Abstract
Abstract. Objectives: The potential of epidermal growth factor receptor (EGFR)‐ and Her2‐targeted antibodies Cetuximab, Pertuzumab and Trastuzumab, used in combination to inhibit cell proliferation of breast cancer cells in vitro, has not been extensively investigated. It is anticipated that there would be differences between specific erbB receptor co‐expression profiles that would affect tumour cell growth. Materials and methods: We have examined the effects of Cetuximab, Pertuzumab and Trastuzumab, applied separately or in combination, on cell proliferation of BT474 and SK‐BR‐3 breast cancer cell lines. Cell cycle progression of BT474 and SK‐BR‐3 cells was statically and dynamically assessed using flow cytometry. In order to discover a potential influence of differential EGFR co‐expression on sensitivity to antibody treatment, EGFR was down‐regulated by siRNA in SK‐BR‐3. An annexinV/propidium iodide assay was used to identify potential induction of apoptosis. Results: Treatment with Pertuzumab and Trastuzumab, both targeted to Her2, resulted in a reduced fraction of proliferating cells, prolongation of G1 phase and a great increase in quiescent BT474 cells. Cetuximab had no additional contribution to the effect of either Pertuzumab or Trastuzumab when administered simultaneously. Treatment with the antibodies did not induce an appreciable amount of apoptosis in either BT474 or SK‐BR‐3 cells. In contrast to SK‐BR‐3, the BT474 cell line appears to be more sensitive to antibody treatment due to low EGFR content besides Her2 overexpression. Conclusion: The extent of decelerated or blocked cell proliferation after antibody treatment that is targeted to EGFR and to Her2 depends both on EGFR and Her2 co‐expression and on antibody combination used in the treatment setting. Cetuximab did not enhance any inhibitory effect of Trastuzumab or Pertuzumab, most probably due to the dominant overexpression of Her2. Cell susceptibility to Trastuzumab/Pertuzumab, both targeted to Her2, was defined by the ratio of EGFR/Her2 co‐expression.
INTRODUCTION
Epidermal growth factor receptor (EGFR) and the related Her2 receptor tyrosine kinase are both subjects of antigen‐specific therapeutic targeting in numerous metastatic malignancies, for example, those of the colon, lung, head and neck (Vokes & Chu 2006), and of breast cancer (Plosker & Keam 2006). Small kinase inhibiting molecules (Shawver et al. 2002) as well as humanized or chimeric antibodies (Adams & Weiner 2005) represent valuable therapeutic agents, used as a supplement to conventional chemotherapy, in clinical trials and practice. They have been proven to augment therapeutic efficiency with respect to recurrence‐free and/or overall survival (Pegram et al. 2004). Immunohistochemical EGFR assessment in colon cancer tissue, independent of absolute expression level, provides the basis for Cetuximab therapy targeted to this receptor. In metastatic breast cancer patients, immunohistochemical identification of Her2 overexpression (usually based on Her2 gene amplification, detectable via in situ hybridization) is a prerequisite for Trastuzumab therapy (Mass et al. 2005). Its usefulness in adjuvant settings is currently being extensively explored (Pritchard et al. 2006). However, there is no obvious correlation between EGFR expression and response to Cetuximab treatment in colorectal cancer patients (Saltz et al. 2004) and in non‐small cell lung cancer (Massarelli & Herbst 2006). Likewise, among breast cancer patients with Her2 overexpression both therapy responders and non‐responders are found – strongly indicating that receptor positivity or overexpression is insufficient to predict individual benefit from antibody therapy, even though detection of EGFR expression and Her2 overexpression provide the diagnostic basis to launch such erbB‐receptor‐based treatment. Rather, additional molecules or a particular molecular signature, seem perhaps to contribute to or even to define antibody treatment responsiveness. An optimal combination of therapeutic reagents, targeted to erbB‐receptors based on the co‐expression pattern of EGFR and Her2, has not been previously identified.
Antibody targeting to EGFR or Her2 is desired to trigger cytostatic or even cytotoxic effects on tumour cells, represented by impeded cell growth and proliferation, or by cell death. Independent and combined application of EGFR‐ and Her2‐targeted immunoglobulins to well‐characterized tumour cells will help to ascertain the most potent treatment mode of erbB‐targeted antibodies, under standardized conditions. In a previous study, we described a different, cell line‐specific sensitivity to Trastuzumab (and to certain growth factors) in BT474 and SK‐BR‐3 breast cancer cell lines (Diermeier et al. 2005). We observed contrasting EGFR and Her2‐receptor interaction and (de)activation upon ligand treatment, and we found some evidence for varied EGFR (not a Trastuzumab target) content in both cell lines to be responsible for specific Trastuzumab responsiveness. Here, we present a bottom‐up study by quantifying the antiproliferative effect of Cetuximab (targeted to EGFR) and Trastuzumab (targeted to Her2) in BT474 and SK‐BR‐3 breast cancer cells. We have included Pertuzumab in the study, targeted to an extracellular Her2 epitope, that is different from the Trastuzumab binding site (Adams et al. 2006). In preclinical studies, Pertuzumab has been shown to be inhibitory to breast, prostate and non‐small cell lung cancer cell lines, expressing Her2 at normal and elevated levels (Nahta et al. 2004). In phase I clinical trials, it has shown activity in a number of human cancers (Agus et al. 2005) and a phase II program investigating the effect of Pertuzumab, in locally advanced and metastatic breast cancer, is in progress (Walshe et al. 2006). BT474 and SK‐BR‐3 breast cancer cell lines are characterized by Her2 receptor protein overexpression (based on gene amplification) but different EGFR content that is 3‐fold higher in SK‐BR‐3 compared to BT474 (Brockhoff et al. 2001). In order to determine the most effective mode of administration of therapeutic agents, in vitro cell treatment was performed using the appropriate antibodies, either separately or in combination of two.
We found that replication of BT474 and SK‐BR‐3 cells was significantly reduced with Trastuzumab treatment, whereas Pertuzumab enhanced this effect only in BT474 cells. Cell cycle deceleration is based on prolongation of the G1 phase and decreased S‐phase fraction (SPF). In addition, a substantial cohort of cells was shown to be driven into quiescence by Trastuzumab and Pertuzumab treatment, whereas SK‐BR‐3 cells appeared less susceptible than BT474 cells. Inhibition of SK‐BR‐3 cell growth by Trastuzumab treatment can be amplified by siRNA‐mediated EGFR‐down‐regulation, indicating a decisive impact of EGFR co‐expression on susceptibility to Her2 targeting. Antibodies targeted to Her2 act complementarily, but Cetuximab had no additional effect on either cell line, indicating that an antiproliferative effect mediated via EGFR‐targeting using Cetuximab can not be achieved in the presence of Her2 overexpression. Although higher EGFR content reduced sensitivity to Trastuzumab, it did not provide sufficient evidence for responsiveness to Cetuximab in the presence of Her2 overexpression. Overall, antiproliferative efficiency of the erbB‐receptor‐targeted therapeutic antibodies varied considerably and was due to the specific EGFR‐Her2 expression ratio. Further investigation, including with additional cell lines and types, will help elucidate the most efficient antibody combination for a given erbB‐receptor profile.
MATERIALS AND METHODS
Cell culture
Human breast cancer cell lines BT474 and SK‐BR‐3 were obtained from the American Type Culture Collection and were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 5% foetal calf serum (FCS) (both from PAN Biotech, Aidenbach, Germany). Cells were seeded at densities of 2 × 105 cells per T75 tissue flask (Greiner Bio‐One, Frickenhausen, Germany) and were incubated in humidified atmosphere containing 5% CO2 at 37 °C. Culture medium was refreshed every 2 days. Cells were routinely grown for 7 days, then were washed with phosphate‐buffered saline (PBS, pH 7.4, Biochrom, Berlin, Germany) and were detached from culture flasks by incubating for 3 min at 37 °C in a PBS solution supplemented with 0.05% trypsin and 0.02% ethylenediamine tetraacetic acid.
