Abstract
Abstract. Objectives: Human amnion is an easy‐to‐obtain novel source of human mesenchymal stem cells, which poses little or no ethical dilemmas. We have previously shown that human amnion‐derived mesenchymal (HAM) cells exhibit certain mesenchymal stem cell‐like characteristics with respect to expression of stem cell markers and differentiation potentials. Materials and methods: In this study, we further characterized HAM cells’ potential for in vivo therapeutic application. Results: Flow cytometric analyses of HAM cells show that they express several stem cell‐related cell surface markers, including CD90, CD105, CD59, CD49d, CD44 and HLA‐ABC, but not CD45, CD34, CD31, CD106 or HLA‐DR. HAM cells at the 10th passage showed normal karyotype. More interestingly, the AbdB‐like HOXA genes HOXA9, HOXA10 and HOXA11 that are expressed in the mesenchyme of the developing female reproductive tract and pregnant uteri are also expressed in HAM cells, suggesting similarities between these two mesenchymal cell types. Progesterone receptor is also highly expressed in HAM cells and expression of genes or proteins in HAM cells could be manipulated with the aid of lentivirus technology or cell‐permeable peptides. To test potentials of HAM cells for in vivo application, we introduced enhanced green fluorescence protein (EGFP)‐expressing HAM cells to mice by intrauterine infusion (into uteri) or by intravenous injection (into the circulation). Presence of EGFP‐expressing cells within the uterine mesenchyme after intrauterine infusion or in lungs after intravenous injection was noted within 1–4 weeks. Conclusions: Collectively, these results suggest that HAM cells are a potential source of mesenchymal stem cells with therapeutic potential.
INTRODUCTION
Embryonic stem cells (ESCs) possess potential for differentiation into a wide range of cell lineages (Thomson et al. 1998). Ethical issues, associated with establishment of human ESC lines as well as questions of undesirable properties such as teratoma formation and immune rejection, need to be resolved prior to taking fully fledged advantage of ESCs (Drukker & Benvenisty 2004). While these questions are being pursued, mesenchymal stem cells (MSCs) obtained from various human adult tissues have been targets of recent interest to substitute ESCs, at least, for some stem cell therapeutics (Mimeault & Batra 2006).
Human mesenchymal stem cells (hMSCs) are capable of differentiating into multiple lineages of cells and thus are considered multipotent (Pittenger et al. 1999). Bone marrow (BM)‐derived hMSCs have been widely investigated for their differentiation potential and immunomodulatory properties in the context of stem cell transplantation (Gregory et al. 2005a,b; Ryan et al. 2005; , Chang et al. 2006). MSCs have been obtained from a variety of sources, including amniotic fluid, amnion, chorion, cord blood and adult tissues (Fukuchi et al. 2004; Tsai et al. 2004; Miki et al. 2005; Zhao et al. 2005; , De Coppi et al. 2007; Guillot et al. 2007; Kim et al. 2007b). Some of these have been shown to possess specific differentiation potentials into, for example, cardiomyocytes and endothelial cells plus other mesenchymal lineages. The human amnion, which can be detached simply from a term placenta, serves as a potential source of hMSCs and poses little in the way of ethical issues. Amniotic epithelial cells have been shown to be able to differentiate into cells of all three germ layers (Miki et al. 2005) and to endothelial cells (Alviano et al. 2007). We have previously established human amnion‐derived mesenchymal (HAM) cells and have examined their differentiational properties into osteogenic, adipogenic and chondrogenic lineages (Kim et al. 2007a). However, in vivo therapeutic potentials of HAM cells are yet to be investigated.
CD34 is generally used to identify haematopoietic stem cells. CD34+ cells obtained from human BM differentiate into multiple lineages of the haematopoietic system and express several HOX genes, such as HOXB4, HOXA9 and HOXA10. These transcription factors are implicated in cell proliferation and differentiation into distinct haematopoietic progenitor lineages. However, in the human BM, CD34− cells, including mesenchymal cells, do not express recognizable levels of HOXA10 (Sauvageau et al. 1994). In mice, AbdB‐like Hoxa genes, namely Hoxa9, Hoxa10 and Hoxa11, are expressed in a nested pattern in the female reproductive tract (FRT) directing differentiation of the oviduct, uterus and vagina (Ma et al. 1998). These genes are all expressed in mesenchymal cells of the reproductive tract during embryonic development and in the adult uterus. Hoxa10 and Hoxa11 are primarily expressed in mouse uterine stromal (mesenchymal) cells during pregnancy and participate in cell proliferation (Lim et al. 1999). It is not yet known whether any human MSCs express AdbB‐like HOXA genes or whether they are involved in self‐renewal or any differentiational potential of MSCs.
In the present investigation, we have characterized cellular properties of HAM cells using multiple experimental approaches. We have confirmed that HAM cells are similar to human BM‐derived MSCs and MSCs from other sources, expressing representative mesenchymal cell surface markers CD105 and CD90. One unique feature of HAM cells reported herein is that they abundantly express HOXA9, HOXA10, HOXA11 and WNT5a, all of which are involved in developmental patterning of the reproductive tract and normal uterine function in adults. Furthermore, utilizing approaches for manipulating gene or protein expression, we provide evidence that HAM cells can serve as a potential source for stem cell therapy.
MATERIALS AND METHODS
Isolation of HAM Cells
Human placentas were obtained with informed consent from patients at Seoul National University Bundang Hospital, with approval of the Institutional Review Board. The fresh amnion was mechanically separated from the chorion and was washed with phosphate‐buffered saline (PBS) to remove cellular debris. Minced amnion pieces were subjected to trypsin digestion [0.25% trypsin in Dulbecco's modified Eagle's medium (DMEM)] to remove epithelial cells. Tissue pieces were then treated with 2 mg/mL collagenase A and 0.05 mg/mL deoxyribonuclease (DNase; Roche, Indianapolis, IN, USA). Dispersed mesenchymal cells were collected by centrifugation at 500 g. After several washes with DMEM, cells were plated in 75‐mL culture flasks containing DMEM supplemented with penicillin (100 U/mL), streptomycin (0.1 mg/mL), sodium carbonate (3.7 mg/mL) and 10% foetal bovine serum (FBS; Invitrogen, Carlsbad, CA, USA). The culture medium was changed once a week until fibroblast‐like cells appeared and thereafter twice a week. Establishment of amniotic fluid (AF)‐derived and umbilical cord (UC)‐derived MSCs was as described previously (Kim et al. 2007b; Kim et al. unpublished).
Flow cytometric analysis for cell surface markers and cell cycle
Trypsinized HAM cells were re‐suspended in DMEM supplemented with 10% FBS and were washed with PBS containing 3% FBS. Cells were incubated with primary antibodies for 1 h at 4 °C, and then unbound antibodies were removed by washing. For stage‐specific embryonic antigen‐4 (SSEA‐4), TRA 1–60, TRA 1–80 and TRA 2–54 staining, FITC‐conjugated secondary antibody was used for labelling. After washing, cells were analysed on a FACSCalibur flow cytometer (BD Biosciences, Franklin Lakes, NJ, USA) equipped with CellQuest software. Monoclonal antibodies for flow cytometric analysis were purchased from the following sources: HLA‐DR, CD31, HLA‐ABC, CD106, CD90, CD59, CD49d, CD45, CD34 and CD44 (BD Biosciences); SSEA‐4, TRA 1–60, TRA 1–80, TRA 2–54 (Chemicon, Temecula, CA, USA); CD105 (R&D System, Minneapolis, MN, USA); HLA‐G (Novus Biologicals, Littleton, CO, USA); CD117 (Miltenyi Biotec, Bergisch Gladbach, Germany). Table 1 shows the list of CD antigens investigated in this study.
