Abstract
Background and objectives: Adipose tissue‐derived stem cells (ASCs) have great potential for regenerative medicine. For molecular understanding of specific functional molecules present in ASCs, we analysed 756 proteins including specific chondrogenic functional factors, using high‐throughput nano reverse‐phase liquid chromatography–electrospray ionization–tandem mass spectrometry.
Materials, methods and results: Of these proteins, 33 were identified as chondrogenic factors or proteins including type 2 collagen, biglycan, insulin‐like growth factor‐binding protein and transforming growth factor‐beta 1 (TGF‐β1). ASCs are a possible cell source for cartilage regeneration as they are able to secrete a number of functional cytokines including chondrogenesis‐inducing molecules such as TGF‐β1 and bone morphogenetic protein 4 (BMP4). The chondrogenic phenotype of cultured ASCs was effectively induced by ASC‐culture media (CM) containing BMP4 and TGF‐β1, and maintained after pre‐treatment for 14 days in vitro and subcutaneous implantation in vivo. Chondrogenic differentiation efficiency of cultured ASCs and cultured mouse skin‐derived progenitor cells (SPCs) depended absolutely on ASC CM‐fold concentration. Cell density was also a very important factor for chondrogenic behaviour development during differentiation of ASCs and SPCs.
Conclusion: ASC CM‐derived TGF‐β1‐induced chondrogenic differentiation of ASCs resulted in significant reduction in chondrogenic activity after inhibition of the p38 pathway, revealing involvement of this MAPK pathway in TGF‐β1 signalling. On the other hand, TGF‐β1 signalling also led to SMAD activation that could directly increase chondrogenic activity of ASCs.
Introduction
Recently, MSCs isolated from human adipose tissue have been identified as another source of multipotent adult stem cells that can differentiate into multiple mesodermal‐derived tissues, and neuron‐like cells expressing neuronal markers have been reported (1, 2, 3). Compared to bone marrow cells, adipose tissue is more easily obtainable. Moreover, adipose tissue‐derived stem cells (ASCs) are a possible cell source of cartilage regeneration as they are able to secrete a number of functional cytokines into the culture medium, including chondrogenesis‐inducing molecules such as transforming growth factor‐beta (TGF‐β), bone morphogenetic protein 4 (BMP4) and fibroblast growth factor (FGF). In this study, the chondrogenic phenotype of cultured ASCs can be induced in culture medium containing BMP4 and TGF‐β; this phenotype can then be maintained after pretreatment for 14 days in vitro then subcutaneous implantation in vivo (4).
We analysed and identified proteins that were completely expressed from ASCs using the Nano RP LC–ESI–MS/MS system to be able to understand functions of ASC‐secreted proteins in detail. Our results demonstrate that ASCs avidly express a number of chondrogenic factors or proteins including type 2 collagen, biglycan, insulin‐like growth factor (IGF)‐binding protein and TGF‐β1. Various types of growth factor including IGF‐1 and members of the TGF‐β superfamily have been demonstrated to regulate cartilage matrix synthesis and to initiate intracellular signal transduction events in articular chondrocytes (5, 6, 7, 8, 9, 10). Besides added cytokines such as TGF‐β, cytokines secreted by ASCs may regulate differentiation of ASCs into chondrocytes. Cytokines produced by cultured cells, other than cytokines added externally to the culture, might affect cell differentiation, especially members of the TGF‐β superfamily, which participate in chondrogenesis (11, 12, 13). Influence of other members of the TGF‐β subfamily on chondrogenic differentiation shows that the mechanism is transduced through two major intracellular signalling pathways. The first pathway involves the SMAD family of signalling molecules (14), and the second involves mitogen‐activated protein kinase (MAPK) signalling (15, 16). Both signalling cascades are operated via a common TGF‐β receptor complex. BMP4 enhances production of articular cartilage matrix by stimulating synthesis of type 2 collagen and aggrecan; the latter is important in maintenance of the articular cartilage phenotype. Exogeneously added BMP4 acts synergistically with TGF‐β in inducing chondrogenesis in ASCs and chondroprogenitor cells; one study has shown that BMP4 functions in a dose‐dependent manner (15). On the other hand, the FGF family of cytokines plays an important role in chondrogenesis via MAPK phosphorylation (17, 18). This signal activation induces expression of Sox9, the master regulator during chondrogenic differentiation. Moreover, regulatory crosstalk between FGF‐activated signalling cascades and the other signalling pathways may occur, initiated by parathyroid hormone‐related peptides and hedgehog (19, 20). In this study, we investigated the role of endogeneously secreted TGF‐β1 and BMP4 in chondrogenic differentiation of cultured ASCs in vitro and in vivo, and clarified the molecular mechanisms involved. Results clearly identify TGF‐β1 and BMP4 as key factors that could be important in regulation and control of chondrogenic phenotype differentiation in cultured ASCs and in engrafted ASCs in vivo, and highlight some of the transduction pathways involved in this process.
Materials and methods
Isolation of ASCs and concentration of conditioned medium
Donor human adipose tissue‐derived stromal cells were obtained under local anaesthesia. The raw adipose tissue was processed according to established methodologies to obtain stromal vascular function (1). Each aliquot of frozen conditioned medium was thawed at 4 °C. As the medium thawed, working volumes of protease inhibitor cocktail solution and PMSF were added to each aliquot. Thawed medium was placed in an Amicon Ultra‐15 centrifugal filter device with 5000 molecular weight cut‐off (MWCO) and was centrifuged at 4000 g; final volumes of conditioned medium were concentrated ∼500‐fold. Total protein content of each extract was quantified using the Bio‐Rad protein assay kit (Milan, Italy).
Mouse skin progenitor cell culture and cell viability assay
Donor ICR mouse (25–30 g) skin tissues for derivation of progenitor cells were obtained under local anaesthesia. To isolate progenitor cells from this adult mouse skin, samples were washed extensively with equal volumes of PBS. Then, the epidermis was separated off, minced and enzyme digested at 37 °C for 30 min with 0.2% trypsin (Sigma, St Louis, MO, USA). Enzyme activity was neutralized with Dulbecco’s Modified Eagle’s Medium (DMEM; Life Technologies) containing 10% FBS, and centrifuged at 1200 g for 10 min to obtain a high‐density cell pellet. This skin progenitor cell pellet was collected by centrifugation as described above, and incubated overnight at 37 °C in 10% FBS containing DMEM. Cell viability was evaluated by visual cell counts in conjunction with trypan blue exclusion. In all viability assays, triplicate wells were used for each condition, and each experiment was repeated at least three times. In all viability assays, triplicate test wells were used for each condition. Raw data from each experiment were analysed using analysis of variance with Fisher’s or t‐tests.