Cell treatment with therapeutic antibodies
Cells were treated with humanized monoclonal 4D5 and 2C4 antibodies Trastuzumab and to Pertuzumab, respectively (both courtesy of Roche Diagnostics, Penzberg, Germany), and with the chimeric monoclonal IMC‐225 antibody Cetuximab (a gift from Merck, Darmstadt, Germany), in different combinations as indicated; untreated cells served as control samples. Therapeutic antibodies were added at a final concentration of 10 µg/mL. Functionality (biological activity) of antibodies was individually verified by treating cells (e.g. human squamous carcinoma cell line A431 and the UROtsa cell line derived from normal urothelium) that both responded on exposure to, for example, Cetuximab (data not shown).
Immunofluorescence staining of BrdU pulse‐labelled cells
To perform anti‐5‐bromo‐2′‐deoxyuridine (BrdU) cell cycle analysis, 1 × 106 cells were seeded (day zero) into T75 culture flasks in DMEM medium supplemented with 5% FCS. After 8 h, medium was replaced with DMEM containing 2% FCS. The next day, cells were treated with 10 µg/mL of therapeutic antibodies. On day 2, cells were pulse‐labelled with 20 µm BrdU (Sigma‐Aldrich, Taufkirchen, Germany). To avoid a potential BrdU‐induced inhibitory effect, 10 µm 2′‐deoxycytidine (Sigma‐Aldrich) was added simultaneously to the growth medium (Diermeier et al. 2004). After 30 min incubation, BrdU was thoroughly removed by three washing steps. According to the procedure described in detail previously (Brockhoff et al. 2001; Diermeier et al. 2005), samples were harvested at 2 h intervals. Over the time range, differences in cell cycle duration became apparent by identifying cohort of cells exiting from G1, then entering S phase of the second cell cycle (between 30 and 40 h after the BrdU pulse). Cells were fixed with 65% MeOH and were stored until further preparation. Samples were run in at least three independent experiments. Cell preparation and antibody staining for detection of BrdU incorporation has been described in detail previously (Brockhoff et al. 2001). In brief, RNA and cytoplasm were degraded and DNA was denatured by subsequent incubation steps with 1 mg/mL RNase (Sigma‐Aldrich) in PBS, supplemented with 0.02% sodium azide (NaN3) and 2% bovine serum albumin (BSA) (PAB2) at 37 °C for 15 min, Pepsin/HCl solution (5 mg enzyme in 15 mL 0.01 m HCl) at 37 °C for 3 min and 2 N HCl at room temperature for 10 min, respectively, with two washing steps using ice‐cooled PAB2 between washings. DNA was stained with 10 µg/mL anti‐BrdU antibody (clone Bu20a, DakoCytomation, Hamburg, Germany) in PAB2. After labelling with fluorescein isothiocyanate (FITC) conjugated to rabbit‐antimouse antibody (54 µg/mL in PAB2) (DakoCytomation) as the second‐step reagent, the entire DNA content of nuclei was stoichiometrically stained with propidium iodide (PI; Sigma‐Aldrich; 50 µg/mL in PAB2). BrdU‐based approach was utilized to ascertain the regulative level at which the antibodies affect cell cycle progress, information that can not be obtained by static DNA analysis.
Quantification of quiescent cells using the BrdU/Hoe‐quenching technique
For BrdU/Hoechst33258 (Hoe)‐quenching measurements, 2 × 105 cells were seeded on day 0 into T75 culture flasks and were incubated for 7 days. On day 3 after cell seeding, samples were treated with therapeutic antibodies as indicated; untreated samples were used as controls. After further 6 h, 120 µm BrdU was added to both antibody‐treated and untreated cell samples as these concentrations resulted in optimal quenching of Hoe fluorescence for the two cell lines. To minimize potential disturbance in the nucleotide pathway due to BrdU treatment, medium was also supplemented with half‐equimolar 2′‐deoxycytidine (Diermeier et al. 2004). In order to protect the cells from irradiation by visible light, culture flasks were wrapped with aluminium foil and cells were harvested at low light intensity. For all samples, cell detachment was performed after 7 days of growth followed by cell storage at a concentration of 106 cells/mL in freezing medium (Rosewell Park Memorial Institute 1640 medium +10% FCS +10% dimethyl sulfoxide [DMSO]) at –20 °C until flow cytometric analysis. For BrdU/Hoe‐quenching cell staining, thawed cells were washed twice with 2 ml of ice cold DNA‐staining buffer (100 mm Tris‐HCl, pH 7.4, 154 mm NaCl, 1 mm CaCl2, 0.5 mm MgCl2, 0.1% IGEPAL CA‐630 [Nonylphenyl‐polyethylenglycol], 0.2% BSA). 5 × 105 cells were resuspended in 1 mL buffer supplemented with 40 µg/mL (2–4 Units/mL) RNase and 1.2 µg/mL Hoe (Sigma‐Aldrich) and incubated for 15 min at 37 °C (Kubbies et al. 1999; Poot et al. 1994). Cellular DNA content was stained with PI (1.5 µg/mL) for 15 min on ice. Samples were passed through a 70‐µm nylon mesh to remove cell aggregates prior to flow cytometric analysis.
Calculation of quiescent G0 cells upon antibody treatment
Because the rate of cell cycle progress through successive cell cycles depends on individual treatment of the respective sample, the calculation of G0 cells requires a correction for the fraction of cells that have divided once, twice or three times, within the period of observation. Proliferating cells were attributed to the first, second and third cell cycle, respectively (Fig. 1). The fraction of dormant cells that did not proceed in the cell cycle was calculated by dividing the number of G0 cells at the time point of the measurement (90 h) by the sum of (1) the number of cells that had incorporated BrdU, but had not yet divided (the first cycle S + G2M), (2) the number of cells that had divided once (the second cycle cells) divided by two (two cells in the second cycle had resulted from one cell in the first cycle), (3) the number of cells that had divided twice (the third cycle cells) divided by four (four cells in the third cycle had resulted from one cell in the first cycle) and (4) the number of G0 cells.
Figure 1.
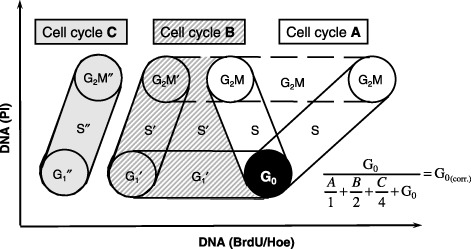
Schematic bivariate diagram of BrdU/Hoe versus propidium iodide fluorescence intensities. The distribution of three successive cell cycles with cell cycle phases G1, S, G2/M of an asynchronous cell population continuously exposed to BrdU is shown. Superscript bars indicate the first (none), the second (′) and the third (″) cell cycle. Figures given in Fig. 4b and 4c are corrected for the cells that divided during the BrdU incubation time of 90 h. In order to calculate the number of cells that have entered G0 and do not proceed in their cell cycle due to treatment, the fraction of G0 cells was divided by the sum of (1) the number of cells that have incorporated BrdU, but have not divided (the first cycle S + G2M), (2) the number of cells that have divided once (the second cycle cells) divided by two (two cells in the second cycle have resulted from one cell in the first cycle), (3) the number of cells that have divided twice (the third cycle cells) divided by four (four cells in the third cycle have resulted from one cell in the first cycle) and (4) the number of G0 cells.
Ki‐67 staining
On day zero 5 × 106 BT474 cells were seeded in T75 cell culture flasks and were treated with antibodies on day 3. Overall, to ensure comparability, cells were treated identically as for G0 quantification with BrdU/Hoe staining. After harvesting cells on day 7, they were centrifuged and washed once in PBS. Subsequently, the pellet was fixed in 4% formalin for 24 h. Sample preparation was performed using the Shandon Cytoblock® Cell Preparation System (Thermo Fisher Scientific, Waltham, MA, USA). In brief, cells were centrifuged, mixed with two drops of reagent 2, and with two drops of reagent 1, in succession. After applying two drops of reagent 1 to a Cytoblock Cassette, cells were transferred and were covered with two drops of reagent 1. The Cytoblock Cassette was then transferred to 70% ethanol for 24 h and was then embedded in paraffin wax. Two‐micrometer sections were cut from formalin‐fixed, paraffin wax‐embedded cell samples and were analysed for Ki‐67 protein staining by immunohistochemistry. The primary antibody anti‐Ki‐67 (rabbit monoclonal clone MIB1, Dako Cytomation GmbH) was applied in at 1 : 10 dilution with final concentration of 5 µg/mL and results were detected using the I‐View 3,3′‐diaminobenzidine detection kit on a Ventana Nexes autostainer (Ventana, Strasbourg, France). Antibody binding was visualized using 3‐amino‐9‐ethylcarbazole (AEC) solution (Dako Cytomation GmbH). As an internal control for Ki‐67 positive expression and also for negativity, paraffin embedded tonsil tissue preparations were used. Percentage of Ki‐67‐positive cells in each sample was determined as described elsewhere (Nocito et al. 2001): nuclei were considered to be Ki‐67 positive if any nuclear staining was seen. Ki‐67 labelling index (percentage of Ki‐67‐positive cells) was determined by assessing the Ki‐67‐positive fraction of 80 cells in a representative area of each sample.
