SUMMARY
Noncoding introns are removed from nuclear precursor messenger RNA (pre-mRNA) in a two-step phosphoryl transfer reaction by the spliceosome, a dynamic multimegadalton enzyme. Cryo-electron microscopy (cryo-EM) structures of the Saccharomyces cerevisiae spliceosome were recently determined in eight key states. Combined with the wealth of available genetic and biochemical data, these structures have revealed new insights into the mechanisms of spliceosome assembly, activation, catalysis, and disassembly. The structures show how a single RNA catalytic center forms during activation and accomplishes both steps of the splicing reaction. The structures reveal how spliceosomal helicases remodel the spliceosome for active site formation, substrate docking, reaction product undocking, and spliceosome disassembly and how they facilitate splice site proofreading. Although human spliceosomes contain additional proteins, their cryo-EM structures suggest that the underlying mechanism is conserved across all eukaryotes. In this review, we summarize the current structural understanding of pre-mRNA splicing.
1. INTRODUCTION
The discovery of “split genes” in adenovirus in 1977 (Berget et al. 1977; Chow et al. 1977) astonished the scientific community and galvanized four decades of intensive research on the underlying mechanism. It became clear early on that the majority of eukaryotic protein-coding genes contain noncoding sequences (introns), which must be removed from the precursor messenger RNA (pre-mRNA) so that the remaining coding sequences (exons) can be ligated to produce the functional mRNA—a process called pre-mRNA splicing. Introns are defined by three conserved sequences, the 5′ splice site (5′SS), the internal branch point (BP) sequence, and the 3′ splice site (3′SS). Early work established that introns are removed from pre-mRNA in two consecutive phosphoryl transfer reactions (Fig. 1A) (reviewed in Will and Lührmann 2011). In the first reaction (branching or step 1), the nucleophilic attack of the BP adenosine 2′-hydroxyl group at the phosphate of the 5′SS produces the free 5′ exon and the lariat intron-3′ exon intermediate. In the second phosphoryl transfer reaction, the 3′-terminal hydroxyl group of the free 5′ exon attacks the phosphate at the 3′SS, producing the ligated exons (mRNA) and a free intron lariat. The intron lariat is a unique structure essential for pre-mRNA splicing, in which the 2′-hydroxyl group of the BP adenosine covalently links to the 5′ phosphate of the first nucleotide of the intron.
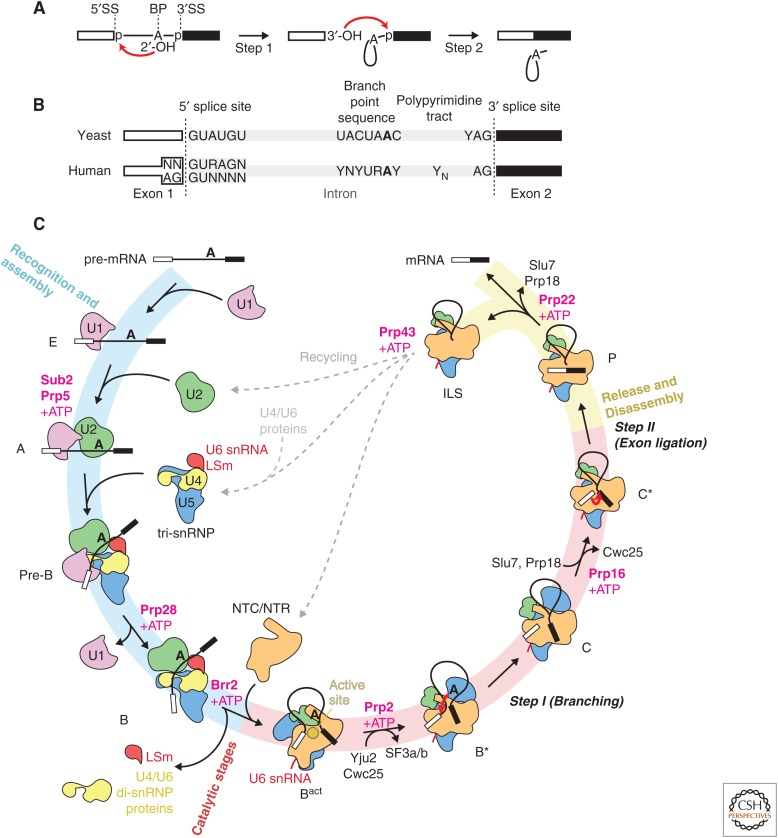
Figure 1.
Schematic overview of the splicing reaction and the cycle. (A) Removal of introns from precursor messenger RNA (pre-mRNA) by the two-step phosphoryl transfer reaction: branching and exon-ligation reactions. (B) Yeast and human introns are identified by short conserved sequences at the 5′ and 3′ splice sites (5′SS and 3′SS) and the internal branch point (BP) sequence. (C) Schematic representation of the yeast splicing cycle. The spliceosome has been isolated in at least 10 distinct states, namely, E, A, pre-B, B, Bact, B*, C, C*, P, and intron lariat spliceosome (ILS) complexes. Transitions between these states are regulated by helicases or ATPases in the Ski2-like (Brr2), DEAD- (Sub2/UAP56, Prp5, Prp28) and DEAH-box (Prp2, Prp16, Prp22, Prp43) families. The 5′SS and BP sequence are recognized by the U1 and U2 small nuclear ribonucleoprotein particles (snRNPs) and the resulting A complex associates with the preassembled U4/U6.U5 tri-snRNP to form the fully assembled pre-B complex. The U1 snRNP dissociates from the pre-B complex by the activity of the DEAD-box helicase Prp28 to form the precatalytic B complex. The Brr2 helicase then unwinds the U4/U6 small nuclear RNA (snRNA) duplex allowing U4 snRNA to dissociate together with the U4/U6 di-snRNP proteins and the LSm proteins. U6 snRNA freed from U4 snRNA folds and associates with part of U2 snRNA to create the group II intron-like active site harboring the catalytic magnesium ions. The nineteen complex (NTC) and nineteen-related (NTR) proteins associate to stabilize the active site RNA structure in Bact complex. The U2 snRNP SF3a and SF3b complexes and the RES complex dissociate by the activity of the Prp2 helicase inducing the binding of the Prp16 helicase and the step 1 (branching) factors, Yju2, Isy1, and Cwc25, which dock the branch helix into the active site. In the B* complex, the BP adenosine 2′-OH group attacks the 5′SS producing the 5′exon and the lariat intron intermediate leading to the formation of the C complex. By its ATPase activity, the Prp16 helicase dissociates together with the step 1 factors, allowing the binding of the Prp22 helicase and step 2 factors, Prp18 and Slu7. In the resulting C* complex, the 3′SS can be docked into the active site enabling the 3′-OH group of the 5′ exon to attack the 3′SS, producing the ligated exons and the lariat intron. The Prp22 helicase activity releases the ligated exon (mRNA) from the postsplicing P complex and the remaining ILS is disassembled by the activity of the Prp43 helicase and the snRNPs are recycled for the next round of splicing.
This simple two-step reaction is catalyzed by a complex molecular machine known as the spliceosome (Brody and Abelson 1985), which comprises five snRNAs and approximately 70 proteins in the yeast Saccharomyces cerevisiae and more than 100 in humans (see Kastner et al. 2019). Each of the five small nuclear RNAs (U1, U2, U4, U5, and U6 snRNAs) organizes with a distinct set of proteins into small nuclear ribonucleoprotein particles (snRNPs) (Lerner and Steitz 1979), whereas other non-snRNP proteins, including the nineteen complex (NTC) and the nineteen-related (NTR) proteins, join during the splicing reaction (Fig. 2) (Will and Lührmann 2011). The RNA active site of the spliceosome is created from U6, U5, and U2 snRNAs after ordered assembly of the snRNPs on the pre-mRNA substrate and extensive conformational and compositional remodeling (Fig. 1B). These events include the dissociation of U1 and U4 snRNAs with their associated proteins, the binding of the NTC and NTR proteins (Chan and Cheng 2005) to form and stabilize the catalytic RNA core, and the binding of specific factors to enable the branching and exon ligation reactions. Eight conserved RNA helicases from the Ski2 (Brr2), DEAD-box (Sub2/UAP56, Prp5, Prp28), and DEAH-box (Prp2, Prp16, Prp22, Prp43) families promote the transitions between the different spliceosome states by remodeling the protein–RNA or RNA–RNA networks and by proofreading the selection of the correct intron sequences (Fig. 1C) (Burgess and Guthrie 1993; Semlow et al. 2016). At least 10 distinct spliceosome states can be defined during the splicing reaction, the E, A, pre-B, B, Bact, B*, C, C*, P, and intron lariat spliceosome (ILS) complexes, which differ in their RNA or protein composition and/or the state of the pre-mRNA substrate (Will and Lührmann 2011).
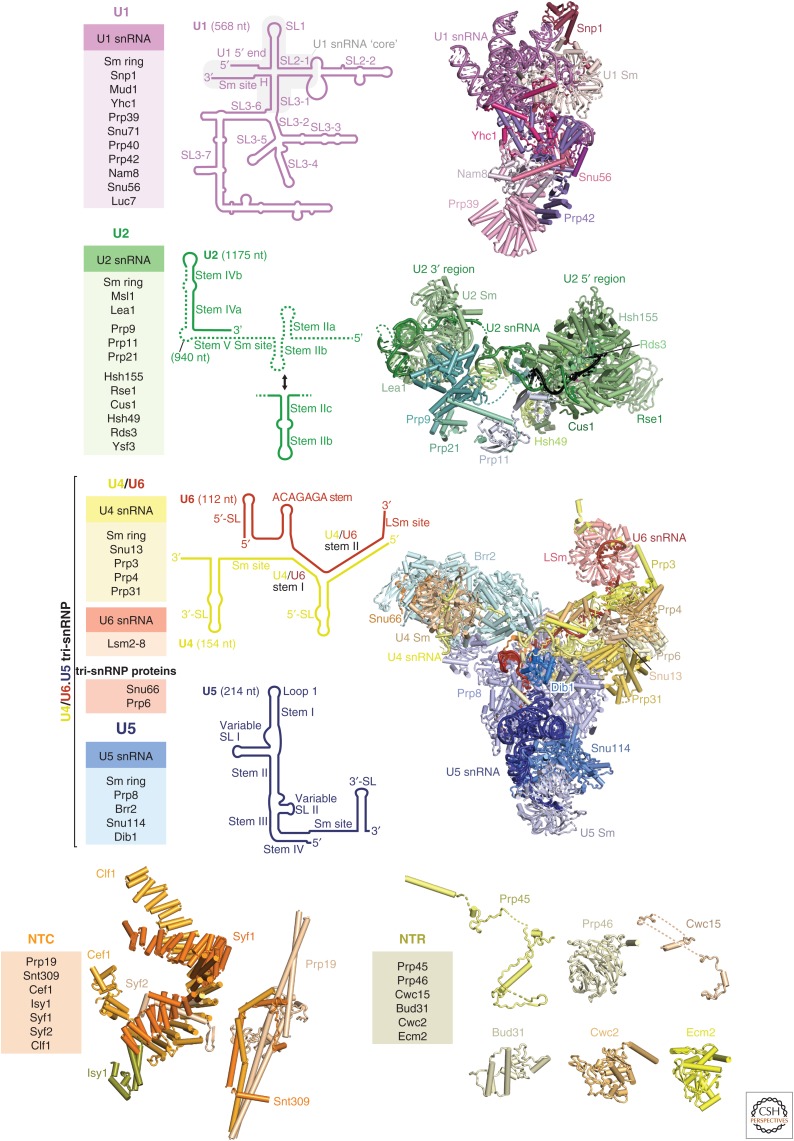
Figure 2.
