistant mRNA fragments protected by initiating ribosomes assembled in vitro. Subsequently, Wolin and Walter (1988, 1989) identified sites of in vitro translational pausing by mapping the relative density of ribosome-protected fragments using a primer extension assay. These studies made fundamental contributions to our understanding of translation through the analysis of ribosome footprints, but they were limited to the study of individual mRNAs translated in vitro.
Historically, studies of in vivo translation relied on analyzing polysomes recovered from cells and tissues (Mathews et al. 2000). These polysomes comprise multiple ribosomes translating a single mRNA template, and they can be fractionated according to the number of ribosomes they contain by ultracentrifugation through a sucrose density gradient. The distribution of an mRNA across these fractions reflects its translational status. Gene-specific and global polysome analyses (Arava et al. 2003) have provided a wealth of information about translation, but their quantitative resolution is limited by the poor separation of heavier polysomes and the difficulty of distinguishing ribosomes on different open reading frames (ORFs) in polycistronic mRNAs and transcripts with regulatory upstream translation. This challenge is exacerbated when comparing translation between different genes, as the number of ribosomes on a transcript scales with the length of the coding sequence as well as the translation level.
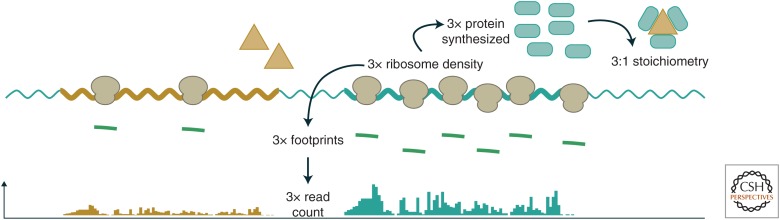
Ribosome profiling circumvents these limitations and precisely measures translation levels by counting discrete ribosome footprints (Ingolia et al. 2009). This quantitative precision revealed a principle of “proportional synthesis” that holds in bacterial and eukaryotic cells: protein subunits of multimeric complexes are synthesized in proportion to their stoichiometry in the assemblies (Li et al. 2014) thus reducing waste of producing unneeded subunits and eliminating the need to dispose of such uncomplexed species (Fig. 3). In bacteria, proteins with differing stoichiometry are often translated from a single polycistronic transcript, and so these differences likely reflect differential translation of the individual reading frames within that RNA. Translation-driven proportional synthesis can even be seen in chloroplasts, in which plastid ribosome profiling revealed that this organelle produces photosystem components in a tight stoichiometric ratio not seen in mRNA abundance (Chotewutmontri and Barkan 2016). More broadly, the fine-tuning seen in proportional synthesis highlights the accuracy and precision of ribosome profiling measurements.
Figure 3.
Quantifying protein synthesis. The number of ribosomes translating a reading frame determines the number of footprints generated in a profiling experiment, and so counting the footprint sequences derived from a reading frame indicates the amount of the encoded protein that is being synthesized. An exemplary polycistronic bacterial transcript is shown, with two open reading frames ([ORFs] A and B) encoding a pair of proteins that assemble with a 1:3 stoichiometric ratio. To achieve this stoichiometry, ORF B is translated threefold more heavily than ORF A, leading to threefold higher ribosome density and threefold more ribosome footprints.
This fine-tuning of stoichiometry also emphasizes how protein abundance is typically the functionally relevant output of gene expression in the cell, and selective constraints arise on protein levels rather than mRNA levels. Protein abundance correlates better with ribosome profiling measurements than with mRNA levels (Liu et al. 2017b; Cheng et al. 2018), and so experiments capture additional, biologically relevant information about gene expression by incorporating profiling data. Combining ribosome profiling with transcriptomic and proteomic data promises the opportunity to learn more about the genetic determinants of protein levels that impact translation as well as transcription. Ribosome profiling has enabled the study of interallelic (Muzzey et al. 2014), interstrain (Albert et al. 2014), and interspecies (Artieri and Fraser 2014b; McManus et al. 2014) translational differences in yeast. It has also been applied to study gene expression variation between individual humans (Battle et al. 2015; Cenik et al. 2015). In some cases, transcriptional divergence is buffered by translational changes to conserve protein levels, whereas in other cases transcriptional and translational changes reinforce each other. Future ribosome profiling studies promise further insight into the roles of translational changes as well as the nature of the polymorphisms that drive these changes.
During dynamic remodeling of cell physiology, global expression profiling has shown how genes are induced just in time to fulfill their functional roles (Brown and Botstein 1999). Ribosome profiling in meiotic yeast has revealed how this principle of just-in-time regulation extends to the translational as well as transcriptional control of the proteins synthesized for each stage of this highly ordered process (Brar et al. 2012). Ribosome profiling in mitosis, too, points to translational control of protein production in concert with cell cycle progression (Stumpf et al. 2013; Tanenbaum et al. 2015).
Subsequently, concerted programs of translational regulation have emerged in many other models, ranging from basal eukaryotic parasites Trypanosoma (Jensen et al. 2014; Vasquez et al. 2014) and Plasmodium (Caro et al. 2014) to circadian cycles in mammalian tissue (Janich et al. 2015). This principle even extends to the distinct mitochondrial translational apparatus, in which profiling of mitoribosomes points to coordinated synthesis of oxidative phosphorylation components translated in the mitochondrion with those produced in the cytosol (Couvillion et al. 2016).
One common theme arising in many studies is the coordinated regulation of ribosomal proteins and ribosome biogenesis factors linked to the rate of cell growth. In metazoa, ribosome production and protein synthesis levels are regulated in part by the protein kinase, mammalian target of rapamycin (mTOR), which serves as a master regulator of growth (Hindupur et al. 2015), in part through controlling the translation of mRNAs encoding ribosomal proteins, but affecting many other transcripts as well (Proud 2018). This mTOR-driven translation supports proliferating cells during normal development and malignant growth in cancer (Dowling et al. 2010; Alain et al. 2012; Robichaud et al. 2018). As mTOR activity drives cell growth downstream from well-known oncogenic signaling pathways, there is great interest in developing active-site mTOR inhibitors as clinically useful anticancer therapies (Bhat et al. 2015). Ribosome profiling studies of cells treated with these inhibitors has revealed a broad range of target transcripts beyond ribosomal proteins that seem to support the cancer cell phenotype (Hsieh et al. 2012; Thoreen et al. 2012). Intriguingly, many of the same genes are translationally repressed in mouse embryonic stem cells induced to differentiate into embryoid bodies rather than continue rapid proliferation (Ingolia et al. 2011).
MOLECULAR MECHANISMS OF TRANSLATIONAL CONTROL
Ribosome profiling has revealed the mechanism underlying the action of other anticancer drugs that target the translational apparatus (Chu and Pelletier 2018). Rocaglate drugs are a class of natural products that target eukaryotic initiation factor 4A (eIF4A), a prototypical DEAD-box RNA helicase, selectively killing cancer cells (Santagata et al. 2013). Ribosome profiling revealed that rocaglates inhibit translation of specific mRNAs, suggesting that this targeted inhibition could explain their selectivity for cancer cells (Wolfe et al. 2014). By measuring the relative sensitivity of different transcripts to rocaglate treatment—which differed greatly from their sensitivity to hippuristanol, a more conventional eIF4A inhibitor—ribosome profiling further elucidated the unique repressive mechanism of these drugs. Rather than mimicking the loss of eIF4A function, rocaglate drugs clamp eIF4A onto certain polypurine RNA sequences, where it serves as a roadblock to translation initiation (Iwasaki et al. 2016). Transcripts with polypurine-rich transcript leaders are thus particularly sensitive to rocaglate treatment.
Similar correspondences between translational changes measured by ribosome profiling and transcript features have provided new insights into the normal function of eIF4A as well as other translation initiation factors, often challenging our current understanding of their roles. Ribosome profiling measurements conducted after inactivation of eIF4A revealed profound but fairly uniform reduction in translation (Sen et al. 2015). Conditional inactivation of yeast Ded1, another DEAD-box translation initiation factor, argued that it is particularly important for translating mRNAs with longer and more structured 5′ untranslated regions (UTRs) (Sen et al. 2015). The scaffolding protein eIF4G recruits eIF4A and stimulates its ATPase activity (Merrick and Pavitt 2018; Sokabe and Fraser 2018). Although eIF4G is typically thought to be recruited to mRNAs through its interactions with the cap-binding protein eIF4E and poly(A)-binding protein, recent studies suggest that in yeast it preferentially binds and promotes the translation of mRNAs with oligo(U) tracts in their 5′UTRs (Zinshteyn et al. 2017). Transcripts that depend on eIF4G also show reduced translation in the absence of the ribosome-associated factor Asc1/RACK1 (Thompson et al. 2016). In plants, poly(A)-binding proteins interact with A-rich motifs directly in the 5′UTR, in which they promote translation in pattern-triggered immune response (Xu et al. 2017).
Global ribosome profiling, combined with mRNA and protein abundance measurements, has also provided critical insights into the mechanism of microRNA-mediated repression (Duchaine and Fabian 2018), making it possible to disentangle the effects of mRNA destabilization from reduced translation. Early ribosome profiling studies measured concordant effects at both stages of expression, with translational repression contributing ∼1/6th of the total reduction in protein abundance at steady state (Guo et al. 2010). Later work in developing zebrafish embryos showed that strong translational repression precedes mRNA decay during the activation of miR-430 in the maternal-to-zygotic transition (Bazzini et al. 2012). It remains unclear whether this reflects a general kinetic pathway for microRNA-mediated repression, or a shift in the mode of action between earlier or later stages of embryogenesis (Subtelny et al. 2014). Similarly, ribosome profiling has distinguished the translational effects of RNA modifications such as adenosine N6 methylation, which seem to influence both translation and mRNA stability (Wang et al. 2015; Zhou et al. 2015; Coots et al. 2017; Slobodin et al. 2017; Vu et al. 2017; Peer et al. 2018).