Table 1.
Immunophenotype of human amnion‐derived mesenchymal (HAM) cells: list of CD antigens examined in this study
| CD antigen | Other names | Function | HAM |
|---|---|---|---|
| CD105 | SH2, Endoglin | Mesenchymal stem cell marker | + |
| CD90 | Thy‐1 | Mesenchymal stem cell marker | + |
| CD59 | Protectin | Negative regulator of complement activation | + |
| CD49d | VLA‐4 | Integrin α4 subunit | + |
| CD44 | H‐CAM | Haematopoietic and non‐haematopoietic stem cell marker, mesenchymal stem cell marker | + |
| CD45 | Leukocyte common antigen | Haematopoietic cell marker | – |
| CD34 | Gp105–120, Mucosialin | Haematopoietic stem cell marker, cell adhesion | – |
| CD31 | PECAM‐1 | Immunoglobulin superfamily marker, cell adhesion | – |
| CD106 | VCAM‐1 | Immunoglobulin superfamily marker | – |
| HLA‐ABC | MHC class I | + | |
| HLA‐DR | MHC class II | – | |
| HLA‐G | Soluble MHC | ± | |
| SSEA4 | EG, EC, ES cell marker | ± | |
| TRA 1–60 | EG, EC, ES cell marker | ± | |
| TRA 1–81 | EG, EC, ES cell marker | ± | |
| TRA 2–54 | EC and ES cell marker | ± | |
| CD117 | c‐kit | Germ cell marker, haematopoietic stem cell marker | ± |
EG, embryonic germ cells; EC, teratocarcinoma stem cells; ES, embryonic stem cells.
±: weakly positive.
RNA isolation and reverse transcription‐PCR analysis
Total RNA was extracted from cultured cells by using TRIzol Reagent (Molecular Research Center Inc., Cincinnati, OH, USA) according to the manufacturer's protocol. The resuspended RNA was then treated with ribonuclease A (RNaseA)‐free DNase (Takara, Otsu, Japan) for 1 h at 37 °C to remove any contaminated genomic DNA. RNA concentration and purity were evaluated by the ratio of optical density (OD)260 : (OD)280 by spectrophotometry. RNA samples (2 µg each) were subjected to reverse transcription (RT) using SuperScript III reverse transcriptase (Invitrogen) for cDNA synthesis. Samples were either used directly for PCR or were stored at –20 °C. PCR was carried out using Prime Taq Premix (2 X) (GeNet Bio, Cheonan, South Korea) and reactions were run on a PCR thermal cycler (Applied Biosystems, Foster City, CA, USA). Following initial denaturation at 95 °C for 5 min, PCR was performed at 95 °C for 30 s at specific annealing temperature (55–62 °C) for 30 s, and at 72 °C for 30–60 s. Amplified products were analysed using 1.5% agarose gel electrophoresis. PCR reaction of hypoxanthine‐guanine phosphoribosyltransferase (HPRT) was performed as internal control. Sequences of primers used for PCR analysis are given in Table 2.
Table 2.
Primers used for RT‐PCR analysis of HAM cells
| Gene name | Primer sequence | Product size (base pair) |
|---|---|---|
| WNT4a | F: CCTTCGTGTACGCCATCTCT R: TCAGAGCATCCTGACCACTG | 142 |
| WNT5a | F: CTGGCAGGACTTTCTCAAGG R: CCTTCGATGTCGGAATTGAT | 123 |
| WNT7a | F: GCCTGGACGAGTGTCAGTTT R: CTGGCCTTGCTTCTCTTTGT | 215 |
| HOXA9 | F: TCGATCCCAATAACCCAGC R: CACTCGTCTTTTGCTCGG | 249 |
| HOXA10 | F: AGAGCAGCAAAGCCTCGCCG R: AAGTTGGCTGTGAGCTCCCGGATCC | 298 |
| HOXA11 | F: CGTGCGCGAAGTGACCTTCAGAGAGTAC R: CCTGCCCACGGTGCTATAGAAATTGGAC | 229 |
| PR | F: AAATCATTGCCAGGTTTTCG R: TGTGAGCTCGACACAACTCC | 404 |
| ERα | F: ACAAGGGAAGTATGGTATG R: CATCTCTCTGGCGCTTGTGT | 309 |
| HPRT | F: TATGGACAGGACTGAACGTCTTGC R: GACACAAACATGATTCAAATCCCTGA | 496 |
F, forward; R, reverse.
Cytogenetic analysis
Chromosome analysis of HAM cells was carried out as follows. To obtain metaphase chromosomes, 0.1 mL of colcemid solution (10 µg/mL; Invitrogen) was added to cell cultures 1 h prior to harvest. Harvested cells were treated with hypotonic KCl (0.075 m) solution for 25 min and were fixed in freshly prepared 3 : 1 methanol‐acetic acid. The metaphase chromosomes were stained using a standard GTG‐banding method and were examined under a Zeiss Axioskop microscope (Zeiss, Jena, Germany). Twenty cells were examined and metaphase images were captured and karyotyped by Cytovision (Applied Imaging Corporation, San Jose, CA, USA).
Immunological characteristics of HAM cells in response to Interferon‐γ
Interferon‐γ (IFN‐γ; LG Life Sciences, Seoul, South Korea) was added to growth media containing HAM cells, at 50–200 U/mL concentrations (100 U = 5 ng). Expression of HLA markers and CD59 were examined by flow cytometry 48 h later.
Preparation of cell‐permeable peptide‐fused recombinant proteins
Full‐length enhanced green fluorescence protein (EGFP) was amplified using pEGFP‐N1 vector (Invitrogen) as a template, and cloned into XhoI and BamH1 sites of the pET15b bacterial expression vector (Novagen, Darmstadt, Germany). MPG peptide (GALFLGWLGAAGSTMGAPKKKRKV) sequence (Morris et al. 1997) was amplified by Klenow reaction and was cloned into NdeI and XhoI sites in pET15b‐EGFP. After sequence verification, the plasmid was transformed into BL21 Escherichia coli strain. TAT‐β‐gal and β‐gal expression plasmids were generous gifts from Dr. S. Dowdy (UCSD, San Diego, CA, USA). Expression and purification of cell‐penetrating peptide (CPP)‐fused recombinant proteins were performed following the protocol provided (Becker‐Hapak et al. 2001).
X‐gal staining
Expression of β‐gal in CPP‐β‐gal‐treated HAM cells was assessed by X‐gal staining as described previously (Nagy et al. 2003). In brief, the cells treated with β‐gal or protein transduction domain (PTD)‐β‐gal were washed twice with PBS and then were fixed in 0.2% glutaraldehyde solution (0.1 m phosphate buffer, pH 7.2, 0.2% glutaraldehyde, 5 mm EGTA, 2 mm MgCl2). Cells were washed twice with PBS containing 2 mm MgCl2 followed by detergent rinse (0.1 m phosphate buffer, pH 7.2, 2 mm MgCl2, 0.01% sodium deoxycholic acid, 0.02% Nonidet P‐40). Substrate solution containing 1 mg/mL X‐gal, 10 mm potassium ferrocyanide, 10 mm potassium ferricyanide, 2 mm MgCl2 in PBS was then added. Cells were incubated at 37 °C until the characteristic blue colouration became visible.