Protein separation and preparation by SDS–polyacrylamide gel electrophoresis and in‐gel digestion
Samples solubilized in sample buffer (62.5 mm Tris–HCl, pH 6.8, 1% (W/V) SDS, 0.1 m DTT and 0.02% bromophenol blue dye) were loaded on to 15% SDS–PAGE. After electrophoresis, gels were stained with colloidal Coomassie blue G250 (ProteomeTech Inc). A 1 DE gel lane was cut in 20 consecutive pieces. Then, each gel piece was digested in‐gel with sequencing grade, modified trypsin (Promega, Madison, WI, USA). In brief, each gel piece was placed in a polypropylene (Eppendorf) tube and washed 4–5 times (until the gel was clear) with 150 μl of 1:1 acetonitrile/25 mm ammonium bicarbonate, pH 7.8. Gel slices were dried in a Speedvac concentrator and then rehydrated in 30 μl of 25 mm ammonium bicarbonate, pH 7.8, containing 20 ng trypsin. After incubation at 37 °C for 20 h, liquid was transferred to a new tube. Tryptic peptides remaining in the gel matrix were extracted for 40 min at 30 °C with 20 μl of 50% (v/v) aqueous acetonitrile containing 0.1% (v/v) formic acid. Combined supernatants were evaporated in a Speedvac concentrator and dissolved in 8 μl of 5% (v/v) aqueous acetonitrile solution containing 0.1% (v/v) formic acid for mass spectrometric analysis.
Identification of proteins by LC‐MS/MS analysis
Resulting tryptic peptides were separated and analysed using reversed phase capillary HPLC, directly coupled to a Finnigan LCQ ion trap mass spectrometer (LC‐MS/MS). Both 0.1 × 20 mm trapping and 0.075 × 130 mm resolving columns were packed with Vydac 218MS low trifluoroacetic acid C18 beads (5 μm in size, 300Å in pore size; Vydac, Hesperia, CA, USA) and placed in‐line. After peptides were bound to the trapping column for 10 min using 5% (v/v) aqueous acetonitrile containing 0.1% (v/v) formic acid, bound peptides were eluted using a 50 min gradient of 5–80% (v/v) acetonitrile containing 0.1% (v/v) formic acid at flow rate of 0.2 μl/min. For tandem mass spectrometry, full mass scan range mode was m/z = 450–2000 Da. After determination of charge states of an ion on zoom scans, product ion spectra were acquired in MS/MS mode with relative collision energy of 55%. Individual spectra from MS/MS were processed using TurboSEQUEST software (Thermo Quest, San Jose, CA, USA). Generated peak list files were used to query either MSDB database or NCBI using the MASCOT program (http://www.matrixscience.com). Modifications of methionine and cysteine, peptide mass tolerance at 2 Da, MS/MS ion mass tolerance at 0.8 Da, allowance of missed cleavage at 2 and charge states (+1, +2 and +3), were taken into account. Only significant hits as defined by MASCOT probability analysis were considered initially. Separation and analyses of tryptic peptides were performed on a Nano RP LC–ESI–MS/MS system, combining Ultimate nanoLC systems including FAMOS autosampler and Switchos column switching valve (LC‐Packings, Amsterdam, The Netherlands) connected to a QSTAR mass spectrometer (Applied Biosystems, Foster city, CA, USA) with nanospray interface with slight modification. For enrichment of peptides, self‐packed precolumn (2 cm × 200 μm I.D) with Zorbax 300SB‐C18, 5 μm (Agilent) and 0.1% formic acid was used at 4 μl/min flow rate for 10 min. Trapped peptides were separated on a self‐packed analytical column (15 cm × 75 μm I. D) with the same resin, and was eluted at flow rate of 0.2 μl/min, using 110 min gradient from 5% ACN to 32% ACN containing 0.1% formic acid. MS/MS spectra were acquired in an automated switching mode tandem mass spectrometer with m/z‐dependent set of collision offset values. Spectra obtained were automatically processed and searched against the NCBI non‐redundant database using MASCOT software package (Matrix Sciences, London, UK). Proteins, whose peptides were matched in statistical significance, were assigned.
Western blot analysis
For confirmation of differentially expressed proteins in experimental samples, cells were pooled and lysed in 500 μl of lysis buffer (20 mm Tris–HCl, pH 7.5, 150 mm NaCl, 1 mm EDTA, 1% Triton X‐100, 2.5 mm sodium pyrophosphate, 1 mm EGTA, 1 mm glycerophosphate, 1 mm Na3VO4 and 1 mm PMSF). Lysates were clarified by centrifugation at 15 000 g for 10 min and total protein content was determined using a Bio‐Rad protein assay kit. For Western blotting, equal amounts (40 μg) of protein extracts in lysis buffer were subjected to 10% SDS–PAGE analysis and transferred on to a nitrocellulose membrane. Anti‐type 2 collagen (1:500, Sigma), anti‐pSAPK/JNK (1:1000, Cell Signaling, Boston, MA, USA), anti‐pERK (1:1000, Cell Signaling), anti‐p38 (1:1000, Cell Signaling), anti‐SMAD2 (1:1000, Cell Signaling) and anti‐β‐actin (1:500, Sigma) antibodies were incubated with membranes. Relative band intensities were determined using Quality‐one 1‐D analysis software (Bio‐Rad, Chicago, IL, USA).
Bisulphite modification and sequencing of genomic DNA
Genomic DNA was purified by phenol/chloroform/isoamyl alcohol extraction, followed by a chloroform extraction, after which DNA was ethanol‐precipitated; DNA was dissolved in distilled water. Bisulphite conversion was conducted using EZ DNA Methylation–Gold Kit (Zymo Research, CA, USA), as described by the manufacturer. Briefly, non‐methylated cytosines in DNA were converted to uracil via heat‐denaturation of DNA, and with specifically designed CT conversion reagent. DNA was then desulphonated and subsequently cleaned and eluted. Bisulphite‐modified DNA was then immediately utilized for PCR or stored at, or below, −20 °C. Converted DNA was amplified by PCR using primers designed with MethPrimer (http://www.urogene.org/methprimer). PCR reactions were conducted in a MyGenie 96 Gradient Thermal Block (Bioneer, Daejeon, South Korea) in accordance with the following protocol: 95 °C for 15 min, 40 cycles of 95 °C for 20 s, 43–58 °C for 40 s and 72 °C for 30 s followed by extension at 72 °C for 10 min, and soaking at 4 °C. After electrophoresis on 1.5% agarose gel, remaining PCR products were cloned into bacteria (DH5α) using pGEM T‐Easy Vector System I (Promega). DNA extracted from bacterial clones was analysed by sequencing with M13 reverse primer, using ABI 3730XL capillary DNA sequencer (Applied Biosystems) and represented as rows of circles, with each circle symbolizing methylation state of one CpG.