Quantitative and comprehensive ribosome profiling measurements broadly offer the insights available from transcriptome profiling, augmented with information about translational regulation. Using these comprehensive measurements as input, modeling approaches have been used to systematically quantify the roles of various transcript features in determining the translational output of an mRNA (Weinberg et al. 2016; Hockenberry et al. 2017). This approach can also be applied to learn about cellular physiology by profiling natural biological processes and to learn about regulatory mechanisms by observing the effects of targeted molecular disruptions, as shown by the examples above. Ribosome profiling contains information about the exact positions of ribosomes as well; however, that goes beyond expression profiling.
DISCOVERY OF NONCANONICAL TRANSLATION EVENTS
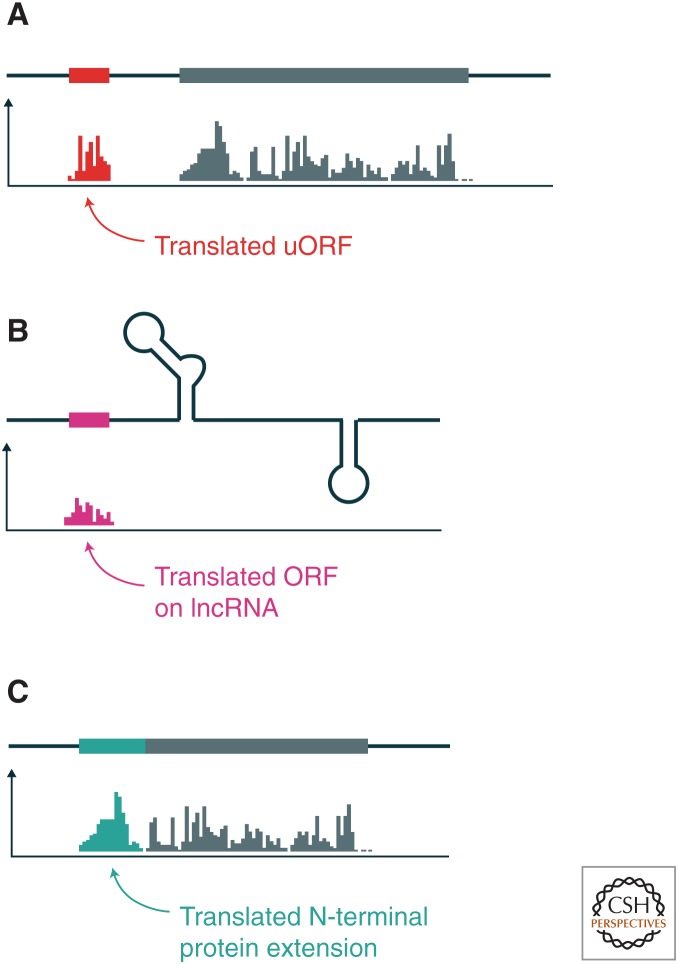
The prevalence of ribosome footprints in 5′UTRs was one of the most striking features we observed in the first ribosome profiling data from yeast and mammalian cells (Fig. 4A) (Ingolia et al. 2009, 2011). Upstream translation itself can serve to repress expression of downstream protein-coding genes when scanning ribosomes initiate at upstream ORFs (uORFs) and translate them instead of proceeding on to the main reading frame (Sonenberg and Hinnebusch 2009). Ribosome profiling data are critical for understanding this mode of regulation, as not all uORFs are translated, and their repressive effects can be modulated by genetic variation between individuals, including polymorphisms that create or destroy uORFs (Calvo et al. 2009; Cenik et al. 2015). Indeed, in some cases, uORFs can promote the translation of the downstream coding sequence (Sonenberg and Hinnebusch 2009). Alternative transcript isoforms can also include or exclude upstream translated regions, thereby modulating translation. In the most extreme cases, totally unproductive transcript isoforms may result from the inclusion of long upstream regions replete with uORFs (Brar et al. 2012; Chen et al. 2017; Cheng et al. 2018).
Figure 4.
Annotating the proteome with ribosome profiling. The figure diagrams mRNAs (top) showing the frequency of ribosome footprints along them (below). (A) Ribosome footprint sequences mapping to the 5′ leader of a transcript indicates the translation of an upstream open reading frame (uORF, red segment) and downstream ORF (gray segment). (B) Likewise, ribosome footprint sequences on a noncoding RNA indicate the presence of a translated region, typically near the 5′ end of the transcript. (C) Alternative protein isoforms translated in addition to or in place of annotated reading frames also appear in ribosome profiling data.
Regulatory uORFs control the paradoxical induction of specific mRNAs such as ATF4 and CHOP during stress-induced translational shutoff, mediated by the phosphorylation of the α subunit of eukaryotic initiation factor 2 (eIF2α) (Sonenberg and Hinnebusch 2009; Wek 2018). Ribosome profiling has now provided a comprehensive list of dozens of genes that show similar up-regulation (Andreev et al. 2015; Sidrauski et al. 2015). Surprisingly, although ribosome profiling confirms the translation of ATF4 and CHOP uORFs, many other targets lack ribosome occupancy in their 5′UTRs, raising the question of how their translation is controlled. Furthermore, translated uORFs appear to be widespread across the transcriptome (Johnstone et al. 2016), not restricted to the small number of phospho-eIF2α-induced genes (Andreev et al. 2015; Sidrauski et al. 2015).
Much of the upstream translation seen in ribosome profiling cannot be attributed to AUG codons, and instead appears to initiate at a near-cognate, non-AUG codon. As the most common non-AUG initiation sites occur at codons that are quite similar to AUG (e.g., CUG), some of this translation results from mispairing of the initiator transfer RNA (tRNA) with the noncanonical start codon. There is also evidence for uORF peptide products that begin with amino acids other than methionine. These alternative start codon products are induced when eIF2 is inhibited by phosphorylation, and depend on the poorly understood initiation factor eIF2A, which is unrelated to the canonical eIF2 complex (Starck et al. 2016; Merrick and Pavitt 2018). Ribosome profiling recently uncovered a shift toward EIF2A-dependent, non-AUG initiation in a SOX2-driven mouse model of squamous cell carcinoma (Sendoel et al. 2017). Many oncogenic transcripts were induced in this translational program, and tumor progression depended on EIF2A, pointing toward a causal link between unconventional 5′UTR translation and cancer. The recent development of translation complex profile sequencing (TCP-Seq) promises a more direct view of the mechanism of translation initiation. TCP-Seq augments the standard ribosome profiling approach with formaldehyde cross-linking that stabilizes preinitiation complexes on mRNAs to profile 40S subunits scanning through 5′UTRs, providing insights into the molecular choreography of the scanning process (Archer et al. 2016).
Ribosome footprints were seen on many presumptively noncoding RNAs in addition to the 5′UTRs of coding genes (Fig. 4B) (Ingolia et al. 2011, 2014; Chew et al. 2013; Ji et al. 2015). The patterns of footprints on long noncoding RNAs (lncRNAs) (Chekulaeva and Rajewsky 2018) matched our expectations for those of translating ribosomes; they fell in AUG-initiated reading frames near the 5′ ends of transcripts and in aggregate showed three-nucleotide periodicity (Ingolia et al. 2011; Calviello et al. 2016). The lack of canonical features of protein-coding sequences in these translated regions motivated a variety of experiments aimed at validating the profiling results. lncRNA footprints copurified with affinity-tagged ribosomes and responded to drugs that targeted the ribosome, and thus represented ribosome-protected footprints rather than nonribosomal background (Ingolia et al. 2014). We have also reported evidence for protein products derived from lncRNA translation (see Chekulaeva and Rajewsky 2018). Viral infection leads to an immunological memory of epitopes derived from lncRNA translation (Stern-Ginossar et al. 2012), similar to the epitopes produced by noncanonical upstream translation (Starck et al. 2012). Certain peptides have also been detected directly (Calviello et al. 2016), although in general these short products are challenging targets for proteomics, and ribosome profiling predictions can greatly aid in finding them (Menschaert et al. 2013).
ANNOTATING THE EXPANDED PROTEOME
The functional impact of pervasive alternative translation remains an important question. In a few cases, ribosome profiling has directly identified short, functional proteins such as Toddler (Pauli et al. 2014). Ribosome profiling points to a remarkable breadth of short, translated ORFs supported by conservation analysis (Menschaert et al. 2013; Aspden et al. 2014; Bazzini et al. 2014; Fields et al. 2015; Calviello et al. 2016) that could add to our growing catalog of functional micropeptides (Anderson et al. 2015; D’Lima et al. 2017). In many cases, however, this translation may be adventitious and subject principally to negative selection to avoid harmful effects from protein products or RNA destabilization (Ulitsky and Bartel 2013). Even nonfunctional proteins can serve as antigens, however (Ingolia 2014), and understanding this cryptic source of antigens (Starck et al. 2012) has implications for cancer immunotherapy (Schumacher and Schreiber 2015) and immunobiology more generally.