Composition of the spliceosome. Protein composition (left), small nuclear RNA (snRNA) secondary structure (middle), and three-dimensional structure (right) of yeast spliceosome components as they assemble on precursor messenger RNA (pre-mRNA) substrates. Each of the five small nuclear ribonucleoprotein particles (snRNPs) is shown with shades of distinct colors (U1, magenta; U2, green, U4, yellow; U5, blue; U6, red). For each snRNA, important secondary structure elements are labeled. The structure of the isolated U2 snRNP remains unknown, and in its stead, we show the yeast U2 snRNP structure bound to pre-mRNA (black) (yeast B complex, PDB 5NRL). For the nineteen complex (NTC) and nineteen-related (NTR), the three-dimensional structures found in the C complex are shown. The NTC forms a complex in isolation (Chan and Cheng 2005), whereas the NTR proteins are likely to join the spliceosome separately. The yeast U1 snRNP (PDB 5UZ5), the yeast U2 snRNP (PDB 5NRL), yeast tri-snRNP (PDB 5NRL), and the NTC and NTR in the Bact complex (PDB 5GM6) structures are shown. SL, stem loop.
A detailed understanding of the molecular mechanism of splicing requires structural knowledge of the spliceosome in its various states. Until recently, structural information on the spliceosome was limited to crystal structures of proteins and RNA–protein complexes (Will and Lührmann 2011), as well as low-resolution electron microscopic images of the snRNPs in specific spliceosome states (reviewed in Stark and Lührmann 2006). Because of recent developments in cryo-electron microscopy (cryo-EM), the structures of biological macromolecules can now often be determined to near-atomic resolution (Fernandez-Leiro and Scheres 2016). This is largely due to the development of direct electron detectors, providing high-quality movies of the biological specimen, and to improved image processing software. In particular, the classification of complex mixtures into structurally homogeneous subsets in programs such as RELION has had a major impact on the determination of spliceosome structures (Wilkinson et al. 2018). The spliceosome is inherently flexible, and some of its domains undergo a continuous motion. Focused classification methods together with partial signal subtraction and focused refinement have enabled us to bring even highly mobile parts of the spliceosome into focus. These new structures are built on a foundation of decades of genetic and biochemical work, that established many of the methods to stall the spliceosome in its various states. The structures of the S. cerevisiae U4/U6.U5 tri-snRNP (Nguyen et al. 2015) and Schizosaccharomyces pombe ILS (Yan et al. 2015), both reported in 2015, presaged a flood of high-resolution cryo-EM structures of yeast and human spliceosomes. The resulting structures have substantially advanced our knowledge of splicing. They not only confirm and explain decades of functional observations, but they also dramatically enhance our understanding of the underlying molecular mechanism, suggesting a high conservation of the splicing mechanism across all eukaryotes. In this review, we will focus on the mechanistic insights gained from structural studies of the S. cerevisiae spliceosome and make only brief references to the human spliceosome, which is discussed in more depth in Kastner et al. (2019) and Yan et al. (2019). For details on cryo-EM and its application to the spliceosome, we refer the interested reader to other recent reviews (e.g., Wilkinson et al. 2018). PyMOL sessions of the yeast spliceosome structures discussed in this review are available online (see www2.mrc-lmb.cam.ac.uk/groups/nagai/resources/).
2. SPLICE SITE RECOGNITION BY THE U1 AND U2 SNRNPS AND PRESPLICEOSOME FORMATION
Much of our current understanding of the mechanism of pre-mRNA splicing is derived from genetic experiments using S. cerevisiae (hereafter termed “yeast,” unless otherwise specified) and biochemical experiments using yeast whole cell extract or HeLa cell nuclear extract (Will and Lührmann 2011). S. cerevisiae contains approximately 5500 protein-coding genes of which approximately 300 contain introns. The human genome contains approximately 20,000 protein-coding genes, and the majority of these genes contain multiple introns, which are often alternatively spliced (Will and Lührmann 2011). In yeast, the 5′SS and BP sequences are stringently conserved to be /GUAUGU (/ is the splice junction) and UACUAAC (BP adenosine is bold) with few exceptions (Fig. 1B) (Parker et al. 1987; Spingola et al. 1999). In humans, most 5′SS sequences fall into two major classes: one with a G/GU(A or G)AG and the other AG/GUA (/ denotes the splice junction) consensus sequence. The metazoan BP sequence is YNCURAC (Will and Lührmann 2011) and precedes the 3′SS by 30–40 nucleotides. In both yeast and humans, the 3′SS is defined as YAG/ in all introns except those processed by the metazoan minor spliceosome (Tarn and Steitz 1996). In metazoans, the YAG is preceded by a poly-pyrimidine tract.
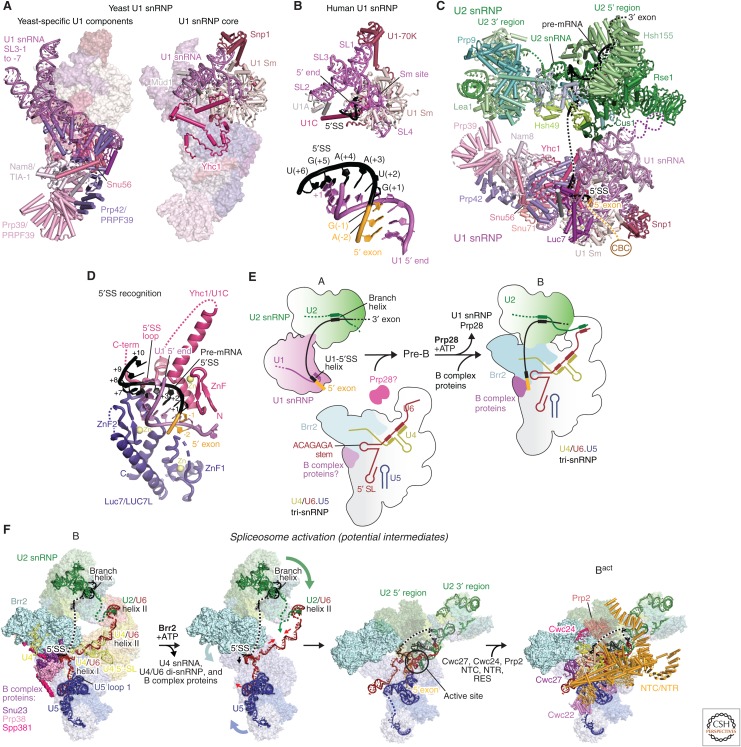
In the first step of spliceosome assembly, the U1 snRNP binds to the pre-mRNA 5′SS by base-pairing with the U1 snRNA 5′end (m32,2,7GpppAUACΨΨACCUG---; Ψ, pseudo-uridine), which is conserved in most species including yeast and humans (see rfam.xfam.org/). The human U1 snRNP contains the seven-membered Sm ring, which also binds the U-rich tract of U2, U4, and U5 snRNAs, and three U1-specific proteins (U1-70k, U1A, and U1C) (Will and Lührmann 2011). In S. cerevisiae, U1 snRNA is expanded and the U1 snRNP contains seven additional yeast-specific proteins (Gottschalk et al. 1998), which, except for Snu56, have homologs in humans, where they function as alternative splicing factors (Fig. 3A) (Gottschalk et al. 1998; Li et al. 2017). The crystal structures of the recombinant U1 snRNP (Pomeranz Krummel et al. 2009) and the chymotrypsin-treated native U1 snRNP (Weber et al. 2010) together provided the complete image of the human U1 snRNP. The crystal structure of the functional core of U1 snRNP bound to a 5′SS RNA oligonucleotide showed that the Zn-finger domain of the U1C protein stabilizes 5′SS binding by making contacts with the sugar-phosphate backbone of the U1–5′SS duplex but not the bases (Fig. 3B) (Kondo et al. 2015). This suggests that 5′SSs are predominantly selected by the stability of the U1–5′SS base-pairing. A recent cryo-EM structure of the free yeast U1 snRNP (Li et al. 2017) shows that its functional core is highly similar to the human U1 snRNP crystal structure, whereas the yeast-specific RNA expansion is stabilized by yeast-specific proteins (Fig. 3A). The 5′ end of U1 snRNA is disordered in the free yeast U1 snRNP but it becomes ordered when it base-pairs with the 5′SS of pre-mRNA. The A complex structure (Plaschka et al. 2018) shows that 5′SS binding induces ordering of Luc7 and flexible parts of the U1C protein by making contacts with the duplex formed between the 5′SS and the 5′ end of U1 snRNA (Fig. 3C). Homologs of Luc7 in humans function as alternative splicing factors and thus the structure of the 5′SS bound yeast U1 snRNP suggests how human homologs of Luc7 stabilize binding of weak 5′SSs (Fig. 3D).
Figure 3.
Spliceosome assembly and activation. (A) The yeast U1 small nuclear ribonucleoprotein particle (snRNP) structure (PDB 5UZ5) contains the conserved core of the U1 snRNP (right) and additionally contains yeast-specific RNA elements and eight proteins (left). (B) 5′ splice site (5′SS) recognition by the U1 small nuclear RNA (snRNA) 5′ end in the human minimal U1 snRNP crystal structure (PDB 4PJO). (C) Yeast A complex assembled on UBC4 precursor messenger RNA (pre-mRNA) (PDB 6G90). (D) The RNA duplex formed between the 5′SS and the 5′ end of U1 snRNA interacts with Yhc1(U1C) and Luc7 in yeast A complex. The 5′ end of U1 snRNA and Luc7, completely disordered in the free U1 snRNP (Li et al. 2017), become ordered when pre-mRNA 5′SS binds to U1 snRNP to form the A complex (Plaschka et al. 2018). (E) Schematic model of early assembly events of the spliceosome. The A complex associates with the U4/U6.U5 tri-snRNP to form the pre-B complex, which is subsequently converted by Prp28 activity into the B complex spliceosome. For more details, see Plaschka et al. (2018). (F) Model for spliceosome activation, the B to Bact complex transition. Possible steps during activation are shown, starting with the cryo-electron microscopy (cryo-EM) structure of the yeast B complex (PDB 5NRL) and ending with the Bact complex (PDB 5GM6). During spliceosome activation, U4 snRNA and at least 22 proteins dissociate, 24 proteins join, and the RNA-based active site containing two catalytic magnesium ions forms. First, the Brr2 helicase unwinds the U4/U6 snRNA duplex to initiate the release of U4 snRNA and U4/U6 di-snRNP-specific, tri-snRNP-specific, and B complex proteins. After U4 snRNA dissociation, U6 snRNA folds to form the internal stem loop (ISL) and associates with part of U2 snRNA to form the catalytic RNA core. The nineteen complex (NTC) and nineteen-related (NTR) proteins and the Bact-specific proteins join to cradle the active site RNA core, which remains largely unchanged during the branching and exon-ligation reactions.