In vitro and in vivo chondrogenesis
For chondrocyte differentiation, a pellet culture system was utilized. Samples of approximately 3 × 106 cells were placed in wells of a 96‐well plate. Pellets were cultured at 37 °C with 5% CO2 in 500 μl of chondrogenic media containing 6.25 g/ml of insulin, 10 ng/ml of TGF‐β1 and 50 ng of ascorbate‐2‐phosphate in control media for 2–3 weeks. Medium was replaced once every 2 days for 15 days. For calcium deposition and chondrocyte analysis in paraffin wax‐embedded tissue, specimens were stained using Masson’s and van Gieson’s methods. To evaluate in vivo differentiation potency, each sample was immobilized in Matrigel (BD Bioscience, Franklin Lakes, NJ, USA). Approximately 3 × 106 cells were mixed with Matrigel and were implanted subcutaneously in 8‐week‐old immunodeficient beige mice (NIH III/bg/nu/xid; Charles River Laboratories, Wilmington, MA, USA). All procedures were conducted in accordance with specifications of the approved protocol. Transplants were recovered 6 weeks later, fixed in 4% formalin and decalcified with 10% EDTA (pH 8.0) for paraffin wax embedding. Sections were cut then deparaffinized and stained in haematoxylin and eosin, and Masson’s and van Gieson’s methods for evaluation of chondrogenic differentiation potency.
Antibody blocking and inhibition assays
For functional study of ASC secreted TGF‐β1 and BMP4 into ASC culture media (CM), we induced antibody blockage to TGF‐β1 and BMP4 chondrogenic factors secreted from the cultured ASCs, using serially diluted anti‐TGF‐β1 and anti‐BMP4 antibodies. We also evaluated chondrogenic differentiation ability of ASCs, and expression and activation of mediated signalling molecules. Moreover, to confirm relevance of p38 and MEK signalling pathways, in terms of controlling chondrogenic differentiation in CM‐treated ASCs, ASCs were seeded in 10 cm dishes at of 5 × 105 density and cultured in 2% FBS containing α‐MEM for 8 h at 37 °C, in a CO2 incubator. Cells were then treated with ASC CM at various fold concentrations. CM‐treated ASCs were treated with p38 MAPK inhibitor, SB203580 (10 μm; Promega), MEK inhibitor, PD98059 (10 μm; Sigma), or were left untreated. Cells were proteins were analysed using Western blotting and RT‐PCR.
Statistical analysis
All data were expressed as mean ± SEM from five or more independent experiments. Statistical significance of difference between groups was calculated using Student’s two‐tailed t‐test.
Results
Characterization of ASC CM proteins by high‐throughput automated LC–MS/MS analysis and meaningful expression of chondrogenesis‐inducing proteins
Cells from three individual donors were combined to identify functional protein expressed by ASCs. As analysis of bands excised from one‐dimensional gels is still cumbersome, we endeavoured to avoid gel electrophoresis altogether by subjecting proteins isolated from cultured ASC extracts to trypsin digestion in solution. Tryptic peptides were fragmented on‐line using mass spectrometry, which provided a larger catalogue of proteins expressed by ASCs. All molecules identified by analysis of individual bands from a one‐dimensional gel were also identified by this approach. A total of 756 individual proteins was identified from cultured ASC extracts by Nano RP LC–ESI–MS/MS. Functionality of these proteins is summarized in Fig. 2. Selected functional categories included development (2%), antioxidation (1%), DNA‐binding protein (1%), cartilage function‐related protein (23%), signal transduction (5%), protein synthesis and processing (1%), metabolism (5%), apoptosis (8%), cell adhesion and migration (10%), cell cycle (7%), neural function (11%) and presently miscellaneous (31%) (Fig. 1a). Among these proteins, 33 were identified as chondrogenic factors or proteins, including type 2 collagen, biglycan, IGF‐binding protein and TGF‐β1. Table 1 presents identity, Moscow hit, molecular weight, scores and accession number of each chondrogenic functional protein. Figure 3a,b show spectra obtained by Nano RP LC–ESI–MS/MS analysis of the peptide mixture. This peptide indicated was matched to the amino acid sequence of TGF‐β1 and type 4 collagen (Fig. 1). Table 1 also shows chondrogenesis related secreted proteins from cultured ASCs, that included collagen alpha 2, extracellular matrix protein, biglycan, IGF binding protein and TGF‐β1.
Figure 2.
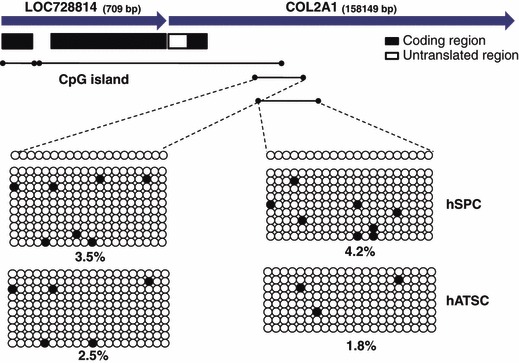
Methylation status of promoter region of type 2 collagen gene of cultured ASCs and SPCs as determined by bisulphite sequencing. Methylation level is presented as percentage of filled circles (denoting methylated cytosines) over total circles (cytosine in CpG) for a given locus in the genomic DNA preparation.
Figure 1.