Ribosome profiling has also revealed alternative translation that extends or truncates classical protein-coding genes (Fig. 4C). This variation can add or remove entire domains, changing or reversing the function of the protein product. For instance, a truncated form of the innate immune signaling protein, mitochondrial antiviral signaling protein (MAVS), called miniMAVS, seems to antagonize the function of full-length MAVS in antiviral gene expression (Brubaker et al. 2014). Our data suggest that such alternative isoforms are widespread (Ingolia et al. 2011; Fields et al. 2015).
The diversity of translation products has spurred adaptations of ribosome profiling optimized for identifying translated regions of the transcriptome. We reported on the use of harringtonine, a drug that immobilizes initiating ribosomes, producing footprints that mark sites of translation initiation (Ingolia et al. 2011). Others showed, independently, that lactimidomycin or pateamine A could likewise trap initiating ribosome footprints (Lee et al. 2012; Gao et al. 2015; Popa et al. 2016), while high doses of the drug puromycin could drive rapid premature termination to produce a similar initiation-specific footprint profile (Fritsch et al. 2012). Joint analysis of lactimidomycin- and harringtonine-treated profiling data promises more robust identification of translational start sites appearing in both data sets by excluding possible artifacts resulting from either of these mechanistically distinct drugs (Stern-Ginossar et al. 2012; Arias et al. 2014). This combined analysis was used to define translated reading frames in human cytomegalovirus, a large herpesvirus with a complex life cycle that expresses a variety of alternative translation products, some of which seem to display specific molecular function (Stern-Ginossar et al. 2012). Novel translated ORFs upstream of, or within, known ORFs have also been detected in other viruses (Stern-Ginossar et al. 2018). More recently, a systematic, regression-based combination of ribosome profiling data generated with these different drugs revealed hundreds of novel coding sequences in mammalian cells along with a wealth of shorter, translated reading frames (Fields et al. 2015).
To draw accurate inferences about in vivo translation from deep-sequencing data, it is essential to know that the RNA fragments being sequenced are ribosome-protected footprints. We have provided several lines of evidence showing that this is true in general (Ingolia et al. 2014), and we and others have shown how straightforward computational approaches can distinguish signatures of translation from footprints left by nonribosomal RNA–protein interactions (Ingolia et al. 2014; Ji et al. 2016). The bulk of these nonribosomal reads derive from abundant structural RNAs, including tRNAs, spliceosomal small nuclear RNAs (snRNAs), and small nucleolar RNAs (snoRNAs) (Ji et al. 2016). Fragments of these RNAs, along with other nonribosomal background, can be identified and excluded from ribosome footprint analysis because they differ in length from ribosome footprints and lack triplet periodicity (Ingolia et al. 2014; Ji et al. 2016).
To equate ribosome footprint density with protein production, it is also important to know that these ribosomes are translating productively. We have followed run-off elongation after blocking new initiation with drugs and shown that coding sequences are quickly depleted of ribosomes (Ingolia et al. 2011), except under conditions in which elongation is arrested (Barry et al. 2017). These basic features seem to hold in most systems, although it does not obviate the need to evaluate them in unusual biological contexts.
TRACKING THE FOOTPRINTS OF TRANSLATION ELONGATION
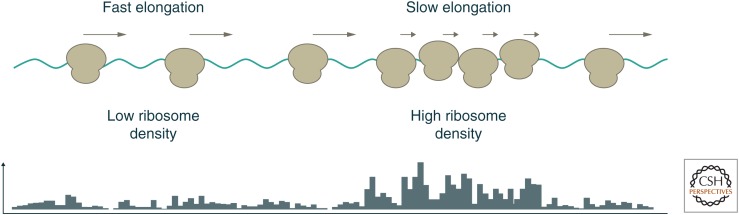
Ribosome profiling has broad applications in annotating genes as well as measuring expression, but its most distinctive contributions may stem from insights into the activities of ribosomes in vivo. We know that the speed of translation elongation can vary across a coding sequence, presumably as a result of variations in the mRNA template and the protein product. Ribosomes will spend more time at positions of slow elongation, and so we will observe a higher density of footprints at these sites (Fig. 5).
Figure 5.
Inferring elongation speed from variations in ribosome footprint density. The lower part of the figure reports the frequency of ribosomal footprints along the mRNA. Regions of slow elongation will accumulate higher ribosome occupancy than regions of faster elongation on the same transcript. These differences in ribosome density are visible in profiling data, and they can be used to infer how codon usage, peptide sequence, and other features control the speed of translation.
Indeed, footprint counts vary substantially across genes and accumulate at specific “pause” sites. There is great interest in understanding the features that correlate with the speed of translation elongation and deconvolving this from biases in capturing and sequencing footprints (Stadler and Fire 2011; Dana and Tuller 2012; Qian et al. 2012; Charneski and Hurst 2013; Lareau et al. 2014; Pop et al. 2014; Liu and Song 2016; O’Connor et al. 2016; Weinberg et al. 2016; Dao Duc et al. 2017). Likewise, there is interest in understanding the factors that drive dramatic ribosome pausing at specific locations, which may reflect a qualitatively different process than the variation in translation speed seen across typical codons (Han et al. 2014; Li et al. 2014; Martens et al. 2015; Mohammad et al. 2016; Zhang et al. 2017). Various measures of codon usage bias correlate with footprint occupancy, suggesting that favored codons are decoded more quickly. However, consensus has not emerged on the exact basis of this effect, which seems to extend beyond the time required for decoding and tRNA recruitment. Elongation rates learned from ribosome profiling nonetheless provide an empirical basis for tuning the translation of a coding sequence, thereby controlling its expression (Tunney et al. 2017).
Translation of even a single codon is a complicated, multistep process (Dever et al. 2018; Rodnina 2018), and ribosome profiling has opened a new window into the operation of the translational machinery by reporting on normal elongation and on the effects of mutations. tRNAs in particular are heavily modified, and disrupting these modifications can change the speed of translation (Zinshteyn and Gilbert 2013) and thereby disrupt protein folding (Nedialkova and Leidel 2015). A broad survey of tRNA modifications revealed diverse effects on specific codons and on gene expression (Chou et al. 2017). In a similar fashion, base modifications on mRNA can affect decoding (Choi et al. 2016; Li et al. 2017), representing another factor that can contribute. In yeast, ribosome profiling provided in vivo confirmation that certain pairs of synonymous codons induce major ribosome pausing only when adjacent and in a particular order, suggesting structural cross talk between tRNAs in the A and P sites (Gamble et al. 2016). Ribosome profiling can distinguish between different phases of the translation elongation cycle, as different ribosome conformations protect footprints of differing length (Lareau et al. 2014). The longer (∼28 nt) footprints, captured in most ribosome profiling experiments, probably reflect unrotated ribosomes. Cycloheximide treatment traps ribosomes in this long-footprint conformation, and yeast ribosome profiling performed without cycloheximide revealed a population of shorter (∼21 nt) footprints that are attributed to rotated ribosomes. Although the abundance of long ribosome footprints correlates with codon usage and tRNA availability, short footprint density correlates with physicochemical amino acid properties instead, likely reflecting effects on translocation rather than decoding. Short footprints accumulate at certain tRNA-dependent stalls (Matsuo et al. 2017), suggesting that this cross talk affects translocation rather than tRNA recruitment (Lareau et al. 2014). Notably, these short footprints differ from the ∼16 nt footprints reflecting a ribosome stalled at the end of a broken mRNA (Guydosh and Green 2014). More generally, these examples show the importance of identifying and quantifying all ribosome footprints regardless of their length (Mohammad et al. 2016).
Translation elongation also slows in response to amino acid limitation, leading to ribosome footprint accumulation on codons encoding the affected amino acids. Footprint density peaks induced by histidine deprivation were used to generate fiduciary marks in ribosome profiling data (Guydosh and Green 2014; Lareau et al. 2014), and inadvertent serine restriction likewise caused a buildup of footprints on serine codons in bacteria (Li et al. 2014). Systematic amino acid starvation coupled with ribosome profiling provided a spectrum of perturbed ribosome occupancy profiles that inform biophysical models of bacterial translation (Subramaniam et al. 2014). Remarkably, ribosome profiling likewise uncovered proline limitations in certain human tumors, based on slowed elongation when decoding proline codons (Loayza-Puch et al. 2016), and may serve more generally to probe for metabolic disruptions in cancer and other diseases.
Ribosome profiling has also facilitated the study of peptide-mediated translational pausing that occurs naturally in cells (Nakatogawa and Ito 2002). We observed ribosome footprint accumulation at certain tandem proline codons in mammalian cells (Ingolia et al. 2011), consistent with the slowed rate of elongation at these sites in vitro and the unfavorable conformation of polyprolyl nascent chains in the ribosome (Huter et al. 2017). The universally conserved elongation factor EF-P/eIF5A is implicated in translation of polyproline peptides (Doerfel et al. 2013; Gutierrez et al. 2013; Ude et al. 2013; Dever et al. 2018; Rodnina 2018) but ribosome profiling after eIF5A depletion reveals broader perturbation of ribosome footprint profiles, supporting a wider role for eIF5A in elongation through many unfavorable peptide sequences and in efficient translation termination (Schuller et al. 2017). Ribosome footprinting of these eIF5A stalls agreed with stalling sites identified by 5PSeq (Pelechano and Alepuz 2017), which focuses on in vivo RNA degradation intermediates whose 5′ terminus marks the trailing edge of the last translating ribosome (Pelechano et al. 2015). In contrast, ribosome profiling showed that Legionella toxins targeting elongation factor 1A (eEF1A) show no such specificity (Barry et al. 2017). Many peptide sequences can block bacterial translation (Woolstenhulme et al. 2013), and this effect is often exploited for biological regulation. Bacterial ribosome profiling has also defined peptide-specific arrest caused by antibiotics targeting the translational machinery (Kannan et al. 2014; Marks et al. 2016). Stalling also occurs at programmed ribosomal frameshifting, which can stand out dramatically in footprint profiles (Michel et al. 2012; Napthine et al. 2017).