Production of EGFP‐expressing HAM cells by lentiviral transduction
The EGFP‐expressing lentiviral vector and packaging plasmids were kindly provided by Dr. Miyoshi (BioResource Center, RIKEN Tsukuba Institute, Tsukuba, Ibaraki, Japan). Packaging plasmids (pCAG‐HIVgp and pCMV‐VSV‐G‐RSV‐Rev) and EGFP‐expressing lentiviral plasmid (CS‐CDF‐EG‐PRE) were transfected into 293T cells using Targetfect F1 transfection reagent (Targeting Systems, San Diego, CA, USA). Lentivirus‐containing conditioned media of 293T cells were concentrated by ultracentrifugation and the pellet was re‐suspended in a small volume of Hank's buffered saline solution (Miyoshi et al. 1998). HAM cells (5 × 104 cells/well) in 6‐well plates were transduced with serially diluted viral stock solution. Four days later, the transduced HAM cells were amplified on new 10‐cm plates; infectivity was determined by using FACS analysis. EGFP+ HAM cells with 98% or more infectivity were maintained and used for intrauterine infusion or intravenous injection.
Intrauterine and intravenous infusions of EGFP‐HAM cells
Six‐week‐old CD‐1 or nude mice were purchased from Orient‐Bio (Kyungki‐do, South Korea) and were used as recipients of HAM cell infusions. All animal procedures were performed in accordance with institutional guidelines of Konkuk University. All mice were kept in an animal care facility located in the College of Veterinary Medicine at Konkuk University. For intrauterine infusions, EGFP‐HAM cells (1 × 105/100 µL PBS) were injected into the uterine lumen of ICR female mice. Total volume of infusion was not more than 10 µL. For intravenous infusion of EGFP‐HAM cells, 5 × 104 cells in 100 µL PBS were injected into tail veins of anaesthetized male nude mice. Mice were killed at indicated days and tissues were processed for observing EGFP fluorescence and for immunofluorescence staining with anti‐human CD59 or anti‐human CD105 antibodies.
Immunofluorescence staining and confocal laser microscopy
Immunofluorescence staining was performed as described previously (Han et al. 2007). Adult mouse tissues were dissected and flash frozen in Histo‐Freeze (Fisher Scientific, Pittsburgh, PA, USA). Frozen sections (12 µm) were mounted on to poly‐L‐lysine‐coated slides (Polysciences Inc., Warrington, PA, USA) and were fixed in cold 4% paraformaldehyde in PBS. To observe EGFP fluorescence, slides were mounted after fixation. For immunofluorescence staining, non‐specific antiserum binding was blocked with 2% bovine serum albumin (BSA)/PBS for 40–60 min. Tissue sections were incubated for 1 h with anti‐human CD59 or anti‐human CD105 antibodies (Santa Cruz Biotechnologies, Santa Cruz, CA, USA) diluted to 1 : 100 in 2% BSA/PBS. Following three washes with 2% BSA/PBS, rabbit anti‐goat Alexa 546 (Molecular Probes, Eugene, OR, USA) secondary antibody (1 : 250 in 2% BSA/PBS) was applied for 30 min. Subsequently, slides were washed with 2% BSA/PBS and nuclei were stained with To‐PRO‐3‐iodide (Molecular Probes; 1 : 500) or Hoechst 33342 (Molecular Probes) for 10–30 min. After a final wash in 2% BSA/PBS, coverslips were mounted using ProFade Gold antifade reagent (Invitrogen). Specimens were examined using Olympus FV1000 spectral confocal microscopy or upright fluorescence microscopy (Olympus, Tokyo, Japan).
RESULTS
Characterization of HAM cells revealed by flow cytometry and cytogenetics
In a previous investigation, we found that HAM cells with fibroblast‐like features expressed several stem cell markers, including Oct‐4, SSEA‐3, SSEA‐4 and Rex‐1. They also exhibit differentiation potential for mesenchymal cell lineages, such as adipocytes, chondrocytes and osteocytes (Kim et al. 2007a). Using flow cytometric analysis, we further characterized and quantified expression of several mesenchymal stem cell markers, HLA antigens and haematopoietic stem cell markers.
HAM cells were similar to human BM‐derived MSCs or MSCs from other sources, expressing representative mesenchymal cell surface markers CD105 (Endoglin) and CD90 (Thy‐1). HAM cells also expressed high levels of CD59 (protectin), CD49d (integrin α4 subunit), and CD44 (H‐CAM), but not CD45 (LCA), CD34 (Mucosialin), CD31 (PECAM‐1) or CD106 (VCAM‐1) (Fig. 1a). In addition, we examined expression of three major histocompatibility complex (MHC) markers, HLA‐ABC (class I), HLA‐DR (class II) and HLA‐G (soluble). HLA‐ABC, but not others, was expressed at a modest level as in other stem cells (Fig. 1a). However, expression of HLA‐G varied among HAM cell lines established from different amnions, ranging from 2% to 90% (data not shown). HAM cells were weakly positive for SSEA‐4, TRA 1–60, TRA 1–81 and TRA 2–54 ranging from 2% to 20% depending on HAM cell lines (data not shown). CD117, a representative stem cell and germ cell marker, was expressed in 2–6% of HAM cells. Overall, similar results were obtained from at least three to five HAM cell lines established from different amnions. It is notable that CD59, a negative regulator of complement activation, was highly expressed. This may indicate that HAM cells possess a mechanism to escape from complement attack by inhibiting formation of membrane attack complex. It has not been reported whether CD59 is highly expressed in MSCs from other sources.
Figure 1.
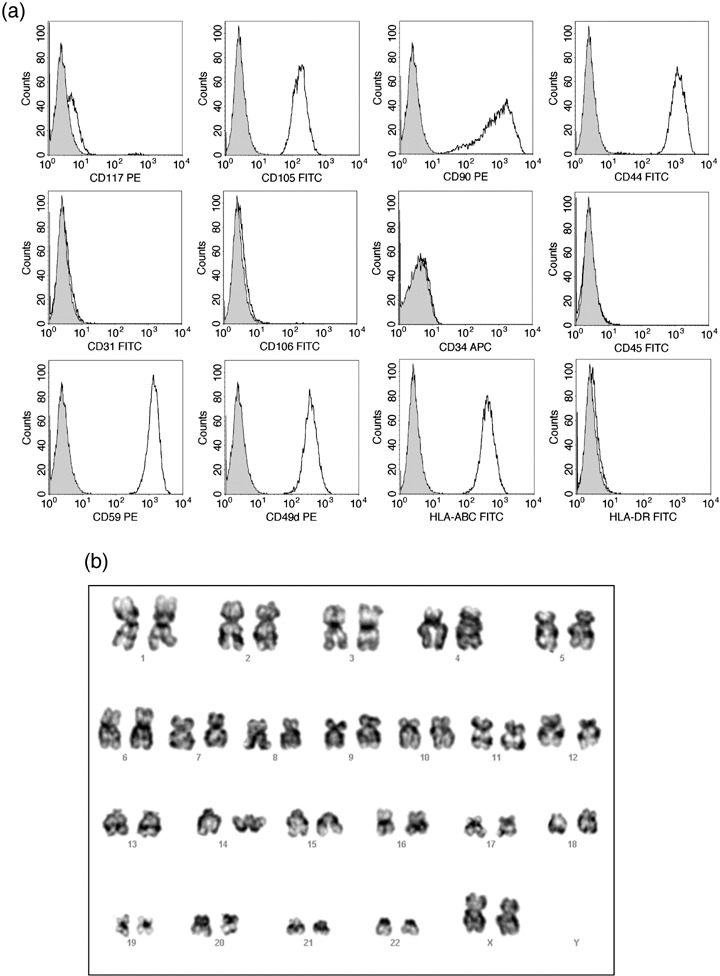
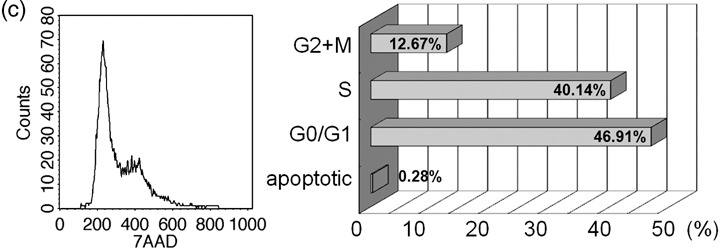
Expression of various cell surface markers in HAM cells. (a) HAM cells were incubated with indicated antibodies and were analysed using flow cytometry. HAM cells were strongly positive for CD105, CD90, CD44, CD59, CD49d and HLA‐ABC but negative for CD31, CD106, CD34, CD45 and HLA‐DR. Expression of CD117 varied from line to line, ranging from 2% to 6%. SSEA‐4 and TRA markers (data not shown) were weakly positive ranging from 2% to 20%. See Table 1 for summary. (b) Normal karyotype of HAM cells at passage 10. (c) Cell cycle phase distribution of HAM cells. Flow cytometric analysis based on DNA content was used to determine cell cycle distribution of asynchronously growing HAM cells. Percentage of cells in each phase is shown as a barogram.
