Abstract
Objectives: In recent years, obesity has become a global epidemic, highlighting the necessity for basic research into mechanisms underlying growth of adipose tissue and differentiation of stem cells into adipocytes, in humans. For better understanding of cell signalling in adipogenesis, the role of DNER (delta/Notch‐like EGF‐related receptor) in adipogenic differentiation of human adipose tissue‐derived mesenchymal stem cells (hAMSC) was investigated.
Materials and methods: To assess the role of DNER in hAMSC adipogenesis, hAMSCs were transfected with DNER small interfering RNA (siDNER). Real‐time quantitative reverse transcriptase polymerase chain reactions to assess expression levels of adipogenesis‐related genes regulated by siDNER, cell cycle and immunoblot analyses were performed.
Results: First, it was determined that DNER mRNA was profoundly expressed in hAMSCs and reduced during adipogenic differentiation. Knockdown of DNER altered cell morphology, inhibited proliferation and increased frequency and efficiency of adipogenesis in hAMSC. Expression of CCAAT/enhancer‐binding protein δ increased and proportion of cells in S phase decreased by knockdown of DNER, using specific siRNA. Moreover, adipocyte‐specific genes including peroxisome proliferator‐activated receptor gamma, fatty acid binding protein 4 and perilipin were up‐regulated in siDNER compared to the siControl group during adipogenesis in hAMSC.
Conclusions: These results indicate that DNER knockdown in hAMSC accelerated onset of adipogenic differentiation by bypassing mitotic clonal expansion during the early stages of adipogenesis.
Introduction
Notch is an indispensable signalling receptor controlling cell fate and pattern formation during embryonic development for invertebrate as well as vertebrate species. The role of Notch in differentiation of mammalian cells is, however, still incompletely understood. Constitutive expression of activated Notch1 inhibits osteoblast differentiation while enhancing adipogenesis in stromal cell lines (1). On the other hand, impaired Notch1 expression prevents adipocyte differentiation (2). Consistent with these findings, over‐expression or down‐regulation of Hes1 (a down‐stream target of Notch) by siRNA, inhibits adipogenesis in 3T3‐L1 preadipocytes (3). This finding suggests that Notch may have roles in both promoting and inhibiting adipogenesis.
DNER (delta/Notch‐like EGF‐related receptor), which is expressed principally in the cerebellum, is a neuron‐specific transmembrane protein with extracellular EGF‐like repeats. Recent studies have shown that DNER mediates Notch signalling via cell–cell interaction.
The primary objective of this study was to determine the effects of DNER on adipogenic differentiation using human adipose tissue‐derived mesenchymal stem cells (hAMSCs).
Up to now, preadipocyte cell lines from rodents, generally, have been employed to gain insight into the molecular mechanisms of adipogenesis and as a tool in development of therapeutic drugs for treatment of human obesity. With regard to currently available human in vitro models of adipogenesis, primary preadipocytes derived from stromal‐vascular cells of human adipose tissue have been utilized extensively. However, after a small number of passages, they undergo dramatic reduction in their ability to differentiate – a phenomenon known as functional senescence – before entering replicative senescence (4). Recently, several studies have shown that hAMSCs are multipotent, that is, they are capable of undergoing differentiation into adipocytes as well as other adult stem cells of mesenchymal lineage in vitro (5, 6). Thus, hAMSC provide a unique in vitro model, which allow for appropriate research into human adipogenesis.
The process of terminal adipocyte differentiation, during which preadipocytes mature into adipocytes, has been well studied in murine models in vitro, such as mouse 3T3‐L1 and 3T3‐F442A cell lines. Transition from preadipocyte to adipocyte in the mouse involves a stage in which an abrupt increase in cell number, referred to as mitotic clonal expansion (MCE), is observed followed by growth arrest and accumulation of lipid in the cytosol (7, 8). Interestingly on the contrary, primary human preadipocytes do not undergo MCE during in vitro adipogenesis (9, 10). Although the adipogenic mechanisms are fairly well understood in a limited number of model systems such as 3T3‐L1 preadipocyte and primary human preadipocyte models, differentiation of human stem cells into adipocytes has been relatively poorly elucidated thus far.