Detailed analyses of ribosome footprint occupancy patterns on individual mRNAs are particularly impacted by technical challenges. Studies in prokaryotes face a unique obstacle: ribonucleases do not degrade unprotected RNA precisely to the edge of the prokaryotic ribosome (Oh et al. 2011), making it challenging to precisely identify functionally relevant positions within each footprint (Woolstenhulme et al. 2013). More universally, all methods for converting ribosome footprints into a deep-sequencing library display biases that over- or underrepresent certain footprints, thereby distorting the apparent ribosome occupancy observed after sequencing (Artieri and Fraser 2014a; Bartholomaus et al. 2016; Lecanda et al. 2016; Tunney et al. 2017). Translation inhibitors used before cell lysis can distort ribosome occupancy profiles more directly (Gerashchenko and Gladyshev 2014; Hussmann et al. 2015). Eukaryotic cells are often treated with cycloheximide before ribosome profiling to immobilize and stabilize ribosomes. In our early studies, we reported that this treatment did not affect overall ribosome occupancy across a coding sequence but did change the pattern of footprints within that sequence (Ingolia et al. 2011). Subsequently, use of cycloheximide varied between studies. Later analysis showed that peaks of ribosome density appear to shift downstream in cycloheximide-treated samples relative to untreated ones (Gerashchenko and Gladyshev 2014; Hussmann et al. 2015).
Interest in a quantitative understanding of elongation has been heightened by recent studies that identified a potential role for elongation rates in dictating mRNA half-lives (Presnyak et al. 2015; Chan et al. 2017), either through direct surveillance of ribosome speed by mRNA decay machinery (Radhakrishnan et al. 2016) or indirectly by inducing ribosome collisions that then trigger decay pathways (Ferrin and Subramaniam 2017; Simms et al. 2017). Ribosome profiling will undoubtedly play a key role in unraveling the molecular mechanisms connecting elongation to decay and in quantifying the role of elongation in determining steady-state mRNA levels.
TRANSLATION TERMINATION AND BEYOND
In most ribosome profiling studies, 3′UTRs are devoid of footprints, in contrast to the surprising abundance of upstream initiation. Stop codon readthrough causes a specific accumulation of in-frame ribosome footprints in 3′UTRs, which are particularly prominent in Drosophila (Dunn et al. 2013). Defects in posttermination ribosome recycling (Hellen 2018) allow unrecycled ribosomes to enter 3′UTRs with no particular reading frame, and then reinitiate translation in some different reading frame, producing 3′UTR footprints out of frame from the coding DNA sequence (CDS) (Young et al. 2015). It appears that the ribosome rescue factors Dom34/Pelota and Hbs1 typically rescue many posttermination, unrecycled ribosomes, as the loss of these factors also causes an accumulation of vacant ribosomes past the stop codon (Guydosh and Green 2014). This distinctive accumulation of 3′UTR footprint patterns arose in reticulocytes and platelets, bringing to light a depletion in normal recycling factors and a disruption of ribosome homeostasis in both of these anucleate blood lineages (Mills et al. 2016). Indeed, translational regulation is pervasive in hematopoiesis (Alvarez-Dominguez et al. 2017), and altered ribosome recycling may underlie the particular sensitivity of red blood cells to defects in the translational machinery (Mills and Green 2017). It seems that the loss of ribosome rescue may serve a positive role in platelets, however. The rescue of ribosomes is linked to quality control processes that degrade aberrant protein products and mRNA templates (Brandman and Hegde 2016) and the loss of ribosome rescue factors seems to stabilize mRNAs that cannot be replaced by transcription (Mills et al. 2017).
PICKING THE RIGHT FOOTPRINTS
Footprinting of purified ribosome subpopulations enables profiling of cotranslational processes that act on proteins but, using a sequencing-based assay. Selective ribosome profiling by purifying ribosomes that are engaged by specific chaperones or targeting factors has revealed the in vivo substrates and engagement patterns in bacteria (Oh et al. 2011) and eukaryotes (Döring et al. 2017). Likewise, profiling of ribosome footprints engaged with the signal recognition particle (SRP) monitors cotranslational secretion and suggests that determinants beyond the classic signal sequence may aid SRP targeting (Chartron et al. 2016). Profiling has even been adapted to profile the folding state of nascent protein chains directly (Han et al. 2014), highlighting the fact that protein folding can be coupled directly to translation (Gloge et al. 2014). Indeed, selective ribosome profiling of ribosomes associated with different members of a multiprotein complex has suggested that complex assembly can begin cotranslationally (Shieh et al. 2015). Such cotranslational assembly could couple with the degradation of monomers that lack partner proteins for complex formation (Ishikawa et al. 2017) to complement proportional synthesis (Li et al. 2014) in maintaining proteome stoichiometry. This selective profiling strategy has been extended to study ribosomal subpopulations with varying composition. After identifying proteins RPL10A and RPL38 as substoichiometric in ribosomes, selective ribosome profiling of only those ribosomes containing these proteins revealed a potential role for heterogeneity between ribosomes in shaping overall translational output (Shi et al. 2017).
Recently, an approach termed proximity-specific ribosome profiling has enabled selective profiling of ribosomes at specific subcellular locations. Subcellular organization is inevitably disrupted by lysis and homogenization, but proximity labeling with a localized biotin ligase can mark ribosomes according to their in vivo localization for subsequent purification and footprinting. This approach was used first to identify the ribosomes that localize near the endoplasmic reticulum and the mitochondria in yeast, providing further insight into protein targeting (Jan et al. 2014; Williams et al. 2014). We have recently combined this method with rapid and specific depletion of SRP to comprehensively characterize the role of SRP in cotranslational localization. This approach uncovered an unexpected class of mRNAs encoding proteins that are normally secreted but become mistargeted to mitochondria in the absence of SRP (Costa et al. 2018). Recent results suggest that subcellular organization of protein synthesis may be widespread, especially in tissues in which cells polarize and form three-dimensional structures (Moor et al. 2017). Localized translational control is particularly prominent in neurons, in which it is implicated in fundamental neural processes such as long-term potentiation and depression (Glock et al. 2017; Biswas et al. 2018; Sossin and Costa-Mattioli 2018).
Ribosome affinity purification has also emerged as a tool for cell type–specific translational profiling in animals, by using translating ribosome affinity purification (TRAP) (Doyle et al. 2008; Heiman et al. 2008) and RiboTag (Sanz et al. 2009). These approaches seem quite complementary to ribosome profiling, and indeed, tissue-specific ribosome profiling was recently shown in Drosophila (Chen and Dickman 2017). TRAP has seen its broadest application in the nervous system, which is characterized by extreme cell type diversity as well as a prominent role for translational control in synaptic plasticity (Sossin and Costa-Mattioli 2018). Further integration of cell type–specific ribosome profiling seems particularly promising in understanding the molecular basis of neuronal functions.
PERSPECTIVE
The translation of mRNA into protein and the folding of the resulting protein into an active form are prerequisites for virtually every cellular process and represent the single largest investment of energy by cells. Ribosome profiling-based approaches have revolutionized our ability to monitor protein synthesis in vivo, making it possible to determine the start, stop, reading frame, chaperone engagement, subcellular targeting, and rate of translation for virtually every mRNA and protein encoded in a cell. The rich and quantitative nature of ribosome profiling data provides an unprecedented opportunity to explore and model complex cellular processes. Finally, by virtue of the precise genomic positional information obtained by ribosome profiling, the protein coding capacity of genomes can now be explored experimentally.
Nonetheless, important technical and conceptual questions remain. For example, the function of the many novel, short, and alternate translated regions identified thus far by ribosome profiling remains an intriguing and largely open question and one whose answer could fundamentally change the way that we believe about information encoding in genomes. Newly available CRISPR-based methods now make it possible to shut down the expression of any transcript (Gilbert et al. 2013, 2014; Liu et al. 2017a) or introduce nonsense mutations into any ORF (Hess et al. 2017). These approaches provide a central tool for efforts to define the functional roles for this broad array of newly identified translation products.
We have already seen demonstrations of specialized alterations to ribosome profiling that will advance its utility in complex systems. These developments include the analysis of molecularly defined subsets of ribosomes, either associated with specific factors or protein modifications, or even specialized ribosomes missing a core ribosomal protein entirely. Similar approaches allow the analysis of localized ribosomes within increasingly specific cell types or subcellular locations. We also know little about how ribosomes are distributed across individual transcripts of the same gene: Is the spacing between ribosomes purely stochastic, or are initiation and elongation “metered” to shape the traffic of ribosomes and minimize collisions? Along this line, understanding the biological roles for the use of synonymous codons remains one of the oldest outstanding questions in the field. Ribosome profiling provides an unprecedented view of their impact by yielding position-specific densities of ribosomes along a message. However, better protocols are needed to ensure that in vivo ribosome positions are captured faithfully and turned into sequencing libraries free of biases or distortions. Finally, transformative advances are likely to emerge from progressively more sophisticated and creative analysis of the rich data sets generated from ribosome profiling experiments, enabling major surprises to be revealed, even in systems that were thought to be well characterized.