Karyotyping HAM cells at the 10th passage showed normal diploid complement of chromosomes (Fig. 1b). FACS analysis based on DNA content was used to determine cell cycle distribution of the cells. As shown in Fig. 1c, around 47% of the cells were in G0/G1 phase and 40% in S phase. We compared cell cycle phase distribution of HEK293 cells in culture (data not shown) with that of HAM cells, and found that a higher percentage of HAM cells was in S phase (~18% in HEK293 versus ~40% in HAM cells). This is typical of stem cells as they undergo rapid self‐renewal (Jirmanova et al. 2002; , Becker et al. 2006).
Interferon‐γ treatment induces expression of MHC class I molecules in HAM cells
Classical MHC class I (HLA‐ABC) proteins are expressed on all cell types and are recognized by cytotoxic T cells or natural killer cells; presentation of ‘non‐self’ moieties generally leads to cell death. MHC class II (HLA‐DR) molecules are restricted to antigen‐presenting cells. HLA‐G, a non‐classical MHC‐I molecule, is thought to protect the foetus from maternal uterine natural killer cells by providing immunological tolerance. This molecule is selectively expressed in lineages of blastocyst trophectoderm. MHC expression in cells is controlled by cytokines, such as IFN‐γ and IFN‐γ normally induces MHC antigen expression, leading to activation of pathways involved in graft rejection (Drukker et al. 2002). It is believed that BM‐MSCs are immune‐privileged in some contexts (Uccelli et al. 2007) but can also be rejected by MHC‐mismatched hosts (Eliopoulos et al. 2005); however, immunological characteristics of HAM cells derived from foetal tissues are not known. To examine whether IFN‐γ treatment increased MHC expression, we treated these cells with 100 U/mL IFN‐γ for 48 h and examined MHC expression by flow cytometry. As shown in Fig. 2, IFN‐γ treatment increased median fluorescence intensity of HLA‐ABC expression (49.58 versus 102.74 in control versus treated) without affecting that of HLA‐DR expression. CD59 also exhibited moderate increases in median fluorescence intensity by IFN‐γ treatment (49.58 versus 72.99, data not shown). These results indicate that HAM cells can express higher levels of MHC class I molecules in response to a cytokine and thus may become targets of immune rejection on encountering immune cells secreting such factor. This result is in contrast to BM‐MSCs as these express high levels of HLA‐DR after IFN‐γ treatment (Le Blanc et al. 2003).
Figure 2.
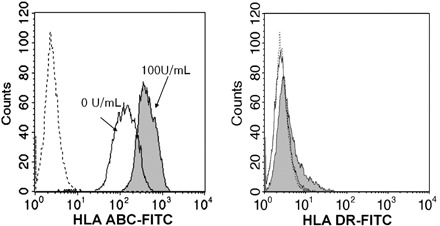
Effect of IFN‐γ on HLA‐ABC and HLA‐DR expression of HAM cells. HAM cells were incubated in the growth media with 100 U/mL of IFN‐γ for 48 h. Induction of HLA‐ABC suggested that HAM cells could be targeted by natural killer cells under in vivo conditions (100 U = 5 ng).
Mesenchymal properties of HAM cells are similar to uterine mesenchymal cells with respect to HOXA, WNT5a and progesterone receptor expression
Hox genes encode closely related transcriptional regulators important for diverse developmental processes. These factors are especially crucial for early embryonic patterning and determination of cell fate during organogenesis (Kmita & Duboule 2003). Among these classical Hox genes, AbdB‐like Hox genes are known for their functions in two distinct cellular contexts. Hoxa9 and Hoxa10 are expressed in CD34+ BM cells and are implicated in normal and malignant haematopoiesis (Sauvageau et al. 1994; , Bjornsson et al. 2001; Lawrence et al. 2005). Hoxa9, Hoxa10 and Hoxa11 all are expressed in developing FRT in a nested pattern and direct proper patterning of the Müllerian duct into oviduct, uterus and vagina (Ma et al. 1998). AbdB‐like Hoxa genes in the uterus are primarily confined to the mesenchymal cell compartment during development and during early pregnancy (Ma et al. 1998; , Lim et al. 1999). These genes are up‐regulated by progesterone (P4) in the uterus via the progesterone receptor and participate in proliferation and differentiation of uterine mesenchymal cells (Lim et al. 1999). Wnt5a is also a regulator of normal patterning of the FRT (Mericskay et al. 2004). Expression of Wnt5a is confined to the mesenchyme of the developing reproductive tract and is implicated in the process of tissue patterning mediated by Hoxa10, Hoxa11 and others (Mericskay et al. 2004). To this end, we examined expression of HOXA9, HOXA10 and HOXA11 in HAM cells to further explore uterine mesenchyme‐like features that are shared by these cells.
By RT‐PCR analysis (Fig. 3), we found that HOXA9, HOXA10 and HOXA11 are all expressed abundantly in HAM cells and while WNT5a is also highly expressed, WNT4a and WNT7a are not expressed in established HAM cell cultures. WNT4a and WNT7a were detected in HAM cells only at passage 1 (P1), possibly because P1 cells contain a mixture of heterogeneous cell types resulting from enzymatic digestion of the amnion. The results that genes crucially involved in patterning of the FRT are highly expressed in HAM cells suggest that these cells may share similar cellular and molecular characteristics with uterine mesenchymal cells. Human ESCs in the undifferentiated state or at 28 days of embryoid body formation (EB28) express very low levels of HOXA9 and HOXA11, but not HOXA10. WNT5a is expressed in the EB28 sample.
Figure 3.