The CCAAT/enhancer‐binding protein (C/EBP) family of transcription factors consists of five different proteins, C/EBPα, C/EBPβ, C/EBPδ, C/EBPγ and CHOP. Each C/EBP protein has unique properties regulating cell type specific growth and differentiation. Sequential expression of these factors is observed during adipocyte differentiation, in which the early expression of C/EBPβ and C/EBPδ promotes expression of C/EBPα and peroxisome proliferator‐activated receptor gamma (PPARγ) (11). C/EBPδ was first characterized as an acute phase inflammatory response gene but it is typically low to undetectable in most cell types and tissues; however, it is rapidly induced by a variety of extracellular stimuli, for example, growth hormone, insulin, interferon‐γ, interleukin (IL)‐1, IL‐6, lipopolysaccharide, tumour necrosis factor‐α, dexamethasone, noradrenaline and glutamate (12, 13). In vitro and in vivo studies have implicated C/EBPδ in proliferation of osteoblasts (14), differentiation of lung epithelial cells (15, 16) but growth arrest of mouse mammary epithelial cells (17, 18) subsequent to immortalization and lack of C/EBPδ promotes proliferation of mouse embryonic fibroblasts (19) yet accelerates adipogenic differentiation in 3T3‐L1 preadipocytes (20). AUTHOR please check/amend the sense of the above sentences.
DNER has recently been identified in the brain as a single‐pass transmembrane protein, consisting of a large extracellular region harbouring 10 EGF repeats, and a short cytoplasmic tail with no enzymatic activity (21, 22). It has been recently reported that DNER promotes maturation of Bergmann glia in the cerebellum via Deltex‐dependent Notch signalling and is essential for precise cerebellar development (23). However, until now, the function of DNER has been assessed only in murine models and the precise mechanisms inherent to DNER signalling remain unknown in the context of hAMSC adipogenesis.
With this in mind, we assessed the effects of DNER on the adipogenesis of hAMSC. Our results demonstrated that the inhibition of DNER resulted in an increase in adipocyte maturation, partly via a reduction of cell proliferation through elevation of C/EBPδ expression. Moreover, unlike the murine cerebellar development model, we observed no connection between DNER and Notch signalling in hAMSC adipogenesis.
Materials and methods
Isolation and culture of hAMSC
hAMSCs (24) were isolated and cultured as described previously. In brief, freshly excised human mammary fat tissue obtained from reduction mammoplasty was minced and digested for 2 h with type I collagenase (1 mg/mL) at 37 °C. After washing with phosphate‐buffered saline (PBS) and 5 min of centrifugation at 1000 rpm, the tissue pellet content was filtered through 100 μm nylon mesh and incubated overnight in DMEM with 10% foetal bovine serum (FBS) at 37 °C with 5% humidified CO2. After 24‐h in culture, unattached cells were removed and culture medium was exchanged for K‐NAC medium supplemented with N‐acetyl‐l‐cysteine (NAC; Sigma, St‐Louis, MO, USA A8199) (2 mm) and l‐ascorbic acid (0.2 mm). All procedures were approved by the institutional review board of Seoul National University (IRB No. 0611/001‐001). Medium was exchanged at 48‐h intervals until cells became 90% confluent or more trypsinized and stored in liquid nitrogen until future use or were immediately subcultured.
In vitro differentiation assay
An in vitro differentiation assay into osteogenic and adipogenic lineage was performed. hAMSCs were initially cultured and propagated in K‐NAC medium with 5% FBS, then changed to adipogenic medium (DMEM supplemented with 5% FBS, 1 μm dexamethasone, 10 μm insulin, 200 μm indomethacin and 0.5 mm isobutylmethylxanthine), or osteogenic medium (DMEM supplemented with 5% FBS, 50 μm l‐ascorbate‐2‐phosphate, 0.1 μm dexamethasone and 10 mm glycerophosphate), which were refreshed every other day. Adipogenic differentiation was evaluated using oil red O staining as indication of intracellular lipid accumulation. Seven days after induction of adipogenic differentiation, cells were washed three times in PBS, fixed for 1 h in 10% formalin and washed a further three times in PBS. The cells were then stained for 30 min with oil red O, washed three times in PBS and photographed after nuclei had been stained with Hoechst 33238 (1 μg/mL; 10 min). Three random fields were counted for each well, triplicate wells counted for each treatment. Osteogenic differentiation was labelle by positive staining with the von Kossa stain, which is specific for calcium.