Footnotes
Editors: Michael B. Mathews, Nahum Sonenberg, and John W.B. Hershey
Additional Perspectives on Translation Mechanisms and Control available at www.cshperspectives.org
REFERENCES
*Reference is also in this collection.
- Alain T, Morita M, Fonseca BD, Yanagiya A, Siddiqui N, Bhat M, Zammit D, Marcus V, Metrakos P, Voyer LA, et al. 2012. eIF4E/4E-BP ratio predicts the efficacy of mTOR targeted therapies. Cancer Res 72: 6468–6476. [DOI] [PubMed] [Google Scholar]
- Albert FW, Muzzey D, Weissman JS, Kruglyak L. 2014. Genetic influences on translation in yeast. PLoS Genet 10: e1004692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Dominguez JR, Zhang X, Hu W. 2017. Widespread and dynamic translational control of red blood cell development. Blood 129: 619–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson DM, Anderson KM, Chang CL, Makarewich CA, Nelson BR, McAnally JR, Kasaragod P, Shelton JM, Liou J, Bassel-Duby R, et al. 2015. A micropeptide encoded by a putative long noncoding RNA regulates muscle performance. Cell 160: 595–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreev DE, O’Connor PB, Fahey C, Kenny EM, Terenin IM, Dmitriev SE, Cormican P, Morris DW, Shatsky IN, Baranov PV. 2015. Translation of 5′ leaders is pervasive in genes resistant to eIF2 repression. eLife 4: e03971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arava Y, Wang Y, Storey JD, Liu CL, Brown PO, Herschlag D. 2003. Genome-wide analysis of mRNA translation profiles in Saccharomyces cerevisiae. Proc Natl Acad Sci 100: 3889–3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archer SK, Shirokikh NE, Beilharz TH, Preiss T. 2016. Dynamics of ribosome scanning and recycling revealed by translation complex profiling. Nature 535: 570–574. [DOI] [PubMed] [Google Scholar]
- Arias C, Weisburd B, Stern-Ginossar N, Mercier A, Madrid AS, Bellare P, Holdorf M, Weissman JS, Ganem D. 2014. KSHV 2.0: A comprehensive annotation of the Kaposi’s sarcoma-associated herpesvirus genome using next-generation sequencing reveals novel genomic and functional features. PLoS Pathog 10: e1003847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artieri CG, Fraser HB. 2014a. Accounting for biases in riboprofiling data indicates a major role for proline in stalling translation. Genome Res 24: 2011–2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artieri CG, Fraser HB. 2014b. Evolution at two levels of gene expression in yeast. Genome Res 24: 411–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aspden JL, Eyre-Walker YC, Phillips RJ, Amin U, Mumtaz MA, Brocard M, Couso JP. 2014. Extensive translation of small open reading frames revealed by Poly-Ribo-Seq. eLife 3: e03528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry KC, Ingolia NT, Vance RE. 2017. Global analysis of gene expression reveals mRNA superinduction is required for the inducible immune response to a bacterial pathogen. eLife 6: e22707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartholomaus A, Del Campo C, Ignatova Z. 2016. Mapping the non-standardized biases of ribosome profiling. Biol Chem 397: 23–35. [DOI] [PubMed] [Google Scholar]
- Battle A, Khan Z, Wang SH, Mitrano A, Ford MJ, Pritchard JK, Gilad Y. 2015. Genomic variation. Impact of regulatory variation from RNA to protein. Science 347: 664–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazzini AA, Lee MT, Giraldez AJ. 2012. Ribosome profiling shows that miR-430 reduces translation before causing mRNA decay in zebrafish. Science 336: 233–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazzini AA, Johnstone TG, Christiano R, Mackowiak SD, Obermayer B, Fleming ES, Vejnar CE, Lee MT, Rajewsky N, Walther TC, et al. 2014. Identification of small ORFs in vertebrates using ribosome footprinting and evolutionary conservation. EMBO J 33: 981–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazzini AA, Del Viso F, Moreno-Mateos MA, Johnstone TG, Vejnar CE, Qin Y, Yao J, Khokha MK, Giraldez AJ. 2016. Codon identity regulates mRNA stability and translation efficiency during the maternal-to-zygotic transition. EMBO J 35: 2087–2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat M, Robichaud N, Hulea L, Sonenberg N, Pelletier J, Topisirovic I. 2015. Targeting the translation machinery in cancer. Nat Rev Drug Discov 14: 261–278. [DOI] [PubMed] [Google Scholar]
- *.Biswas J, Liu Y, Singer RH, Wu B. 2018. Fluorescence imaging methods to investigate translation in single cells. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a032722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandman O, Hegde RS. 2016. Ribosome-associated protein quality control. Nat Struct Mol Biol 23: 7–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brar GA, Weissman JS. 2015. Ribosome profiling reveals the what, when, where and how of protein synthesis. Nat Rev Mol Cell Biol 16: 651–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brar GA, Yassour M, Friedman N, Regev A, Ingolia NT, Weissman JS. 2012. High-resolution view of the yeast meiotic program revealed by ribosome profiling. Science 335: 552–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown PO, Botstein D. 1999. Exploring the new world of the genome with DNA microarrays. Nat Genet 21: 33–37. [DOI] [PubMed] [Google Scholar]
- Brubaker SW, Gauthier AE, Mills EW, Ingolia NT, Kagan JC. 2014. A bicistronic MAVS transcript highlights a class of truncated variants in antiviral immunity. Cell 156: 800–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calviello L, Mukherjee N, Wyler E, Zauber H, Hirsekorn A, Selbach M, Landthaler M, Obermayer B, Ohler U. 2016. Detecting actively translated open reading frames in ribosome profiling data. Nat Methods 13: 165–170. [DOI] [PubMed] [Google Scholar]
- Calvo SE, Pagliarini DJ, Mootha VK. 2009. Upstream open reading frames cause widespread reduction of protein expression and are polymorphic among humans. Proc Natl Acad Sci 106: 7507–7512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caro F, Ahyong V, Betegon M, DeRisi JL. 2014. Genome-wide regulatory dynamics of translation in the Plasmodium falciparum asexual blood stages. eLife 3: e04106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cenik C, Cenik ES, Byeon GW, Grubert F, Candille SI, Spacek D, Alsallakh B, Tilgner H, Araya CL, Tang H, et al. 2015. Integrative analysis of RNA, translation, and protein levels reveals distinct regulatory variation across humans. Genome Res 25: 1610–1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan LY, Mugler CF, Heinrich S, Vallotton P, Weis K. 2017. Non-invasive measurement of mRNA decay reveals translation initiation as the major determinant of mRNA stability. bioRxiv 10.1101/214775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charneski CA, Hurst LD. 2013. Positively charged residues are the major determinants of ribosomal velocity. PLoS Biol 11: e1001508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chartron JW, Hunt KC, Frydman J. 2016. Cotranslational signal-independent SRP preloading during membrane targeting. Nature 536: 224–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Chekulaeva M, Rajewsky N. 2018. Roles of long noncoding RNAs and circular RNAs in translation. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a032680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Dickman D. 2017. Development of a tissue-specific ribosome profiling approach in Drosophila enables genome-wide evaluation of translational adaptations. PLoS Genet 13: e1007117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Tresenrider A, Chia M, McSwiggen DT, Spedale G, Jorgensen V, Liao H, van Werven FJ, Unal E. 2017. Kinetochore inactivation by expression of a repressive mRNA. eLife 6: e27417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z, Otto GM, Powers E, Keskin A, Mertins P, Carr S, Jovanovic M, Brar GA. 2018. Pervasive, coordinated protein level changes driven by transcript isoform switching during meiosis. Cell 172: 910–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chew GL, Pauli A, Rinn JL, Regev A, Schier AF, Valen E. 2013. Ribosome profiling reveals resemblance between long non-coding RNAs and 5′ leaders of coding RNAs. Development 140: 2828–2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J, Ieong KW, Demirci H, Chen J, Petrov A, Prabhakar A, O’Leary SE, Dominissini D, Rechavi G, Soltis SM, et al. 2016. N6-methyladenosine in mRNA disrupts tRNA selection and translation-elongation dynamics. Nat Struct Mol Biol 23: 110–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chotewutmontri P, Barkan A. 2016. Dynamics of chloroplast translation during chloroplast differentiation in maize. PLoS Genet 12: e1006106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou HJ, Donnard E, Gustafsson HT, Garber M, Rando OJ. 2017. Transcriptome-wide analysis of roles for tRNA modifications in translational regulation. Mol Cell 68: 978–992 e974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Chu J, Pelletier J. 2018. Translating therapeutics. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a032995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coots RA, Liu XM, Mao Y, Dong L, Zhou J, Wan J, Zhang X, Qian SB. 2017. m6A facilitates eIF4F-independent mRNA translation. Mol Cell 10.1016/j.molcel.2017.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa EA, Subramanian K, Nunnari J, Weissman JS. 2018. Defining the physiological role of SRP in protein targeting efficiency and specificity. Science 359: 689–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couvillion MT, Soto IC, Shipkovenska G, Churchman LS. 2016. Synchronized mitochondrial and cytosolic translation programs. Nature 533: 499–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dana A, Tuller T. 2012. Determinants of translation elongation speed and ribosomal profiling biases in mouse embryonic stem cells. PLoS Comput Biol 8: e1002755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dao Duc K, Saleem ZH, Song YS. 2018. Theoretical analysis of the distribution of isolated particles in the TASEP: Application to mRNA translation rate estimation. Phys Rev E 97: 012106. [DOI] [PubMed] [Google Scholar]