After 5′SS recognition, the BP sequence is recognized by the Msl5(BBP)–Mud2(U2AF65) heterodimer in yeast, whereby Msl5 binds to the branch site and interacts with U1 snRNP, bridging the interaction between the 5′SS and the branch site (Berglund et al. 1998). The DEAD-box helicases Sub2 and Prp5 then replace Msl5-Mud2 with the U2 snRNP and promote the interaction between U1 and U2 snRNPs to form the A complex (Shao et al. 2012). The region of U2 snRNA between stems I and IIa forms the branchpoint-interacting stem loop (BSL), in which the BP binding sequence is located in the loop (Perriman and Ares 2010). Prp5 associates with U2 snRNP by interacting with the BSL and is released upon the base-pairing of U2 with the branch site to allow the recruitment of the tri-snRNP (Liang and Cheng 2015). The stability of the BSL is fine-tuned such that mutations in the stem affect the binding of suboptimal BP sequences. In S. pombe, Prp5 has been shown to interact directly with the SF3b HEAT repeat protein SF3B1 (Hsh155) and similar interactions may occur in other species. Mutations in SF3B1 are found in human myelodysplastic syndromes (MDSs) and other cancers. The equivalent mutations in Hsh155 in S. cerevisiae specifically change splicing of pre-mRNA substrates with suboptimal BP sequence (Tang et al. 2016). Mutations in Hsh155 and Prp5 change the direct physical interaction between Hsh155 and Prp5 and influence the loading (i.e., “fidelity”) of the BS–U2 duplex into the SF3B complex during prespliceosome formation, thereby influencing branch site selection. The complete U2 snRNP structure with the bound BP sequence is now available as part of the A and B complexes (Plaschka et al. 2017, 2018). Consistent with negative stain EM analysis of the U2 snRNP, it has a bipartite structure consisting of the 5′ domain comprising SF3b (Rauhut et al. 2016; Yan et al. 2016; Plaschka et al. 2017) and the U2 snRNA 5′ end, as well as the 3′ region comprising the 3′ end of U2 snRNA, the SF3a subcomplex, the U2 Sm ring, and the U2B˝(Msl1 in yeast)-U2A′(Lea1 in yeast) dimer (Plaschka et al. 2017). The BP sequence pairs with U2 snRNA to form the branch helix, which wedges into the ends of the horseshoe-shaped Hsh155 HEAT repeats, as discussed in more detail below.
3. U4/U6.U5 TRI-SNRNP
The U4/U6.U5 tri-snRNP is the largest preassembled spliceosomal complex and comprises the U5 snRNP, the U4/U6 di-snRNP, and tri-snRNP-specific proteins. In the U4/U6 di-snRNP, the U4 and U6 snRNAs are extensively base-paired to form a three-way junction, stabilized in a compact structure by di-snRNP proteins (Snu13, Prp31, Prp3, and Prp4) (Fig. 2) (Nguyen et al. 2015; 2016; Wan et al. 2016a). Only the U5 snRNP and U6 snRNA remain from the tri-snRNP after spliceosome activation and become part of the catalytic state of the spliceosome. The U5 snRNP comprises U5 snRNA, Prp8, Brr2, Snu114, and the U5 Sm ring.
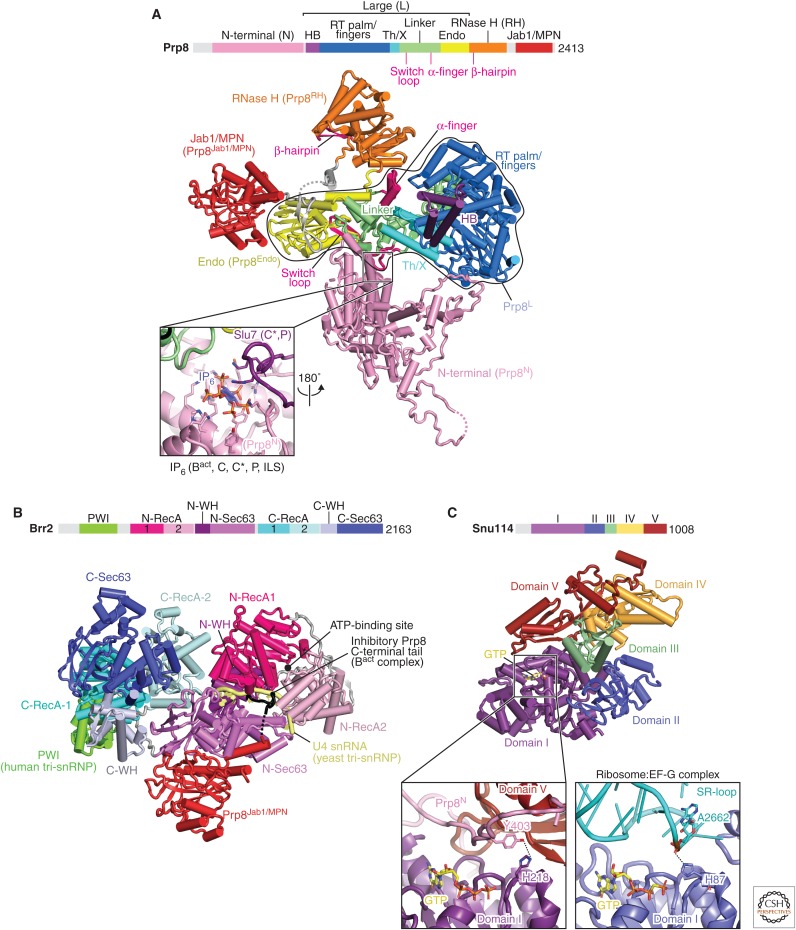
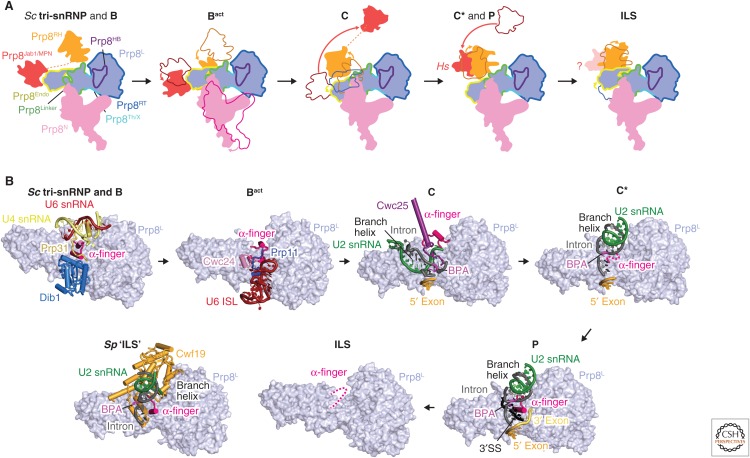
Prp8 is the largest and most highly conserved protein in the spliceosome and contains four structurally distinct domains (amino-terminal, Large, RNaseH and Jab1/MPN domains), connected by flexible linkers (Fig. 4A) (Galej et al. 2013; Nguyen et al. 2015; 2016; Yan et al. 2015). These four domains extensively rearrange at different stages of splicing to organize the catalytic RNA core and regulate the docking and undocking of the substrate pre-mRNA (Fig. 5A,B) (reviewed in Fica and Nagai 2017; Shi 2017). The Prp8 Large domain consists of the helix bundle (HB), reverse transcriptase, Linker and endonuclease (Endo) domains, reminiscent of the group II intron encoded protein (maturase) and suggesting their common evolutionary origin (Galej et al. 2013; Zhao and Pyle 2016). Prp8 cross-links with each of the three splicing reactants in the pre-mRNA substrate (5′SS, BP, and 3′SS) and hence was thought to be at “the heart of the spliceosome” (Grainger and Beggs 2005). This has now been confirmed by the spliceosome structures. The active site cavity of the spliceosome was mapped to the Linker domain in the Prp8 crystal structure based on the Prp8–pre-mRNA crosslinks (Galej et al. 2013). The pre-mRNA branch point (BP+2) nucleotide was cross-linked to the disordered loop in the Linker domain, between residues 1575 and 1598, pinpointing the location of the active site (Galej et al. 2013). This region forms a helix-turn-helix in the yeast U4/U6.U5 tri-snRNP and was hence named the “α-finger” (Nguyen et al. 2016), although the α-finger region alters its structure in different states of the active site.
Figure 4.
Details of key spliceosome protein components Prp8, Brr2, and Snu114. (A) Prp8 comprises four structural units, the amino-terminal (N-terminal), Large, RNase H, and Jab1 domains, which are connected by flexible linkers. These domains change relative positions during the splicing cycle. The Large domain (encircled in black) contains the helical bundle (HB), reverse transcriptase (RT), thumb/X, linker, and endonuclease (En) domains and is reminiscent of the group II intron-encoded protein (IEP), which, however, lacks the amino-terminal HB domain. The catalytic RNA core of the spliceosome forms in the space between the Linker and HB domains. The inset shows the binding site for an IP6 molecule, which is present in all catalytic stage spliceosome structures determined to date (yeast and human). In C* and P complexes (yeast), Slu7 is in part stabilized by binding the IP6. (B) The Brr2 helicase comprises an amino-terminal region with a PWI domain and two Ski2-like helicase cassettes, where only the amino-terminal cassette is catalytically active. The Prp8 Jab1 domain wedges between the two helicase cassettes, anchoring Brr2 to the spliceosome. The Prp8 carboxyl terminus (red) forms an extended peptide, which can bind the amino-terminal helicase cassette and has been proposed to inhibit the binding of RNA substrates (Mozaffari-Jovin et al. 2013). (C) Snu114 is homologous to the translation translocases EF-G (bacterial) and eEF2 (eukaryotic). The inset shows the GTPase active sites of Snu114 and EF-G (PDB 4JUW), respectively. The catalytic His218 in Snu114 is hydrogen-bonded to Tyr403 in the amino-terminal domain of Prp8 and, unlike its EF-G counterpart, is not poised for GTP hydrolysis. Further, the Snu114 His218 is not essential for yeast viability (Nguyen et al. 2016) and its configuration is unchanged in all spliceosome structures determined, suggesting that if GTP hydrolysis occurs, it may instead play a role in spliceosome biogenesis. C-terminal, carboxy-terminal.
Figure 5.
Prp8 rearrangements throughout the splicing cycle. (A) The four Prp8 domains (amino-terminal, Large, RNase H, and Jab) rearrange throughout the splicing cycle. All available spliceosome cryo-electron microscopy (cryo-EM) structures were aligned on the Prp8 Large domain and relative movements of the other domains are shown with a cartoon schematic. Although the amino-terminal domain moves only during the B to Bact transition, to clamp the 5′ exon, the RNase H and Jab1 domains move extensively between states. Thereby they contribute to the stabilization of exchanging spliceosome proteins (RNase H, Jab1) and movements of the branch helix (RNase H). See Figures 6 and 7. (B) Structural remodeling of the Prp8 α-finger throughout the splicing cycle. The Prp8 α-finger lies at the heart of the active site, where it changes its conformation to help organize the active site during multiple steps of splicing. See Figures 6 and 7.