Cell staining for univariate DNA analysis
For univariate DNA staining, fixed cells were washed twice with PBS supplemented with 0.2% BSA followed by incubation in the same buffer, supplemented with 10 U/mL RNase (20 min, 37 °C). Subsequently, PI was added to a final concentration of 25 µg/mL. Samples were incubated for 15 min and were analysed by flow cytometry. Cells were kept in the dark during preparation.
Flow cytometry instrumentation and data analysis
Both static DNA measurements as well as anti‐BrdU measurements were performed on a standard FACSCalibur flow cytometer (BD Biosciences, San Jose, CA, USA). PI was measured on channel 3 with a 670 nm long‐pass filter and anti‐BrdU FITC on channel 1 with a 530/30 band‐pass filter. An LSR‐I three laser flow cytometer (Becton Dickinson Biosciences, San Jose, CA, USA) was used for BrdU/Hoe‐quenching measurements. A 325‐nm ultraviolet laser line was used for Hoe excitation. PI fluorescence was excited with an air‐cooled 488 nm argon laser. Fluorescence emission was detected via 425/44 band‐pass and 630/22 band‐pass filter optic, respectively. Sample measurements and data analysis were performed with CellQuest software (Becton Dickinson Biosciences) using a Macintosh G3 computer. Cell debris and aggregates were excluded from analysis using pulse processing (DNA width signal against DNA area signal). By drawing regions around the initial G0 cell fraction (1, 4), we identified cells that remained BrdU negative within a period of 90 h of BrdU incubation time, indicating quiescent cells. Quantification of cell cycle distribution in static DNA analysis was performed with WinCycle software (Phoenix Flow Systems, San Diego, CA, USA).
Figure 4.
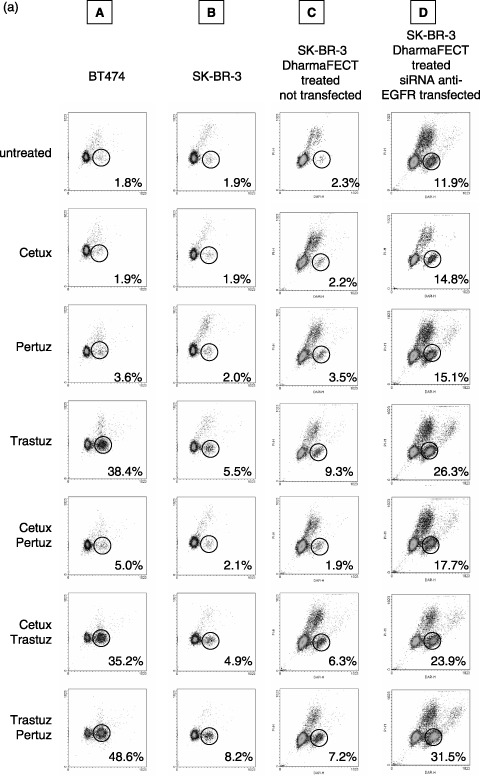
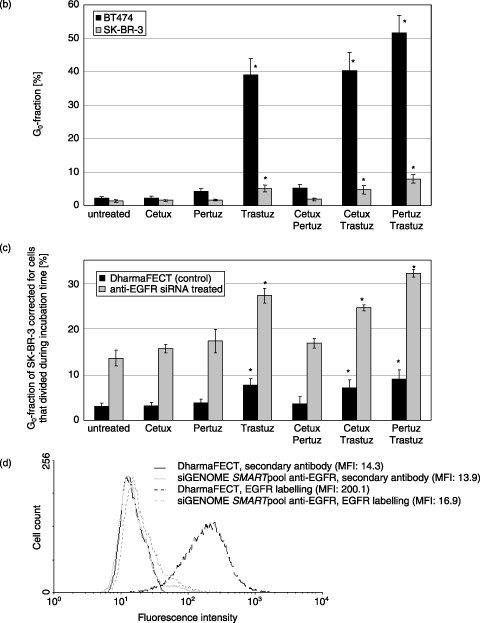
(a) Quantification of quiescent BT474 and SK‐BR‐3 cells without (columns A, B, C) and after (column D) siRNA mediated EGFR knockdown (column C = control). Cells were treated for 96 h with Cetuximab, Trastuzumab and Pertuzumab, applied separately or in pairwise combinations as indicated. Dot plots representative for a single experiment are shown. Without antibody treatment only 1.8% of BT474 and 1.9% of SK‐BR‐3 cells were found without BrdU incorporation within 90 h. The fraction of quiescent cells is strongly elevated in BT474 after Trastuzumab treatment (38.4%) and this effect is further enhanced if cells are treated with Trastuzumab and Pertuzumab simultaneously (48.6%). The fraction of quiescent SK‐BR‐3 cells is 5.5% and 8.2% induced by Trastuzumab treatment alone or in combination with Pertuzumab, respectively. Overall, Cetuximab does not increase the fraction of dormant cells; only a minor effect is seen in BT474 cells when Cetuximab is applied in combination with Pertuzumab (5% quiescent cells). The effect of antibody treatment in SK‐BR‐3 cells was evaluated after EGFR knockdown (column D). Cells treated with DharmaFECT without siRNA served as control (column C). The basal level of dormant cells in siRNA treated SK‐BR‐3 cells is higher (11.9%) than in untreated cells (1.9%), probably due to the lower EGFR content. Overall, the sensitivity to each antibody used in this study is increased after EGFR knockdown compared to normal SK‐BR‐3 cells. The most pronounced effect is observed in Trastuzumab/Pertuzumab treated cells showing 31.5% quiescent cells compared to 7.2% in the control sample. Data overview is given in Fig. 4b and 4c. Efficiency of EGFR knockdown in SK‐BR‐3 cells is demonstrated in Fig. 4d. (b) Quantification of quiescent G0‐cells in BT474 and SK‐BR‐3 cell lines after antibody treatment. Cells were treated for 96 h with Cetuximab, Trastuzumab and Pertuzumab separately or in pairwise combinations. The values for G0 cell fractions were corrected for the cell number that has divided during the BrdU labelling period as described in Materials and Methods. The data represent mean values of three independent experiments. Error bars indicate the standard deviation of the mean. * indicates statistical significance (P < 0.05) related to control. (c) G0‐fraction of SK‐BR‐3 cells without and after siGENOME SMARTpool anti‐EGFR siRNA treatment. The G0 fraction is presented in absolute numbers and corrected for the number of cells that have divided during incubation time as described in Materials and Methods. Overall, SK‐BR‐3 cells are sensitized to antibody treatment by EGFR knockdown. All treatment modalities are more effective after EGFR knockdown, represented by a larger cell fraction that is driven into quiescence by antibody treatment. The most significant effect is evident after Trastuzumab treatment (alone or in combination): The G0‐fraction after siRNA transfection is increased from 7.8% to 27.3% (by Trastuzumab treatment), from 7.2% to 24.8% (by Trastuzumab, Cetuximab treatment) and from 9.3% to 32.4% (by Trastuzumab, Pertuzumab treatment). Three independent experiments were performed. Error bars indicate the standard deviation of the mean. * indicates statistical significance (P < 0.05) related to the control. (d) EGFR knockdown in SK‐BR‐3 cells by siGENOME SMARTpool anti‐EGFR siRNA 3 days after first transfection. Mean fluorescent intensity (MFI) of EGFR staining in non‐transfected cells is 200.1 compared to DharmaFECT treated control (14.3). 96% of EGFR was homogeneously knocked down in the SK‐BR‐3 cell population by siGENOME SMARTpool anti‐EGFR transfection (MFI 16.9).
siRNA‐mediated EGFR‐down‐regulation
Eighty thousands SK‐BR‐3 cells were seeded per well in a 6‐well plate in DMEM supplemented with 5% FCS on day 0. The next day, medium was removed and 1.6 mL fresh DMEM/5% FCS was added. The transfection mix was prepared by incorporation of 4 µL DharmaFECT1 (Dharmacon, Lafayette, CO, USA) with 196 µL Opti‐MEM (Invitrogen, Karlsruhe, Germany) in tube 1 and 5 µL of 20 µm siRNA (siGENOME SMARTpool anti‐EGFR, Dharmacon) and 195 µL Opti‐MEM in tube 2. After 5 min at room temperature, tube 1 was added to tube 2 and was thoroughly mixed. After a further incubation step of 20 min at room temperature, the 400 µL transfection mix was added per well to give a final siRNA concentration of 50 nm. On day 2 cells were treated with 10 µg/mL Trastuzumab, Pertuzumab and Cetuximab alone or in combinations of two. Transfection was repeated on day 3 by addition of 1.8 mL of fresh DMEM/5% FCS and half the amount of transfection mix (200 µL) per well. Antibodies were added in the same concentration as used on day 2.
