Abstract
Objective
This study aimed to investigate the role of the TGFβ signalling pathway in angiogenesis in a three‐dimensional (3D) collagen gel model, with co‐culture between adipose‐derived stromal cells (ASCs) and endothelial cells (ECs).
Materials and methods
A 3D collagen gel, implanted with green fluorescent protein‐labelled mouse ASCs and red fluorescent protein‐labelled mouse ECs, was established in vitro. Phenomena of angiogenesis with or without type I TGFβ receptor inhibitor (LY2157299) treatment, were observed 7 days post‐implantation, using confocal laser scanning microscopy. To detect expression of angiogenesis‐related genes, semi‐quantitative PCR and quantitative real‐time PCR were conducted. Zymography was performed to explore secretion of matrix metalloproteinase 2 (MMP‐2) and matrix metalloproteinase 9 (MMP‐9) after treatment with LY2157299 of 5, 10, 20 to 50 μm concentrations, for 24 h.
Results
Angiogenesis was found to be attenuated in co‐culture gels after ASC and EC treatment with LY2157299. Genes VEGF‐A,VEGF‐B,VE‐ca,FGF‐1,FGF‐2,PDGF,HGF,BMP‐4 were significantly reduced in the presence of LY2157299 in both mono‐cultured and co‐cultured ECs. Furthermore, reduction in co‐cultured ECs was prominent relative to mono‐cultured ECs, while the same results did not occur to ASCs. We further confirmed that gelatinases secreted by ECs were reduced in a dose‐dependent manner, after treatment with LY2157299.
Conclusions
These results indicate that in ASC/EC co‐culture, the TGFβ signalling pathway regulated angiogenesis via EC activity. Co‐cultured ECs were regulated more significantly than mono‐cultured ECs suggesting that inhibition of TGFβRI may regulate paracrine secretion of ASCs to further modulate EC angiogenesis.
Introduction
In tissue regeneration and wound repair processes, angiogenesis plays a pivotal role by allowing blood supply of oxygen and nutrients and removing metabolic products. During formation of blood vessels, ECs elongate filopodia and form network structures. Previous studies have shown that ASCs promote survival, proliferation, endothelial tubulogenesis and vascularization of ECs and stable vascular assembly 1. Combination of ASCs and ECs further promote formation of vascular networks 2. In addition, co‐implantation of mesenchymal stromal cells ((MSCs), similar to ASCs) and endothelial cells, markedly expedite formation of functional microvascular beds and provides possible methods for cell‐based revascularization therapies to treat various diseases, such as in the peripheral vasculature and ischaemic heart disease 3, 4.
TGFβ signal transduction is mediated through a heterodimeric receptor complex formed by type I (TGFβRI) and type II (TGFβRII) 5 receptors. Typically, TGFβ binds to TGFβ receptor II, which recruits TGFβ receptor I by phosphorylation 6, with subsequent activation of Smad transcription factors. Smads then move into the nucleus and regulate transcription of relevant genes 7, 8.
With reference to angiogenesis, TGFβ is thought to modulate EC properties by balancing their pro‐proliferative and pro‐differentiational signalling pathways through TGFβ receptor II (TGFβRII) and two distinct TGFβRIs, activin receptor‐like kinase‐1(ALK1) and ALK5 9, 10, 11.
In this study, we have investigated whether TGFβ signalling modulated angiogenesis of co‐implantation ASCs and ECs, by inhibition of TGFβ receptor I, and we further explored influence of crosstalk between ASCs and ECs on the angiogenetic process.
Materials and methods
Cell culture
Animal cells and tissues for this study were obtained according to governing ethical principles, and the experiental protocol was reviewed and approved by the Institutional Review Board (IRB) of our hospital.
ASCs were isolated from subcutaneous adipose tissue from groins of 3‐week female mice. Collected tissues were cut into small pieces and digested in 0.075% type I collagenase solution in a shaking incubator for 60 min. This was then neutralized 1:1 (v/v) with 10% heat‐activated foetal bovine serum (FBS) α‐MEM (0.1 mm non‐essential amino acids, 4 mm L‐glutamine, 1% penicillin–streptomycin solution) and suspensions were centrifuged at 1000 rpm for 5 min. After removal of supernatants, remaining ASCs were resuspended in 10% FBS α‐MEM and seeded into plates or T25 flasks, then incubated at 37 °C in a 5% CO2 incubator. Cells were subcultured to passage 2 for further experimentation 12.
Brain microvascular tissue was collected from neonatal mice to obtain ECs. This was cut into small pieces and digested in 0.25% trypsin solution for 30 min. Supernatant was then removed and replaced with 0.5% type II collagenase for 3 h. Type II collagenase was neutralized 1:1 (v/v) with fresh 10% heat‐activated foetal bovine serum (FBS) DMEM (high‐glucose DMEM, 0.1 mm non‐essential amino acids, 4 mm L‐glutamine, 1% penicillin–streptomycin solution) and suspensions were centrifuged at 1000 rpm for 5 min. After removal of supernatants, remaining ECs were resuspended with 10% FBS DMEM, seeded into plates or T25 flasks, then incubated at 37 °C in a 5% CO2 incubator.
To obtain green fluorescent protein (GFP)‐positive ASCs and DsRed‐Express‐positive ECs, subcutaneous adipose tissue and brain microvascular tissues were collected from enhanced GFP transgenic mice (The Centre of Genetically Engineered Mice, West China Hospital, Sichuan University, Chengdu, China), and DsRed‐Express transgenic mice (The Genetic Centre of Institute of Laboratory Animal Sciences, Chinese Academy of Medical Sciences and Centre of Comparative Medicine, Peking Union Medical College, Beijing, China) respectively. Cell isolation was the same as described above.