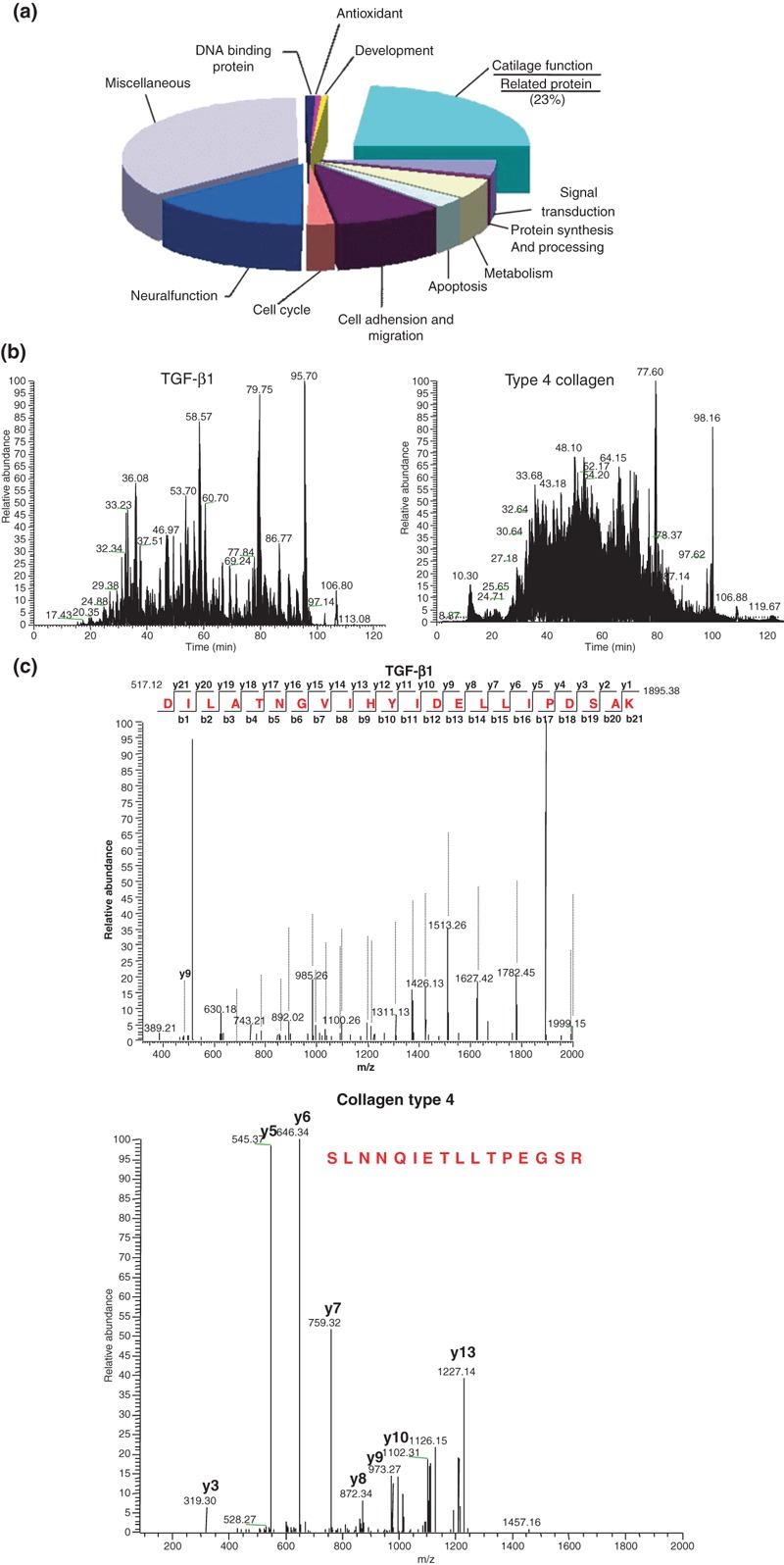
Functional annotation of total proteins secreted in ASC media and Nano RP LC–ESI–MS/MS analysis of TGB‐β1 and type 2 collagen proteins. (a) Proteins classified by biological processes. Values represent number of proteins classified into the indicated functional categories. (b) Nano RP LC–ESI–MS/MS analysis of peptide mixture of TGB‐β1 and type 4 collagen. (c) Total ion chromatogram of reverse‐phase LC separation of peptide mixture and MS spectrum of peptides eluted and detected at a dotted line of TGB‐β1 and type 4 collagen.
Table 1.
Major protein list in ASC CM
| Acc num | Protein name |
|---|---|
| gi|115351 | Collagen alpha 2(V) chain precursor |
| gi|1418930 | Prepro‐alpha2(I) collagen [Homo sapiens] |
| gi|1421278 | Extracellular Matrix Protein Mol_id: 1; Molecule: Sparc; Chain: Null; Fragment: |
| gi|15011913 | Collagen, type VI, alpha 1 precursor [Homo sapiens] |
| gi|16197601 | Type III preprocollagen alpha 1 chain [Homo sapiens] |
| gi|179433 | Biglycan |
| gi|179596 | Pre‐pro‐alpha‐2 type I collagen [Homo sapiens] |
| gi|180414 | Alpha‐1 type III collagen |
| gi|180671 | Collagenase type IV precursor |
| gi|54696462 | Decorin |
| gi|183894 | Insulin‐like growth factor binding protein 6 |
| gi|20380052 | COL3A1 protein [Homo sapiens] |
| gi|219510 | Collagen alpha 1(V) chain precursor [Homo sapiens] |
| gi|2388555 | Alpha2(I) collagen [Homo sapiens] |
| gi|25989621 | Collagen triple helix repeat‐containing protein 1 [Homo sapiens] |
| gi|27769056 | SERPINE2 protein [Homo sapiens] |
| gi|30030 | Alpha‐1 collagen VI (AA 574‐1009) [Homo sapiens] |
| gi|30054 | Alpha1 (III) collagen [Homo sapiens] |
| gi|30102 | Type I collagen [Homo sapiens] |
| gi|30582147 | Insulin‐like growth factor binding protein 7 [Homo sapiens] |
| gi|30582531 | Cofilin 1 (non‐muscle) [Homo sapiens] |
| gi|32879983 | Transforming growth factor, beta‐induced, 68kDa [Homo sapiens] |
| gi|33150528 | Osteoglycin OG [Homo sapiens] |
| gi|339992 | Tumour necrosis factor |
| gi|398164 | Insulin‐like growth factor binding protein 3 [Homo sapiens] |
| gi|4502945 | Alpha 1 type I collagen preproprotein [Homo sapiens] |
| gi|4507467 | Transforming growth factor, beta‐induced, 68kDa [Homo sapiens] |
| gi|5453834 | Periostin, osteoblast specific factor [Homo sapiens] |
| gi|55743106 | Alpha 3 type VI collagen isoform 5 precursor [Homo sapiens] |
| gi|57162640 | Collagen, type V, alpha 1 [Homo sapiens] |
| gi|6165881 | Collagen type XI alpha‐1 [Homo sapiens] |
| gi|62087268 | Alpha 1 type XI collagen isoform C preproprotein variant [Homo sapiens] |
| gi|984956 | Connective tissue growth factor |
Figure 3.
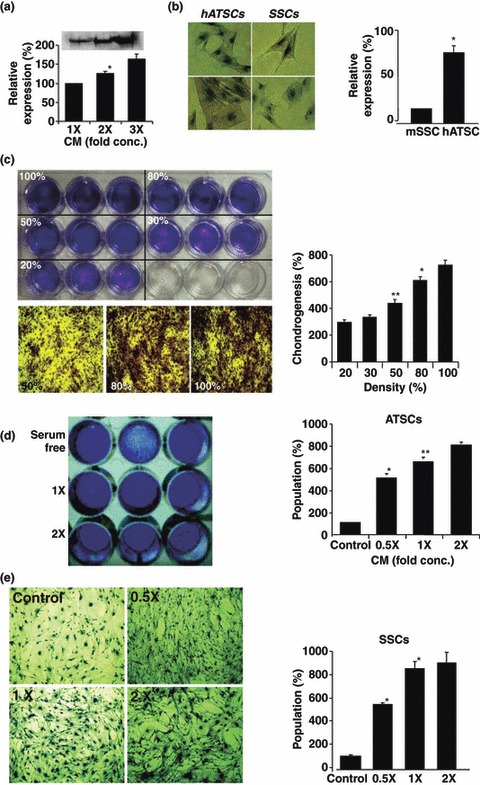
Effects of cell density and concentration of ASC CM on chondrogenic potency in cultured ASCs and SPCs. (a) Western blot analysis of secreted type 2 collagen at various‐fold concentrations in ASC CM. (b) Cell density effects on chondrogenic activity in ASCs and SPCs. Toluidine blue‐positive cells enumerated after chondrogenic differentiation and expressed as percentage relative to total number of cells. (c) Effect of density of ASCs on chondrogenic activity. At various densities of cultured ASCs, different efficiencies of chondrogenic behaviour were evident. Cell density of 100% resulted in most efficient chondrogenic behaviour. (d–e) Effect of ASC CM concentration on chondrogenesis of ASCs (d) and SPCs (e) (*P < 0.05, **P < 0.01).