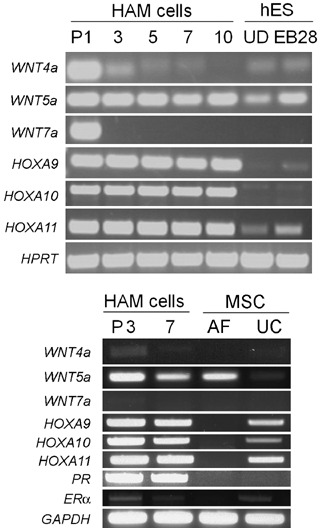
Expression of AbdB‐like HOXA and WNT genes in HAM cells, amniotic fluid (AF)‐mesenchymal stem cells (MSCs) and umbilical cord (UC)‐MSCs. RT‐PCR assays were performed using cDNAs prepared from HAM cell RNAs of indicated passages. RNA samples of human embryonic stem (hES) cells in the undifferentiated state (UD) and 28 days after embryoid body formation (EB28) were used as controls. AF‐MSC and UC‐MSC samples were both prepared at passage 5. Primers are shown in Table 2.
We then compared gene expression profiles of HAM cells with AF‐ and UC‐derived MSCs to examine whether uterine ‘mesenchymeness’ is unique to HAM cells. As shown in Fig. 3, AF‐MSCs express WNT5a but not HOXA genes. In contrast, UC‐MSCs express HOXA9, HOXA10 and HOXA11, but none of the WNT genes. Notably, progesterone receptor was also expressed abundantly in HAM cells while it was below detectable levels in both AF‐ and UC‐MSCs. Collectively, these results led us to hypothesize that HAM cells could be applied as a therapy for replenishing uterine cells when scarcity or low proliferation of uterine endometrial cells is associated with pregnancy failure.
Short‐ or long‐term regulation of gene expression achieved in HAM cells
Devising methods for manipulating gene expression in HAM cells is critical in exploring in vivo transplantation potential of these cells, thus, we tested two methods for introducing gene or protein. Cell‐permeable or cell‐penetrating peptides (CPPs) are defined by their ability to enter live cells across the lipid bilayer (this is by unknown mechanisms). These peptides, commonly with stretches of basic amino acids, are capable of mediating cellular uptake of macromolecules of substantial sizes (Joliot & Prochiantz 2004). Among many known CPPs, we chose MPG and PTD of the HIV TAT protein, both of which are effective in intracellular delivery of various macromolecules (Morris et al. 1997; Nagahara et al. 1998).
We produced CPP‐fusion proteins by using a bacterial expression system (Becker‐Hapak et al. 2001). GFP alone, MPG‐EGFP, β‐gal alone and PTD‐β‐gal recombinant proteins were purified and added to culture media containing HAM cells at indicated concentrations. In contrast to purified β‐gal protein without PTD fusion, PTD‐β‐gal fusion protein was readily penetrable into HAM cells with high efficiency, as visualized by X‐gal staining (Fig. 4). The incubation time for PTD‐β‐gal was 19 h. MPG‐EGFP and EGFP proteins were added to HAM cells for 1 h, and EGFP fluorescence was observed using an inverted microscope with an epifluorescence attachment (Fig. 4). MPG‐EGFP was readily visible within HAM cells, but EGFP was not confined to the nucleus but was also present in a patchy pattern in the cytoplasm. These results show that the CPP system is an effective method for achieving transient protein expression in HAM cells.
Figure 4.
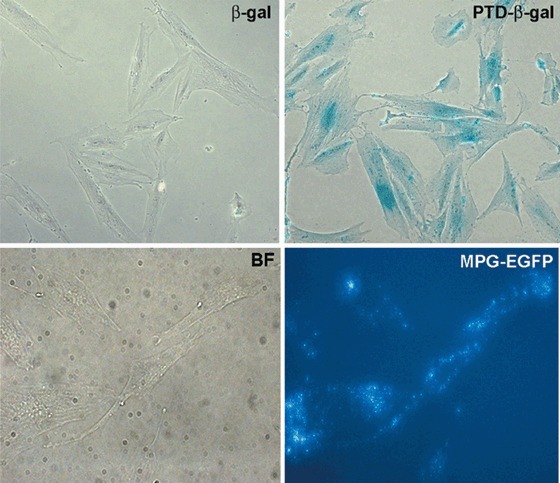
Efficacy of cell‐permeable peptides for short‐term gene regulation in HAM cells. Purified PTD‐β‐gal or β‐gal protein was added to the culture media of HAM cells at 100 nm 19 h later, cells were washed with PBS containing 2 mm MgCl2 twice, fixed with glutaraldehyde, and rinsed with detergent rinse solution. X‐gal‐containing substrate solution was added and colour was developed for 1 h. Note blue X‐gal staining mainly in the nuclear region (upper panel). EGFP or MPG‐EGFP protein was added to culture media of HAM cells at 40 µg/mL and incubated for 1 h. Cells were washed twice with HBSS and observed using inverted microscopy with an epifluorescence attachment (lower panel). Note the patched pattern of EGFP expression throughout the cytoplasm of MPG‐EGFP‐treated HAM cells. Control EGFP treatment did not show any fluorescence after washing (data not shown). BF, bright field.
We then tested the HIV‐1‐based lentiviral vector system to see if it would be effective in transducing HAM cells for non‐transient genomic manipulation. Lentivirus has been used successfully in several stem cell systems (Chan et al. 2005; Clements et al. 2006; Van Damme et al. 2006). In the lentiviral vector used, the EGFP reporter gene is under the control of the human elongation factor 1 promoter (Miyoshi et al. 1998). Transduction conditions were optimized by using different concentrations of the viral stock and the efficiency of transduction was analysed using FACS for EGFP fluorescence. Generally, more than 98% of HAM cells exhibited EGFP fluorescence (Fig. 5a); EGFP expression in the virus‐infected HAM cells was also confirmed by fluorescence microscopy (Fig. 5b). The results confirmed that HAM cells are easy to manipulate for gene expression and in vivo cell trafficking by both CPP and lentiviral systems.
Figure 5.