Brr2 contains two Ski2-type helicase domains. The crystal structure of human Brr2 showed how the enzymatically active amino-terminal helicase domain forms a tight complex with the enzymatically inactive carboxy-terminal domain (Fig. 4B) (Mozaffari-Jovin et al. 2013). Brr2 forms a stable complex with the Jab1 domain of Prp8 and is therefore a constituent subunit of the U5 snRNP. Brr2 remains a part of every spliceosome complex, unlike all other spliceosome helicases that bind only transiently. The crystal structures of human and yeast Brr2 in complex with the Prp8 Jab1 domain show that the Jab1 domain is wedged between the amino- and carboxy-terminal helicase domains (Mozaffari-Jovin et al. 2013; Nguyen et al. 2013). Brr2 unwinds the U4/U6 snRNA duplex and plays a crucial role in spliceosome activation (Laggerbauer et al. 1998; Raghunathan and Guthrie 1998). Consistent with this role, in the yeast tri-snRNP (Nguyen et al. 2015; 2016; Wan et al. 2016a; Plaschka et al. 2017) the single-stranded region of U4 snRNA is loaded into the active site of the amino-terminal helicase cassette, ready to unwind the U4/U6 snRNA duplex to activate the spliceosome (Laggerbauer et al. 1998; Raghunathan and Guthrie 1998).
The U5 snRNP additionally contains Snu114, which is homologous to the ribosome translocases EF-G (prokaryotes) and EF-2 (eukaryotes) (Fig. 4C) (Fabrizio et al. 1997). Because of this homology, it was suggested that Snu114 induces conformational changes in the spliceosome upon GTP hydrolysis, as EF-G does during ribosome translocation. Biochemical experiments suggested that Snu114 may trigger conformational changes of the spliceosome on GTP binding (Small et al. 2006) or hydrolysis (Bartels et al. 2003) to activate Brr2. However, in all the available spliceosome cryo-EM structures GTP is bound to Snu114, except for the S. pombe intron lariat spliceosome (ILS) structure, in which GDP was modelled (Yan et al. 2015). However, close inspection of the density suggests GTP may be a better fit. In EF-G the catalytic His places a water molecule near the GTP γ-phosphate. The equivalent His-218 in Snu114 is hydrogen-bonded to Tyr-403 in the Prp8 amino-terminal domain. Neither mutation of His-218 nor Tyr-403 affect yeast viability or growth, showing that Snu114-bound GTP is not hydrolyzed during the splicing cycle (Nguyen et al. 2016). In contrast, mutations in the P-loop of Snu114 which abolish GTP binding are lethal (Bartels et al. 2003) showing GTP binding is important. The ternary complex between Prp8, Snu114, and Aar2, coexpressed in yeast, also contains GTP (L Strittmatter, W Galej, and K Nagai, unpubl.) and it remains possible that GTP binding or hydrolysis plays a role during U5 snRNP biogenesis.
In the yeast tri-snRNP (Nguyen et al. 2016), the U6 ACAGAGA sequence, which pairs with the 5′SS sequence during activation (Lesser and Guthrie 1993; Kandels-Lewis and Séraphin 1993), is occluded by forming a short stem loop (ACAGAGA stem) to prevent premature interactions with the 5′SS (Nguyen et al. 2016). The structure of the human U4/U6.U5 tri-snRNP (Agafonov et al. 2016) is substantially different from its yeast counterpart and the Brr2–Prp8 Jab1 domain complex is anchored near Snu114 by Sad1, in which the conserved Brr2 PWI domain contacts the Sad1 protein (Agafonov et al. 2016). This interaction keeps the Brr2 helicase away from its U4 snRNA substrate to prevent premature unwinding of the U4/U6 snRNA duplex. A similar tri-snRNP conformation may exist in yeast, in which Sad1 transiently interacts with the tri-snRNP and is essential for splicing (Huang et al. 2014). The Prp28 DEAD-box helicase facilitates the disruption of the U1 snRNP–5′SS interaction (Staley and Guthrie 1999; Chen et al. 2001) and is bound to the Prp8 amino-terminal domain in the human tri-snRNP (Agafonov et al. 2016), whereas in the yeast tri-snRNP the B complex proteins replace Prp28 at the same binding site.
4. SPLICEOSOME ASSEMBLY
The U4/U6.U5 tri-snRNP associates with the A complex to form the fully assembled spliceosome, the pre-B complex (Fig. 3C) (Boesler et al. 2016). The U2, U5, and U6 snRNAs, which form the catalytic RNA core during activation, as well as the branching substrates (5′SS and BP) are thus brought together into one assembly. The pre-B complex forms in an ATP-independent manner and is held together predominantly by an RNA–RNA (U2/U6 helix II) interaction (Fig. 3C) (Will and Lührmann 2011). In the subsequent step, the helicase Prp28 facilitates 5′SS transfer from the U1 snRNP to the U6 snRNA ACAGAGA sequence, forming the so-called ACAGAGA helix (Kandels-Lewis and Séraphin 1993; Lesser and Guthrie 1993). In the presence of an inactive Prp28 mutant, the human splicing reaction stalls after pre-B complex formation and just before 5′SS transfer. Negative-stain electron microscopy of the Prp28-stalled human pre-B complex revealed a morphology distinct from the B complex, which forms after 5′SS transfer (Boesler et al. 2016). Thus, the U1 and U2 snRNPs may be dynamically associated with tri-snRNP in the pre-B complex. On addition of a 5′SS RNA oligonucleotide, the pre-B complex is transformed into a B complex-like structure, suggesting that dissociation of the U1 snRNP or pairing of the U6 ACAGAGA region with the 5′SS leads to a large conformational rearrangement in the human pre-B complex (Boesler et al. 2016). In the human tri-snRNP, Brr2 bound to the Prp8 Jab1 domain is kept inactive through its location near Snu114. In the yeast tri-snRNP, the Brr2 helicase is similarly bound to the Prp8 Jab1 domain (Mozaffari-Jovin et al. 2013; Nguyen et al. 2013) but is located near the Prp8 Endo domain and the single-stranded region of U4 snRNA is already loaded in the active site of the amino-terminal Brr2 helicase domain (Nguyen et al. 2015; 2016; Wan et al. 2016a). Several different mechanisms have been proposed to prevent premature unwinding of the U4/U6 snRNA duplex in the yeast tri-snRNP (Mozaffari-Jovin et al. 2013). In both the yeast and human B-complex structures (Bertram et al. 2017a; Plaschka et al. 2017), Brr2 is found in a similar location on the U4 snRNA substrate as in the isolated yeast tri-snRNP (Nguyen et al. 2015; 2016; Wan et al. 2016a), suggesting that spliceosome activation proceeds through similar mechanisms. The timing and mechanism of Sad1 release from human pre-B complex are currently unknown.
The pre-B complex is transformed into the B complex, following Prp28 helicase activity and U1 snRNP release. In the human B complex structure, which was stalled with low Mg2+ ion concentration, the U6 ACAGAGA box is paired with the 5′SS to accommodate the 5′ exon in the “exon channel” (Bertram et al. 2017a). In contrast, in the yeast B complex structure, stalled at low ATP concentration, the 5′SS instead interacts with U6 snRNA upstream of the ACAGAGA box, in agreement with cross-linking results (Sawa and Abelson 1992; Chan and Cheng 2005). This suggests that the yeast B complex was stalled at a slightly earlier stage than its human counterpart. In both B complex structures and in the yeast tri-snRNP (Nguyen et al. 2016) the so-called B complex proteins (Snu23, Prp38, and Spp381/human MFAP1) bind at the Prp8 amino-terminal domain to stabilize the 5′SS–U6 snRNA interactions and to guide Brr2 onto its U4 snRNA substrate (Fig. 3D). In the yeast B complex structure (Plaschka et al. 2017), the Snu23 amino terminus reaches near the active amino-terminal Brr2 helicase cassette, and this may couple the productive 5′SS-U6 snRNA interaction with Brr2-mediated spliceosome activation.
5. SPLICEOSOME ACTIVATION
The cryo-EM structures of yeast B and Bact complexes provide important insights into the mechanism of spliceosome activation, which results in the formation of the RNA catalytic core of the spliceosome (Fig. 3D) (Rauhut et al. 2016; Yan et al. 2016; Plaschka et al. 2017). During activation, U4 snRNA and its associated U4/U6 di-snRNP-specific and tri-snRNP-specific proteins dissociate, and the active spliceosome forms around the remaining U5 snRNP components together with the newly associating NTC and NTR proteins. Spliceosome activation begins with translocation of Brr2 along U4 snRNA in a 3′ to 5′ direction. This disrupts the U4/U6 stem I, and U4/U6 stem II may also be unwound, although this remains unclear (Laggerbauer et al. 1998; Raghunathan and Guthrie 1998). During this process, at least 24 proteins are released (U4 snRNP proteins, the LSm ring, Dib1, Snu66, and Prp6) and 22 proteins associate (RES, NTC and NTR complexes, and the Bact-specific proteins Cwc24, Cwc27). U6 snRNA, freed from U4 snRNA, forms an internal stem loop (ISL), whereas the adjacent U6 snRNA sequence pairs with U2 snRNA to form U2/U6 helix Ia and Ib (Madhani and Guthrie 1992), producing the active center of the spliceosome (Figs. 3D, 6A–D) (Madhani and Guthrie 1992; Steitz and Steitz 1993; Fica et al. 2013; Rauhut et al. 2016; Yan et al. 2016). Three consecutive triple base pairs (catalytic triplex) in the RNA-based active site coordinate the two catalytic magnesium ions (Fig. 6D) (Steitz and Steitz 1993; Fica et al. 2013; Yan et al. 2015). This RNA structure is reminiscent of the group II intron active site, consistent with the idea that both splicing systems arose from a common ancestor (Toor et al. 2008).
Figure 6.
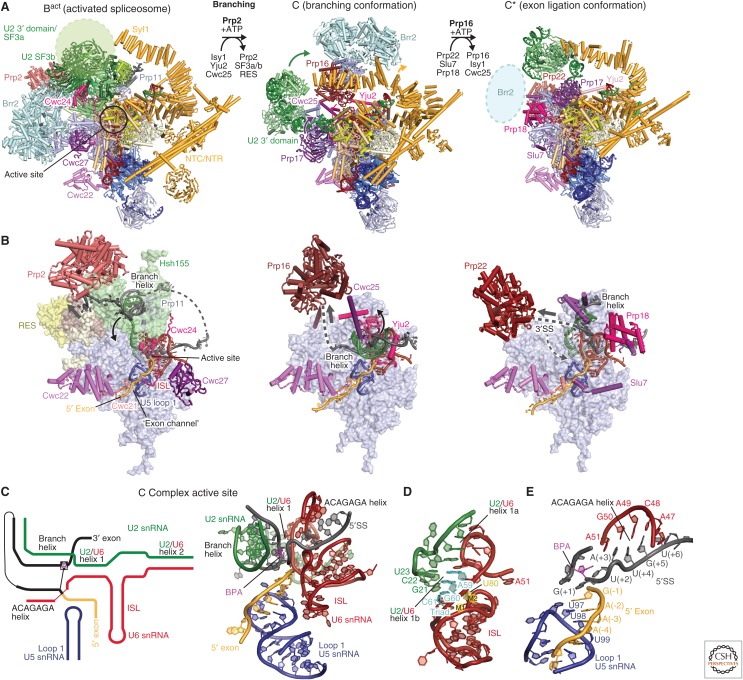
Branching and spliceosome remodeling. (A) Yeast Bact, C, and C* complex spliceosomes and step-specific factors are required to remodel the spliceosome between these states to move along the splicing pathway. The helicases Prp2 (salmon) and Prp16 (maroon) facilitate the remodeling from Bact to B* for branching and C to C* for exon ligation, respectively. Prp22 (light maroon) facilitates 3′SS proofreading in C* before exon ligation. The three structures, Bact (PDB 5GM6), C (PDB 5JL5), and C* (PDB 5MPS), are aligned using their Prp8 Large domains. (B) The location of the spliceosome active site, step-specific factors and the helicase-mediated remodeling in the Bact, C, and C* complex structures. Although the Bact and C complex structures show Prp2 (red) and Prp16 (dark red), respectively, poised for spliceosome remodeling, the C* structure, which lacks a “docked” 3′SS, reveals instead how Prp22 may contribute to 3′SS proofreading. Prp2 releases the branch helix from the U2 small nuclear ribonucleoprotein particle (snRNP) SF3b subcomplex, allowing it to bind in the active site for branching to occur. Prp16 subsequently undocks the branch helix together with step-specific factors Cwc25 and the Yju2 amino terminus, to allow docking of the 3′SS in C*. An arrow indicates the direction of RNA “translocation” (3′-5′ direction) of the helicases, which may remain bound to their respective locations and act at a distance (Semlow et al. 2016). (C) Secondary structure diagram (left) and cryo-electron microscopy (cryo-EM) structure (right) of the RNA-based active site in C complex (PDB 5JL5). This active site conformation is characteristic of all catalytic stage complexes, except for the precise position of the branch helix and remodeling of the ACAGAGA helix. (D) Structure of the U6 small nuclear RNA (snRNA) internal stem loop (ISL) and U2/U6 helix I in C complex. The U2/U6 “catalytic” triad (cyan) and the U6 residue U80 (yellow) coordinate the two catalytic metal ions, which are essential for splicing (M1 and M2, not modeled). (E) Recognition of the precursor messenger RNA (pre-mRNA) 5′ splice site (5′SS) and 5′ exon in the C complex (PDB 5JL5). These interactions remain almost unchanged throughout the catalytic splicing stages.