RNA interference
Transfection of siRNA into the cells was conducted when they had reached 70% confluence. The small interfering RNAs of DNER (siDNER, L‐018708‐01) and the non‐targeting control (siControl #1, D‐001810‐01) were purchased from Dharmacon (Chicago, IL, USA). Experiments were conducted using DharmaFECT1 (Dharmacon) as transfection agent and siRNA at concentration of 25 nmol/L. For mRNA analysis, the cells were transfected with target gene siRNA or control non‐targeting siRNA using DharmaFECT1. After 48 h, culture medium was exchanged and the cells were cultured with or without adipogenic medium.
Cell counts
To evaluate cell proliferation, hAMSC were differentiated in six‐well plates for 7 days. Then cells were rinsed gently in PBS and were harvested by trypsinization before reconstitution in PBS for counting using a haemocytometer; counts were performed on days 0, 1, 3 and 7. At each time point, each well was counted twice, and at least three wells were counted per each sample.
Measurement of cell population growth potential and cell cycle distribution
The effect of siDNER‐transfection on hAMSC proliferation was assessed by 3‐(4,5‐dimethylthiazol‐2‐yl)‐2,5‐diphenyltetrazolium bromide (MTT) assay as described previously (25). In brief, cells were seeded in six‐well microplates at a density of 5 × 104/mL, and were incubated for 24 h. They were then transfected for 48 h with siDNER or siControl. After culture for the indicated periods, 100 μL of MTT stock solution (5 mg/mL, Sigma) was added to each well and incubated for an additional 4 h at 37 °C. The supernatant was removed and 500 μL of dimethyl sulphoxide was added to each well to dissolve the water‐insoluble purple formazan crystals, and then transferred to 96‐well microplates for reading. Absorbance at a wavelength of 500 nm was measured using an EL800 microplate reader (Bio‐Tek Instrument, Winooski, VT, USA). All measurements were performed in triplicate. Flow cytometry cell cycle analysis using propidium iodide (PI) staining was performed, also as described previously (26). In brief, cells were transfected for 48 h with siDNER or siControl and then harvested by trypsinization at the indicated time. After washing in ice‐cold PBS, cells were fixed in 70% ethanol at −20 °C and stained with 50 μg/mL of PI in the presence of 100 μg/mL RNase A for 30 min. Cell cycle distribution was analysed using a FACSCalibur flow cytometer (Becton&Dickinson, San Jose, CA, USA).
RT‐PCR and real‐time quantitative PCR
Total RNA was isolated using Trizol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. cDNA synthesis was accomplished by adding the purified RNA and oligo‐dT primers to Accupower RT premix (Bioneer, Daejon, Korea) according to the manufacturer’s instructions. PCR was conducted using Accupower PCR premix (Bioneer). All PCR products were analysed by gel electrophoresis on 1.5% agarose gels with ethidium bromide staining, followed by fluorescence digitization using a Bio‐Rad GelDoc XR system (Bioneer, Daejon, Korea). Primers were designed using the Primer3 input software (http://frodo.wi.mit.edu/primer3/) and specificity of each primer was controlled using BLAST software. Primer sets used in the RT‐PCR are listed in Table S1. Real‐time quantitative PCR was conducted using an SYBR green PCR kit (ABI, Foster City, CA, USA) and validated using an Applied Biosystem 7500 (ABI) real‐time quantitative PCR system. Primers employed in this study are indicated in Table S2. The comparative method of relative quantification (2−ΔΔcCt) was employed to calculate expression levels of each target gene (normalized to β‐actin).