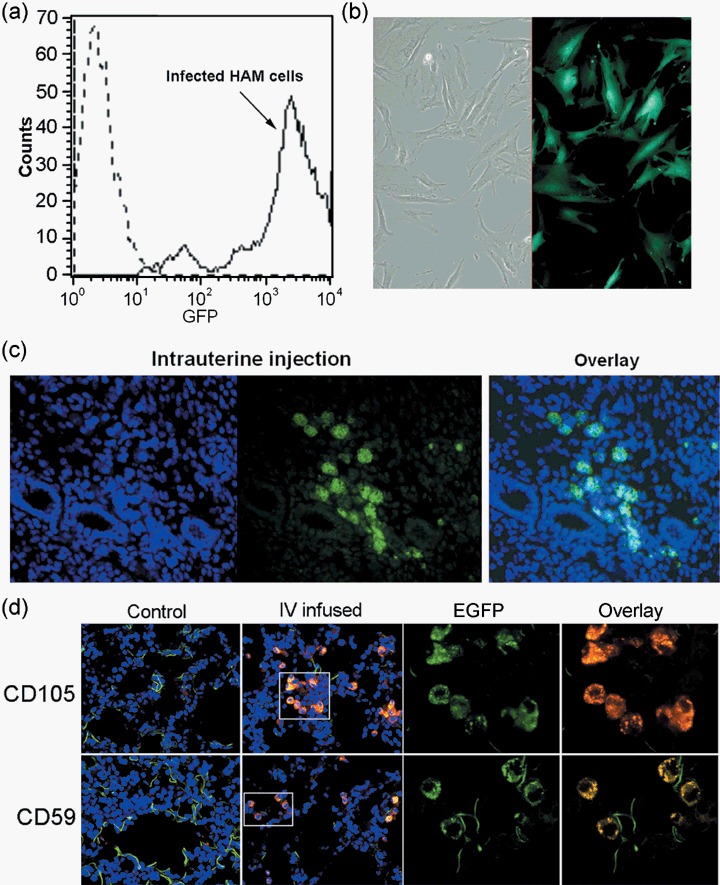
Transplantation of lentivirus‐transduced HAM cells into mice. (a) Flow cytometry confirmed high transduction rate of EGFP lentivirus in HAM cells. More than 98% of cells show EGFP expression. (b) Fluorescence microscopic image of HAM cells expressing EGFP. (c) Transplantation of EGFP expressing HAM cells into immunocompetent CD‐1 mice by intrauterine infusion. Small volume of EGFP‐HAM cells at 1 × 105 cells/0.1 mL were infused into the uterine lumen. Uteri were taken after 10 days and were frozen for cryosectioning. Sections were stained with Hoechst dye after fixation. Image on the far left shows nuclear staining (blue) and green fluorescence in the middle indicates EGFP fluorescence. These images are overlaid and shown on the right. (d) Intravenous infusion of EGFP‐HAM cells was performed via tail veins of anaesthetized nude mice. About 200 µL of cells were given at 5 × 104 cells/0.1 mL. Tissues were collected for up to 4 weeks at 1‐week interval and frozen for cryosectioning. This representative figure is obtained from lung sections from a nude mouse at 4 weeks after infusion. Presence of EGFP‐HAM cells was again confirmed by immunofluorescence imaging using human specific anti‐CD59 or anti‐CD105 antibodies. Control is a lung section from a non‐injected mouse. Laser confocal microscopic images are shown at 60×. Insets are shown enlarged in the right panels [EGFP alone, EGFP (green) + CD marker (red) overlay].
Intrauterine and intravenous transplantations of HAM cells
Lentivirally engineered HAM cells retain EGFP expression for many passages (data not shown). Thus, we used EGFP‐expressing HAM cells for in vivo transplantation. Two strategies for in vivo delivery were taken. First, to examine whether HAM cells sharing similar HOXA gene profiles and characteristics with uterine mesenchymal cells would contribute to the uterine endometrium, we infused HAM cells directly into the uterine lumen of immunocompetent mice. The presence of HAM cells was examined 7–10 days later on cryosections of the infused uterus. As shown in Fig. 5c, EGFP‐HAM cells with large nuclei were noted in some uterine sections; however, the results were somewhat variable between several mice, possibly due to inconsistent infusions. Nonetheless, the presence of live HAM cells with EGFP expression was observed within several mice uteri.
We next examined whether a systemic injection of undifferentiated HAM cells into mice leads to a contribution of these cells into multiple organs. We gave intravenous injections of EGFP‐HAM cells into tail veins of nude mice. The presence of EGFP‐expressing cells within tissues and organs was examined at weekly intervals up to 4 weeks. As reported previously, transplanted cells are often found in lungs (Anjos‐Afonso et al. 2004; Schrepfer et al. 2007), and we also observed the presence of large EGFP‐positive cells in the lung of injected mice. Identity of these cells was confirmed by immunofluorescence staining with anti‐human CD59 or anti‐human CD105 antibodies. EGFP and CD marker expression were observed in same cells as shown in Fig. 5d.
DISCUSSION
Hypoplastic endometrium is one cause of female infertility. Moreover, uterine insufficiency resulting from chemotherapy or radiation therapy in cancer patients leaves them with little hope for normal fertility. Thus, there is a need for rejuvenating uterine insufficiency by stem cell therapy. Multipotent mesenchymal cells have high potential as a source of generating specific cell lineages. They are relatively easy to handle and do not provoke ethical issues. The highlights of this investigation are that HAM cells derived from the term placenta possess characteristics as multipotent MSCs and that HAM cells can be engineered to express genes or proteins of interest, by a lentiviral or CPP system. Based on gene expression studies, we show that HAM cells share similar molecular characteristics with uterine mesenchymal cells.
CD90 (Thy‐1) is a well‐known surface marker for MSCs (In 't Anker et al. 2003). It is interesting to note that CD90 is also expressed in a subset of human uterine stromal fibroblasts, thus serving as a uterine stromal cell marker (Gargett 2004). CD90+ uterine cells are functionally diverse from CD90− cells with respect to cytokine profiles and basic morphology (Koumas et al. 2001). A further striking similarity between HAM cells and uterine mesenchymal cells is that they share similar expression patterns for HOXA and WNT5a genes. Among the 39 HOX genes identified that are involved in embryonic patterning, HOXA9, HOXA10 and HOXA11 are specifically involved in the Müllerian duct cell patterning into oviduct, uterus and vagina (Ma et al. 1998) and also in uterine function during early pregnancy (Benson et al. 1996; Lim et al. 1999). It is interesting to note that CD34− HAM cells express high levels of HOXA9 and HOXA10, pointing towards two possibilities. First, CD34− HAM cells, when properly engineered to express CD34, may develop characteristics resembling haematopoietic cell progenitors (Sauvageau et al. 1994; , Bjornsson et al. 2001; Lawrence et al. 2005). Alternatively, high expression of HOXA9, HOXA10 and HOXA11 in HAM cells may be useful for uterine cell therapy to replenish endometrial cells, when poor endometrial proliferation is associated with infertility or poor pregnancy outcome. Indeed, HOXA10 is implicated in regulation of proliferation of both haematopoietic and endometrial cells (Lim et al. 1999; , Bjornsson et al. 2001). Thus, it is interesting to observe that after intrauterine infusion, HAM cells remain viable in the uterus for many days even in immunocompetent mice (Fig. 5c). However, it remains to be determined whether these cells are in fact incorporated into endometrial cell population. We also show that HAM cells express high levels of progesterone (P4) receptor, while AF‐ or UC‐MSCs do not. Therefore, it will be interesting to examine whether P4 supplementation with intrauterine infusion of HAM cells would have a better outcome with respect to cell proliferation and incorporation. Overall, cellular and molecular similarities of HAM cells to uterine mesenchymal cells are not observed in AF‐ or UC‐MSCs. The results strongly suggest that HAM cells possess unique similarities to the uterine mesenchyme and that they may be a potential source for stem cell therapy associated with uterine insufficiency.
Immunological characteristics of MSCs are controversial, generating mixed observations of immune privilege and immune rejection (Uccelli et al. 2007). These reports are mostly observations derived from studies using BM‐MSCs and thus need to be validated more systematically in HAM cells. HAM cells show heightened levels of MHC class I expression on IFN‐γ treatment (Fig. 2). However, our findings that intraluminally infused HAM cells survived in the uterus indicate that these cells may become immune‐privileged in the uterine milieu. It has long been considered that the uterus is an immune‐privileged site that can circumvent maternal immune surveillance when certain cellular conditions are met (Niederkorn 2006). Several mechanisms are operative in the trophoblasts lining the maternal–foetal interface, to avert immunological attack. Expression of CD59 (protectin), is associated with inhibiting activation of the complement pathway in trophoblast cells (Girardi et al. 2006). Thus, it is plausible that high expression of CD59 (Fig. 1a) helps protect HAM cells from complement‐mediated immune attack in immunocompetent mice uteri. Indeed, expression of human CD59 and other associated factors of the complement pathway in swine xenografts contribute to delaying allograft rejection (McCurry et al. 1995). At present, whether other MSCs or stem cells express CD59 is not known.