In the Bact structure, the catalytic RNA core forms and is stabilized by the NTR/NTC proteins Cwc2, Bud31, Cef1, Ecm2, Clf1, and Syf2 (Rauhut et al. 2016; Yan et al. 2016). The structure of the catalytic RNA core “enzyme” remains unchanged throughout the catalytic phase of the splicing cycle (Fica and Nagai 2017; Shi 2017). The ACAGAGA helix, formed between the U6 snRNA ACAGAGA box and the GUAUGU sequence at the 5′ end of the intron, positions the exon–intron junction in a curved conformation to expose the scissile phosphate for nucleophilic attack by the BP adenosine (Fig. 6E). In the Bact structure (Rauhut et al. 2016; Yan et al. 2016), the BP adenosine is located 50 Å away from the 5′SS, and the SF3a subunit Prp11 and Bact-specific protein Cwc24 together protect the scissile bond at the 5′SS (Fig. 6A,B). The 5′ exon base-pairs with the conserved U5 snRNA loop 1 (Newman and Norman 1992; Sontheimer and Steitz 1993). The adjacent 5′ exon is loaded in the “exon channel” between the Prp8 amino-terminal and Large domains of Prp8, where it is stabilized by Cwc21 and Cwc22 and clamped by the Prp8 amino-terminal and Large domains.
The gap between the Prp8 amino-terminal and Large domains is wider in the B complex such that the 5′ exon can slide into this gap to replace the 5′ end of U6 snRNA (Plaschka et al. 2017). In the Bact complex, the gap between the two Prp8 domains closes, presumably as the result of NTC and NTR protein binding, and the switch loop of Prp8 (residues 1615–1639) together with Cwc21 fills the gap to clamp the 5′ exon between the two Prp8 domains (Rauhut et al. 2016; Yan et al. 2016) until the exon-ligation reaction is complete (Figs. 3D, 5A, 6B) (Bai et al. 2017; Liu et al. 2017; Wilkinson et al. 2017). In yeast, the GUAUGU sequence at the 5′ end of the intron is stringently conserved and the UGU within this sequence forms Watson–Crick base pairs with ACA of the ACAGAGA box (Kandels-Lewis and Séraphin 1993; Lesser and Guthrie 1993) and nucleates formation of the flanking non-Watson–Crick base pairs (Galej et al. 2016; Wan et al. 2016b). In humans, the 5′SS sequence is much more degenerate and only the G/GU sequence is stringently conserved. Therefore, in humans the pairing between the 5′SS and the ACAGAGA box (Will and Lührmann 2011) is unlikely to be driven by Watson–Crick pairs. This suggests that the U1 snRNP delivers the bound 5′SS directly to its site of interaction with U6, to load the 5′ exon into the gap between the amino-terminal and Large domains of Prp8.
The structure of the complete U2 snRNP was determined as part of the yeast B complex (Plaschka et al. 2017), comprising the SF3a/b subcomplexes and the U2 3′ domain, and this organization is unchanged in the Bact structure (Fig. 2). The U2 snRNP SF3b subcomplex consists of Rse1, containing three β-propeller domains, and the HEAT-repeat protein Hsh155, which together surround Ysf3 (Rcp10) and the Zn-containing Rds3 protein. Cus1 and Hsh49 associate on the periphery, to form the complete SF3b complex (Plaschka et al. 2017). The branch helix, which initially forms in the A complex between the pre-mRNA BP sequence and U2 snRNA, wedges into the split ends of the circular Hsh155 HEAT repeats within SF3b (Rauhut et al. 2016; Yan et al. 2016; Plaschka et al. 2017). The nucleotide base of the BP adenosine is flipped out from the branch helix and makes contact with Tyr35 of Rds3 and a pocket formed by Hsh155 HEAT repeats H15–H17.
Although the organization of the U2 snRNP SF3b complex in the B and Bact complexes is unchanged, the U2 snRNP undergoes a major repositioning during spliceosome activation. Comparisons between the B and Bact structures suggest that this large movement of the U2 snRNP is mediated by the energetically favorable folding of the U6 snRNA and remodeling of the U2 snRNA from the stem IIa/b to IIb/c conformation (Fig. 3D) (Hilliker et al. 2007; Perriman and Ares 2007). In the B complex, the U2 and U5 snRNPs are linked primarily through U6 snRNA, in particular through U2/U6 helix II. The U2 snRNA region that forms U2/U6 helix Ia and Ib as well as the Prp11 loop that folds over the 5′SS are disordered in the B complex, but become ordered in Bact. This suggests that as the RNA core forms during spliceosome activation, the U2 snRNP is pulled via the branch helix toward the active site, where it is held in place by association of the NTC and NTR complexes. Hence, although the helicase Brr2 is needed to overcome an initial barrier for spliceosome activation, the subsequent conformational remodeling is driven by U6 snRNA folding and new protein–RNA contacts within the spliceosome. The U2 3′ domain is held at one end of the Syf1/Clf1 arch and the other end is anchored by Syf2 near the U2/U6 stem 1. The solenoid structure of Syf1/Clf1 allows a large movement of the U2 3′ domain during different catalytic stages of the spliceosome while it is suspended above the active site (Fica and Nagai 2017).
After formation of the RNA-based active site in Bact, the BP adenosine is held 50 Å away from the active site. The branch helix must subsequently be inserted into the active site to enable the nucleophilic attack of the 2′ hydroxyl group of the BP adenosine at the scissile phosphate of the 5′SS. The Prp2 helicase is required to remodel Bact into this branching conformation, characteristic of the B* and C complexes. In the Bact structure, the intron sequence downstream from the BP winds through the SF3b and RES complexes to reach Prp2 (Warkocki et al. 2015; Rauhut et al. 2016; Yan et al. 2016). As Prp2 translocates along the intron in a 3′ to 5′ direction, the SF3a/b and RES complexes dissociate from the branch helix. This allows the Prp16 helicase and the branching factors Yju2, Cwc25, and Isy1 to bind (Fica and Nagai 2017).
6. STEP I (LARIAT FORMATION)
The structure of the yeast C complex was determined to high-resolution, revealing the spliceosome just after the branching reaction (Fig. 6A,B) (Galej et al. 2016; Wan et al. 2016b). Although the B* complex structure has not been determined so far, it is likely to closely resemble the C complex conformation. In Bact, the 3′ domain of U2 snRNP, comprising the Sm ring and the Msl1/Lea1 complex, binds the SF3a subcomplex, but in the C complex SF3a has dissociated and the U2 3′ domain bridges between the Prp8 RNaseH domain and the amino-terminal end of the Syf1 HAT domain. Thus, the U2 3′ domain may also be a sensor for spliceosome conformation. Upon release of the SF3b complex from Bact, U2 snRNA adopts the stem IIb/c configuration (Hilliker et al. 2007; Perriman and Ares 2007). Remarkably, the products of the branching reaction, the free 5′ exon and the lariat intron intermediate, remain close to the catalytic metal ions in the C complex structure (Galej et al. 2016; Wan et al. 2016b). This arrangement is consistent with the catalytic magnesium ion coordination inferred from metal rescue experiments (Fica et al. 2013). The 5′ phosphate of the first intron nucleotide (G+1) forms a 2′−5′ phosphodiester linkage with the 2′-hydroxyl group of the branch point adenosine. The branch helix is accommodated in the active site cleft together with the step 1 factors, Cwc25, Yju2, and Isy1. Although the branch helix is in the canonical A-form in B and Bact complexes, in C complex the branch helix is highly distorted by the associated branching factors and thereby fits into the active site cleft (Galej et al. 2016). A long amino-terminal α-helix of Cwc25 interacts with the Prp8 RNaseH domain and the now unfolded Prp8 α-finger region and its amino terminus penetrates into the widened major groove of the branch helix. The Yju2 amino terminus wraps around the branch helix and makes intimate contacts with the 5′ exon. The Isy1 amino terminus also extends into the active site and interacts with the branch helix and U(+2) of the intron. In B and Bact complexes, the BP adenosine base is flipped out from the branch helix to interact with the surrounding SF3b proteins, whereas in the C complex the BP adenosine base is hydrogen bonded to intron nucleotide U68 (−2 from the BP), creating an unusual backbone conformation that allows the 2′-hydroxyl group of the BP adenosine to project toward the 5′SS for branching to occur.
The ATPase activity of Prp16 is essential for remodeling of the spliceosome for exon ligation (step 2). The structure by Wan et al. (2016b) lacks Prp16 whereas the position of Prp16 and Brr2 was revealed by focused classification in the C complex structure by Galej et al. (2016) providing structural insight into the Prp16-induced remodeling of the spliceosome. The sixth nucleotide downstream from the BP emerges from the spliceosome and the distance between the last ordered nucleotide and the Prp16 active site is consistent with Prp16–pre-mRNA crosslinks eighteen nucleotides downstream from the BP (Semlow et al. 2016). Prp16 is positioned near Prp8, Cwc25, and Yju2, in which the intron downstream from the BP emerges from the spliceosome. Prp16 translocates along the intron in a 3′ to 5′ direction on ATP hydrolysis, to undock the branch helix from the active site and to release the branching factors. The structure of human C complex was reported recently and reveals a similar arrangement of Prp16 showing that the remodeling mechanism is conserved in the human spliceosome (Zhang et al. 2018).