Immunoblot analysis
Cells were lysed with 50 mm Tris–HCl buffer containing 0.1% Triton X100, freshly supplemented with 1 mm phenylmethylsulphonyl fluoride, 1 mm aprotinin, 1 mm leupeptin, 1 mm anti‐pain and 0.1 mm sodium orthovanadate. Protein content was determined using the DC assay kit (Bio‐Rad) and separated using 15% sodium dodecyl sulphate–polyacrylamide gel electrophoresis. Samples were then transferred to nitrocellulose membranes at 40 V, 350 mA for 5 h, and incubated with antibody to fatty acid binding protein 4 (FABP4; Abcam, Cambridge, UK) and β‐actin (Sigma). All antibodies were used according to manufacturers’ instructions and protein bands were detected using an enhanced chemiluminescence detection kit (Amersham Pharmacia Biotech, Buckinghamshire, UK).
Statistical analysis
All experiments were conducted at least in triplicate (n = 3) and results were expressed as mean ± SD. Statistical analyses of these data were conducted using Student’s t‐test. A value of P < 0.05 was considered significant.
Results
DNER expressed in hAMSC decreased dramatically during adipogenesis
To assess expression of DNER in hAMSC, as an initial step, we performed real‐time quantitative PCR. mRNA expression level of DNER was remarkably high in hAMSC and hUCB (human umbilical cord blood)‐MSC, suggesting that DNER was up‐regulated and might have roles particular to MSCs; on the other hand, DNER expression levels in most human cancer cell lines are lower than in hAMSC or undetectable in real‐time quantitative PCR (Fig. 1a). During hAMSC adipogenesis, DNER was down‐regulated. As shown in Fig. 1b, expression level of DNER began to decrease in early stages of adipogenic differentiation (5 h post‐induction) and down‐regulation continued up to the later stages of adipogenesis (7 days post‐induction). As mentioned previously, DNER is a well‐known neuron‐specific transmembrane protein, which as a ligand of Notch, facilitates morphological differentiation of Bergmann glia via Deltex‐dependent Notch signalling (23). With this in mind, we attempted to determine whether DNER functioned as a Notch ligand during adipogenesis in hAMSC. Real‐time quantitative PCR was conducted to determine expression levels of Notch‐related genes such as Notch1, Hey1, Deltex1 and Hes1. Interestingly, mRNA expression level of DNER was reduced, whereas the Notch‐related genes were up‐regulated during adipogenesis (Fig. 1c). These results were contrary to those previously reported on the function of DNER as a Notch ligand in the mouse cerebellum. Our data indicate that DNER might not be involved in Notch signalling in adipogenesis of hAMSC.
Figure 1.
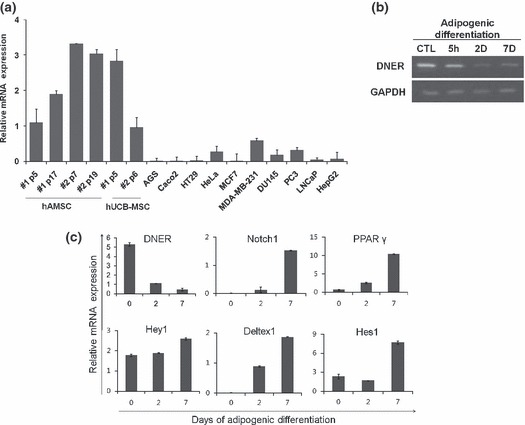
mRNA expression of DNER and Notch‐related genes during hAMSC adipogenesis. (a) Real‐time quantitative PCR analysis of DNER expression in a number of human cancer cell lines, hAMSCs and hUCB‐MSCs at different passages. (b) RT‐PCR results for DNER mRNA expression during adipogenesis. DNER decreased throughout the course of adipogenesis. hAMSCs were induced to differentiate and total RNA was isolated from the cell extracts at the indicated time points. (c) hAMSC were induced to undergo adipogenic differentiation and the mRNA was used for real‐time quantitative PCR analysis on DNER, Notch1, PPARγ, Hey1, Deltex1 and Hes1. DNER levels were reduced during adipogenesis; however, the level of Notch‐related genes increased during adipogenesis. The indicated values are expressed as the mean ± SD for three independent experiments.