Co‐culture
GFP‐positive ASCs from fat tissues and DsRed‐Express‐positive BMECs from brain microvascular tissues, from transgenic mice, were used to establish a 3D collagen gel model. ASCs and ECs were mixed at 1:1 ratio and suspended in DMEM‐HG (Hyclone, Logan, UT, USA), and rat tail tendon type I collagen (Shengyou Biotechnology, Hangzhou, China) with 10 μM LY2157299 (Selleckchem, Houston, TX, USA) [concentration was adopted as described in previous research articles 13, 14] or PBS (blank group). Seeded cells were transferred to 96‐well plates to form gel samples at 37 °C, and cultured for 7 day. Morphology of multiple vascular‐like structures was captured using a Leica DMIRE2 confocal laser scanning microscope (TCS SP2, Leica Microsystems, Wetzlar, Germany, parameter: 20×, Leica Microsystems original image: 1024 × 1024, 100 μm) equipped with a 60× oil immersion objective lens. Image analysis software Imaris 7.0.0 (Bitplane, Zurich, Switzerland) was used for three‐dimensional reconstruction.
To detect gene profiles of growth factors of ASCs and ECs at different concentrations of TGFβRI inhibitor LY 2157299, we used transwell co‐culture. ECs were seeded into six‐well plates, 2.5 × 106 cells per well (85–95% confluence) and ASCs were seeded at 2 × 106 cells per well, in transwell chambers. After 24 h equilibration, culture media were replaced with 2% FBS DMEM for 12 h starvation. Then, ECs were split into two groups. For mono‐cultured ECs, media were replaced with 1% FBS DMEM with concentration gradients of LY2157299 from 0, 5, 10, 20 to 50 μm; meanwhile, transwell chambers with no ASCs were filled with 1% FBS α‐MEM with concentration gradients corresponding to mono‐cultured ECs of LY2157299 (culture media control). For co‐culture ECs and ASCs, media were respectively replaced with 1% FBS DMEM and 1% FBS α‐MEM, both with concentration gradients the same as mono‐culture groups.
Semi‐quantitative polymerase chain reaction (PCR) and quantitative real‐time PCR (qRT‐PCR)
Total RNA was extracted from ASCs and ECs using RNeasy Plus Mini Kit (Qiagen, Shanghai, China) with a genomic DNA eliminator. Dissolved in RNase‐free water, extracted RNA samples were quantified by measuring absorbance at 260 nm using a spectrophotometer. Then, with treatment of DNase I (Mbi, Glen Burnie, MD, USA), around 0.5 μg of each total RNA was reverse transcribed using a first‐strand cDNA synthesis kit (Mbi).
To evaluate expression levels of growth factors in different treated groups, semi‐quantitative PCR and quantitative real‐time PCR (qPCR) were performed. Selected sets of primers are listed in Table 1. All primers were determined through BLAST and amplification of β‐Actin was used as control for assessing PCR efficiency. Semi‐quantitative PCR reactions were performed using a PCR kit (Mbi) plus thermo‐cycler (Bio‐Rad, Hercules, CA, USA) in 25 μl volume containing 1 μl cDNA sample. PCR programme was composed of 30 s denaturation cycle at 94 °C, 30 s annealing cycle at 55–65 °C and 72 °C, 30 s elongation cycle and 28–34 amplification cycles. Products were electrophoresed on 2% agarose gels, stained with ethidium bromide and visualized using Quantity One software (Bio‐Rad).
Table 1.
Primer sequences of β‐Actin and target genes
| Target gene (mouse)k | Primer pairs |
|---|---|
| β‐ACTIN (266bp) |
Forward: GTCCCTCACCCTCCCAAAAG Reverse: GCTGCCTCAACACCTCAACCC |
| VEGF‐A (106bp) |
Forward: CTGCTGTGGACTTGTGTTGG Reverse: AAAGGACTTCGGCCTCTCTC |
| VEGF‐B (128bp) |
Forward: GCAACACCAAGTCCGAATG Reverse: CTGGCTTCACAGCACTCTCC |
| VEGF‐C (76bp) |
Forward: AGGCTCGTACCTGACACCCGG Reverse: CGGCGGCTCGCAACTTTTGC |
| VE‐ca (102bp) |
Forward: CATCGCAGAGTCCCTCAGTT Reverse: TCAGCCAGCATCTTGAACCT |
| PDGF (107bp) |
Forward: CCGATAAAGACAGCCAACATC Reverse: CATTCACAGAGCACATCCTGA |
| FGF‐1(124bp) |
Forward: CCACAGCCCAGCAGTTATC Reverse: CTCCTACGCCCACTCTTCAG |
| FGF‐2(138bp) |
Forward: AGGAAGATGGACGGCTGCT Reverse: GCCCAGTTCGTTTCAGTGC |
| BMP‐4(103bp) |
Forward: TCTTCAACCTCAGCAGCATC Reverse: AAGCCCTGTTCCCAGTCAG |
| HGF(103bp) |
Forward: GGTGTTTCACAAGCAATCCA Reverse: GGACCTCTGTAGCTTTCACCA |
Quantitative real‐time PCR (qPCR) amplification of target message RNA was performed using SYBR Green I PCR master mix (Takara, Tokyo, Japan). Reactions were carried out on an ABI 7300 (Applied Biosystems, Shanghai, China) under the following procedures: Denaturation 3 min at 94 °C, followed by 40 cycles, consisting 5 s at 94 °C and 34 s at 60 °C. For each reaction, a melting curve was generated to test for primer dimmer formation and false priming.
Zymography
MMP‐2 and MMP‐9 secreted by ECs were assayed from culture media samples by zymography. Briefly, protein concentrations were determined using the BCA kit (Beyotime, Nanjing, China). Samples were prepared in standard, non‐reducing loading buffer (62.5 mm Tris‐HCl, pH6.8, 25% glycerol, 2% sodium dodecyl sulphate (SDS), 0.01% bromophenol blue, no β‐mercaptoethanol) for SDS‐PAGE (in 10% SDS‐polyacrylamide gel electrophoresis (PAGE) gel copolymerized with 0.05% gelatin). Following electrophoresis, SDS was removed from gels by incubation in 2.5% Triton X‐100, three times, for 1.5 h, followed by incubation in proteolysis buffer (50 mm CaCl2, 0.5 m NaCl, 50 mm Tris, pH 7.8), incubation at 37 °C for 12–16 h. Zymograms were subsequently stained with Coomassie brilliant blue for 1 h on a rotator, and areas of digestion appeared as clear bands against Coomassie blue background, where substrate had been degraded by the enzyme.