7. SPLICEOSOME REMODELING
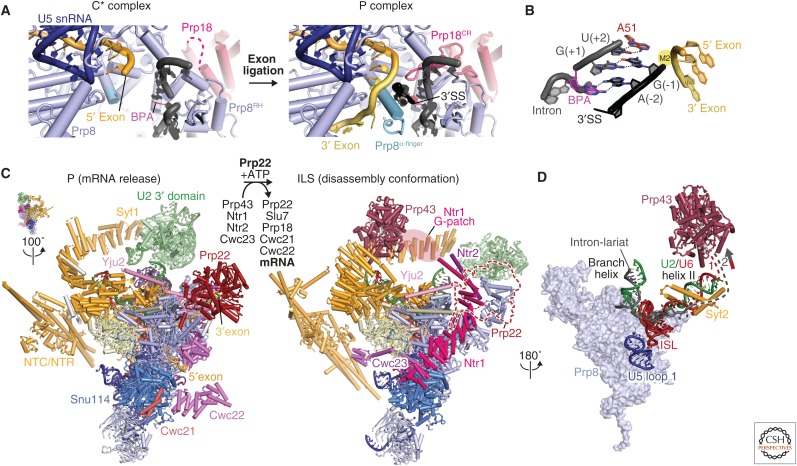
Prp16 remodels the spliceosome from the branching (C complex) to the exon-ligation conformation (C* complex). The yeast C* structures (Fig. 6A,B) (Fica et al. 2017; Yan et al. 2017) confirm earlier observations that the spliceosome indeed harbors a single RNA-based active site (Fica et al. 2013). The 5′ exon participates in both branching and exon ligation and is held firmly in the exon channel for both catalytic steps. In contrast, the branch helix has to vacate the active site after branching, removing the BP, to allow the 3′SS to enter the active site for exon ligation. In the C* complex structure, Prp16 and the branching factors have dissociated, allowing the distorted branch helix to spring back into the canonical A form and to rotate 75° away from the active site. The step 2 (exon-ligation) factors, Prp18 and Slu7, bind to the Prp8 RNaseH domain and associate with the spliceosome together with the Prp22 helicase. The C to C* transition is accompanied by a rotation of the Prp8 RNaseH domain and a large movement of the Prp17 WD40 domain. The β-finger of the rotated RNaseH domain extends toward Cef1, and Prp17 bridges their interaction to lock the branch helix in its new orientation. This conformational change creates a binding site for the incoming 3′ exon. The conserved U5 snRNA loop 1 was shown genetically and biochemically (Newman and Norman 1992; Sontheimer and Steitz 1993) to align the 5′ and 3′ exons during exon ligation. As in the C complex the Prp8 RNaseH domain interacts with the 3′ domain of the U2 snRNP, which in turn interacts with one end of the HAT domain of Syf1. Hence, the rotation of the Prp8 RNaseH domain is accompanied by a large movement of the U2 3′ domain and the HAT domains of Syf1 and Clf1. The C* complex structures do not reveal how the 3′ exon docks into the active site before exon ligation. 3′ exon docking may be very transient and coincide with exon ligation, making its structural observation challenging. However, insights into 3′SS selection have been gained from the postsplicing P complex structure (see below). The binding of an inositol hexaphosphate molecule (IP6) to Prp8 was first documented in the C* complex (PDB 5MPS) but putative IP6 density is present in all yeast and human catalytic stage spliceosome structures (Bact, C, C*, and P and ILS complexes), but not in free Prp8, tri-snRNP, or B complex. In the C* and P complexes, Slu7 contacts the IP6 molecule, suggesting a role in the stabilization of this exon-ligation factor (Fig. 4A).
8. STEP II (EXON LIGATION)
After 3′SS docking in C*, the exons are ligated and the product mRNA released. This depends on the ATPase activity of the DEAH-box helicase Prp22, which carries out 3′SS proofreading as well as mRNA release (Company et al. 1991; Schwer 2008). The cryo-EM structure of the postsplicing complex (P complex) was recently determined by three groups, yielding new insights into the exon-ligation reaction (Fig. 7A–C). All three groups used ATPase-deficient mutants of Prp22, which are unable to release the ligated exon (Bai et al. 2017; Liu et al. 2017; Wilkinson et al. 2017). In the structure, the ligated mRNA and the lariat intron, cleaved at the 3′SS, are bound in the active site (Fig. 7A). The 3′-hydroxyl group of the last intron nucleotide is close to the phosphorus atom at the exon junction, showing that these atoms have not moved significantly after exon ligation. The invariant intron nucleotides (YAG where Y denotes pyrimidine) immediately preceding the 3′ splice junction dock into the active site and are specifically recognized (Fig. 7B). The Hoogsteen edge of the 3′SS G(−1) forms a base pair with the Watson–Crick edge of the first intron nucleotide G(+1) consistent with the analysis of Parker and Siliciano (1993). In two of the P complex structures (Liu et al. 2017; Wilkinson et al. 2017), the 3′SS A(−2) forms a symmetric Hoogsteen–Hoogsteen base pair with the BP adenosine, which is linked via its 2′-oxygen to the 5′SS G(+1). This is consistent with the stringent conservation of the penultimate A(−2) at the 3′SS. In contrast, in the P complex structure reported by Bai et al. (2017), A(−2) has a different geometry and does not appear to form a Hoogsteen–Hoogsteen base pair with the BP adenosine. In these structures, all invariant nucleotides at the 5′SS and 3′SS and BP come together and hydrogen bond with each other for exon ligation. The exon-ligation reaction could not occur unless the 5′SS and 3′SS substrates are precisely positioned by this network of hydrogen bonds, accounting for the stringent conservation of the GU and AG dinucleotides at the 5′SS and 3′SS and the identity of the branch nucleotide. In the P complex structures of Liu et al. (2017) and Wilkinson et al. (2017), the highly conserved Gln1594 forms a hydrogen bond with the O2 carbonyl group of U(−3). Cytosine at the −3 position could form an equivalent hydrogen bond accounting for the conservation of pyrimidines at −3 of the 3′SS. In the spliceosome, the intron folds back to bring the 3′SS and branch site into the active site. Interestingly, Liu et al. (2017) show that in a large number of pre-mRNAs, part of the intron between the BP sequence and the 3′SS may base-pair with itself, which may facilitate the insertion of the 3′ exon into the active site.
Figure 7.
Exon ligation and spliceosome disassembly. (A) Comparison of the C* and P complex active sites. The extended Prp8 α-finger (light blue) and the Prp18 conserved region (CR) (pink) stabilize 3′SS docking in the active site. This positions the 3′-OH group of the 5′ exon to attack at the 3′SS for exon ligation (PDB 6EXN). (B) Recognition of the conserved YAG sequence at the 3′SS. The last intron nucleotide, G(−1), forms a non-Watson–Crick base pair with the first intron nucleotide G(+1) and the penultimate intron nucleotide A(–2) forms a symmetric Hoogsteen–Hoogsteen base pair with the branch point adenosine (PDB 6EXN). The third last pyrimidine (Y) is recognized by Gln-1594 of Prp8. (C) Transition from the P (PDB 6EXN) to ILS (intron lariat spliceosome) (PDB 5Y88). Prp22-mediated ligated-exon release facilitates the binding of Prp43/Ntr1/Ntr2/Cwc23 and the release of the step 2 factors (Prp18 and Slu7) and Cwc21 and Cwc22, which interact with 5′ exon. (D) Prp43 induces disassembly of ILS by pulling on either the intron or U6 small nuclear RNA (snRNA) (PDB 5Y88).
A network of protein–RNA interactions cradles the P complex active site, facilitating the exon-ligation reaction. Helix 8 in the α-finger region (residues 1570–1615) of the Prp8 Linker domain is extended in the P complex compared with the C* complex (Fica et al. 2017; Yan et al. 2017) and before exon ligation the intron-3′ exon nucleotides wrap around this extended α-helix to project the scissile phosphate toward the 3′ terminal hydroxyl group of the 5′ exon. The α-finger residue Arg1604 contacts the phosphate backbone of the 3′SS residues −3 and −4 and the mutation of this residue to Ala abolishes the exon-ligation reaction showing that Arg1604 is crucial in positioning the 3′SS residues in the active site. The step 2 factors, Prp18 and Slu7, are essential for exon ligation, but only the conserved region of Prp18 inserts into the active site and stabilizes the binding of the 3′SS (Liu et al. 2017; Wilkinson et al. 2017). Slu7 extends over a large surface of the spliceosome, and although its precise role in exon ligation is still unclear, it was proposed to assist with guiding the 3′SS into the active site when the distance between the branch point and the 3′SS exceeds nine nucleotides (Schwer 2008; Ohrt et al. 2013; Fica et al. 2017).
The study by Wilkinson et al. (2017) revealed an “undocked” state of the 3′SS in the P complex, in which the exons are ligated but the 3′SS is not docked in the active site. This provides insights into the mechanism of 3′SS discrimination and kinetic proofreading by the Prp22 helicase. Cognate and noncognate 3′SS sequences could potentially both enter the spliceosome active site but only the cognate 3′SS will induce the “docked” conformation, where the 3′SS, BP and 5′SS cooperatively pair with each other to align the 3′SS for nucleophilic attack by the 3′-hydroxyl group of the 5′ exon. This allows the exon ligation reaction to occur and Prp22 activity would subsequently release the ligated mRNA (Schwer 2008). However, if a noncognate 3′SS docks and does not induce the docked conformation to ligate within a “proofreading” window, then Prp22 would pull on the intron to remove the noncognate 3′SS from the active site and allow sampling of a new 3′SS (Mayas et al. 2010).
9. SPLICEOSOME DISASSEMBLY AND RECYCLING
After exon ligation, the ligated exon (mRNA) is released from the spliceosome and the remaining ILS is disassembled to recycle its snRNA and protein components for subsequent splicing rounds. The DEAH-box helicase Prp43 participates in this process (Arenas and Abelson 1997) and forms a stable complex with two NTC interacting proteins, Ntr1 and Ntr2 (Tsai et al. 2005). The purified Prp43-Ntr1-Ntr2 complex can catalyze the disassembly of ILS into U2, U5, U6, and NTC in an ATP-dependent manner. Prp43 interacts with the Ntr1 amino-terminal G-patch domain and this interaction is required to stimulate Prp43 ATPase activity (Tsai et al. 2005; Tanaka et al. 2007). It was suggested that Brr2 helicase is also involved in the disassembly of ILS (Small et al. 2006), yet recent in vitro experiments suggest that ILS disassembly does not depend on Brr2 helicase activity (Fourmann et al. 2016).