There are reports suggesting the contribution of BM‐derived cells to endometrial cells in mice (Du & Taylor 2007) and in female patients receiving BM transplantation (Taylor 2004). While these studies require further validation, they suggest that stem cells of different origins could differentiate into endometrial cells. Further investigation of uterine cell type‐specific differentiation pathways of HAM cells will provide new information and tools for therapeutic applications.
We have shown herein that two approaches to manipulate gene or protein expression, namely lentivirus and CPP, are applicable to HAM cells. CPP systems have a wide range of applications in cell biology, delivering diverse macromolecules into cells (Joliot & Prochiantz 2004). Among many known CPPs, we tested two types of CPP system, PTD and MPG, and they exhibited distinct subcellular localization on uptake of fusion proteins. Thus, these two types of CPP may target distinct subcellular compartments and can be used to deliver cargoes to distinct subcellular locations to direct specific differentiation pathways in HAM cells. Our work also demonstrates for the first time that MPG‐fused recombinant protein can be delivered into the cells.
ACKNOWLEDGEMENT
This work was supported by the Korean Science and Engineering Foundation grant funded from the Korean Government (MOST) (No. R01‐2006‐000‐10501‐0).
REFERENCES
- Alviano F, Fossati V, Marchionni C, Arpinati M, Bonsi L, Franchina M, Lanzoni G, Cantoni S, Cavallini C, Bianchi F, Tazzari PL, Pasquinelli G, Foroni L, Ventura C, Grossi A, Bagnara GP (2007) Term Amniotic membrane is a high throughput source for multipotent mesenchymal stem cells with the ability to differentiate into endothelial cells in vitro . BMC Dev. Biol. 7, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anjos‐Afonso F, Siapati EK, Bonnet D (2004) In vivo contribution of murine mesenchymal stem cells into multiple cell‐types under minimal damage conditions. J. Cell Sci. 117, 5655–5664. [DOI] [PubMed] [Google Scholar]
- Becker KA, Ghule PN, Therrien JA, Lian JB, Stein JL, Van Wijnen AJ, Stein GS (2006) Self‐renewal of human embryonic stem cells is supported by a shortened G1 cell cycle phase. J. Cell. Physiol. 209, 883–893. [DOI] [PubMed] [Google Scholar]
- Becker‐Hapak M, McAllister SS, Dowdy SF (2001) TAT‐mediated protein transduction into mammalian cells. Methods 24, 247–256. [DOI] [PubMed] [Google Scholar]
- Benson GV, Lim H, Paria BC, Satokata I, Dey SK, Maas RL (1996) Mechanisms of reduced fertility in Hoxa‐10 mutant mice: uterine homeosis and loss of maternal Hoxa‐10 expression. Development 122, 2687–2696. [DOI] [PubMed] [Google Scholar]
- Bjornsson JM, Andersson E, Lundstrom P, Larsson N, Xu X, Repetowska E, Humphries RK, Karlsson S (2001) Proliferation of primitive myeloid progenitors can be reversibly induced by HOXA10. Blood 98, 3301–3308. [DOI] [PubMed] [Google Scholar]
- Chan J, O'Donoghue K, De La Fuente J, Roberts IA, Kumar S, Morgan JE, Fisk NM (2005) Human fetal mesenchymal stem cells as vehicles for gene delivery. Stem Cells 23, 93–102. [DOI] [PubMed] [Google Scholar]
- Chang CJ, Yen ML, Chen YC, Chien CC, Huang HI, Bai CH, Yen BL (2006) Placenta‐derived multipotent cells exhibit immunosuppressive properties that are enhanced in the presence of interferon‐gamma. Stem Cells 24, 2466–2477. [DOI] [PubMed] [Google Scholar]
- Clements MO, Godfrey A, Crossley J, Wilson SJ, Takeuchi Y, Boshoff C (2006) Lentiviral manipulation of gene expression in human adult and embryonic stem cells. Tissue Eng. 12, 1741–1751. [DOI] [PubMed] [Google Scholar]
- De Coppi P, Bartsch G Jr, Siddiqui MM, Xu T, Santos CC, Perin L, Mostoslavsky G, Serre AC, Snyder EY, Yoo JJ, Furth ME, Soker S, Atala A (2007) Isolation of amniotic stem cell lines with potential for therapy. Nat. Biotechnol. 25, 100–106. [DOI] [PubMed] [Google Scholar]
- Drukker M, Benvenisty N (2004) The immunogenicity of human embryonic stem‐derived cells. Trends Biotechnol. 22, 136–141. [DOI] [PubMed] [Google Scholar]
- Drukker M, Katz G, Urbach A, Schuldiner M, Markel G, Itskovitz‐Eldor J, Reubinoff B, Mandelboim O, Benvenisty N (2002) Characterization of the expression of MHC proteins in human embryonic stem cells. Proc. Natl. Acad. Sci. USA 99, 9864–9869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du H, Taylor HS (2007) Contribution of bone marrow‐derived stem cells to endometrium and endometriosis. Stem Cells 25, 2082–2086. [DOI] [PubMed] [Google Scholar]
- Eliopoulos N, Stagg J, Lejeune L, Pommey S, Galipeau J (2005) Allogeneic marrow stromal cells are immune rejected by MHC class I‐ and class II‐mismatched recipient mice. Blood 106, 4057–4065. [DOI] [PubMed] [Google Scholar]
- Fukuchi Y, Nakajima H, Sugiyama D, Hirose I, Kitamura T, Tsuji K (2004) Human placenta‐derived cells have mesenchymal stem/progenitor cell potential. Stem Cells 22, 649–658. [DOI] [PubMed] [Google Scholar]
- Gargett CE (2004) Stem cells in gynaecology. Aust. N. Z. J. Obstet. Gynaecol. 44, 380–386. [DOI] [PubMed] [Google Scholar]
- Girardi G, Bulla R., Salmon JE, Tedesco F (2006) The complement system in the pathophysiology of pregnancy. Mol. Immunol. 43, 68–77. [DOI] [PubMed] [Google Scholar]
- Gregory CA, Prockop DJ, Spees JL (2005a) Non‐hematopoietic bone marrow stem cells: molecular control of expansion and differentiation. Exp. Cell Res. 306, 330–335. [DOI] [PubMed] [Google Scholar]
- Gregory CA, Ylostalo J, Prockop DJ (2005b) Adult bone marrow stem/progenitor cells (MSCs) are preconditioned by microenvironmental ‘niches’ in culture: a two‐stage hypothesis for regulation of MSC fate. Sci. STKE 2005, pe37. [DOI] [PubMed] [Google Scholar]
- Guillot PV, Gotherstrom C, Chan J, Kurata H, Fisk NM (2007) Human first‐trimester fetal MSC express pluripotency markers and grow faster and have longer telomeres than adult MSC. Stem Cells 25, 646–654. [DOI] [PubMed] [Google Scholar]
- Han K, Song H, Moon I, Augustin R, Moley K, Rogers M, Lim H (2007) Utilization of DR1 as true RARE in regulating the Ssm, a novel retinoic acid‐target gene in the mouse testis. J. Endocrinol. 192, 539–551. [DOI] [PubMed] [Google Scholar]
- In 't Anker PS, Scherjon SA, Kleijburg‐van der Keur C, Noort WA, Claas FH, Willemze R, Fibbe WE, Kanhai HH (2003) Amniotic fluid as a novel source of mesenchymal stem cells for therapeutic transplantation. Blood 102, 1548–1549. [DOI] [PubMed] [Google Scholar]