- *.Dever TE, Dinman JD, Green R. 2018. Translation elongation and recoding in eukaryotes. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a032649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Lima NG, Ma J, Winkler L, Chu Q, Loh KH, Corpuz EO, Budnik BA, Lykke-Andersen J, Saghatelian A, Slavoff SA. 2017. A human microprotein that interacts with the mRNA decapping complex. Nat Chem Biol 13: 174–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerfel LK, Wohlgemuth I, Kothe C, Peske F, Urlaub H, Rodnina MV. 2013. EF-P is essential for rapid synthesis of proteins containing consecutive proline residues. Science 339: 85–88. [DOI] [PubMed] [Google Scholar]
- Döring K, Ahmed N, Riemer T, Suresh HG, Vainshtein Y, Habich M, Riemer J, Mayer MP, O’Brien EP, Kramer G, et al. 2017. Profiling Ssb-nascent chain interactions reveals principles of Hsp70-assisted folding. Cell 170: 298–311e220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowling RJ, Topisirovic I, Alain T, Bidinosti M, Fonseca BD, Petroulakis E, Wang X, Larsson O, Selvaraj A, Liu Y, et al. 2010. mTORC1-mediated cell proliferation, but not cell growth, controlled by the 4E-BPs. Science 328: 1172–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle JP, Dougherty JD, Heiman M, Schmidt EF, Stevens TR, Ma G, Bupp S, Shrestha P, Shah RD, Doughty ML, et al. 2008. Application of a translational profiling approach for the comparative analysis of CNS cell types. Cell 135: 749–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Duchaine TF, Fabian MR. 2018. Mechanistic insights into microRNA-mediated gene silencing. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a032771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn JG, Foo CK, Belletier NG, Gavis ER, Weissman JS. 2013. Ribosome profiling reveals pervasive and regulated stop codon readthrough in Drosophila melanogaster. eLife 2: e01179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrin MA, Subramaniam AR. 2017. Kinetic modeling predicts a stimulatory role for ribosome collisions at elongation stall sites in bacteria. eLife 6: e23629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields AP, Rodriguez EH, Jovanovic M, Stern-Ginossar N, Haas BJ, Mertins P, Raychowdhury R, Hacohen N, Carr SA, Ingolia NT, et al. 2015. A regression-based analysis of ribosome-profiling data reveals a conserved complexity to mammalian translation. Mol Cell 60: 816–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritsch C, Herrmann A, Nothnagel M, Szafranski K, Huse K, Schumann F, Schreiber S, Platzer M, Krawczak M, Hampe J, et al. 2012. Genome-wide search for novel human uORFs and N-terminal protein extensions using ribosomal footprinting. Genome Res 22: 2208–2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamble CE, Brule CE, Dean KM, Fields S, Grayhack EJ. 2016. Adjacent codons act in concert to modulate translation efficiency in yeast. Cell 166: 679–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Wan J, Liu B, Ma M, Shen B, Qian SB. 2015. Quantitative profiling of initiating ribosomes in vivo. Nat Methods 12: 147–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerashchenko MV, Gladyshev VN. 2014. Translation inhibitors cause abnormalities in ribosome profiling experiments. Nucleic Acids Res 42: e134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, Stern-Ginossar N, Brandman O, Whitehead EH, Doudna JA, et al. 2013. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 154: 442–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert LA, Horlbeck MA, Adamson B, Villalta JE, Chen Y, Whitehead EH, Guimaraes C, Panning B, Ploegh HL, Bassik MC, et al. 2014. Genome-scale CRISPR-mediated control of gene repression and activation. Cell 159: 647–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingold H, Tehler D, Christoffersen NR, Nielsen MM, Asmar F, Kooistra SM, Christophersen NS, Christensen LL, Borre M, Sorensen KD, et al. 2014. A dual program for translation regulation in cellular proliferation and differentiation. Cell 158: 1281–1292. [DOI] [PubMed] [Google Scholar]
- Glock C, Heumuller M, Schuman EM. 2017. mRNA transport and local translation in neurons. Curr Opin Neurobiol 45: 169–177. [DOI] [PubMed] [Google Scholar]
- Gloge F, Becker AH, Kramer G, Bukau B. 2014. Co-translational mechanisms of protein maturation. Curr Opin Struct Biol 24: 24–33. [DOI] [PubMed] [Google Scholar]
- Guo H, Ingolia NT, Weissman JS, Bartel DP. 2010. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 466: 835–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez E, Shin BS, Woolstenhulme CJ, Kim JR, Saini P, Buskirk AR, Dever TE. 2013. eIF5A promotes translation of polyproline motifs. Mol Cell 51: 35–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guydosh NR, Green R. 2014. Dom34 rescues ribosomes in 3′ untranslated regions. Cell 156: 950–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y, Gao X, Liu B, Wan J, Zhang X, Qian SB. 2014. Ribosome profiling reveals sequence-independent post-initiation pausing as a signature of translation. Cell Res 24: 842–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiman M, Schaefer A, Gong S, Peterson JD, Day M, Ramsey KE, Suarez-Farinas M, Schwarz C, Stephan DA, Surmeier DJ, et al. 2008. A translational profiling approach for the molecular characterization of CNS cell types. Cell 135: 738–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Hellen CUT. 2018. Translation termination and ribosome recycling in eukaryotes. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a032656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess GT, Tycko J, Yao D, Bassik MC. 2017. Methods and applications of CRISPR-mediated base editing in eukaryotic genomes. Mol Cell 68: 26–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindupur SK, Gonzalez A, Hall MN. 2015. The opposing actions of target of rapamycin and AMP-activated protein kinase in cell growth control. Cold Spring Harb Perspect Biol 7: a019141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockenberry AJ, Pah AR, Jewett MC, Amaral LA. 2017. Leveraging genome-wide datasets to quantify the functional role of the anti-Shine–Dalgarno sequence in regulating translation efficiency. Open Biol 7: 160239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh AC, Liu Y, Edlind MP, Ingolia NT, Janes MR, Sher A, Shi EY, Stumpf CR, Christensen C, Bonham MJ, et al. 2012. The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature 485: 55–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussmann JA, Patchett S, Johnson A, Sawyer S, Press WH. 2015. Understanding biases in ribosome profiling experiments reveals signatures of translation dynamics in yeast. PLoS Genet 11: e1005732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huter P, Arenz S, Bock LV, Graf M, Frister JO, Heuer A, Peil L, Starosta AL, Wohlgemuth I, Peske F, et al. 2017. Structural basis for polyproline-mediated ribosome stalling and rescue by the translation elongation factor EF-P. Mol Cell 68: 515–527.e516. [DOI] [PubMed] [Google Scholar]
- Ingolia NT. 2014. Ribosome profiling: New views of translation, from single codons to genome scale. Nat Rev Genet 15: 205–213. [DOI] [PubMed] [Google Scholar]
- Ingolia NT. 2016. Ribosome footprint profiling of translation throughout the genome. Cell 165: 22–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. 2009. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324: 218–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Lareau LF, Weissman JS. 2011. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 147: 789–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Brar GA, Rouskin S, McGeachy AM, Weissman JS. 2012. The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nat Protoc 7: 1534–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Brar GA, Stern-Ginossar N, Harris MS, Talhouarne GJ, Jackson SE, Wills MR, Weissman JS. 2014. Ribosome profiling reveals pervasive translation outside of annotated protein-coding genes. Cell Rep 8: 1365–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa K, Makanae K, Iwasaki S, Ingolia NT, Moriya H. 2017. Post-translational dosage compensation buffers genetic perturbations to stoichiometry of protein complexes. PLoS Genet 13: e1006554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki S, Floor SN, Ingolia NT. 2016. Rocaglates convert DEAD-box protein eIF4A into a sequence-selective translational repressor. Nature 534: 558–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan CH, Williams CC, Weissman JS. 2014. Principles of ER cotranslational translocation revealed by proximity-specific ribosome profiling. Science 346: 1257521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janich P, Arpat AB, Castelo-Szekely V, Lopes M, Gatfield D. 2015. Ribosome profiling reveals the rhythmic liver translatome and circadian clock regulation by upstream open reading frames. Genome Res 25: 1848–1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen BC, Ramasamy G, Vasconcelos EJ, Ingolia NT, Myler PJ, Parsons M. 2014. Extensive stage-regulation of translation revealed by ribosome profiling of Trypanosoma brucei. BMC Genomics 15: 911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Z, Song R, Regev A, Struhl K. 2015. Many lncRNAs, 5′UTRs, and pseudogenes are translated and some are likely to express functional proteins. eLife 4: e08890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Z, Song R, Huang H, Regev A, Struhl K. 2016. Transcriptome-scale RNase-footprinting of RNA–protein complexes. Nat Biotechnol 34: 410–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnstone TG, Bazzini AA, Giraldez AJ. 2016. Upstream ORFs are prevalent translational repressors in vertebrates. EMBO J 35: 706–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannan K, Kanabar P, Schryer D, Florin T, Oh E, Bahroos N, Tenson T, Weissman JS, Mankin AS. 2014. The general mode of translation inhibition by macrolide antibiotics. Proc Natl Acad Sci 111: 15958–15963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami K, Pande S, Faiola B, Moore DP, Boeke JD, Farabaugh PJ, Strathern JN, Nakamura Y, Garfinkel DJ. 1993. A rare tRNA-Arg(ccu) that regulates Ty1 element ribosomal frameshifting is essential for Ty1 retrotransposition in Saccharomyces cerevisiae. Genetics 135: 309–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimchi-Sarfaty C, Oh JM, Kim IW, Sauna ZE, Calcagno AM, Ambudkar SV, Gottesman MM. 2007. A “silent” polymorphism in the MDR1 gene changes substrate specificity. Science 315: 525–528. [DOI] [PubMed] [Google Scholar]