Statistical analysis
All experiments in vitro involving gene profiling analyses and zymography, from collected cell samples, were all obtained from at least three different female mice and experiments were performed in triplicate and reproduced at least three separate times. Statistical analysis of data was performed using SPSS 19.0 (IBM, Silicon Valley, CA, USA) and one‐way ANOVA, to compare means of all groups. Student–Newman–Keuls (SNK‐q) testing was used to compare means of each two groups. Data were considered significantly different if two‐tailed P value was <0.05.
Results
LY2157299 attenuated formation of vessel‐like structures in the co‐culture 3D collagen gel model
To explore whether the TGFβ signalling pathway plays an important role in angiogenesis, we used 3D collagen gel moulds with co‐culture of RFP‐mECs and GFP‐mASCs, in vitro. By 7 days post‐implantation, co‐culture with normal conditions significantly expedited formation of extended capillary branches and vessel‐like structures (Fig. 1a, control). However, formation of these structures was suppressed by treatment of LY2157299 (10 μm), and most ECs remained as single cells (Fig. 1a, LY2157299 (10 μm)). Furthermore, after treatment with LY2157299 (10 μm), average length and width of vessel‐like structures was shorter (Fig. 1c,d), and sprout‐ like structures and connections were fewer (Fig. 1b).
Figure 1.
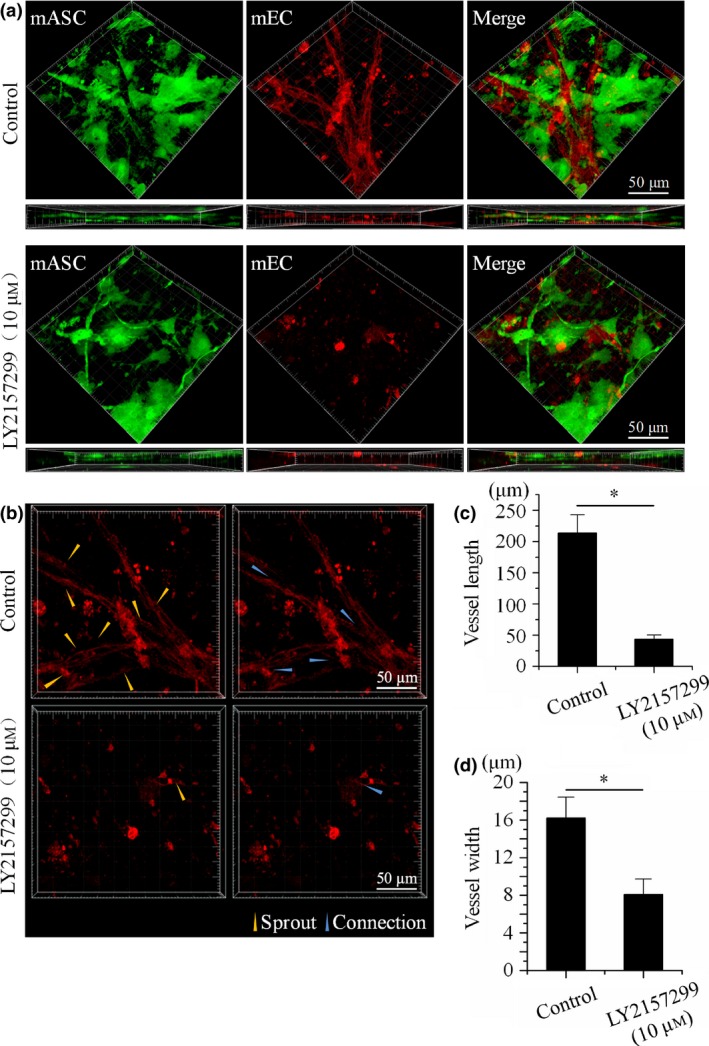
LY 2157299 attenuated angiogenesis in co‐culture 3D collagen gels. (a) Angiogenesis in 3D gels after 7 days co‐culture of ASCs and ECs under normal conditions and with LY2157299, in vitro. 3D construction morphologies and horizontal views reveal that co‐culture with LY2157299 significantly reduced formation of vessel‐like structures (n = 3). (b) Formation of sprouts and connections in co‐culture control groups and inhibitor‐treated groups (n = 3). In control groups, sprouts and connections existed and are shown; whereas in LY2157299‐treated groups, few were seen. (c) Different lengths of vessel‐like structures in examples of co‐culture control groups and inhibitor‐treated groups. Vessel‐like structure lengths were analysed using Image‐Pro Plus Software 6.0. Data expressed as mean of three different experiments (n = 3). (d) Different lumen diameters in co‐culture control group and inhibitor‐treated group. Vessel‐like structure widths were analysed using Image‐Pro Plus Software 6.0. Data expressed as mean of three different experiments (n = 3).
VEGF and VE‐ca were down‐regulated in ECs after treatment with LY2157299
To explore expressions of angiogenesis‐related genes in the co‐culture 3D gels, transwell chambers were used to achieve gene detection both in ECs and ASCs after treatments of concentration gradients of LY2157299 from 0 (normal), 5, 10, 20 to 50 μm for 24 h. Results of semi‐quantitative PCR (Fig. 2a) indicate that expressions of VEGF‐A, VEGF‐B and VE‐ca in ECs were down‐regulated after transwell co‐culture with LY2157299 from 5, 10, 20 to 50 μm for 24 h (Fig. 2a EC group), compared to corresponding mono‐cultured ECs. Considering the possibility that high concentrations such as 20 and 50 μm LY2157599 might affect survival and adhesion of cells, 10 μm was chosen as moderated concentration for explorations such as 3D co‐culture and quantitative real‐time PCR (qRT‐PCR). QRT‐PCR was performed to further investigate expression of VEGF‐A, VEGF‐B and VE‐ca in ECs. As for VEGF‐A (Fig. 2b), in the normal group, co‐cultured ECs expressed more VEGF‐A compared to mono‐cultured ECs. In ECs treated with LY2157299 (10 μm) (Fig. 2b), down‐regulated expression of VEGF‐A was found in mono‐cultured ECs and co‐cultured ECs after treatment with LY2157299 (10 μm). Furthermore, expressions of VEGF‐A in the co‐culture group were lower than in the corresponding mono‐culture group. The same tendency was also discovered in expressions of VEGF‐B (Fig. 2c) and VE‐ca (Fig. 2d). Concerning ASCs, quantification of semi‐quantitative PCR results by the OD method showed that there were no significant differences in expressions of VEGF‐A and VEGF‐B (see Table 4 for mono‐cultured ASCs and co‐cultured ASCs). VE‐ca was not expressed in ASCs.