Inhibition of DNER decelerated proliferation potential of hAMSC
To evaluate the role of DNER in hAMSC adipogenesis, we attempted to knockdown DNER using specific siRNA. As shown in Fig. 2a, after seven days treatment, adipogenic media successfully induced adipogenesis in which cells grew to confluence in the control and siControl‐transfected groups. However, inhibition of DNER in hAMSC caused growth retardation and change in cell morphology into cobblestone‐shape, similar to that of preadipocytes. As mentioned previously, MCE occurs in 3T3‐L1 preadipocytes, but not in primary human preadipocytes. Although controversial, this process may prove necessary for efficient differentiation (8, 27). We further attempted to determine whether DNER influenced MCE in the early phases of adipogenic differentiation. MCE was assessed by cell counting at the indicated time points during hAMSC adipogenesis. The number of cells had increased by approximately 2‐fold after 7 days of adipogenesis in the siControl‐transfected group. On the other hand, we noted no significant increase in cell numbers in the siDNER‐transfected group (Fig. 2b). In addition, in attempt to confirm whether siDNER inhibits cell proliferation, we conducted an MTT cell proliferation assay. The results demonstrated that the cell proliferation level was lower in siDNER‐transfected group as compared with the siControl‐transfected group (Fig. S1). Collectively, our results showed that inhibition of DNER induced a morphological change and might shorten early stages of adipogenesis by induced retardation of cell proliferation.
Figure 2.
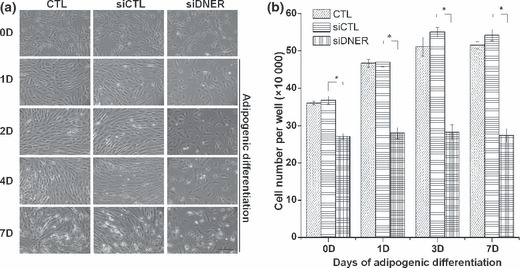
Knockdown of DNER induced growth arrest during the adipogenic differentiation of hAMSC. (a) Phenotypical changes of hAMSC by inhibition of DNER. The cells were transfected with siDNER or siControl. After transfection, the cells were induced to adipogenic differentiation and photomicrographs of hAMSC were taken at the indicated time points. The bar represents 100 μm. (b) To assess cell proliferation, the cells were harvested at the indicated time points of adipogenic differentiation after transfection, and then, at least three wells per sample were counted. The values indicated are expressed as the mean ± SD for three independent experiments. Asterisks indicate statistically significant differences (*P < 0.05).
Inhibition of DNER induced cell cycle arrest
To evaluate whether DNER regulates cell growth of hAMSC populations, we attempted to analyse cell cycle distribution after siDNER transfection, using PI. Knockdown of DNER led to a decrease in S‐phase cells (3‐fold) as compared to siControl‐transfected cells in early phase (Fig. 3a). We also examined expression levels of CDK2, CDK4 and cyclin D1 (CCND1). Expression of CDK2, CDK4 and CCND1 mRNA decreased in the siDNER‐transfected group compared to siControl‐transfected cells (Fig. 3b,c).
Figure 3.
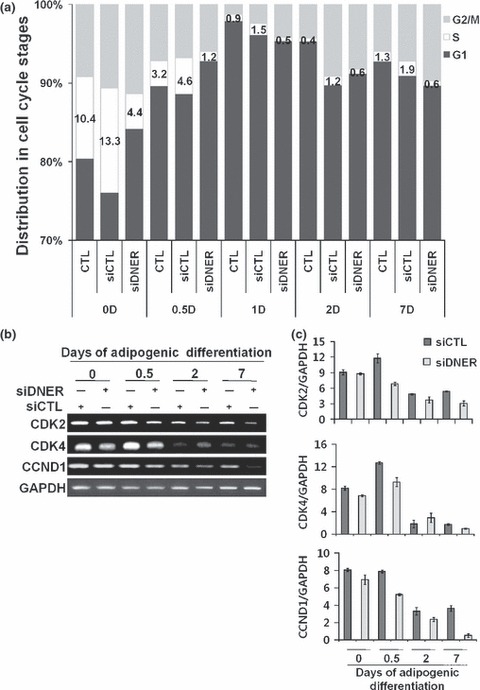
Cell cycle arrest in hAMSC after transfection with siDNER. (a) Representative diagrams of cell cycle distribution induced by siDNER or siControl‐transfected in hAMSC. Cell cycle distribution of hAMSC was analysed by flow cytometry as described in Materials and methods. The percentage of cells in each phase of the cell cycle was indicated as a bar graph. (b) Modulation of expression levels of CDK2, CDK4 and CCND1 upon siDNER transfection. Decrease in mRNA expression was detected in siDNER‐transfected group. RT‐PCR was conducted in triplicate for each transfectant. GAPDH was used as a loading control. (c) Densitometric analyses were obtained using the NIH ImageJ software for CDK2, CDK4 and CCND1 and the expression levels were normalized to GAPDH. The indicated values were expressed as the mean ± SD for three independent experiments.