For univariate DNA analysis, cells were harvested on day 4, as described above, resulting in an incubation time for antibody treatment of 48 h. For quantification of G0 cell fraction, medium was changed on day 4. On the same day antibodies and, after an additional 6 h incubation period, BrdU were added as described above for the BrdU/Hoe‐quenching technique. On day 7, medium was again changed and antibodies and BrdU were replaced. Cells were harvested on day 8 resulting in an incubation time for antibodies and BrdU treatment of 96 h and 90 h, respectively.
AnnexinV/propidium iodide assay/evaluation of apoptosis and necrosis
For evaluation of potential apoptotic and necrotic cells induced by antibody treatment, 2.6 × 104 cells were seeded per well in a 24‐well plate in DMEM supplemented with 5% FCS on day 0; next day, serum concentration was reduced to 2%. Therapeutic antibody treatment was performed on day 2. Cells were harvested on day 4 and were stained using the TACS™ annexinV‐FITC kit (R & D Systems, Wiesbaden‐Nordenstadt, Germany). According to Koopman et al. (1994), cells in early apoptosis are annexinV‐FITC positive but remain impenetrable to PI at low concentrations, whereas late apoptotic cells are both annexinV‐FITC and PI positive. In contrast, live cells are annexinV‐FITC and PI negative.
As positive apoptotic control, SK‐BR‐3 cells were seeded and harvested as described above, but were treated with 10 µm Camptothecin for 19.5 h. As BT474 cells were unaffected by Camptothecin treatment, NaN3 was used to induce apoptosis in this cell line according to Spiridon et al. (2002). Cells were seeded in a 24‐well plate at a cell density of 5 × 104 cells per well. After 24 h, NaN3 was added to a final concentration of 180 mm and cells were incubated for additional 4 h. Untreated cells served as negative controls for both cell lines. After detachment with trypsin, cells were washed twice with PBS supplemented with 2% BSA and were stained according to the manufacturer's instructions. PI was added 1 min before flow cytometric analysis.
Statistical analysis
Student's t‐test for independently acquired samples was applied to calculate mean values +/− standard deviation. Each measurement was performed in at least triplicate (n = 3).
RESULTS
Cell cycle progress upon antibody treatment quantified via anti‐BrdU pulse labelling
Figure 2a shows dynamic assessment of cell proliferation of BT474 and SK‐BR‐3 cell upon treatment with Cetuximab, Pertuzumab and Trastuzumab either applied alone or in combination two‐at‐a‐time. Cell proliferation was analysed using the flow cytometric anti‐BrdU technique. Corresponding quantification of cell cycle progress of the BrdU‐positive cell cohort at 38 h after BrdU pulse labelling is shown in Fig. 2b.
Figure 2.
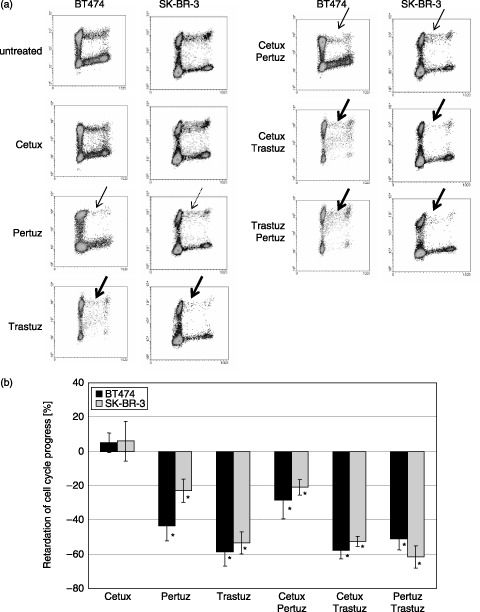
(a) Dynamic proliferation assessment of BT474 and SK‐BR‐3 cells after antibody treatment. Dotted, thin and thick lined arrows indicate low, moderate and strong inhibition of cell cycle progress compared to untreated control cells. Cetuximab treatment does not affect proliferation of both cell lines, whereas Pertuzumab slightly (SK‐BR‐3) and moderately (BT474) detains cell cycle progress. Trastuzumab is the most efficient inhibitor in both cell lines. BT474 and SK‐BR‐3 cells in G1 phase of the second cell cycle do not reenter into S phase within 50 h in the presence of Trastuzumab (data not shown), representing a significant prolongation of G1 by Trastuzumab. Simultaneous administration of Trastuzumab and Pertuzumab to BT474 and SK‐BR‐3 cells results in an additive inhibition of cell proliferation, whereas Cetuximab application does not show any inhibitory effect even in combination with Pertuzumab or Trastuzumab. (b) Cell cycle progress in BT474 and SK‐BR‐3 cells affected by antibody treatment. S/G2/M fractions in BrdU positive cells were calculated in relation to the total BrdU positive cell fraction 38 h after BrdU incubation. Cetuximab does not affect cell cycle progression in both cell lines. However, Pertuzumab and Trastuzumab decelerate the cell cycle progress by 43.4% and 58.7% (BT474) and 23% and 53.3% (SK‐BR‐3), respectively. This indicates first that Trastuzumab is more effective than Pertuzumab and second that BT474 cells are more susceptible to antibody treatment than SK‐BR‐3 cells. Treatment with a combination of two antibodies did not augment the inhibitory effect of antibodies administered separately. * indicates statistical significance (P < 0.05) related to control.
Compared to untreated cells, BT474 and SK‐BR‐3 cell proliferation was markedly inhibited by Trastuzumab treatment. Cell counts of BrdU‐positive cells proceeding from G1 into S phase 38 h after BrdU pulse labelling were diminished by 58.7% in BT474 and 53.3% in SK‐BR‐3 compared to the control (Fig. 2b). Even at the 50 h time point, no BrdU‐positive cells moved forwards from G1 into S phase of the second cell cycle (data not shown) revealing a significant prolongation of G1 phase by Trastuzumab treatment that was more pronounced in SK‐BR‐3 than in BT474 cells. Inhibition of cell cycle progression was similar for the combination treatment of Trastuzumab with Pertuzumab or with Cetuximab in both cell lines.
Cetuximab treatment did not affect cell cycle progression of BT474 and SK‐BR‐3. In contrast, both cell lines were retarded in cell cycle transition from G1 into S phase by 43.4% (BT474) and 23% (SK‐BR‐3) when treated with Pertuzumab. This identified BT474 as the more sensitive cell line for Pertuzumab treatment compared to SK‐BR‐3 cells. Combination treatment with Cetuximab and Pertuzumab did not further increase the retarding effect of Pertuzumab treatment alone. Hence, reduction in the cell fraction moving from G1 into S phase of BrdU‐labelled BT474 by 28.5% and SK‐BR‐3 by 21% must be exclusively attributed to Pertuzumab treatment.