- Jirmanova L, Afanassieff M, Gobert‐Gosse S, Markossian S, Savatier P (2002) Differential contributions of ERK and PI3‐kinase to the regulation of cyclin D1 expression and to the control of the G1/S transition in mouse embryonic stem cells. Oncogene 21, 5515–5528. [DOI] [PubMed] [Google Scholar]
- Joliot A, Prochiantz A (2004) Transduction peptides: from technology to physiology. Nat. Cell Biol. 6, 189–196. [DOI] [PubMed] [Google Scholar]
- Kim J, Kang HM, Kim H, Kim MR, Kwon HC, Gye MC, Kang SG, Yang HS, You J (2007a) Ex vivo characteristics of human amniotic membrane‐derived stem cells. Cloning Stem Cells 9, 581–594. [DOI] [PubMed] [Google Scholar]
- Kim J, Lee Y, Kim H, Hwang KJ, Kwon HC, Kim SK, Cho DJ, Kang SG, You J (2007b) Human amniotic fluid‐derived stem cells have characteristics of multipotent stem cells. Cell Prolif. 40, 75–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kmita M, Duboule D (2003) Organizing axes in time and space; 25 years of colinear tinkering. Science 301, 331–333. [DOI] [PubMed] [Google Scholar]
- Koumas L, King AE, Critchley HO, Kelly RW, Phipps RP (2001) Fibroblast heterogeneity: existence of functionally distinct Thy 1(+) and Thy 1(–) human female reproductive tract fibroblasts. Am. J. Pathol. 159, 925–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence HJ, Christensen J, Fong S, Hu YL, Weissman I, Sauvageau G, Humphries RK, Largman C (2005) Loss of expression of the Hoxa‐9 homeobox gene impairs the proliferation and repopulating ability of hematopoietic stem cells. Blood 106, 3988–3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Blanc K, Tammik C, Rosenthal K, Zetterberg E, Ringden O (2003) HLA expression and immunologic properties of differentiated and undifferentiated mesenchymal stem cells. Exp. Hematol. 31, 890–896. [DOI] [PubMed] [Google Scholar]
- Lim H, Ma L, Ma WG, Maas RL, Dey SK (1999) Hoxa‐10 regulates uterine stromal cell responsiveness to progesterone during implantation and decidualization in the mouse. Mol. Endocrinol. 13, 1005–1017. [DOI] [PubMed] [Google Scholar]
- Ma L, Benson GV, Lim H, Dey SK, Maas RL (1998) Abdominal B (AbdB) Hoxa genes: regulation in adult uterus by estrogen and progesterone and repression in mullerian duct by the synthetic estrogen diethylstilbestrol (DES). Dev. Biol. 197, 141–154. [DOI] [PubMed] [Google Scholar]
- McCurry KR, Kooyman DL, Alvarado CG, Cotterell AH, Martin MJ, Logan JS, Platt JL (1995) Human complement regulatory proteins protect swine‐to‐primate cardiac xenografts from humoral injury. Nat. Med. 1, 423–427. [DOI] [PubMed] [Google Scholar]
- Mericskay M, Kitajewski J, Sassoon D (2004) Wnt5a is required for proper epithelial–mesenchymal interactions in the uterus. Development 131, 2061–2072. [DOI] [PubMed] [Google Scholar]
- Miki T, Lehmann T, Cai H, Stolz. DB , Strom SC (2005) Stem cell characteristics of amniotic epithelial cells. Stem Cells 23, 1549–1559. [DOI] [PubMed] [Google Scholar]
- Mimeault M, Batra SK (2006) Concise review: recent advances on the significance of stem cells in tissue regeneration and cancer therapies. Stem Cells 24, 2319–2345. [DOI] [PubMed] [Google Scholar]
- Miyoshi H, Blomer U, Takahashi M, Gage FH, Verma IM (1998) Development of a self‐inactivating lentivirus vector. J. Virol. 72, 8150–8157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris MC, Vidal P, Chaloin L, Heitz F, Divita G (1997) A new peptide vector for efficient delivery of oligonucleotides into mammalian cells. Nucleic Acids Res. 25, 2730–2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagahara H, Vocero‐Akbani AM, Snyder EL, Ho A, Latham DG, Lissy NA, Becker‐Hapak M, Ezhevsky SA, Dowdy SF (1998) Transduction of full‐length TAT fusion proteins into mammalian cells: TAT‐p27Kip1 induces cell migration. Nat. Med. 4, 1449–1452. [DOI] [PubMed] [Google Scholar]
- Nagy A, Gertsenstein M, Vintersten K, Behringer R (2003) Manipulation the Mouse Embryo. New York: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Niederkorn JY (2006) See no evil, hear no evil, do no evil: the lessons of immune privilege. Nat. Immunol. 7, 354–359. [DOI] [PubMed] [Google Scholar]
- Pittenger MF, Mackay AM, Beck SC, Jaiswal RK, Douglas R, Mosca JD, Moorman MA, Simonetti DW, Craig S, Marshak DR (1999) Multilineage potential of adult human mesenchymal stem cells. Science 284, 143–147. [DOI] [PubMed] [Google Scholar]
- Ryan JM, Barry FP, Murphy JM, Mahon BP (2005) Mesenchymal stem cells avoid allogeneic rejection. J. Inflamm. (Lond.) 2, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauvageau G, Lansdorp PM, Eaves CJ, Hogge DE, Dragowska WH, Reid DS, Largman C, Lawrence HJ, Humphries RK (1994) Differential expression of homeobox genes in functionally distinct CD34+ subpopulations of human bone marrow cells. Proc. Natl. Acad. Sci. USA 91, 12223–12227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrepfer S, Deuse T, Reichenspurner H, Fischbein MP, Robbins RC, Pelletier MP (2007) Stem cell transplantation: the lung barrier. Transplant. Proc. 39, 573–576. [DOI] [PubMed] [Google Scholar]
- Taylor HS (2004) Endometrial cells derived from donor stem cells in bone marrow transplant recipients. JAMA 292, 81–85. [DOI] [PubMed] [Google Scholar]
- Thomson JA, Itskovitz‐Eldor J, Shapiro SS, Waknitz. MA , Swiergiel JJ, Marshall VS, Jones JM (1998) Embryonic stem cell lines derived from human blastocysts. Science 282, 1145–1147. [DOI] [PubMed] [Google Scholar]
- Tsai MS, Lee JL, Chang YJ, Hwang SM (2004) Isolation of human multipotent mesenchymal stem cells from second‐trimester amniotic fluid using a novel two‐stage culture protocol. Hum. Reprod. 19, 1450–1456. [DOI] [PubMed] [Google Scholar]
- Uccelli A, Pistoia V, Moretta L (2007) Mesenchymal stem cells: a new strategy for immunosuppression? Trends Immunol. 28, 219–226. [DOI] [PubMed] [Google Scholar]
- Van Damme A, Thorrez L, Ma L, Vandenburgh H, Eyckmans J, Dell’Accio F, De BC, Luyten F, Lillicrap D, Collen D, Vandendriessche T, Chuah MK (2006) Efficient lentiviral transduction and improved engraftment of human bone marrow mesenchymal cells. Stem Cells 24, 896–907. [DOI] [PubMed] [Google Scholar]
- Zhao P, Ise H, Hongo M, Ota M, Konishi I, Nikaido T (2005) Human amniotic mesenchymal cells have some characteristics of cardiomyocytes. Transplantation 79, 528–535. [DOI] [PubMed] [Google Scholar]