Epigenetic priority of ASCs in chondrogenic potency
To study epigenetic roles of type 2 collagen genes in chondrogenic activity of ASCs and skin progenitor cells (SPCs), we evaluated DNA methylation intensity in functionally active type 2 collagen gene regulatory site of promoter regions (Fig. 2). Bisulphate sequencing analysis was also carried out to establish 5′‐3′ CpG methylation profiles across each test gene proximal promoter, proximal enhancer and early transcription start site. In the case of ASCs, two amplicons were assessed, which harboured meaningfully demethylated CpG dinucleotides relative to the transcriptional start site (Fig. 2).
ASCs effectively differentiate in a cartilage‐, CM concentration‐ and ASC density‐dependent manner
The effect of CM concentration on chondrogenic differentiation potency of SPCs and ASCs was assessed by inducing their differentiation, using serially (1‐, 2‐ and 3‐fold) concentrated CM. Results were quantitatively evaluated and chondrogenic potencies were compared against anti‐type 2 collagen by Western blotting. High dose of concentrated CM was more effective in inducing chondrogenesis in both ASCs and SPCs of constant density (Fig. 3a,b). To demonstrate that autocrine TGF‐β1 signalling was necessary for their self‐renewal, ASCs and mouse SPCs cultured at various cell densities (20–100%) were evaluated for chondrogenic differentiation potentials. When seeded at high clonal density, both types of cell more effectively stimulated chondrogenic differentiation than those at low clonal density (Fig. 3c). In particular, 100% cell density resulted in almost 4‐fold increase in chondrogenic activity compared to that evident at 20% cell density (Fig. 3c). Compared to ASCs and SPCs, those cells showed similar patterns of chondrogenic potency at a range of cell density.
ASC‐cultured CM induces SMAD2, SAPK/JUNK and ERK1/2 activation in ASCs and SPCs in a dose‐dependent manner
Treatment of ASCs and SPCs with CM for 14 days induced chondrogenic differentiation in monolayers, as assessed by increase in type 2 collagen immunoreactivity. Quantification of alizarin red staining demonstrated significant increase in proteoglycan synthesis (Fig. 6b). During CM treatment of cultured ASCs and SPCs, SMAD2, SAPK/JUNK and ERK1/2 signalling molecules were prominently phosphorylated at specific time points. Moreover, CM treatment in the presence of SMAD signal inhibitor SB431542, resulted in significant reduction in chondrogenic differentiation potency along with inhibition of SMAD phosphorylation (Fig. 4a). Moreover, SB431542 treatment resulted in substantial inhibition of ERK1/2 activation at high concentration (over 20 μm). High concentrations of CM effectively induced SMAD, SAPK/JUNK and ERK1/2 activation (Fig. 4b). Thus, ASCs CM‐induced chondrogenesis actively involves SMAD, SAPK/JUNK and ERK1/2 pathways.
Figure 6.
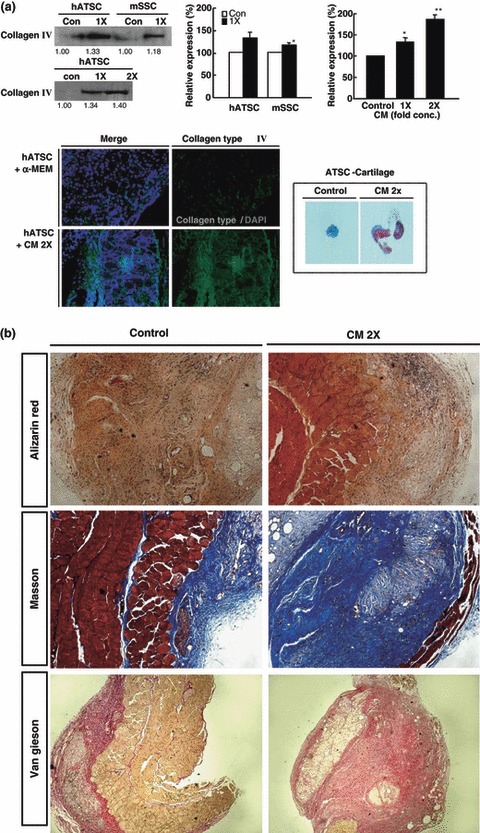
ASC CM effectively induces chondrogenesis of cultured ASCs and SPCs in NOD/SCID mice. (a) Western blot and immunohistochemistry revealed ASC CM effectively promotes chondrogenesis of ASCs and SPCs in vivo in a CM concentration‐dependent manner (*P < 0.05, **P < 0.01). (b) Confirmation of chondrogenic activity of ASC CM combined ASCs in NOD/SCID mice by histological treatment with alizarin red, Masson’s trichrome, and van Gieson’s stain.
Figure 4.
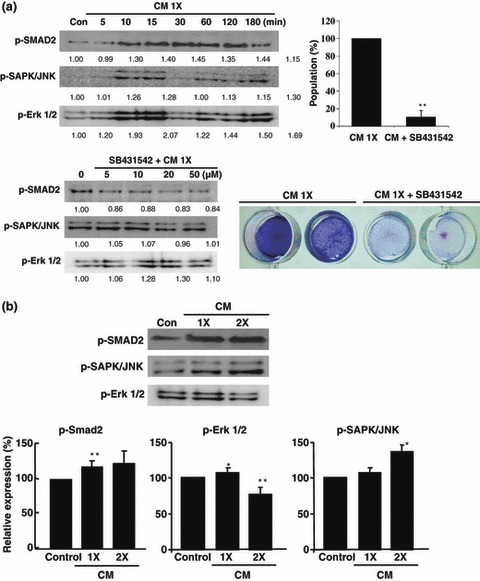
Differential expressions of TGF‐β1‐mediated signal pathway mediators at various times. (a) Involvement of p38/JUNK, ERK1/2 and SMAD2‐mediated signal pathways in ASC CM induction of chondrogenic differentiation of cultured ASCs. Inhibition of SMAD2 activation using the specific inhibitor SB431542l resulted in blocked chondrogenic activity in ASCs. (b) p38/JUNK, ERK1/2 and SMAD2 activation gradually increased in a CM concentration‐dependent manner (*P < 0.05, **P < 0.01).