Figure 2.
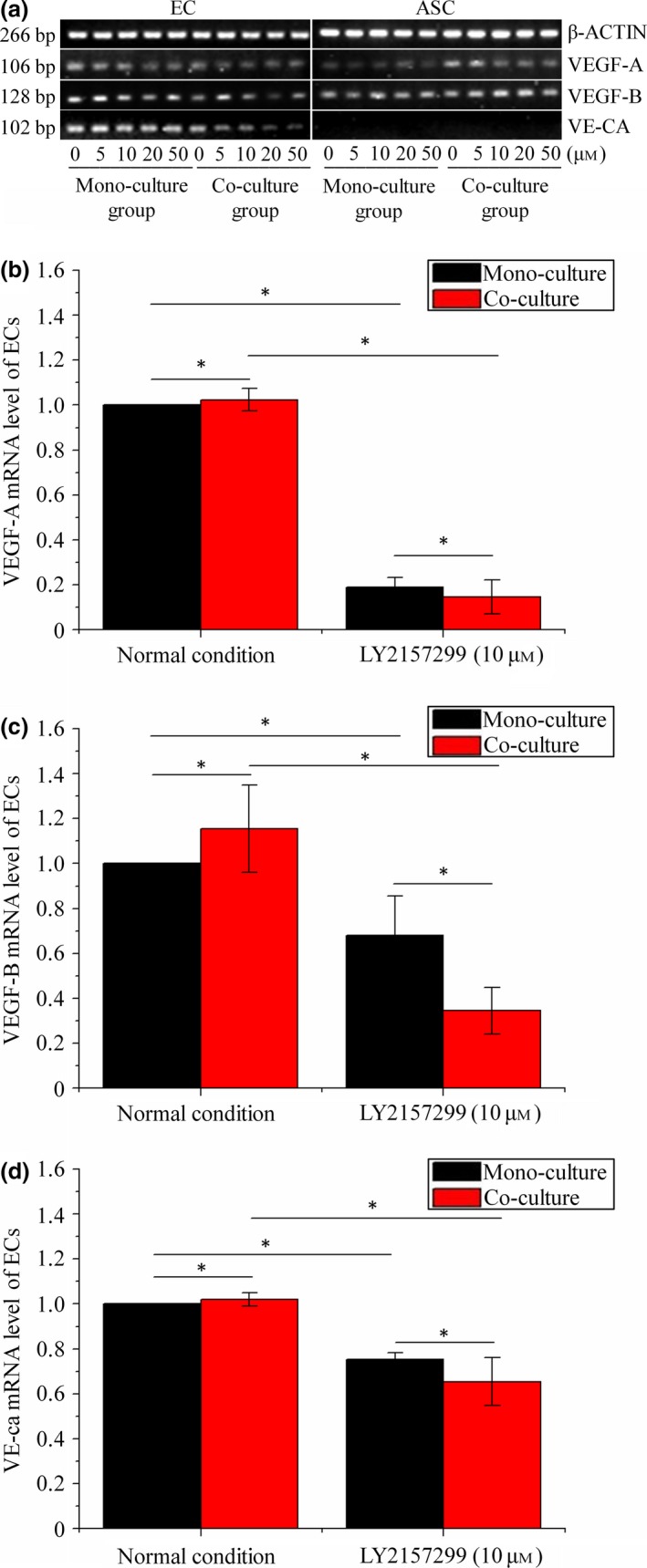
Examples of gene profiles of VEGF ‐A/B and VE ‐ca after 24 h in mono‐culture control groups, co‐culture control groups and LY 2157299‐treated groups by semi‐quantitative PCR and qPCR . (a) Gene variations of VEGF‐A,VEGF‐B and VE‐ca in ECs and ASCs in mono‐culture control group, co‐culture control group and treated group with LY2157299 from 0, 5, 10, 20 to 50 μm, by semi‐quantitative PCR. Gels shown are representative of three different experiments (n = 3). (b–d) Gene variations of VEGF‐A (b), VEGF‐B (c) and VE‐ca (d) in ECs in mono‐culture control group, co‐culture control group and treated group with LY2157299 (10 μm) by qPCR. β‐Actin levels set as internal normalized control. Results shown are representative of three different experiments (n = 3). Data presented as means ± SD, *P < 0.05.
Table 4.