S‐phase fraction of antibody‐treated SK‐BR‐3 and BT474
In addition to calculating proliferating BrdU‐positive cells, we analysed the total cell population by labelling their DNA with PI and flow cytometrically assessing cell cycle parameters upon antibody treatment. Trastuzumab was revealed to be the most effective agent for reducing SPF in BT474 and SK‐BR‐3 (Fig. 3). SPF was significantly reduced in both BT474 (from 26.1% to 8.1%) and in SK‐BR‐3 (from 19.9% to 15.1%) cells, indicating BT474 as the more sensitive cell line. Likewise, the SPF in BT474 (not in SK‐BR‐3) decreased upon Pertuzumab treatment (down to 20.4%). Cetuximab alone or in combination had no significant effect on SPF for either BT474 or SK‐BR‐3. In contrast, Pertuzumab and Trastuzumab administered in combination were capable of significantly intensifying the effect of corresponding single antibody‐treatments only in BT474 (8.1% in Trastuzumab treated cells versus 4.9% for the combination treatment) but not in SK‐BR‐3 (15.1% versus 13.7%) cells.
Figure 3.
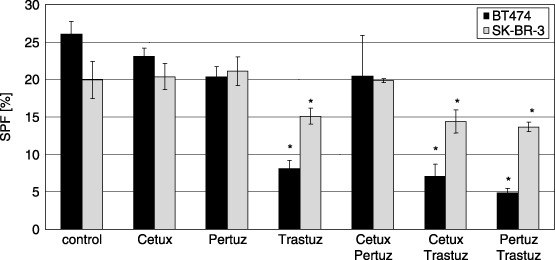
S‐phase fraction (SPF) in BT474 and SK‐BR‐3 cells after antibody treatment. SPF is significantly reduced in both cell lines in the presence of Trastuzumab. This effect is more pronounced in BT474 cells (8.1% versus 26.1%) than in SK‐BR‐3 cells (15.1% versus 19.9%). Pertuzumab reduces SPF in BT474 slightly but significantly (20.4% versus 26.1%) but not in SK‐BR‐3. Cetuximab has no significant effect on SPF in both cell lines. The effect of Trastuzumab is enhanced in both cell lines by the simultaneous administration of Pertuzumab. However, this observation is significant only in BT474 cells (4.9% versus 8.1%) but not in SK‐BR‐3 cells (13.7% versus 15.1%). Cetuximab has no additional effect on SPF when added either together with Trastuzumab or in combination with Pertuzumab. * indicates statistical significance (P < 0.05) related to control.
Quantification of quiescent cells upon antibody‐treatment quantified via BrdU/Hoe staining
Cells were treated at 96 h with Cetuximab, Trastuzumab and Pertuzumab for 96 h and were continuously labelled with BrdU for 90 h. Antibodies were applied alone or in combination of two (Fig. 4a–c). The cell fraction transferred into quiescence (G0) by antibody treatment was flow cytometrically determined by PI/Hoe staining (Fig. 4a) and was quantified as described in the Materials and Methods section (see also Fig. 4b,c).
Antibody‐untreated control samples had only 1.8% (BT474) and 1.9% (SK‐BR‐3) cells in the G0 compartment (Fig. 4a,b). The fraction of quiescent cells was quite high in BT474 cells upon Trastuzumab treatment (38.4%) and was further elevated if Trastuzumab and Pertuzumab were combined (48.6%). The fraction of quiescent SK‐BR‐3 cells was 5.5% for Trastuzumab treatment alone and 8.2% in combination with Pertuzumab. Combining Cetuximab with Trastuzumab for treatment of BT474 or SK‐BR‐3 had no further impact on increasing the G0 cell fraction compared to treatment with Trastuzumab alone.
Cetuximab did not increase the amount of dormant cells in either SK‐BR‐3 or in BT474. Likewise, the SK‐BR‐3 G0 cell fraction remained at the same value with Pertuzumab treatment (2.0%) compared to the control (1.9%). Even combining Cetuximab and Pertuzumab for treatment did not change the G0 fraction of SK‐BR‐3 cells.
A slight increase in the number of dormant cells was induced in BT474 cells when cells were treated with Pertuzumab alone (3.6%). If Pertuzumab and Cetuximab were combined for treatment of BT474 cells a slight, but non‐significant increase in the dormant cell count (5.0% quiescent cells) was found compared to either of the treatments, alone (Cetuximab: 1.9%; Pertuzumab: 3.6%).
Quantification of quiescent cells upon antibody treatment in SK‐BR‐3 after siRNA mediated EGFR‐down‐regulation
Both SK‐BR‐3 and BT474 cells overexpress HER2/neu but differentially express EGFR, the level of EGFR protein being around 3‐fold in SK‐BR‐3 compared to BT474 (Brockhoff et al. 2001; Diermeier et al. 2005). Because BT474 cells show higher sensitivity to antibody‐induced cell cycle exit, the role of the level of EGFR co‐expression on sensitivity to induction of transferring cells into quiescence, was determined by siRNA‐mediated down‐regulation of EGFR in SK‐BR‐3 cells. As shown in Fig. 4d, the level of EGFR expression was effectively reduced by 96% 3 days after transfection.
When EGFR is down‐regulated in SK‐BR‐3 cells the fraction of dormant cells is increased to 11.9% compared to 2.3% in DharmaFECT‐treated controls, without EGFR‐down‐regulation. DharmaFECT treatment alone does not change the G0 fraction in SK‐BR‐3 cells compared to untreated cells (1.9%). However, if cells are treated with Trastuzumab, the G0 cell fraction seems to rise by the presence of DharmaFECT (9.3% versus 5.5%). The reason for this effect remains unknown.
G0 cell fractions for the different antibody treatment modalities of DharmaFECT controls and siRNA tranfected cells are represented as absolute numbers (Fig. 4c) and are corrected for the number of cells that have divided during the incubation of BrdU period continuous labelling. Increase in the G0 cell fraction of siRNA transfected SK‐BR‐3 cells is higher for any of the tested antibody treatments compared to DharmaFECT controls. Interestingly, this effect was most efficient with cells that treated with Trastuzumab in combination with Pertuzumab, followed by Trastuzumab alone, and with its combination with Cetuximab. In contrast to untreated SK‐BR‐3 cells (which are insensitive to Pertuzumab), and Cetuximab treatment (Fig. 4b), EGFR‐down‐regulation established sensitivity of SK‐BR‐3 to Pertuzumab and Cetuximab shown by an increase in G0 cell fraction.
Ki‐67 staining of antibody‐treated SK‐BR‐3 and BT474 cells
In order to differentiate between prolongation of G1, and real cell cycle exit into quiescence, we applied immunohistochemical Ki‐67 staining to paraffin‐embedded preparations of BT474 and SK‐BR‐3 cells. Samples treated with Trastuzumab and Pertuzumab in combination contained fewer cells that were positive for the proliferation marker Ki‐67 compared to control cells (Fig. 5). Table 1 shows the calculation of the fraction of Ki‐67 positive cells for the examples given in Fig. 5 and for additional treatment modalities. Cells were deprived of serum for a positive control. Table 1 shows that Trastuzumab decreased the fraction of Ki‐67‐positive cells both alone (BT474: 31.2% versus 56.9%; SK‐BR‐3: 33.3% versus 50.0%) and in combination with Cetuximab (BT474: 35.2%; SK‐BR‐3: 38.5%) or Pertuzumab (BT474: 13.7%; SK‐BR‐3: 27.7%). The combination Trastuzumab/Pertuzumab was the most effective antibody treatment modality; BT474 and SK‐BR‐3 cells showed a reduction in Ki‐67‐positive cell fractions by 75.9% in BT474 cells and 44.6% in SK‐BR‐3 cells.
Figure 5.
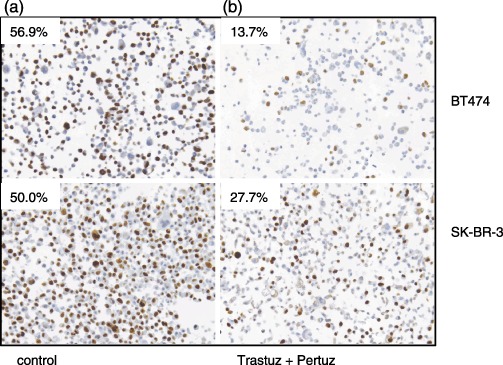
Ki‐67 staining by IHC in paraffin embedded preparations of BT474 and SK‐BR‐3 cells. One example for cell treatment (Tratuzumab/Pertuzumab) representing the largest change in Ki‐67 positivity according to Table 1 is shown for each cell line and compared to untreated cells. The numbers represent the fraction of Ki‐67 positive cells. Samples treated with Trastuzumab and Pertuzumab in combination (b) show fewer Ki‐67‐positive cells compared to control cells (a).