Expression of C/EBP genes during adipogenesis after transfection with siDNER.
C/EBPβ and C/EBPδ of the C/EBP gene family are expressed prior to C/EBPα during adipogenesis. Therefore, the principal pathway of adipogenesis involves induction of C/EBPβ and C/EBPδ, which then facilitates expression of C/EBPα (20). Subsequent to immortalization, lack of C/EBPδ protein promotes proliferation of mouse embryonic fibroblasts (19). We hypothesized that inhibition of cell division and subsequent enhancement of adipogenesis by siDNER transfection might have been caused by C/EBPδ. To confirm this hypothesis, we assessed C/EBPs mRNA levels using real‐time quantitative PCR. C/EBPβ mRNA expression did not produce any significant differences during adipogenesis. On the other hand, C/EBPδ mRNA expression was transiently up‐regulated dramatically in the siDNER‐transfected group as compared to the siControl group in early adipogenesis. C/EBPα, a late adipogenic differentiation stage marker, was higher after 2 days of adipogenic differentiation in the siDNER‐transfected group, as shown using real‐time quantitative PCR (Fig. 4).
Figure 4.
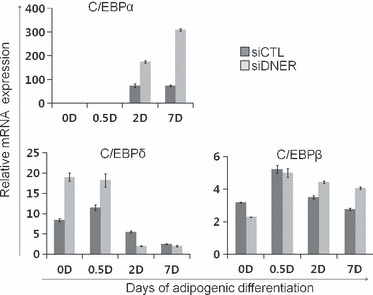
Knockdown of DNER increases expression of C/EBP family during adipogenesis. (a) The cells were transfected with siDNER or siControl. After transfection, the cells were induced to differentiate into adipocyte and total RNA was isolated from the cell extracts at the indicated time points. C/EBPδ mRNA was up‐regulated in siDNER‐transfected group during the early stage of adipogenesis. Real‐time quantitative PCR was conducted in triplicate for each transfectant. The indicated values were expressed as the mean ± SD for three independent experiments.
Knockdown of DNER accelerated adipogenic differentiation in hAMSC
We demonstrated that siDNER transfection reduced cell proliferation during adipogenic differentiation. To determine whether inhibition of DNER (using specific siRNA) would directly influence efficiency of adipogenic differentiation of hAMSC, we stained lipid droplets using oil Red O at 7 days after adipogenic differentiation. The siDNER‐transfected group had higher adipocyte frequency and lipid droplet size compared to the siControl‐transfected group (Fig. 5). These results showed that inhibition of DNER enhanced hAMSC adipogenesis. On the 7th day, effects of DNER on expression of genes critical for adipocyte phenotypes, including PPARγ, FABP4 and Perilipin, were analysed using real‐time quantitative PCR with mRNA prepared from siRNA‐transfected or non‐transfected cells, prior to and after induction of adipogenic differentiation. hAMSCs transfected with siDNER had higher adipogenic efficiency compared to the siControl group, as shown by the earlier appearance and higher level of adipocyte‐specific gene expression. Levels of PPARγ, FABP4 and Perilipin expression increased substantially in all groups throughout adipogenesis, for up to 7 days of the adipogenic induction period. The siDNER‐transfected group, however, demonstrated a significant increase in PPARγ, FABP4 and Perilipin expression levels compared to the siControl group as shown in Fig. 6a. We also demonstrated, using immunoblot analysis, that the protein level of FABP4, adipose specific fatty acid binding protein, was higher in the siDNER‐transfected group after 3 days of adipogenic induction (Fig. 6b). In summary, siDNER‐transfected hAMSCs differentiated into mature adipocytes to a greater extent than was observed in the control group, as indicated by the observed higher levels of PPARγ, FABP4 and Perilipin expression, and by observation of increases in size and number of adipocytes. These results might indicate that DNER knockdown enhances adipogenesis, presumably, via omission of MCE, and ultimately inhibits population growth of hAMSCs.