ASC CM‐derived TGF‐β1 and BMP4 were directly involved in ASCs CM‐induced chondrogenesis via SMAD2/JUNK activation
CM‐induced chondrogenic activity in ASCs and SPCs was dramatically inhibited by treatment with anti‐TGF‐β1 and anti‐BMP4 antibodies. They separately blocked CM during chondrogenesis induction in both ASCs and SPCs and promptly decreased chondrogenic activity by 50% and 60%, respectively, compared to control CM‐treated cells (Fig. 5a). Anti‐TGF‐β1 and anti‐BMP4 antibodies markedly blocked SMAD, SAPK/JUNK and ERK1/2 phosphorylation at low concentration of antibody (Fig. 5a). Moreover, treatment of ASCs with cultured CM induced stable gene expression of BMP4, Runx2 and Sox‐9 in differentiating ASCs; these genes could be important in ASC CM‐induced ASC chondrogenesis (Fig. 5b). Finally, ASCs producing CM effectively induced TGF‐β1 and BMP4 to cooperatively promote chondrogenesis in vitro via SMAD2 and SAPK/JUNK signal pathways and also BMP4‐induced ERK1/2 phosphorylation (Fig. 5a,c). When BMP4 present in ASC CM was blocked, chondrogenic activity of ASC CM was markedly inhibited along with ERK1/2 phosphorylation (Fig. 5a,c).
Figure 5.
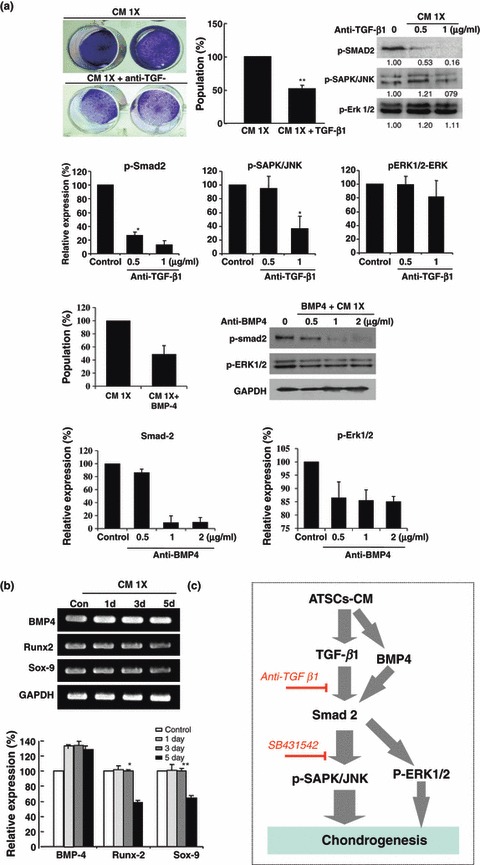
TGF‐β1 and BMP4 being crucial in ASC CM‐induced chondrogenesis in cultured ASCs and nature of mechanism involved. (a) Blockage of CM‐derived TGF‐β1 using anti‐TGF‐β1 antibody resulted in prominent decrease of chondrogenic activity of CM and also inhibited TGF‐β1‐mediated activation and expression of signalling molecules such as SMAD, SAPK/JUNK and ERK1/2. (b) Effective blockage of CM‐derived BMP4 using anti‐BMP4 antibody resulted in prominent decrease of chondrogenic activity of CM and also blocked related activation of signalling molecules such as SMAD, SAPK/JUNK and ERK1/2. (c) Several chondrogenenesis‐related transcription factors and BMP4 expression before and after treatment with ASC CM. (d) Schematic flowchart of major signalling pathways important in CM‐stimulated chondrogenesis in cultured ASCs (*P < 0.05, **P < 0.01).
In vivo chondrogenesis acts in an ASC CM concentration‐dependent manner
The effect of ASC CM on chondrogenic potency of cultured ASCs and SPCs was evaluated in vivo in NOD/SCID mice. An ASC mixture that combined ASC CM (with or without control) and ASCs mixed with Matrigel were injected into 5‐ to 6‐week‐old NOD/SCID mice to induce chondrogenic differentiation. After 4 weeks engraftment, CM‐treated ASCs effectively generated type 2 collagen and proteoglycan‐expressing cartilage, compared to mice in which untreated ASCs were engrafted; this shows that the CM effect was dose‐dependent (Fig. 6a). At higher concentration, CM‐mixed ASCs more effectively differentiated into cartilage that expressed high amounts of type 2 collagen. Moreover, the amount of differentiated cartilage was higher than that achieved after injection of untreated ASCs. To confirm chondrogenic activity of CM‐treated ASCs in vivo, differentiated cartilage tissue was examined after alizarin red, Masson’s trichrome, and van Gieson staining. These results also revealed that doubly concentrated CM‐mixed ASCs markedly produced cartilage in vivo (Fig. 6b).
Discussion
Several types of articular cartilage defect caused by ageing, chondral injury or degenerative disease are seldom repaired, causing long‐term pain and restricted mobility. Therapy is usually restricted to surgical intervention including cell‐based therapeutic options of autologous chondrocyte transfer; regenerative therapy for specific chondroprogenitor cells is urgently required. The discovery of adult stem cells such as adipose stromal cells is germane to cartilage regenerative medicine and also paves the way for identification of molecular events characterizing cell number expansion and differentiation, with the aim of generating stable articular chondrocytes. In general, cytokines are produced by cultured cells, apart from those added externally to culture medium, and these might influence cell differentiation into several lineages, including chondrogenesis. Thus, cell density could have some affect on cell differentiation as cytokine production rate depends on cell density. Our study also revealed that ASC and SPC densities were very important for differentiation. Moreover, high cell densities showed more effective chondrogenic potency. Besides externally added cytokines, cytokines secreted by ASCs have been considered to regulate differentiation of ASCs themselves. But it remains unclear how density of ASCs influences their differentiation into chondrocytes. To confirm presence of soluble factors produced by cells during differentiation and acceleration of ASC chondrogenesis, CM harvested from cultured ASCs were concentrated into 1‐, 2‐ and 3‐fold concentrated CM, and chondrogenic potency was evaluated after application of different CM concentrations to cultured ASCs. High concentrations of CM effectively induced chondrogenic differentiation in both cultured ASCs and SPCs. ASC CM might contain soluble factors accelerating increase in expression level of type 2 collagen. Concentration of these possible soluble factors might be higher in cultures inoculated with increased numbers of cells. Moreover, ASCs are a possible cell source for cartilage regeneration because they can differentiation into cartilage‐like cells. Optimal culture conditions and proper stimulation can induce differentiation of ASCs into chondrocytes. Regulatory mechanisms involved in chondrogenic lineage signalling cascades are not sufficiently understood, but do involve numerous growth factors, proteases and cytokines, in a strictly controlled time‐dependent manner (1, 21, 22, 23, 24, 25, 26, 27). Chondrogenesis is controlled by several transcription factors, in particular, members of the Sox and Cbfa families. Sox9 has been characterized as a master transcription factor with essential direct or indirect regulatory effects in the entire chondrogenic process (28, 29, 30). We observed that BMP4, Runx2 and Sox9 were also prominently expressed after induction of chondrogenesis by ASC CM treatment. Expression of the major osteogenic transcription factor Cbfa1 or Runx2 is necessary for inducing gene expression of collagen X in terminally differentiated chondrocytes (28, 29, 31, 32).