Table 1.
Quantification of Ki‐67‐positive cells by IHC in paraffin embedded preparations of BT474 and SK‐BR‐3 cells. Eighty cells of each sample within a representative area were counted and the fraction of Ki‐67‐positive cells was calculated. As positive control, cells were deprived of serum.
| BT474 | SK‐BR‐3 | |
|---|---|---|
| untreated | 56.9% | 50.0% |
| 0% FCS | 37.5% | 18.6% |
| Trastuzumab | 31.2% | 33.3% |
| Trastuzumab + Cetuximab | 35.2% | 38.5% |
| Trastuzumab + Pertuzumab | 13.7% | 27.7% |
S‐phase fraction of antibody‐treated SK‐BR‐3 cells after siRNA mediated EGFR‐down‐regulation
Figure 6 shows the SPF of siGENOME SMARTpool anti‐EGFR transfected SK‐BR‐3 cells compared to DharmaFECT‐only treated controls upon antibody treatment. SPFs for transfected and non‐transfected cells were not significantly changed by Pertuzumab or Cetuximab treatment, either when applied alone or in combination. EGFR‐down‐regulation caused a decrease in SPF in untreated cells (SPF = 16.6%) by 13.6% compared to Trastuzumab‐treated cells (SPF = 14.3%). In DharmaFECT controls, Trastuzumab lead to reduction in SPF from 23.2% in untreated cells to 17.9% in Trastuzumab‐treated cells, which was equivalent to reduction of 22.9%. Therefore, when Trastuzumab is present, the SPF compared to untreated cells is reduced less effectively in EGFR‐down‐regulated SK‐BR‐3 cells (13.6%) than in DharmaFECT control cells (22.9%). When cells were treated with Cetuximab and Trastuzumab in combination, EGFR‐down‐regulation did not lead to any change in SPF. Although the efficiency of Trastuzumab on reduction of SPF was weaker if the EGFR is down‐regulated, the correlation is reversed for combination treatment with Trastuzumab and Pertuzumab. Reduction in EGFR expression level increased the efficiency of SPF reduction 2‐fold when cells were treated with Trastuzumab and Pertuzumab, as represented by additional decrease in SPF by 32.0% in EGFR‐down‐regulated cells (from 16.6% in untreated cells to 11.25% in Trastuzumab/Pertuzumab‐treated cells) compared to 16.4% in DharmaFECT controls (from 23.2% in untreated cells to 19.4% in Trastuzumab/Pertuzumab‐treated ones).
Figure 6.
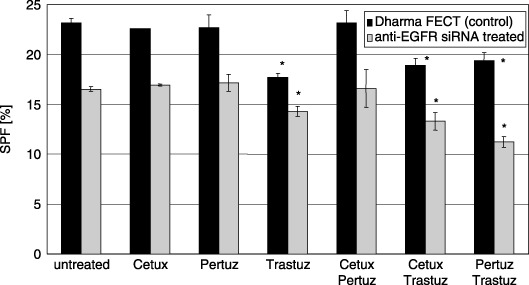
S‐phase fraction (SPF) of SK‐BR‐3 cells after antibody treatment without and after siGENOME SMART pool anti‐EGFR siRNA transfection. Cetuximab and Pertuzumab have no effect on SPF in SK‐BR‐3 cells either with or without EGFR down‐regulation. In DharmaFECT treated control cells (black bars) the SPF is reduced from 23.2% to 17.9% with Trastuzumab treatment, to 18.9% with Cetuximab/Trastuzumab treatment and to 19.4% with Pertuzumab/Trastuzumab treatment. In EGFR‐down‐regulated cells (grey columns), the SPF of untreated cells is reduced from 16.6% to 14.3% with Trastuzumab treatment, to 13.3% with Cetuximab/Trastuzumab treatment and to 11.3% with Pertuzumab/Trastuzumab treatment. The decrease in SPF upon Trastuzumab treatment compared to untreated cells is 22.9% in DharmaFECT control cells and 13.6% in EGFR‐down‐regulated cells. The correlation is reversed for combination treatment of Trastuzumab and Pertuzumab. SPF is decreased by 16.4% in DharmaFECT control cells and by 32% in EGFR‐down‐regulated SK‐BR‐3. For Cetuximab/Trastuzumab treatment the decrease in SPF compared to untreated cells is similar for DharmaFECT control cells (18.5%) and EGFR‐down‐regulated cells (19.6%). * indicates statistical significance (P < 0.05) related to control.
Quantification of apoptosis after antibody treatment
Potential induction of apoptosis, mediated by antibody treatment, was analysed using annexinV labelling and was measured by flow cytometry (Fig. 7a); early and late apoptotic cell fractions were quantified by quadrant analysis (Fig. 7b). In positive controls (NaN3 for BT474 and Camptothecin for SK‐BR‐3), the apoptotic cell fraction was significantly increased. While most antibodies did not induce any apoptosis, combination treatment of Pertuzumab/Cetuximab and Pertuzumab/Trastuzumab induced a statistically significant increase in apoptosis in BT474 cells compared to controls. The slight increase in apoptosis in SK‐BR‐3 cells upon Trastuzumab treatment (both when applied alone and in combination) was not significant. Neither antibody treatment modality induced a level of apoptosis approximating that of positive controls; even 100 µg/mL Cetuximab did not increase the number of apoptotic cells (data not shown). In agreement with these results, caspase activation assays have shown a slight increase in apoptotic cell fraction in BT474 cells when two antibodies were combined for treatment (data not shown), but the level of apoptosis was far below the amount of apoptosis induced in positive controls.
Figure 7.
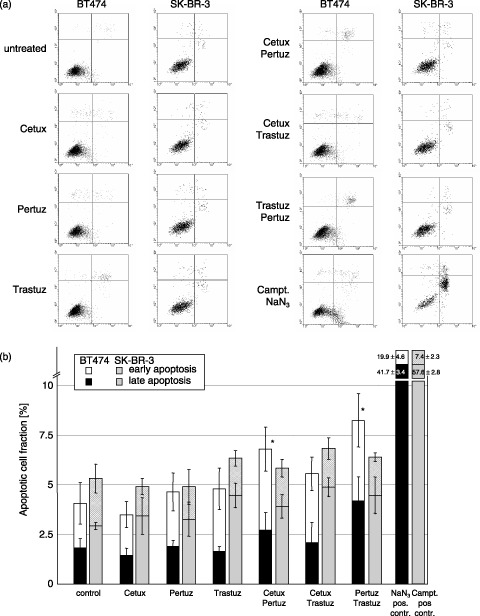
(a) Quantification of apoptotic cells by annexinV/propidium iodide (PI) staining. Compared to untreated cells, NaN3 or Camptothecin treatment (positive controls for BT474 and SK‐BR‐3, respectively) induced a significant amount of early (annexinV positive) and late (annexinV/PI double positive) apoptotic cells. However, none of the antibodies used in this study was able to induce apoptosis in BT474 or SK‐BR‐3 cells when administered individually. Only after Cetuximab/Pertuzumab (7.0% versus 4%) and Pertuzumab/Trastuzumab (7.9% versus 4%) treatment of BT474 cells, a small but significant increase in the apoptotic cell fraction could be observed. (b) Apoptotic cell fractions in BT474 and SK‐BR‐3 cell lines after antibody treatment. * indicates statistical significance (P < 0.05) related to control.
DISCUSSION
Cell proliferation analysis of BT474 and SK‐BR‐3 breast cancer cell lines revealed that both Trastuzumab and Pertuzumab, both targeted to Her2, decelerate cell cycle progress and drive cells into quiescence, and Trastuzumab is more effective than Pertuzumab. Nevertheless, Pertuzumab potentiates the effect of Trastuzumab. However, Cetuximab, targeted to EGFR conveyed no antiproliferative effect in either BT474 or in SK‐BR‐3 cells and was also ineffective when used in combination with Trastuzumab or Pertuzumab.