OD values of angiogenesis‐related growth factor in mono‐cultured and co‐cultured ASCs
| Mono‐cultured ASCs | Co‐cultured ASCs | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 5 μm | 10 μm | 20 μm | 50 μm | 0 | 5 μm | 10 μm | 20 μm | 50 μm | |
| VEGF‐A | ||||||||||
| OD | 442 ± 44 | 466 ± 33 | 461 ± 35 | 501 ± 17 | 523 ± 17 | 671 ± 27 | 561 ± 48 | 563 ± 67 | 480 ± 55 | 528 ± 48 |
| P | 0.1053 | 0.3703 | 0.0973 | 0.1466 | P | 0.1374 | 0.1196 | 0.0523 | 0.0816 | |
| VEGF‐B | ||||||||||
| OD | 1058 ± 24 | 1033 ± 26 | 1042 ± 13 | 1035 ± 22 | 1049 ± 30 | 1291 ± 66 | 1242 ± 54 | 1332 ± 53 | 1244 ± 18 | 1262 ± 55 |
| P | 0.4724 | 0.0836 | 0.4644 | 0.5405 | P | 0.286 | 0.1888 | 0.2688 | 0.5607 | |
| VEGF‐C | ||||||||||
| OD | 459 ± 25 | 451 ± 38 | 432 ± 14 | 455 ± 24 | 429 ± 23 | 682 ± 21 | 656 ± 16 | 655 ± 18 | 652 ± 27 | 633 ± 36 |
| P | 0.8297 | 0.7359 | 0.8593 | 0.3761 | P | 0.3385 | 0.1144 | 0.1746 | 0.2693 | |
| HGF | ||||||||||
| OD | 1173 ± 86 | 1149 ± 55 | 1250 ± 51 | 1218 ± 22 | 1222 ± 85 | 1104 ± 48 | 1063 ± 48 | 1161 ± 55 | 1080 ± 67 | 1075 ± 44 |
| P | 0.7762 | 0.1281 | 0.5314 | 0.6584 | P | 0.4096 | 0.2901 | 0.7343 | 0.631 | |
| FGF2 | ||||||||||
| OD | 554 ± 44 | 560 ± 33 | 578 ± 34 | 603 ± 23 | 633 ± 13 | 593 ± 18 | 559 ± 16 | 605 ± 8 | 615 ± 15 | v572 ± 11 |
| P | 0.798 | 0.3691 | 0.1931 | 0.1359 | P | 0.0931 | 0.1717 | 0.1739 | 0.3035 | |
Relevant growth factors down‐regulated in ECs after treatment with LY 2157299
Gene expressions of growth factors related to angiogenesis in both ECs and ASCs after transwell co‐culture were further investigated by semi‐quantitative PCR (Fig. 3). These results (Fig. 3) show that expressions of PDGF, HGF, FGF‐1, FGF‐2 and BMP‐4 were down‐regulated in both mono‐cultured ECs and co‐cultured ECs after treatment with LY2157299 from 5, 10, 20 to 50 μm for 24 h (see Table 2 for mono‐cultured ECs and co‐cultured ECs); moreover, expressions of PDGF, HGF, FGF‐1, FGF‐2 and BMP‐4 in ECs reduced in a dose‐dependent manner (see Table 2 for mono‐cultured ECs and co‐cultured ECs). Furthermore, expressions of these gene products in co‐culture groups were lower than corresponding mono‐culture groups (see Table 3 for comparison between treated mono‐cultured ECs and co‐cultured ECs). Regarding ASCs, after treatment with LY2157299, there were no significant differences in expression of VEGF‐C, HGF and FGF‐2 either in mono‐cultured ASCs or in co‐cultured ASCs (see Table 4 for mono‐cultured ASCs and co‐cultured ASCs). BMP‐4 and FGF‐1 were not expressed in ASCs (Fig. 3). PDGF in ASCs was not expressed when treated with LY2157299.
Figure 3.
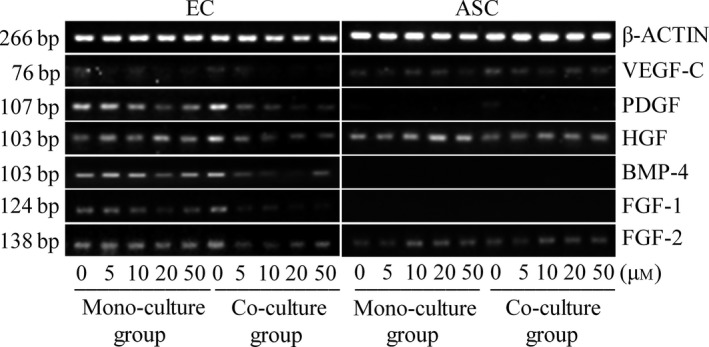
Different gene variations of growth factor profiles induced by LY 2157299 in EC s and ASC s using semi‐quantitative PCR . After screening by semi‐quantitative PCR, the variety of genes, including VEGF‐C,PDGF,HGF,FGF‐1,FGF‐2 and BMP‐4 in ECs and ASCs are shown. Gels shown are representative of three different experiments (n = 3).
Table 2.
Gene changes of angiogenesis‐related growth factors in mono‐cultured and co‐cultured ECs
| Mono‐cultured ECs | Co‐cultured ECs | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 5 μm | 10 μm | 20 μm | 50 μm | 0 | 5 μm | 10 μm | 20 μm | 50 μm | |
| VEGF‐C | 100% | 84.94% | 79.51% | 59.40% | 54.58% | 100% | 66.25% | 33.59% | 30.31% | 26.67% |
| P | 0.04726 | 0.0291 | 0.00206 | 0.00175 | P | 0.00094 | 0.00068 | 0.00027 | 0.00012 | |
| PDGF | 100% | 96.69% | 61.41% | 52.35% | 49.01% | 100% | 31.24% | 21.21% | 14.79% | 13.95% |
| P | 0.00137 | 0.00116 | 0.00009 | 0.00064 | P | 0.00007 | 0.00009 | 0.00006 | 0.00008 | |
| HGF | 100% | 95.93% | 91.81% | 84.05% | 75.37% | 100% | 48.16% | 29.68% | 28.12% | 26.54% |
| P | 0.00363 | 0.0168 | 0.01275 | 0.00705 | P | 0.00493 | 0.00168 | 0.00128 | 0.00163 | |
| BMP4 | 100% | 96.83% | 91.71% | 80.98% | 46.89% | 100% | 39.29% | 33.38% | 23.93% | 23.18% |
| P | 0.00791 | 0.00746 | 0.00512 | 0.00065 | P | 0.00043 | 0.00004 | 0.00073 | 0.00003 | |
| FGF1 | 100% | 94.25% | 69.85% | 60.08% | 58.57% | 100% | 35.58% | 33.33% | 15.27% | 12.57% |
| P | 0.00217 | 0.00044 | 0.00085 | 0.00038 | P | 0.00182 | 0.00209 | 0.00055 | 0.00087 | |
| FGF2 | 100% | 91.14% | 82.63% | 62.98% | 60.48% | 100% | 47.60% | 36.92% | 34.07% | 33.40% |
| P | 0.02356 | 0.00549 | 0.00178 | 0.00167 | P | 0.00011 | 0.00039 | 0.00003 | 0.00001 | |
Table 3.