The cryo-EM structure of the S. cerevisiae ILS (Wan et al. 2017) has provided mechanistic insight into the disassembly process (Fig. 7C,D). In agreement with biochemical data (Fourmann et al. 2016), Cef1, Clf1, and Syf2 (NTC proteins) and Cwc2, Cwc15, Bud31, Ecm2, Prp45, and Prp46 (NTR proteins) remain bound to the U5 snRNP in the ILS, cradling the catalytic RNA core as in Bact (Rauhut et al. 2016; Yan et al. 2016), C (Galej et al. 2016; Wan et al. 2016b), C* (Fica et al. 2017; Yan et al. 2017), and P (Bai et al. 2017; Liu et al. 2017; Wilkinson et al. 2017) complexes. In contrast, Prp22, Cwc21, and Cwc22 have dissociated from the spliceosome. Prp22 likely dissociates together with the mRNA from the P complex, and this would release Cwc21 and Cwc22 from the exon channel. In metazoans, the exon-junction complex (EJC) assembles on Cwc22 to bind the mRNA, and also dissociates with the mRNA in P complex. This frees several spliceosome surfaces that allow the Prp43-Ntr1-Ntr2 complex to associate. In the cryo-EM structure of the S. cerevisiae ILS, Prp43 is bound to Syf1, near U2/U6 snRNA helix II. Ntr1 is anchored to Snu114 with its carboxy-terminal domain and the Ntr1 super-helical domain spans almost the entire length of the spliceosome, bridging between Prp43 and Snu114, which may serve as a marker for a disassembling spliceosome. The Ntr1 amino-terminal G patch domain, which binds Prp43 (Tsai et al. 2005), was not identified in the cryo-EM map. Although there is no RNA density bound to Prp43, it is located near the U6 snRNA 3′ end and the intron, suggesting that either of these RNAs may be the functional substrate of Prp43 (Fig. 7D). Similar to Prp16 in C complex, and Prp22 in C*, Prp43 is likely to pull on U6 snRNA or the intron to disassemble the ILS. No density was observed for Brr2 in the ILS structure, suggesting that it is mobile.
Unlike its S. cerevisiae counterpart, whole-cell extract from S. pombe does not support in vitro splicing, apparently because an endogenous U2/U6.U5 complex accumulates. This complex may correspond to another ILS-like intermediate. The cryo-EM structure of the S. pombe ILS (Yan et al. 2015) shows that its catalytic RNA core is very similar to its yeast counterpart, except that it lacks the Prp43-Ntr1-Ntr2 complex and Brr2, in agreement with earlier proteomic studies. Interestingly, the carboxy-terminal domain of Cwf19 (Cwf19L2 in human) is wedged between the Prp8 Large and RNaseH domains in the S. pombe ILS. The carboxy-terminal domain of Cwf19 shows significant similarity to the carboxy-terminal domain of S. cerevisiae protein Drn1 whose amino-terminal domain is known to interact with the debranching enzyme Dbr1. It is possible that Drn1 binds to Prp8 in P complex and recruits Dbr1 to the lariat intron after the mRNA and step 2 factors dissociate from the complex. Alternatively, the S. pombe ILS may represent a novel and distinct dead-end complex, which explains why S. pombe splicing extract is inactive.
10. CONCLUSION
Since 2015, cryo-EM studies of the spliceosome have provided snapshots of splicing in its key steps. These have advanced our structural and mechanistic knowledge of the underlying reaction, and build on years of genetic, biochemical, and structural studies of the spliceosome, confirming the pioneering work by many laboratories, and leading to new molecular insights that can be tested in the future. The most important conclusions can be summarized in the following points.
The active site of the spliceosome is an RNA-based metalloenzyme cradled by many proteins.
A single active site performs the two trans-esterification reactions: branching and exon ligation.
The catalytic RNA core forms during the transition from the B to Bact complex and remains unchanged until the spliced mRNA is released.
RNA helicases play crucial roles not only in generating the active RNA core but also in docking and undocking the substrates (BP adenosine and the 3′SS) together with step-specific factors. By doing so, these helicases play a crucial role in proofreading the splice sites.
The spliceosome shows a remarkable structural and mechanistic conservation between yeast and human systems (Bertram et al. 2017b; Fica et al. 2017; Yan et al. 2017; Zhang et al. 2017).
ACKNOWLEDGMENTS
We thank the past and present members of the Nagai group for their contribution to our spliceosome project and critical reading of the manuscript. We are particularly grateful to Kelly Nguyen, Wojtek Galej, Sebastian Fica, Pei-Chun Lin, Max Wilkinson, Chris Oubridge, Lisa Strittmatter, Clément Charenton, and Chris Norman. We apologize for the omission of many important references because of the restriction on the number of references. Our work was supported by the Medical Research Council (MC_U105184330) and European Research Council Advanced Grant (AdG-693087-SPLICE3D). C.P. was supported by a European Molecular Biology Organization (EMBO) fellowship.
Footnotes
Editors: Thomas R. Cech, Joan A. Steitz, and John F. Atkins
Additional Perspectives on RNA Worlds available at www.cshperspectives.org
REFERENCES
* Reference is in this collection.
- Agafonov DE, Kastner B, Dybkov O, Hofele RV, Liu WT, Urlaub H, Lührmann R, Stark H. 2016. Molecular architecture of the human U4/U6.U5 tri-snRNP. Science 351: 1416–1420. [DOI] [PubMed] [Google Scholar]
- Arenas JE, Abelson JN. 1997. Prp43: An RNA helicase-like factor involved in spliceosome disassembly. Proc Natl Acad Sci 94: 11798–11802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai R, Yan C, Wan R, Lei J, Shi Y. 2017. Structure of the post-catalytic spliceosome from Saccharomyces cerevisiae. Cell 171: 1589–1598. [DOI] [PubMed] [Google Scholar]
- Bartels C, Urlaub H, Lührmann R, Fabrizio P. 2003. Mutagenesis suggests several roles of Snu114p in pre-mRNA splicing. J Biol Chem 278: 28324–28334. [DOI] [PubMed] [Google Scholar]
- Berget SM, Moore C, Sharp PA. 1977. Spliced segments at the 5′ terminus of adenovirus 2 late mRNA. Proc Natl Acad Sci 74: 3171–3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berglund JA, Abovich N, Rosbash M. 1998. A cooperative interaction between U2AF65 and mBBP/SF1 facilitates branchpoint region recognition. Genes Dev 12: 858–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram K, Agafonov DE, Dybkov O, Haselbach D, Leelaram MN, Will CL, Urlaub H, Kastner B, Lührmann R, Stark H. 2017a. Cryo-EM structure of a pre-catalytic human spliceosome primed for activation. Cell 170: 701–713. [DOI] [PubMed] [Google Scholar]
- Bertram K, Agafonov DE, Liu WT, Dybkov O, Will CL, Hartmuth K, Urlaub H, Kastner B, Stark H, Lührmann R. 2017b. Cryo-EM structure of a human spliceosome activated for step 2 of splicing. Nature 542: 318–323. [DOI] [PubMed] [Google Scholar]
- Boesler C, Rigo N, Anokhina MM, Tauchert MJ, Agafonov DE, Kastner B, Urlaub H, Ficner R, Will CL, Lührmann R. 2016. A spliceosome intermediate with loosely associated tri-snRNP accumulates in the absence of Prp28 ATPase activity. Nat Commun 7: 11997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brody E, Abelson J. 1985. The “spliceosome”: Yeast pre-messenger RNA associates with a 40S complex in a splicing-dependent reaction. Science 228: 963–967. [DOI] [PubMed] [Google Scholar]
- Burgess SM, Guthrie C. 1993. A mechanism to enhance mRNA splicing fidelity: The RNA dependent ATPase Prp16 governs usage of a discard pathway for aberrant lariat intermediates. Cell 73: 1377–1391. [DOI] [PubMed] [Google Scholar]
- Chan SP, Cheng SC. 2005. The Prp19-associated complex is required for specifying interactions of U5 and U6 with pre-mRNA during spliceosome activation. J Biol Chem 280: 31190–31199. [DOI] [PubMed] [Google Scholar]
- Chen JY, Stands L, Staley JP, Jackups RR Jr, Latus LJ, Chang TH. 2001. Specific alterations of U1-C protein or U1 small nuclear RNA can eliminate the requirement of Prp28p, an essential DEAD box splicing factor. Mol Cell 7: 227–232. [DOI] [PubMed] [Google Scholar]
- Chow LT, Gelinas RE, Broker TR, Roberts RJ. 1977. An amazing sequence arrangement at the 5′ ends of adenovirus 2 messenger RNA. Cell 12: 1–8. [DOI] [PubMed] [Google Scholar]
- Company M, Arenas J, Abelson J. 1991. Requirement of the RNA helicase-like protein PRP22 for release of messenger RNA from spliceosomes. Nature 349: 487–493. [DOI] [PubMed] [Google Scholar]
- Fabrizio P, Laggerbauer B, Lauber J, Lane WS, Lührmann R. 1997. An evolutionarily conserved U5 snRNP-specific protein is a GTP-binding factor closely related to the ribosomal translocase EF-2. EMBO J 16: 4092–4106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Leiro R, Scheres SH. 2016. Unravelling biological macromolecules with cryo-electron microscopy. Nature 537: 339–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fica SM, Nagai K. 2017. Cryo-electron microscopy snapshots of the spliceosome: Structural insights into a dynamic ribonucleoprotein machine. Nat Struct Mol Biol 24: 791–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fica SM, Tuttle N, Novak T, Li NS, Lu J, Koodathingal P, Dai Q, Staley JP, Piccirilli JA. 2013. RNA catalyses nuclear pre-mRNA splicing. Nature 503: 229–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fica SM, Oubridge C, Galej WP, Wilkinson ME, Bai XC, Newman AJ, Nagai K. 2017. Structure of a spliceosome remodelled for exon ligation. Nature 542: 377–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fourmann JB, Dybkov O, Agafonov DE, Tauchert MJ, Urlaub H, Ficner R, Fabrizio P, Lührmann R. 2016. The target of the DEAH-box NTP triphosphatase Prp43 in Saccharomyces cerevisiae spliceosomes is the U2 snRNP-intron interaction. eLife 5: e15564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galej WP, Oubridge C, Newman AJ, Nagai K. 2013. Crystal structure of Prp8 reveals active site cavity of the spliceosome. Nature 493: 638–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galej WP, Wilkinson ME, Fica SM, Oubridge C, Newman AJ, Nagai K. 2016. Cryo-EM structure of the spliceosome immediately after branching. Nature 537: 197–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottschalk A, Tang J, Puig O, Salgado J, Neubauer G, Colot HV, Mann M, Séraphin B, Rosbash M, Lührmann R, et al. 1998. A comprehensive biochemical and genetic analysis of the yeast U1 snRNP reveals five novel proteins. RNA 4: 374–393. [PMC free article] [PubMed] [Google Scholar]
- Grainger RJ, Beggs JD. 2005. Prp8 protein: At the heart of the spliceosome. RNA 11: 533–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilliker AK, Mefford MA, Staley JP. 2007. U2 toggles iteratively between the stem IIa and stem IIc conformations to promote pre-mRNA splicing. Genes Dev 21: 821–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YH, Chung CS, Kao DI, Kao TC, Cheng SC. 2014. Sad1 counteracts Brr2-mediated dissociation of U4/U6.U5 in tri-snRNP homeostasis. Mol Cell Biol 34: 210–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandels-Lewis S, Séraphin B. 1993. Involvement of U6 snRNA in 5′ splice site selection. Science 262: 2035–2039. [DOI] [PubMed] [Google Scholar]