The complementary effect of Trastuzumab and Pertuzumab can be explained by different mechanisms of action on the receptor level. Pertuzumab, which binds to the extracellular Her2 subdomain II, disrupts (ligand‐induced) heterointeraction of Her2 with other erbB‐receptors (lateral signalling) that most probably would interrupt subsequent intracellular downstream signalling (Sliwkowski et al. 1999; Cho et al. 2003; Badache & Hynes 2004; Mass et al. 2005; Adams et al. 2006; Bernard et al. 2006; Kumar & Pegram 2006). In contrast, binding of Trastuzumab to subdomain IV of Her2 does not interfere with the receptor dimerization loop, involved in receptor association, thus not interrupting cross‐signalling activity (Badache et al. 2004). In a previous study, we have shown rather that Trastuzumab induces and stabilizes Her2 homodimerization, which is observed even without any ligand stimulation, if the Her2 expression is high (Diermeier et al. 2005). We have previously revealed that the Trastuzumab‐enhanced Her2 homodimerization was associated with Y877 and Y1248 phosphorylation in SK‐BR‐3 cells’ and additionally with Y1112 phosphorylation in BT474 cells’ recruiting pathways with phosphatase and tensin homolog deleted on chromosome ten (PTEN), Chk‐kinase and Cbl E3 ubiquitin ligase involvement, thereby reducing cell survival (PTEN), cell cycle progression (Chk) and inducing receptor degradation (Cbl), respectively (Diermeier et al. 2005). The Trastuzumab complementary effect caused by Pertuzumab might be mediated via obstruction of Her2 interaction with Her3 or Her4 receptors, albeit we have previously quantified fairly low Her3 and Her4 expression both in BT474 and SK‐BR‐3 cells (Brockhoff et al. 2001). Our data are in agreement with other studies showing enhanced sensitivity to Trastuzumab by treating BT474 cells with Pertuzumab, in addition to demonstrating that the efficiency of erbB‐receptor targeting can be elevated by a dual treatment of cells using antibodies interacting with different epitopes of the same receptor and thereby addressing different molecular mechanisms (Massarelli et al. 2006). However, insensitivity of SK‐BR‐3 cells to additional Pertuzumab treatment reveals that this phenomenon is not universally valid in breast cancer cells and is determined – most likely among other factors – by EGFR co‐expression.
In vitro cell susceptibility to Her2‐specific antibody treatment is substantially determined by EGFR‐Her2 co‐expression and is additionally modulated by activity of growth factors. As shown in SK‐BR‐3 cells, EGFR‐down‐regulation raises the sensitivity to Trastuzumab. siRNA‐mediated EGFR‐down‐regulation could provoke a shift from Her2‐EGFR heterointeraction in favour of Her2‐Her2 homodimerization and thereby provide the basis for Trastuzumab to operate more efficiently, because its primary impact is on Her2 homodimerization. In order to prove this hypothesis, specific fluorescence resonance energy transfer experiments are currently underway. The finding that susceptibility to Trastuzumab, targeted to Her2, directly depends on the level of co‐expressed EGFR, strongly suggests that erbB‐receptor‐based strategies, dedicated to inhibition of tumour cell proliferation, have to account (at least) for both EGFR and Her2 receptors, and most probably for other erbB‐receptors as well (Emlet et al. 2006). Boosting the therapeutic efficiency of Trastuzumab might be achieved by simultaneous EGFR‐down‐regulation, an auspicious extension of currently practiced antibody treatment, that appears well worth being systematically evaluated in preclinical trials.
It is important to note that a larger part of antibody‐induced cell proliferation inhibition must be attributed to cells that are transferred into quiescence (as shown by two independent experiments) rather than to cell cycle prolongation of the same cells. This observation suggests two strategies for supplementing antibody treatment: first, cytostatic agents may be used that complementarily target remaining proliferating cells that are not efficiently affected by antibody treatment and second, substances can be applied that drive antibody‐induced dormant cells into apoptosis.
That growth factors can partially (EGF) or even completely (HRG) compensate for antiproliferative effects induced by Trastuzumab (Diermeier et al. 2005) and Pertuzumab (data not shown) is an important observation. Assessing erbB‐related growth factors in tumour tissues should be considered to be included into prognosis because their presence or absence could have a significant impact on the therapeutic efficiency of erbB‐receptor targeted antibodies.
Antibody triggered cell death by apoptosis does not play a significant role in the system investigated in this study. Minimal appearance of apoptotic BT474 cells upon Cetuximab‐Pertuzumab and Pertuzumab‐Trastuzumab treatment was negligible. In contrast, Nahta et al. (2004) found apoptosis in BT474 breast cancer cells upon Trastuzumab‐Pertuzumab treatment. However, this discrepancy might well be attributed to the exceptionally high and non‐physiological antibody concentrations this group used to treat the cells. Their dose‐response assay ranged from 0.1 to 100 µg/mL Trastuzumab, and it revealed a significant fraction of apoptotic BT474 cells only upon 100 µg/mL Trastuzumab treatment, that is 10‐fold higher than the concentration used in this investigation.
Cetuximab failed to show any significant or additional inhibitory effect in BT474 or SK‐BR‐3 cell proliferation, which is most probably due to overexpression of Her2 in these cell lines. In contrast to our observation, Ye et al. (1999) have demonstrated an augmentation of Trastuzumab‐induced growth inhibition by Cetuximab in ovarian OVCA420 cells. However, in OVCA420 cells EGFR content is higher than Her2 expression as shown in their and other studies. Therefore, co‐expression ratio of EGFR/Her2 would be converse compared to that in BT474 or SK‐BR‐3 cells, thus eliminating the apparent contradiction between their results and ours. In vitro targeting overexpressed Her2 with Trastuzumab depends on the absolute EGFR content, whereas the EGFR density seems to be negligible if targeted with Cetuximab in the presence of Her2 overexpression. As seen in many other studies, this observation emphasizes the dominant malignant potential of Her2 (Graus‐Porta et al. 1997). If overexpressed Her2 has been shown to be constitutively activated (Yuste et al. 2005), persistent Her2 signalling might be dominant over any inhibitory effect, resulting from attacking the EGFR. Furthermore, factors other than absolute EGFR content have been recently proposed to have more impact on responsiveness to Cetuximab, for example, basal activation state of EGFR (Balin‐Gauthier et al. 2006) and the ratio of high‐ and low‐affinity receptors (Italiano 2006). Continued functional investigations (ligand‐induced receptor interaction, cross‐activation, intracellular trafficking) are needed to elucidate the individual mechanism of erbB‐receptors to develop their malignant potential within the context of co‐expression. In view of clinical utilization, simultaneous quantification of EGFR and Her2 in the same tumour sample might render individual diagnosis and facilitate the elevation of erbB‐based therapy efficiency more precisely.
In summary, blocking or inhibiting uncontrolled cell proliferation of breast cancer cells can be achieved by treatment with antibodies targeted to erbB‐receptors. However, treatment efficiency varies considerably and the antiproliferative impact using antibodies targeted to EGFR (Cetuximab) and Her2 (Trastuzumab, Pertuzumab) strictly depends on co‐expression ratios of both receptors. Moreover, treatment efficiency is known to be lower in the presence of erbB‐receptor‐related growth factors EGF and HRG (Diermeier et al. 2005). The search for a molecular signature with predictive value for responsiveness to antibody treatment must particularly take into account individual co‐expression profiles of erbB‐receptors.
ACKNOWLEDGEMENTS
This work was supported by the Else Kroener‐Fresenius‐Stiftung (eksvrt161). The authors thank Rosi Kromas for her highly dependable assistance.