Gene changes in ECs after co‐culture
| 5 μm | 10 μm | 20 μm | 50 μm | |
|---|---|---|---|---|
| VEGF‐C | ||||
| Down to | 87.29% | 47.28% | 57.10% | 54.68% |
| P | 0.03651 | 0.00204 | 0.00024 | 0.00637 |
| PDGF | ||||
| Down to | 41.27% | 44.13% | 36.08% | 36.36% |
| P | 0.00004 | 0.00092 | 0.00002 | 0.0046 |
| HGF | ||||
| Down to | 74.26% | 47.82% | 49.49% | 52.09% |
| P | 0.02073 | 0.00006 | 0.00154 | 0.00335 |
| BMP4 | ||||
| Down to | 44.21% | 39.65% | 32.19% | 53.85% |
| P | 0.00098 | 0.00214 | 0.00013 | 0.00099 |
| FGF1 | ||||
| Down to | 42.96% | 54.30% | 28.93% | 24.43% |
| P | 0.00011 | 0.00195 | 0.00114 | 0.00071 |
| FGF2 | ||||
| Down to | 55.94% | 47.87% | 57.94% | 59.16% |
| P | 0.00007 | 0.00063 | 0.00002 | 0.00049 |
Activation of MMP‐2 and MMP‐9 with and without inhibition of TGFβRI
After quantification using the OD method (with Quantity One 4.6.3 software (Bio‐Rad)), we found that secretions of MMP‐2 and MMP‐9 were attenuated both in mono‐cultured (Fig. 4b) and co‐cultured ECs (Fig. 4c) after treatment of LY2157299.
Figure 4.
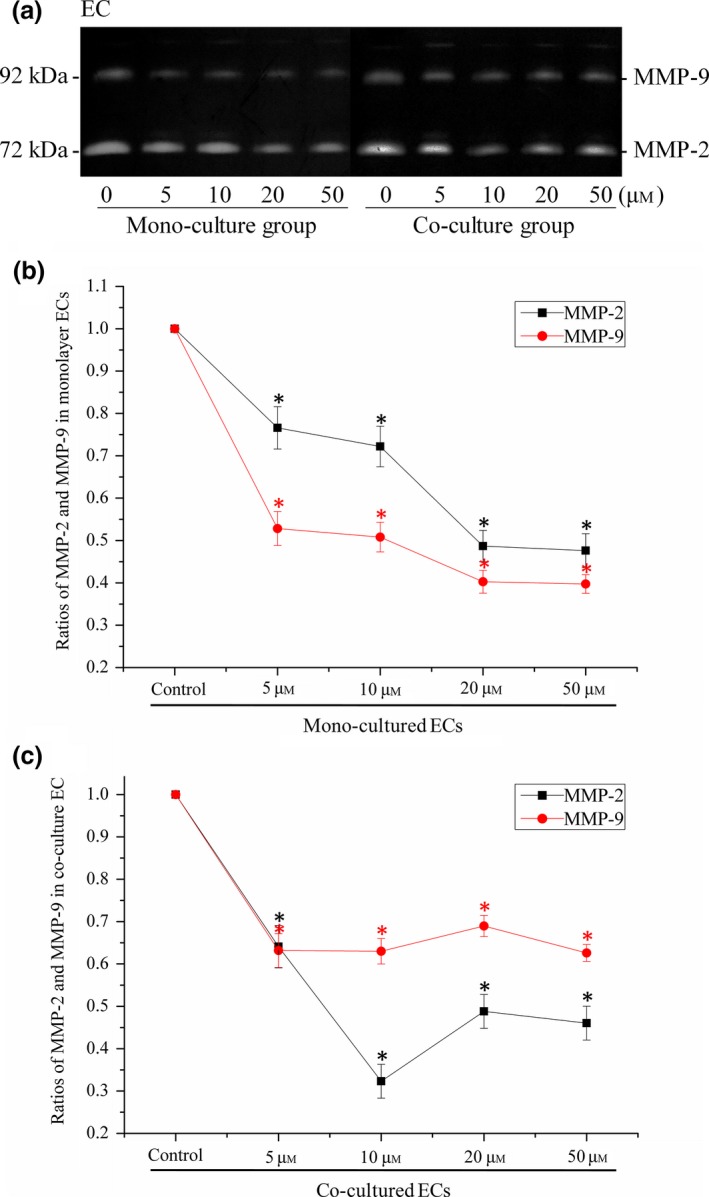
LY 2157299 reduced MMP ‐2 and MMP ‐9 secretions by EC s. (a) Zymography showed MMP‐2 and MMP‐9 secreted in mono‐cultured ECs and co‐cultured ECs induced by LY2157299 from 0, 5, 10, 20 to 50 μm. Gels shown are representative of three different experiments (n = 3). (b) Quantification of MMP‐2 and MMP‐9 secreted in mono‐cultured ECs was performed using the OD method. Secretions of MMP‐2 and MMP‐9 were lower after treatment with LY2157299. *Significant difference with respect to normal controls (P < 0.05). (c) Quantification of MMP‐2 and MMP‐9 secreted in co‐cultured ECs was performed using the OD method. Secretions of MMP‐2 and MMP‐9 were lower after treatment with LY2157299. *Significant difference with respect to normal controls (P < 0.05).
Discussion
ASCs, one of the important post‐natal types of adult stem cell, are thought to be one of the most promising stem cell populations for therapeutic use, as human adipose tissue is ubiquitous and easily obtained in large quantities, with little donor site morbidity or patient discomfort. Thus, use of autologous ASCs as both research tools and as cellular therapeutics is feasible, and has been shown to be both safe and efficacious in pre‐clinical and clinical studies of injury and disease 15, 16.