Although potency of bone marrow stromal cells for chondrogenesis and cartilage application has been well reported as a stem cell source (33, 34, 35), little is known of how such soluble factors affect cartilage differentiation potency, their molecular pathways and integration processes of cells. In this study, the chondrogenic phenotype has been induced by TGF‐β1‐containing differentiation medium and can be maintained in vitro after pre‐treatment, and in vivo after subcutaneous engraftment. Presently, TGF‐β1‐induced chondrogenic differentiation of ASCs and inhibition of the p38 pathway resulted in significant reduction in chondrogenic activity, revealing involvement of the MAPK pathway in TGF‐β1 signalling, whereas inhibition of ERK1/2 and PI3K pathways did not affect chondrogenic differentiation (data not shown). The present results also demonstrated involvement of the p38 pathway in chondrogenic gene expression in human ASCs treated with ASC CM prominently containing TGF‐β1, during in vitro chondrogenesis. On the other hand, activation of TGF‐β1 signaling may lead to SMAD activation that might directly increase chondrogenic activity of ASCs. TGF‐β1 upregulated Sox9 gene expression in differentiating cells. Sox9 protein binds to promoter regions of type 2 collagen and aggrecan, enhancing transcription of type 2 collagen and aggrecan mRNA. The SMAD signalling pathway also interacted with various other TGF‐β1‐related signalling pathways including p38, ERK1/2 and JUNK phosphorylation. Yet, hydrostatic pressure and BMP6 effectively compensated for altered TGF beta receptor and BMP profile inducing reduced chondrogenic potential of ASCs (36, 37).
In conclusion, our study has identified ASC derived as a valuable chondrogenic effector for chondrogenesis of autologously available ASCs or skin‐derived progenitor cells. We have identified a definitive molecular mechanism involving chondrogenic behaviour of ASC CM. Finally, cultured ASCs’ CM, in combination with ASCs may initiate a promising means of treating cartilage defects.
Acknowledgements
This study was supported by the 21st Century Frontier/Stem Cell Research Committee (SC5112) in South Korea.
References
- 1. Kang SK, Putnam L, Dufour J, Ylostalo J, Jung JS, Bunnell BA (2004) Expression of telomerase extends the lifespan and enhances osteogenic differentiation of adipose tissue‐derived stromal cells. Stem Cells 22, 1356–1372. [DOI] [PubMed] [Google Scholar]
- 2. Kang SK, Putnam LA, Ylostalo J, Popescu IR, Dufour J, Belousov A et al. (2004) Neurogenesis of Rhesus adipose stromal cells. J. Cell Sci. 117, 4289–4299. [DOI] [PubMed] [Google Scholar]
- 3. Pittenger MF, Mackay AM, Beck SC, Jaiswal RK, Douglas R, Mosca JD et al. (1999) Multilineage potential of adult human mesenchymal stem cells. Science 284, 143–147. [DOI] [PubMed] [Google Scholar]
- 4. Erickson GR, Gimble JM, Franklin DM, Rice HE, Awad H, Guilak F (2002) Chondrogenic potential of adipose tissue‐derived stromal cells in vitro and in vivo. Biochem. Biophys. Res. Commun. 290, 763–769. [DOI] [PubMed] [Google Scholar]
- 5. Frenkel SR, Saadeh PB, Mehrara BJ, Chin GS, Steinbrech DS, Brent B et al. (2000) Transforming growth factor beta superfamily members: role in cartilage modeling. Plast. Reconstr. Surg. 105, 980–990. [DOI] [PubMed] [Google Scholar]
- 6. Grimaud E, Heymann D, Redini F (2002) Recent advances in TGF‐beta effects on chondrocyte metabolism. Potential therapeutic roles of TGF‐beta in cartilage disorders. Cytokine Growth Factor Rev. 13, 241–257. [DOI] [PubMed] [Google Scholar]
- 7. Heng BC, Cao T, Lee EH (2004) Directing stem cell differentiation into the chondrogenic lineage in vitro. Stem Cells 22, 1152–1167. [DOI] [PubMed] [Google Scholar]
- 8. Miyazono K (2000). Positive and negative regulation of TGF‐beta signaling. J. Cell Sci.. 113(Pt 7), 1101–1109. [DOI] [PubMed] [Google Scholar]
- 9. Roelen BA, Dijke P (2003) Controlling mesenchymal stem cell differentiation by TGFBeta family members. J. Orthop. Sci. 8, 740–748. [DOI] [PubMed] [Google Scholar]
- 10. Starkman BG, Cravero JD, Delcarlo M, Loeser RF (2005) IGF‐I stimulation of proteoglycan synthesis by chondrocytes requires activation of the PI 3‐kinase pathway but not ERK MAPK. Biochem. J. 389, 723–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hall FL, Benya PD, Padilla SR, Carbonaro‐Hall D, Williams R, Buckley S et al. (1996). Transforming growth factor‐beta type‐II receptor signalling: intrinsic/associated casein kinase activity, receptor interactions and functional effects of blocking antibodies. Biochem. J.. 316(Pt 1), 303–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mackay AM, Beck SC, Murphy JM, Barry FP, Chichester CO, Pittenger MF (1998) Chondrogenic differentiation of cultured human mesenchymal stem cells from marrow. Tissue Eng. 4, 415–428. [DOI] [PubMed] [Google Scholar]
- 13. Oh CD, Chun JS (2003) Signaling mechanisms leading to the regulation of differentiation and apoptosis of articular chondrocytes by insulin‐like growth factor‐1. J. Biol. Chem. 278, 36563–36571. [DOI] [PubMed] [Google Scholar]
- 14. Massague J, Wotton D (2000) Transcriptional control by the TGF‐beta/Smad signaling system. EMBO J. 19, 1745–1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nakayama N, Duryea D, Manoukian R, Chow G, Han CY (2003) Macroscopic cartilage formation with embryonic stem‐cell‐derived mesodermal progenitor cells. J. Cell Sci. 116, 2015–2028. [DOI] [PubMed] [Google Scholar]