- Lareau LF, Hite DH, Hogan GJ, Brown PO. 2014. Distinct stages of the translation elongation cycle revealed by sequencing ribosome-protected mRNA fragments. eLife 3: e01257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecanda A, Nilges BS, Sharma P, Nedialkova DD, Schwarz J, Vaquerizas JM, Leidel SA. 2016. Dual randomization of oligonucleotides to reduce the bias in ribosome-profiling libraries. Methods 107: 89–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Liu B, Lee S, Huang SX, Shen B, Qian SB. 2012. Global mapping of translation initiation sites in mammalian cells at single-nucleotide resolution. Proc Natl Acad Sci 109: E2424–E2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li GW, Burkhardt D, Gross C, Weissman JS. 2014. Quantifying absolute protein synthesis rates reveals principles underlying allocation of cellular resources. Cell 157: 624–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Xiong X, Zhang M, Wang K, Chen Y, Zhou J, Mao Y, Lv J, Yi D, Chen XW, et al. 2017. Base-resolution mapping reveals distinct m1A methylome in nuclear- and mitochondrial-encoded transcripts. Mol Cell 68: 993–1005.e1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu TY, Song YS. 2016. Prediction of ribosome footprint profile shapes from transcript sequences. Bioinformatics 32: i183–i191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu SJ, Horlbeck MA, Cho SW, Birk HS, Malatesta M, He D, Attenello FJ, Villalta JE, Cho MY, Chen Y, et al. 2017a. CRISPRi-based genome-scale identification of functional long noncoding RNA loci in human cells. Science 355: aah7111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu TY, Huang HH, Wheeler D, Xu Y, Wells JA, Song YS, Wiita AP. 2017b. Time-resolved proteomics extends ribosome profiling-based measurements of protein synthesis dynamics. Cell Syst 4: 636–644.e639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loayza-Puch F, Rooijers K, Buil LC, Zijlstra J, Oude Vrielink JF, Lopes R, Ugalde AP, van Breugel P, Hofland I, Wesseling J, et al. 2016. Tumour-specific proline vulnerability uncovered by differential ribosome codon reading. Nature 530: 490–494. [DOI] [PubMed] [Google Scholar]
- Marks J, Kannan K, Roncase EJ, Klepacki D, Kefi A, Orelle C, Vazquez-Laslop N, Mankin AS. 2016. Context-specific inhibition of translation by ribosomal antibiotics targeting the peptidyl transferase center. Proc Natl Acad Sci 113: 12150–12155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens AT, Taylor J, Hilser VJ. 2015. Ribosome A and P sites revealed by length analysis of ribosome profiling data. Nucleic Acids Res 43: 3680–3687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews MB, Nahum S, John WBH. 2000. Origins and principles of translational control. In Cold Spring Harbor monograph archive; Volume 39: Translational control of gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Matsuo Y, Ikeuchi K, Saeki Y, Iwasaki S, Schmidt C, Udagawa T, Sato F, Tsuchiya H, Becker T, Tanaka K, et al. 2017. Ubiquitination of stalled ribosome triggers ribosome-associated quality control. Nat Commun 8: 159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGlincy NJ, Ingolia NT. 2017. Transcriptome-wide measurement of translation by ribosome profiling. Methods 126: 112–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McManus CJ, May GE, Spealman P, Shteyman A. 2014. Ribosome profiling reveals post-transcriptional buffering of divergent gene expression in yeast. Genome Res 24: 422–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menschaert G, Van Criekinge W, Notelaers T, Koch A, Crappe J, Gevaert K, Van Damme P. 2013. Deep proteome coverage based on ribosome profiling aids mass spectrometry-based protein and peptide discovery and provides evidence of alternative translation products and near-cognate translation initiation events. Mol Cell Proteomics 12: 1780–1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Merrick WC, Pavitt GD. 2018. Protein synthesis initiation in eukaryotic cells. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a033092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel AM, Baranov PV. 2013. Ribosome profiling: A hi-def monitor for protein synthesis at the genome-wide scale. Wiley Interdiscip Rev RNA 4: 473–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel AM, Choudhury KR, Firth AE, Ingolia NT, Atkins JF, Baranov PV. 2012. Observation of dually decoded regions of the human genome using ribosome profiling data. Genome Res 22: 2219–2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills EW, Green R. 2017. Ribosomopathies: There’s strength in numbers. Science 358: eaan2755. [DOI] [PubMed] [Google Scholar]
- Mills EW, Wangen J, Green R, Ingolia NT. 2016. Dynamic regulation of a ribosome rescue pathway in erythroid cells and platelets. Cell Rep 17: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills EW, Green R, Ingolia NT. 2017. Slowed decay of mRNAs enhances platelet specific translation. Blood 129: e38–e48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammad F, Woolstenhulme CJ, Green R, Buskirk AR. 2016. Clarifying the translational pausing landscape in bacteria by ribosome profiling. Cell Rep 14: 686–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moor AE, Golan M, Massasa EE, Lemze D, Weizman T, Shenhav R, Baydatch S, Mizrahi O, Winkler R, Golani O, et al. 2017. Global mRNA polarization regulates translation efficiency in the intestinal epithelium. Science 357: 1299–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzzey D, Sherlock G, Weissman JS. 2014. Extensive and coordinated control of allele-specific expression by both transcription and translation in Candida albicans. Genome Res 24: 963–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatogawa H, Ito K. 2002. The ribosomal exit tunnel functions as a discriminating gate. Cell 108: 629–636. [DOI] [PubMed] [Google Scholar]
- Napthine S, Ling R, Finch LK, Jones JD, Bell S, Brierley I, Firth AE. 2017. Protein-directed ribosomal frameshifting temporally regulates gene expression. Nat Commun 8: 15582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedialkova DD, Leidel SA. 2015. Optimization of codon translation rates via tRNA modifications maintains proteome integrity. Cell 161: 1606–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor PB, Andreev DE, Baranov PV. 2016. Comparative survey of the relative impact of mRNA features on local ribosome profiling read density. Nat Commun 7: 12915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E, Becker AH, Sandikci A, Huber D, Chaba R, Gloge F, Nichols RJ, Typas A, Gross CA, Kramer G, et al. 2011. Selective ribosome profiling reveals the cotranslational chaperone action of trigger factor in vivo. Cell 147: 1295–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauli A, Norris ML, Valen E, Chew GL, Gagnon JA, Zimmerman S, Mitchell A, Ma J, Dubrulle J, Reyon D, et al. 2014. Toddler: An embryonic signal that promotes cell movement via Apelin receptors. Science 343: 1248636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pechmann S, Chartron JW, Frydman J. 2014. Local slowdown of translation by nonoptimal codons promotes nascent-chain recognition by SRP in vivo. Nat Struct Mol Biol 21: 1100–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Peer E, Moshitch-Moshkovitz S, Rechavi G, Dominissini D. 2018. The epitranscriptome in translation regulation. Cold Spring Harb Perspect Biol 10.110l/cshperspect.a032623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelechano V, Alepuz P. 2017. eIF5A facilitates translation termination globally and promotes the elongation of many non polyproline-specific tripeptide sequences. Nucleic Acids Res 45: 7326–7338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelechano V, Wei W, Steinmetz LM. 2015. Widespread co-translational RNA decay reveals ribosome dynamics. Cell 161: 1400–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotkin JB, Kudla G. 2011. Synonymous but not the same: The causes and consequences of codon bias. Nat Rev Genet 12: 32–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pop C, Rouskin S, Ingolia NT, Han L, Phizicky EM, Weissman JS, Koller D. 2014. Causal signals between codon bias, mRNA structure, and the efficiency of translation and elongation. Mol Syst Biol 10: 770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popa A, Lebrigand K, Barbry P, Waldmann R. 2016. Pateamine A-sensitive ribosome profiling reveals the scope of translation in mouse embryonic stem cells. BMC Genomics 17: 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presnyak V, Alhusaini N, Chen YH, Martin S, Morris N, Kline N, Olson S, Weinberg D, Baker KE, Graveley BR, et al. 2015. Codon optimality is a major determinant of mRNA stability. Cell 160: 1111–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Proud CG. 2018. Phosphorylation and signal transduction pathways in translational control. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a033050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian W, Yang JR, Pearson NM, Maclean C, Zhang J. 2012. Balanced codon usage optimizes eukaryotic translational efficiency. PLoS Genet 8: e1002603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radhakrishnan A, Chen YH, Martin S, Alhusaini N, Green R, Coller J. 2016. The DEAD-Box protein Dhh1p couples mRNA decay and translation by monitoring codon optimality. Cell 167: 122–132 e129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Robichaud N, Sonenberg N, Ruggero D, Schneider RJ. 2018. Translational control in cancer. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a032896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Rodnina MV. 2018. Translation in prokaryotes. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a032664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell JB, Cook GM. 1995. Energetics of bacterial growth: Balance of anabolic and catabolic reactions. Microbiol Rev 59: 48–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santagata S, Mendillo ML, Tang YC, Subramanian A, Perley CC, Roche SP, Wong B, Narayan R, Kwon H, Koeva M, et al. 2013. Tight coordination of protein translation and HSF1 activation supports the anabolic malignant state. Science 341: 1238303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz E, Yang L, Su T, Morris DR, McKnight GS, Amieux PS. 2009. Cell-type-specific isolation of ribosome-associated mRNA from complex tissues. Proc Natl Acad Sci 106: 13939–13944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuller AP, Wu CC, Dever TE, Buskirk AR, Green R. 2017. eIF5A functions globally in translation elongation and termination. Mol Cell 66: 194–205 e195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher TN, Schreiber RD. 2015. Neoantigens in cancer immunotherapy. Science 348: 69–74. [DOI] [PubMed] [Google Scholar]