In the process of angiogenesis by ECs, expressions of angiogenesis‐related genes such as VEGF‐A, VEGF‐B, FGF‐1 and FGF‐2 are essential 17. After inhibition of TGFβRI by LY2157299 (LY2157299 dissolved in DMSO then diluted to 5, 10, 20 and 50 μm by culture medium, LY2157299 inhibiting TGFβ signal pathway by inhibiting the TGFβ receptor I) for 24 h, marked attenuation of related genes such as VEGF‐A, VEGF‐B, VE‐ca, PDGF, HGF, FGF‐1 and FGF‐2. VEGF‐A and VEGF‐B was found in ECs; these are critical mediators of angiogenesis and vasculogenesis. VEGF‐A has two major biological activities, the capacity to stimulate vascular endothelial cell proliferation 18, and the ability to increase vascular permeability 19. VEGF‐A also promotes survival and migration of ECs 20. VE‐cadherin (VE‐ca) regulates activity of high affinity receptors for vascular endothelial growth factor (VEGF), transforming growth factor (TGF)‐β and fibroblast growth factor (FGF) 21. PDGF family members have been reported to play key pro‐angiogenic roles in model studies in vivo 22; PDGFB from endothelial cells and its receptor (PDGFRβ) from pericytes, play important roles in pericyte recruitment to blood vessels 23, 24. HGF promotes angiogenesis through up‐regulation of VEGF and also via down‐regulating thrombospondin‐1 (a potent angiogenic inhibitor) expression 25, 26. FGF2 has been attributed to be a key player in inducing different angiogenic and lymphangiogenic pathways. FGFs induce angiogenesis by promoting endothelial cell proliferation 27, 28, extracellular matrix degradation, altering intercellular adhesion and communication by affecting cadherins, gap junctions and by modulating integrin expression 29. BMP‐4 has been shown to stimulate angiogenesis by production of VEGF‐A in osteoblasts 30. Suppression of angiogenesis‐related genes suppresses survival, migration and proliferation of ECs.
When non‐activated vascular endothelial cells are activated, they have higher vessel permeability and integrin on cell surfaces and secrete proteolytic enzymes such as MMPs for remodelling extracellular matrix 31, 32. By degrading surrounding proteins, MMPs assist movement of vascular endothelial cells into surrounding sites for formation of micro‐vessels. Most of them, MMP‐2 and MMP‐9, play key roles in extracellular matrix remodelling 33, 34 and providing new space for vascular endothelial cells to form new micro‐vessels.
From zymography results that secretions of MMP‐2 and MMP‐9 wereattenuated after treatment with LY2157299, we drew the conclusion that inhibition of the TGFβ signalling pathway attenuated secretion of MMPs, limited remodelling of extracellular matrix, reduced space for formation of new micro‐vessels and finally inhibited angiogenesis.
Based on indistinctive expressions of angiogenic genes other than PDGF in ASCs after inhibition of TGFβRI, we suppose that in co‐culture of ASCs and ECs, the TGFβ signalling pathway regulated angiogenesis via ECs. The result that inhibition affected angiogenesis in co‐cultured ECs, regulated more significantly than mono‐cultured ECs, suggests that inhibition of TGFβRI regulated paracrine secretion of ASCs, to further modulate angiogenesis of ECs.
We recognise that there are some limitations to the study. First, angiogenesis in 3D gel requires cell–cell contact crosstalk. To isolate gene expressions in the ASCs and ECs, non‐contact transwell co‐culture was used. This may be somewhat inaccurate due to gaps between contact and non‐contact co‐cultures. Secondly, we screened the growth factor profile, based on a common genebank; other unscreened growth factors may also play a vital role in angiogenesis. Thirdly, exact mechanisms of ASC factor secretion to further suppress EC angiogenesis, needs to be further explored.
Acknowledgements
This work was funded by National Natural Science Foundation of China (81470721, 81321002, 31170929), Sichuan Science and Technology Innovation Team (2014TD0001).
Shiyu Lin and Jing Xie contributed equally to this work.
References
- 1. Kolonin MG, Evans KW, Mani SA, Gomer RH (2012) Alternative origins of stroma in normal organs and disease. Stem Cell Res. 8, 312–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Park IS, Kang JA, Kang J, Rhie JW, Kim SH (2014) Therapeutic effect of human adipose‐derived stromal cells cluster in rat hind‐limb ischemia. Anat. Rec. (Hoboken) 297, 2289–2298. [DOI] [PubMed] [Google Scholar]
- 3. Abdollahi H, Harris LJ, Zhang P, McIlhenny S, Srinivas V, Tulenko T et al (2011) The role of hypoxia in stem cell differentiation and therapeutics. J. Surg. Res. 165, 112–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cao Y, Sun Z, Liao L, Meng Y, Han Q, Zhao RC (2005) Human adipose tissue‐derived stem cells differentiate into endothelial cells in vitro and improve postnatal neovascularization in vivo. Biochem. Biophys. Res. Commun. 332, 370–379. [DOI] [PubMed] [Google Scholar]
- 5. Shen J, Li S, Chen D (2014) TGF‐beta signaling and the development of osteoarthritis. Bone Res. 2, 14002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rahman MS, Akhtar N, Jamil HM, Banik RS, Asaduzzaman SM (2015) TGF‐β/BMP signaling and other molecular events: regulation of osteoblastogenesis and bone formation. Bone Res. 3, 15005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Massague J, Seoane J, Wotton D (2005) Smad transcription factors. Genes Dev. 19, 2783–2810. [DOI] [PubMed] [Google Scholar]