- *.Kastner B, Will CL, Stark H, Lührmann R. 2019. Structural insights into nuclear pre-mRNA splicing in higher eukaryotes. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a032417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo Y, Oubridge C, van Roon AM, Nagai K. 2015. Crystal structure of human U1 snRNP, a small nuclear ribonucleoprotein particle, reveals the mechanism of 5′ splice site recognition. eLife 4 10.7554/eLife.04986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laggerbauer B, Achsel T, Lührmann R. 1998. The human U5-200kD DEXH-box protein unwinds U4/U6 RNA duplices in vitro. Proc Natl Acad Sci 95: 4188–4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner MR, Steitz JA. 1979. Antibodies to small nuclear RNAs complexed with proteins are produced by patients with systemic lupus erythematosus. Proc Acad Natl Sci 76: 5495–5499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesser CF, Guthrie C. 1993. Mutations in U6 snRNA that alter splice site specificity: Implications for the active site. Science 262:1982–1988. [DOI] [PubMed] [Google Scholar]
- Li X, Liu S, Jiang J, Zhang L, Espinosa S, Hill RC, Hansen KC, Zhou ZH, Zhao R. 2017. CryoEM structure of Saccharomyces cerevisiae U1 snRNP offers insight into alternative splicing. Nat Commun 8: 1035–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang WW, Cheng SC. 2015. A novel mechanism for Prp5 function in prespliceosome formation and proofreading the branch site sequence. Genes Dev 29: 81–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Li X, Zhang L, Jiang J, Hill RC, Cui Y, Hansen KC, Zhou ZH, Zhao R. 2017. Structure of the yeast spliceosomal postcatalytic P complex. Science 358: 1278–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhani HD, Guthrie C. 1992. A novel base-pairing interaction between U2 and U6 snRNAs suggests a mechanism for the catalytic activation of the spliceosome. Cell 71: 803–817. [DOI] [PubMed] [Google Scholar]
- Mayas RM, Maita H, Semlow DR, Staley JP. 2010. Spliceosome discards intermediates via the DEAH box ATPase Prp43p. Proc Natl Acad Sci 107: 10020–10025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozaffari-Jovin S, Wandersleben T, Santos KF, Will CL, Lührmann R, Wahl MC. 2013. Inhibition of RNA helicase Brr2 by the C-terminal tail of the spliceosomal protein Prp8. Science 341: 80–84. [DOI] [PubMed] [Google Scholar]
- Newman AJ, Norman C. 1992. U5 snRNA interacts with exon sequences at 5′ and 3′ splice sites. Cell 68: 743–754. [DOI] [PubMed] [Google Scholar]
- Nguyen TH, Li J, Galej WP, Oshikane H, Newman AJ, Nagai K. 2013. Structural basis of Brr2-Prp8 interactions and implications for U5 snRNP biogenesis and the spliceosome active site. Structure 21: 910–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen TH, Galej WP, Bai XC, Savva CG, Newman AJ, Scheres SH, Nagai K. 2015. The architecture of the spliceosomal U4/U6.U5 tri-snRNP. Nature 523: 47–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen THD, Galej WP, Bai XC, Oubridge C, Newman AJ, Scheres SHW, Nagai K. 2016. Cryo-EM structure of the yeast U4/U6.U5 tri-snRNP at 3.7 Å resolution. Nature 530: 298–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohrt T, Odenwälder P, Dannenberg J, Prior M, Warkocki Z, Schmitzová J, Karaduman R, Gregor I, Enderlein J, Fabrizio P, et al. 2013. Molecular dissection of step 2 catalysis of yeast pre-mRNA splicing investigated in a purified system. RNA 19: 902–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker R, Siliciano PG. 1993. Evidence for an essential non-Watson–Crick interaction between the first and last nucleotides of a nuclear pre-mRNA intron. Nature 361: 660–662. [DOI] [PubMed] [Google Scholar]
- Parker R, Siliciano PG, Guthrie C. 1987. Recognition of the TACTAAC box during mRNA splicing in yeast involves base pairing to the U2-like snRNA. Cell 49: 229–239. [DOI] [PubMed] [Google Scholar]
- Perriman RJ, Ares M Jr. 2007. Rearrangement of competing U2 RNA helices within the spliceosome promotes multiple steps in splicing. Genes Dev 21: 811–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perriman R, Ares M Jr. 2010. Invariant U2 snRNA nucleotides form a stem loop to recognize the intron early in splicing. Mol Cell 38: 416–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaschka C, Lin PC, Nagai K. 2017. Structure of a pre-catalytic spliceosome. Nature 546: 617–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaschka C, Lin PC, Charenton C, Nagai K. 2018. Prespliceosome structure provides insights into spliceosome assembly and regulation. Nature doi: 10.1038/nature. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomeranz Krummel DA, Oubridge C, Leung AK, Li J, Nagai K. 2009. Crystal structure of human spliceosomal U1 snRNP at 5.5 Å resolution. Nature 458: 475–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghunathan PL, Guthrie C. 1998. RNA unwinding in U4/U6 snRNPs requires ATP hydrolysis and the DEIH-box splicing factor Brr2. Curr Biol 8: 847–855. [DOI] [PubMed] [Google Scholar]
- Rauhut R, Fabrizio P, Dybkov O, Hartmuth K, Pena V, Chari A, Kumar V, Lee CT, Urlaub H, Kastner B, Lührmann R, et al. 2016. Molecular architecture of the Saccharomyces cerevisiae activated spliceosome. Science 353: 1399–1405. [DOI] [PubMed] [Google Scholar]
- Sawa H, Abelson J. 1992. Evidence for a base-pairing interaction between U6 small nuclear RNA and 5′ splice site during the splicing reaction in yeast. Proc Natl Acad Sci 89: 11269–11273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwer B. 2008. A conformational rearrangement in the spliceosome sets the stage for Prp22-dependent mRNA release. Mol Cell 30: 743–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semlow DR, Blanco MR, Walter NG, Staley JP. 2016. Spliceosomal DEAH-Box ATPases remodel pre-mRNA to activate alternative splice sites. Cell 164: 985–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao W, Kim HS, Cao Y, Xu YZ, Query CC. 2012. A U1-U2 snRNP interaction network during intron definition. Mol Cell Biol 32: 470–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y. 2017. Mechanistic insights into precursor messenger RNA splicing by the spliceosome. Nat Rev Mol Cell Biol 18: 655–670. [DOI] [PubMed] [Google Scholar]
- Small EC, Leggett SR, Winans AA, Staley JP. 2006. The EF-G-like GTPase Snu114p regulates spliceosome dynamics mediated by Brr2p, a DExD/H box ATPase. Mol Cell 23: 389–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sontheimer EJ, Steitz JA. 1993. The U5 and U6 small nuclear RNAs as active site components of the spliceosome. Science 262:1989–1996. [DOI] [PubMed] [Google Scholar]
- Spingola M, Grate L, Haussler D, Ares M Jr. 1999. Genome-wide bioinformatic and molecular analysis of introns in Saccharomyces cerevisiae. RNA 5: 221–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staley JP, Guthrie C. 1999. An RNA switch at the 5′ splice site requires ATP and the DEAD box protein Prp28p. Mol Cell 3: 55–64. [DOI] [PubMed] [Google Scholar]
- Stark H, Lührmann R. 2006. Cryo-electron microscopy of spliceosomal components. Annu Rev Biophys Biomol Struct 35: 435–457. [DOI] [PubMed] [Google Scholar]
- Steitz TA, Steitz JA. 1993. A general two-metal-ion mechanism for catalytic RNA. Proc Natl Acad Sci 90: 6498–6502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka N, Aronova A, Schwer B. 2007. Ntr1 activates the Prp43 helicase to trigger release of lariat-intron from the spliceosome. Genes Dev 21: 2312–2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Q, Rodriguez-Santiago S, Wang J, Pu J, Yuste A, Gupta V, Moldón A, Xu YZ, Query CC. 2016. SF3B1/Hsh155 HEAT motif mutations affect interaction with the spliceosomal ATPase Prp5, resulting in altered branch site selectivity in pre-mRNA splicing. Genes Dev 30: 2710–2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarn WY, Steitz JA. 1996. A novel spliceosome containing U11, U12, and U5 snRNPs excises a minor class (AT–AC) intron in vitro. Cell 84: 801–811. [DOI] [PubMed] [Google Scholar]
- Toor N, Keating KS, Taylor SD, Pyle AM. 2008. Crystal structure of a self-spliced group II intron. Science 320: 77–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai RT, Fu RH, Yeh FL, Tseng CK, Lin YC, Huang YH, Cheng SC. 2005. Spliceosome disassembly catalyzed by Prp43 and its associated components Ntr1 and Ntr2. Genes Dev 19: 2991–3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan R, Yan C, Bai R, Wang L, Huang M, Wong CC, Shi Y. 2016a. The 3.8 Å structure of the U4/U6.U5 tri-snRNP: Insights into spliceosome assembly and catalysis. Science 351: 466–475. [DOI] [PubMed] [Google Scholar]
- Wan R, Yan C, Bai R, Huang G, Shi Y. 2016b. Structure of a yeast catalytic step I spliceosome at 3.4 Å resolution. Science 353: 895–904. [DOI] [PubMed] [Google Scholar]
- Wan R, Yan C, Bai R, Lei J, Shi Y. 2017. Structure of an intron lariat spliceosome from Saccharomyces cerevisiae. Cell 171: 120–132. [DOI] [PubMed] [Google Scholar]
- Warkocki Z, Schneider C, Mozaffari-Jovin S, Schmitzová J, Höbartner C, Fabrizio P, Lührmann R. 2015. The G-patch protein Spp2 couples the spliceosome-stimulated ATPase activity of the DEAH-box protein Prp2 to catalytic activation of the spliceosome. Genes Dev 29: 94–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber G, Trowitzsch S, Kastner B, Lührmann R, Wahl MC. 2010. Functional organization of the Sm core in the crystal structure of human U1 snRNP. EMBO J 29: 4172–4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson ME, Fica SM, Galej WP, Norman CM, Newman AJ, Nagai K. 2017. Postcatalytic spliceosome structure reveals mechanism of 3′-splice site selection. Science 358: 1283–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson ME, Lin PC, Plaschka C, Nagai K. 2018. Cryo-EM studies of pre-mRNA splicing: From sample preparation to model visualization. Annu Rev Biophys 10.1146/annurev-biophys-070317-033410. [DOI] [PubMed] [Google Scholar]
- Will CL, Lührmann R. 2011. Spliceosome structure and function. Cold Spring Harb Perspect Biol 3: a003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan C, Hang J, Wan R, Huang M, Wong CCL, Shi Y. 2015. Structure of a yeast spliceosome at 3.6-Angstrom resolution. Science 349: 1182–1191. [DOI] [PubMed] [Google Scholar]
- Yan C, Wan R, Bai R, Huang G, Shi Y. 2016. Structure of a yeast activated spliceosome at 3.5 Å resolution. Science 353: 904–911. [DOI] [PubMed] [Google Scholar]
- Yan C, Wan R, Bai R, Huang G, Shi Y. 2017. Structure of a yeast step II catalytically activated spliceosome. Science 355: 149–155. [DOI] [PubMed] [Google Scholar]
- *.Yan C, Wan R, Shi Y. 2019. Molecular mechanisms of pre-mRNA splicing through structural biology of the spliceosome. Cold Spring Harb Perspect Biol 10.1101/cshperspect.032409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Yan C, Hang J, Finci L, Lei J, Shi Y. 2017. An atomic structure of the human spliceosome. Cell 169: 1–12. [DOI] [PubMed] [Google Scholar]
- Zhang X, Yan C, Zhan X, Li L, Lei J, Shi Y. 2018. Structure of the human activated spliceosome in three conformational states. Cell Res 28: 307–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Pyle AM. 2016. Crystal structures of a group II intron maturase reveal a missing link in spliceosome evolution. Nat Struct Mol Biol 23: 558–565. [DOI] [PMC free article] [PubMed] [Google Scholar]