- Sen ND, Zhou F, Ingolia NT, Hinnebusch AG. 2015. Genome-wide analysis of translational efficiency reveals distinct but overlapping functions of yeast DEAD-box RNA helicases Ded1 and eIF4A. Genome Res 25: 1196–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sendoel A, Dunn JG, Rodriguez EH, Naik S, Gomez NC, Hurwitz B, Levorse J, Dill BD, Schramek D, Molina H, et al. 2017. Translation from unconventional 5′ start sites drives tumour initiation. Nature 541: 494–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Z, Fujii K, Kovary KM, Genuth NR, Rost HL, Teruel MN, Barna M. 2017. Heterogeneous ribosomes preferentially translate distinct subpools of mRNAs genome-wide. Mol Cell 67: 71–83.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shieh YW, Minguez P, Bork P, Auburger JJ, Guilbride DL, Kramer G, Bukau B. 2015. Operon structure and cotranslational subunit association direct protein assembly in bacteria. Science 350: 678–680. [DOI] [PubMed] [Google Scholar]
- Sidrauski C, McGeachy AM, Ingolia NT, Walter P. 2015. The small molecule ISRIB reverses the effects of eIF2α phosphorylation on translation and stress granule assembly. eLife 4: 05033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simms CL, Yan LL, Zaher HS. 2017. Ribosome collision is critical for quality control during no-go decay. Mol Cell 68: 361–373.e365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slobodin B, Han R, Calderone V, Vrielink J, Loayza-Puch F, Elkon R, Agami R. 2017. Transcription impacts the efficiency of mRNA translation via co-transcriptional N6-adenosine methylation. Cell 169: 326–337.e312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Sokabe M, Fraser CS. 2018. Toward a kinetic understanding of eukaryotic translation. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a032706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenberg N, Hinnebusch AG. 2009. Regulation of translation initiation in eukaryotes: Mechanisms and biological targets. Cell 136: 731–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Sossin WS, Costa-Mattioli M. 2018. Translational control in the brain in health and disease. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a032912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler M, Fire A. 2011. Wobble base-pairing slows in vivo translation elongation in metazoans. RNA 17: 2063–2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starck SR, Jiang V, Pavon-Eternod M, Prasad S, McCarthy B, Pan T, Shastri N. 2012. Leucine-tRNA initiates at CUG start codons for protein synthesis and presentation by MHC class I. Science 336: 1719–1723. [DOI] [PubMed] [Google Scholar]
- Starck SR, Tsai JC, Chen K, Shodiya M, Wang L, Yahiro K, Martins-Green M, Shastri N, Walter P. 2016. Translation from the 5′ untranslated region shapes the integrated stress response. Science 351: aad3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steitz JA. 1969. Nucleotide sequences of the ribosomal binding sites of bacteriophage R17 RNA. Cold Spring Harb Symp Quant Biol 34: 621–630. [DOI] [PubMed] [Google Scholar]
- Stern-Ginossar N, Weisburd B, Michalski A, Le VT, Hein MY, Huang SX, Ma M, Shen B, Qian SB, Hengel H, et al. 2012. Decoding human cytomegalovirus. Science 338: 1088–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Stern-Ginossar N, Thompson SR, Mathews MB, Mohr I. 2018. Translational control in virus-infected cells. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a033001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpf CR, Moreno MV, Olshen AB, Taylor BS, Ruggero D. 2013. The translational landscape of the mammalian cell cycle. Mol Cell 52: 574–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramaniam AR, Zid BM, O’Shea EK. 2014. An integrated approach reveals regulatory controls on bacterial translation elongation. Cell 159: 1200–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subtelny AO, Eichhorn SW, Chen GR, Sive H, Bartel DP. 2014. Poly(A)-tail profiling reveals an embryonic switch in translational control. Nature 508: 66–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanenbaum ME, Stern-Ginossar N, Weissman JS, Vale RD. 2015. Regulation of mRNA translation during mitosis. eLife 4: 07957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson MK, Rojas-Duran MF, Gangaramani P, Gilbert WV. 2016. The ribosomal protein Asc1/RACK1 is required for efficient translation of short mRNAs. eLife 5: e11154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoreen CC, Chantranupong L, Keys HR, Wang T, Gray NS, Sabatini DM. 2012. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature 485: 109–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tunney RJ, McGlincy NJ, Graham ME, Naddaf N, Pachter L, Lareau L. 2017. Accurate design of translational output by a neural network model of ribosome distribution. bioRxiv 10.1101/201517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ude S, Lassak J, Starosta AL, Kraxenberger T, Wilson DN, Jung K. 2013. Translation elongation factor EF-P alleviates ribosome stalling at polyproline stretches. Science 339: 82–85. [DOI] [PubMed] [Google Scholar]
- Ulitsky I, Bartel DP. 2013. lincRNAs: Genomics, evolution, and mechanisms. Cell 154: 26–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasquez JJ, Hon CC, Vanselow JT, Schlosser A, Siegel TN. 2014. Comparative ribosome profiling reveals extensive translational complexity in different Trypanosoma brucei life cycle stages. Nucleic Acids Res 42: 3623–3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verduyn C, Stouthamer AH, Scheffers WA, van Dijken JP. 1991. A theoretical evaluation of growth yields of yeasts. Antonie Van Leeuwenhoek 59: 49–63. [DOI] [PubMed] [Google Scholar]
- Vogel C, Marcotte EM. 2012. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat Rev Genet 13: 227–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu LP, Pickering BF, Cheng Y, Zaccara S, Nguyen D, Minuesa G, Chou T, Chow A, Saletore Y, MacKay M, et al. 2017. The N6-methyladenosine (m6A)-forming enzyme METTL3 controls myeloid differentiation of normal hematopoietic and leukemia cells. Nat Med 23: 1369–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, Weng X, Chen K, Shi H, He C. 2015. N6-methyladenosine modulates messenger RNA translation efficiency. Cell 161: 1388–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg DE, Shah P, Eichhorn SW, Hussmann JA, Plotkin JB, Bartel DP. 2016. Improved ribosome-footprint and mRNA measurements provide insights into dynamics and regulation of yeast translation. Cell Rep 14: 1787–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Wek RC. 2018. Role of eIF2α kinases in translational control and adaptation to cellular stresses. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a032870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams CC, Jan CH, Weissman JS. 2014. Targeting and plasticity of mitochondrial proteins revealed by proximity-specific ribosome profiling. Science 346: 748–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe AL, Singh K, Zhong Y, Drewe P, Rajasekhar VK, Sanghvi VR, Mavrakis KJ, Jiang M, Roderick JE, Van der Meulen J, et al. 2014. RNA G-quadruplexes cause eIF4A-dependent oncogene translation in cancer. Nature 513: 65–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolin SL, Walter P. 1988. Ribosome pausing and stacking during translation of a eukaryotic mRNA. EMBO J 7: 3559–3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolin SL, Walter P. 1989. Signal recognition particle mediates a transient elongation arrest of preprolactin in reticulocyte lysate. J Cell Biol 109: 2617–2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolstenhulme CJ, Parajuli S, Healey DW, Valverde DP, Petersen EN, Starosta AL, Guydosh NR, Johnson WE, Wilson DN, Buskirk AR. 2013. Nascent peptides that block protein synthesis in bacteria. Proc Natl Acad Sci 110: E878–E887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G, Greene GH, Yoo H, Liu L, Marques J, Motley J, Dong X. 2017. Global translational reprogramming is a fundamental layer of immune regulation in plants. Nature 545: 487–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young DJ, Guydosh NR, Zhang F, Hinnebusch AG, Green R. 2015. Rli1/ABCE1 recycles terminating ribosomes and controls translation reinitiation in 3′UTRs in vivo. Cell 162: 872–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Hubalewska M, Ignatova Z. 2009. Transient ribosomal attenuation coordinates protein synthesis and co-translational folding. Nat Struct Mol Biol 16: 274–280. [DOI] [PubMed] [Google Scholar]
- Zhang S, Hu H, Zhou J, He X, Jiang T, Zeng J. 2017. Analysis of ribosome stalling and translation elongation dynamics by deep learning. Cell Syst 5: 212–220. e216. [DOI] [PubMed] [Google Scholar]
- Zhou J, Wan J, Gao X, Zhang X, Jaffrey SR, Qian SB. 2015. Dynamic m6A mRNA methylation directs translational control of heat shock response. Nature 526: 591–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinshteyn B, Gilbert WV. 2013. Loss of a conserved tRNA anticodon modification perturbs cellular signaling. PLoS Genet 9: e1003675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinshteyn B, Rojas-Duran MF, Gilbert WV. 2017. Translation initiation factor eIF4G1 preferentially binds yeast transcript leaders containing conserved oligo-uridine motifs. RNA 23: 1365–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]