- 8. Lu Q (2008) Transforming growth factor‐beta1 protects against pulmonary artery endothelial cell apoptosis via ALK5. Am. J. Physiol. Lung Cell. Mol. Physiol. 295, L123–L133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Oh SP, Seki T, Goss KA, Imamura T, Yi Y, Donahoe PK et al (2000) Activin receptor‐like kinase 1 modulates transforming growth factor‐beta 1 signaling in the regulation of angiogenesis. Proc. Natl Acad. Sci. USA 97, 2626–2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Seki T, Yun J, Oh SP (2003) Arterial endothelium‐specific activin receptor‐like kinase 1 expression suggests its role in arterialization and vascular remodeling. Circ. Res. 93, 682–689. [DOI] [PubMed] [Google Scholar]
- 11. Lutty GA, Merges C, Threlkeld AB, Crone S, McLeod DS (1993) Heterogeneity in localization of isoforms of TGF‐beta in human retina, vitreous, and choroid. Invest. Ophthalmol. Vis. Sci. 34, 477–487. [PubMed] [Google Scholar]
- 12. Yue Y, Yang X, Wei X, Chen J, Fu N, Fu Y et al (2013) Osteogenic differentiation of adipose‐derived stem cells prompted by low‐intensity pulsed ultrasound. Cell Prolif. 46, 320–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Steinway SN, Zanudo JG, Ding W, Rountree CB, Feith DJ, Loughran TP Jr. et al (2014) Network modeling of TGFbeta signaling in hepatocellular carcinoma epithelial‐to‐mesenchymal transition reveals joint sonic hedgehog and Wnt pathway activation. Cancer Res. 74, 5963–5977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li CY, Wood DK, Huang JH, Bhatia SN (2013) Flow‐based pipeline for systematic modulation and analysis of 3D tumor microenvironments. Lab Chip 13, 1969–1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gimble JM, Katz AJ, Bunnell BA (2007) Adipose‐derived stem cells for regenerative medicine. Circ. Res. 100, 1249–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tobita M, Orbay H, Mizuno H (2011) Adipose‐derived stem cells: current findings and future perspectives. Discov. Med. 11, 160–170. [PubMed] [Google Scholar]
- 17. Salgado AJ, Reis RL, Sousa NJ, Gimble JM (2010) Adipose tissue derived stem cells secretome: soluble factors and their roles in regenerative medicine. Curr. Stem Cell Res. Ther. 5, 103–110. [DOI] [PubMed] [Google Scholar]
- 18. Leung DW, Cachianes G, Kuang WJ, Goeddel DV, Ferrara N (1989) Vascular endothelial growth factor is a secreted angiogenic mitogen. Science 246, 1306–1309. [DOI] [PubMed] [Google Scholar]
- 19. Keck PJ, Hauser SD, Krivi G, Sanzo K, Warren T, Feder J et al (1989) Vascular permeability factor, an endothelial cell mitogen related to PDGF. Science 246, 1309–1312. [DOI] [PubMed] [Google Scholar]
- 20. Takahashi H, Shibuya M (2005) The vascular endothelial growth factor (VEGF)/VEGF receptor system and its role under physiological and pathological conditions. Clin. Sci. (Lond.) 109, 227–241. [DOI] [PubMed] [Google Scholar]
- 21. Giannotta M, Trani M, Dejana E (2013) VE‐cadherin and endothelial adherens junctions: active guardians of vascular integrity. Dev. Cell 26, 441–454. [DOI] [PubMed] [Google Scholar]
- 22. Kilvaer TK, Valkov A, Sorbye SW, Smeland E, Bremnes RM, Busund LT et al (2011) Fibroblast growth factor 2 orchestrates angiogenic networking in non‐GIST STS patients. J. Transl. Med. 9, 104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Armulik A, Abramsson A, Betsholtz C (2005) Endothelial/pericyte interactions. Circ. Res. 97, 512–523. [DOI] [PubMed] [Google Scholar]
- 24. Saik JE, Gould DJ, Watkins EM, Dickinson ME, West JL (2011) Covalently immobilized platelet‐derived growth factor‐BB promotes angiogenesis in biomimetic poly(ethylene glycol) hydrogels. Acta Biomater. 7, 133–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sengupta S, Gherardi E, Sellers LA, Wood JM, Sasisekharan R, Fan TP (2003) Hepatocyte growth factor/scatter factor can induce angiogenesis independently of vascular endothelial growth factor. Arterioscler. Thromb. Vasc. Biol. 23, 69–75. [DOI] [PubMed] [Google Scholar]
- 26. Xin X, Yang S, Ingle G, Zlot C, Rangell L, Kowalski J et al (2001) Hepatocyte growth factor enhances vascular endothelial growth factor‐induced angiogenesis in vitro and in vivo. Am. J. Pathol. 158, 1111–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Potente M, Gerhardt H, Carmeliet P (2011) Basic and therapeutic aspects of angiogenesis. Cell 146, 873–887. [DOI] [PubMed] [Google Scholar]
- 28. Lang I, Hoffmann C, Olip H, Pabst MA, Hahn T, Dohr G et al (2001) Differential mitogenic responses of human macrovascular and microvascular endothelial cells to cytokines underline their phenotypic heterogeneity. Cell Prolif. 34, 143–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cao Y, Cao R, Hedlund EM (2008) R Regulation of tumor angiogenesis and metastasis by FGF and PDGF signaling pathways. J. Mol. Med. (Berl) 86, 785–789. [DOI] [PubMed] [Google Scholar]
- 30. Kim JM, Kang SW, Shin SM, Su Kim D, Choi KK, Kim EC et al (2014) Inhibition of matrix metalloproteinases expression in human dental pulp cells by all‐trans retinoic acid. Int. J. Oral. Sci. 6, 150–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Deregibus MC, Cantaluppi V, Calogero R, Lo Iacono M, Tetta C, Biancone L et al (2007) Endothelial progenitor cell derived microvesicles activate an angiogenic program in endothelial cells by a horizontal transfer of mRNA. Blood 110, 2440–2448. [DOI] [PubMed] [Google Scholar]
- 32. Mahabeleshwar GH, Feng W, Reddy K, Plow EF, Byzova TV (2007) Mechanisms of integrin‐vascular endothelial growth factor receptor cross‐activation in angiogenesis. Circ. Res. 101, 570–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Page‐McCaw A, Ewald AJ, Werb Z (2007) Matrix metalloproteinases and the regulation of tissue remodelling. Nat. Rev. Mol. Cell Biol. 8, 221–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Schultz GS, Wysocki A (2009) Interactions between extracellular matrix and growth factors in wound healing. Wound Repair Regen. 17, 153–162. [DOI] [PubMed] [Google Scholar]