REFERENCES
- Adams CW, Allison DE, Flagella K, Presta L, Clarke J, Dybdal N, McKeever K, Sliwkowski MX (2006) Humanization of a recombinant monoclonal antibody to produce a therapeutic HER dimerization inhibitor, Pertuzumab. Cancer Immunol. Immunother. 55, 717–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams GP, Weiner LM (2005) Monoclonal antibody therapy of cancer. Nat Biotechnol. 23, 1147–1157. [DOI] [PubMed] [Google Scholar]
- Agus DB, Gordon MS, Taylor C, Natale RB, Karlan B, Mendelson DS, Press MF, Allison DE, Sliwkowski MX, Lieberman G, Kelsey SM, Fyfe G (2005) Phase I clinical study of Pertuzumab, a novel HER dimerization inhibitor, in patients with advanced cancer. J. Clin. Oncol. 23, 2534–2543. [DOI] [PubMed] [Google Scholar]
- Badache A, Hynes NE (2004) A new therapeutic antibody masks ErbB2 to its partners. Cancer Cell 5, 299–301. [DOI] [PubMed] [Google Scholar]
- Balin‐Gauthier D, Delord JP, Rochaix P, Mallard V, Thomas F, Hennebelle I, Bugat R, Canal P, Allal C (2006) In vivo and in vitro antitumor activity of oxaliplatin in combination with Cetuximab in human colorectal tumor cell lines expressing different level of EGFR. Cancer Chemother Pharmacol. 57, 709–718. [DOI] [PubMed] [Google Scholar]
- Bernard‐Marty C, Lebrun F, Awada A, Piccart MJ (2006) Monoclonal antibody‐based targeted therapy in breast cancer: current status and future directions. Drugs 66, 1577–1591. [DOI] [PubMed] [Google Scholar]
- Brockhoff G, Heiss P, Schlegel J, Hofstaetter F, Knuechel R (2001) Epidermal growth factor receptor, c‐erbB2 and c‐erbB3 receptor interaction, and related cell cycle kinetics of SK‐BR‐3 and BT474 breast carcinoma cells. Cytometry 44, 338–348. [DOI] [PubMed] [Google Scholar]
- Cho HS, Mason K, Ramyar KX, Stanley AM, Gabelli SB, Denney DW Jr, Leahy DJ (2003) Structure of the extracellular region of HER2 alone and in complex with the Herceptin Fab. Nature 421, 756–760. [DOI] [PubMed] [Google Scholar]
- Diermeier S, Horvath G, Knuechel‐Clarke R, Hofstaedter F, Szollosi J, Brockhoff G (2005) Epidermal growth factor receptor coexpression modulates susceptibility to Herceptin in HER2/neu overexpressing breast cancer cells via specific erbB‐receptor interaction and activation. Exp. Cell Res. 304, 604–619. [DOI] [PubMed] [Google Scholar]
- Diermeier S, Schmidt‐Bruecken E, Kubbies M, Kunz‐Schughart LA, Brockhoff G (2004) Exposure to continuous bromodeoxyuridine (BrdU) differentially affects cell cycle progression of human breast and bladder cancer cell lines. Cell Prolif. 37, 195–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emlet DR, Schwartz. R , Brown KA, Pollice AA, Smith CA, Shackney SE (2006) HER2 expression as a potential marker for response to therapy targeted to the EGFR. Br. J. Cancer 94, 1144–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graus‐Porta D, Beerli RR, Daly JM, Hynes NE (1997) ErbB‐2, the preferred heterodimerization partner of all ErbB receptors, is a mediator of lateral signaling. EMBO J. 16, 1647–1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Italiano A (2006) Targeting the epidermal growth factor receptor in colorectal cancer: advances and controversies. Oncology 70, 161–167. [DOI] [PubMed] [Google Scholar]
- Koopman G, Reutelingsperger CP, Kuijten GA, Keehnen RM, Pals ST, Van Oers MH (1994) AnnexinV for flow cytometric detection of phosphatidylserine expression on B cells undergoing apoptosis. Blood 84, 1415–1420. [PubMed] [Google Scholar]
- Kubbies M (1999) High‐resolution cell cycle analysis: the flow cytometric bromodeoxyuridine‐Hoechst quenching technique. In: Radbruch A, ed. Flow Cytometry and Cell Sorting, 2nd edn.
- Kumar Pal S, Pegram M (2006) Targeting HER2 epitopes. Semin Oncol. 33, 386–391. [DOI] [PubMed] [Google Scholar]
- Mass RD, Press MF, Anderson S, Cobleigh MA, Vogel CL, Dybdal N, Leiberman G, Slamon DJ (2005) Evaluation of clinical outcomes according to HER2 detection by fluorescence in situ hybridization in women with metastatic breast cancer treated with Trastuzumab. Clin. Breast Cancer 6, 240–246. [DOI] [PubMed] [Google Scholar]
- Massarelli E, Herbst RS (2006) Use of novel second‐line targeted therapies in non‐small cell lung cancer. Semin. Oncol. 33 (Suppl. 1), S9–S16. [DOI] [PubMed] [Google Scholar]
- Ye D, Mendelsohn J, Fan Z (1999) Augmentation of a humanized anti‐HER2 mAb 4D5 induced growth inhibition by a human‐mouse chimeric anti‐EGF receptor mAb C225. Oncogene 18, 731–738. [DOI] [PubMed] [Google Scholar]
- Nahta R, Hung MC, Esteva FJ (2004) The HER‐2‐targeting antibodies Trastuzumab and Pertuzumab synergistically inhibit the survival of breast cancer cells. Cancer Res. 64, 2343–2346. [DOI] [PubMed] [Google Scholar]
- Nocito A, Bubendorf L, Maria Tinner E et al (2001) Microarrays of bladder cancer tissue are highly representative of proliferation index and histological grade. J. Pathol. 194, 349–357. [DOI] [PubMed] [Google Scholar]
- Pegram MD, Konecny GE, O’Callaghan C, Beryt M, Pietras R, Slamon DJ (2004) Rational combinations of Trastuzumab with chemotherapeutic drugs used in the treatment of breast cancer. J. Natl. Cancer Inst. 96, 739–749. [DOI] [PubMed] [Google Scholar]
- Plosker GL, Keam SJ (2006) Trastuzumab: a review of its use in the management of HER2‐positive metastatic and early‐stage breast cancer. Drugs 66, 449–475. [DOI] [PubMed] [Google Scholar]
- Poot M, Hoehn H, Kubbies M, Grossmann A, Chen Y, Rabinovitch PS (1994) Cell‐cycle analysis using continuous bromodeoxyuridine labeling and Hoechst 33358‐ethidium bromide bivariate flow cytometry. Methods Cell Biol. 41, 327–340. [DOI] [PubMed] [Google Scholar]
- Pritchard KI, Shepherd LE, O'Malley FP, Andrulis IL, Tu D, Bramwell VH, Levine MN (2006) National Cancer Institute of Canada Clinical Trials Group HER2 and responsiveness of breast cancer to adjuvant chemotherapy. N. Engl. J. Med. 354, 2103–2111. [DOI] [PubMed] [Google Scholar]
- Saltz. LB , Meropol NJ, Loehrer PJ Sr, Needle MN, Kopit J, Mayer RJ (2004) Phase II trial of Cetuximab in patients with refractory colorectal cancer that expresses the epidermal growth factor receptor. J. Clin. Oncol. 22, 1201–1208. [DOI] [PubMed] [Google Scholar]
- Shawver LK, Slamon D, Ullrich A (2002) Smart drugs: tyrosine kinase inhibitors in cancer therapy. Cancer Cell 1, 117–123. [DOI] [PubMed] [Google Scholar]
- Sliwkowski MX, Lofgren JA, Lewis GD, Hotaling TE, Fendly BM, Fox JA (1999) Non‐clinical studies addressing the mechanism of action of Trastuzumab (Herceptin). Semin. Oncol. 26 (Suppl. 12), 60–70. [PubMed] [Google Scholar]
- Spiridon CI, Ghetie MA, Uhr J, Marches R, Li JL, Shen GL, Vitetta ES (2002) Targeting multiple Her‐2 epitopes with monoclonal antibodies results in improved antigrowth activity of a human breast cancer cell line in vitro and in vivo . Clin. Cancer Res. 8, 1720–1730. [PubMed] [Google Scholar]
- Vokes EE, Chu E (2006) Anti‐EGFR therapies: clinical experience in colorectal, lung, and head and neck cancers. Oncology (Williston Park) 20 (Suppl. 2), 15–25. [PubMed] [Google Scholar]
- Walshe JM, Denduluri N, Berman AW, Rosing DR, Swain SM (2006) A phase II trial with Trastuzumab and Pertuzumab in patients with HER2‐overexpressed locally advanced and metastatic breast cancer. Clin. Breast Cancer 6, 535–539. [DOI] [PubMed] [Google Scholar]
- Yuste L, Montero JC, Esparis‐Ogando A, Pandiella A (2005) Activation of ErbB2 by overexpression or by transmembrane neuregulin results indifferential signaling and sensitivity to Herceptin. Cancer Res. 65, 6801–6810. [DOI] [PubMed] [Google Scholar]