- 16. Tuli R, Seghatoleslami MR, Tuli S, Howard MS, Danielson KG, Tuan RS (2002) p38 MAP kinase regulation of AP‐2 binding in TGF‐beta1‐stimulated chondrogenesis of human trabecular bone‐derived cells. Ann. N Y Acad. Sci. 961, 172–177. [DOI] [PubMed] [Google Scholar]
- 17. Ornitz DM, Marie PJ (2002) FGF signaling pathways in endochondral and intramembranous bone development and human genetic disease. Genes Dev. 16, 1446–1465. [DOI] [PubMed] [Google Scholar]
- 18. Murakami S, Kan M, McKeehan WL, De Crombrugghe B (2000) Up‐regulation of the chondrogenic Sox9 gene by fibroblast growth factors is mediated by the mitogen‐activated protein kinase pathway. Proc. Natl. Acad. Sci. USA 97, 1113–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chen L, Li C, Qiao W, Xu X, Deng C (2001) A Ser(365)‐‐>Cys mutation of fibroblast growth factor receptor 3 in mouse downregulates Ihh/PTHrP signals and causes severe achondroplasia. Hum. Mol. Genet. 10, 457–465. [DOI] [PubMed] [Google Scholar]
- 20. Naski MC, Ornitz DM (1998) FGF signaling in skeletal development. Front Biosci. 3, d781–d794. [DOI] [PubMed] [Google Scholar]
- 21. Chen P, Carrington JL, Hammonds RG, Reddi AH (1991) Stimulation of chondrogenesis in limb bud mesoderm cells by recombinant human bone morphogenetic protein 2B (BMP‐2B) and modulation by transforming growth factor beta 1 and beta 2. Exp. Cell Res. 195, 509–515. [DOI] [PubMed] [Google Scholar]
- 22. Kramer J, Hegert C, Guan K, Wobus AM, Muller PK, Rohwedel J (2000) Embryonic stem cell‐derived chondrogenic differentiation in vitro: activation by BMP‐2 and BMP‐4. Mech. Dev. 92, 193–205. [DOI] [PubMed] [Google Scholar]
- 23. Kuroda R, Usas A, Kubo S, Corsi K, Peng H, Rose T et al. (2006) Cartilage repair using bone morphogenetic protein 4 and muscle‐derived stem cells. Arthritis Rheum. 54, 433–442. [DOI] [PubMed] [Google Scholar]
- 24. Mehlhorn AT, Niemeyer P, Kaschte K, Muller L, Finkenzeller G, Hartl D et al. (2007) Differential effects of BMP‐2 and TGF‐beta1 on chondrogenic differentiation of adipose derived stem cells. Cell Prolif. 40, 809–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Miljkovic ND, Cooper GM, Marra KG (2008) Chondrogenesis, bone morphogenetic protein‐4 and mesenchymal stem cells. Osteoarthritis Cartilage 16, 1121–1130. [DOI] [PubMed] [Google Scholar]
- 26. Mouw JK, Connelly JT, Wilson CG, Michael KE, Levenston ME (2007) Dynamic compression regulates the expression and synthesis of chondrocyte‐specific matrix molecules in bone marrow stromal cells. Stem Cells 25, 655–663. [DOI] [PubMed] [Google Scholar]
- 27. Shea CM, Edgar CM, Einhorn TA, Gerstenfeld LC (2003) BMP treatment of C3H10T1/2 mesenchymal stem cells induces both chondrogenesis and osteogenesis. J. Cell. Biochem. 90, 1112–1127. [DOI] [PubMed] [Google Scholar]
- 28. Bi W, Deng JM, Zhang Z, Behringer RR, De Crombrugghe B (1999) Sox9 is required for cartilage formation. Nat. Genet. 22, 85–89. [DOI] [PubMed] [Google Scholar]
- 29. Lefebvre V, Behringer RR, De Crombrugghe B (2001) L‐Sox5, Sox6 and Sox9 control essential steps of the chondrocyte differentiation pathway. Osteoarthritis Cartilage 9(Suppl. A), 69–75. [DOI] [PubMed] [Google Scholar]
- 30. Semba I, Nonaka K, Takahashi I, Takahashi K, Dashner R, Shum L et al. (2000) Positionally‐dependent chondrogenesis induced by BMP4 is co‐regulated by Sox9 and Msx2. Dev. Dyn. 217, 401–414. [DOI] [PubMed] [Google Scholar]
- 31. De Crombrugghe B, Lefebvre V, Behringer RR, Bi W, Murakami S, Huang W (2000) Transcriptional mechanisms of chondrocyte differentiation. Matrix Biol. 19, 389–394. [DOI] [PubMed] [Google Scholar]
- 32. Mundlos S, Olsen BR (1997) Heritable diseases of the skeleton. Part I: molecular insights into skeletal development‐transcription factors and signaling pathways. FASEB J. 11, 125–132. [DOI] [PubMed] [Google Scholar]
- 33. Mehlhorn AT, Niemeyer P, Kaiser S, Finkenzeller G, Stark GB, Sdkamp NP et al. (2006) Differential expression pattern of extracellular matrix molecules during chondrogenesis of mesenchymal stem cells from bone marrow and adipose tissue. Tissue Eng. 12, 2853. [DOI] [PubMed] [Google Scholar]
- 34. Knippenberg M, Helder MN, Zandieh Doulabi B, Wuisman PI, Klein‐Nulend J (2006) Osteogenesis versus chondrogenesis by BMP‐2 and BMP‐7 in adipose stem cells. Biochem. Biophys. Res. Commun. 14, 902–908. [DOI] [PubMed] [Google Scholar]
- 35. Afizah H, Yang Z, Hui JH, Ouyang HW, Lee EH (2007) A comparison between the chondrogenic potential of human bone marrow stem cells (BMSCs) and adipose‐derived stem cells (ADSCs) taken from the same donors. Tissue Eng. 13, 659–667. [DOI] [PubMed] [Google Scholar]
- 36. Ogawa R, Mizuno S, Murphy GF, Orgill DP. (2009). The effect of hydrostatic pressure on 3‐D chondroinduction of human adipose‐derived stem cells. Tissue Eng. 15, 2937–2945. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hennig T, Lorenz H, Thiel A, Goetzke K, Dickhut A, Geiger F et al. (2007) Reduced chondrogenic potential of adipose tissue derived stromal cells correlates with an altered TGFbeta receptor and BMP profile and is overcome by BMP‐6. J. Cell. Physiol. 211, 682–691. [DOI] [PubMed] [Google Scholar]