Figure 5.
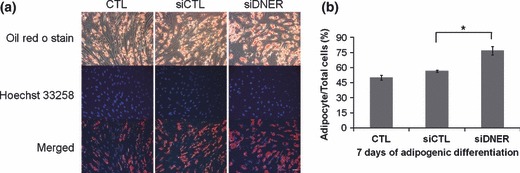
Inhibition of DNER accelerated the onset of adipogenic differentiation. (a) Cells were transfected with siDNER or siControl. After transfection, the cells were induced to undergo adipogenic differentiation for 7 days and were stained with Oil Red O and Hoechst 33258 (nuclei marker, blue). Bar represents 100 μm. (b) Percentage of the number of adipocytes derived from hAMSC to total cells.
Figure 6.
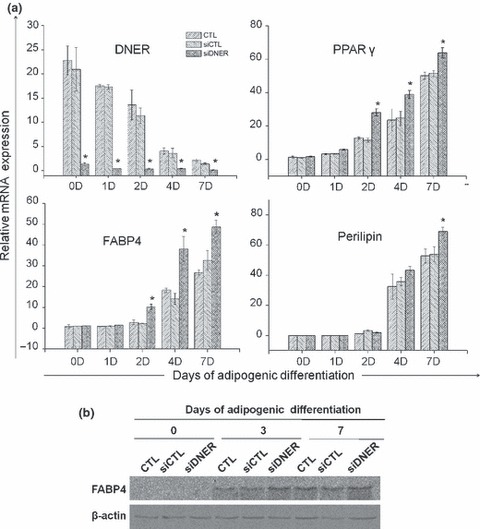
Knockdown of DNER partially increased adipogenesis in hAMSC. (a) Real‐time quantitative PCR results for the DNER, PPARγ, FABP4 and Perilipin gene. The cells were transfected with siDNER or siControl. After transfection, the cells underwent adipogenic differentiation and total RNA was isolated from the cell extracts at the indicated time points. The siDNER‐transfected group evidenced increased numbers of adipocytic genes. β‐actin was used as an internal control for normalization. The values indicated are expressed as the mean ± SD for three independent experiments. Asterisks indicate statistically significant differences (*P < 0.05). (b) Expression of FABP4 protein was detected using Immunoblot analysis. Cells were transfected with siDNER or siControl. After transfection, the cells were differentiated into adipocytes and the whole cell lysates were acquired at the indicated time points. β‐actin was used as loading controls. The results are representative of at least three independent experiments. Twenty micrograms of protein was loaded into each well.
Cell population growth arrest specifically by knockdown of DNER enhances adipogenic differentiation
The aforementioned data suggested that DNER performs a regulatory function in human adipogenesis. Inhibition of DNER retarded cell proliferation and accelerated adipogenesis, as shown by the up‐regulation of adipocytic markers, as well as an increase in size and frequency of adipocytes. On the basis of these observations, we hypothesized that loss of MCE was a cause of this enhancement of adipogenesis in DNER‐knockdown hAMSCs. In effort to determine whether the growth arrest specifically induced by DNER‐knockdown promoted adipogenic differentiation, we administered a non‐cytocidal level of mitomycin C, which induced cell cycle distribution similar to siDNER as a cell population growth inhibitory signal (28) and compared adipogenic efficiency of our hAMSCs with that of siDNER (Fig. S2). The cells were pre‐treated for 2 h with mitomycin C at a concentration of 10 μg/mL in culture medium, prior to adipogenic induction. Interestingly, as is shown in Fig. 7, mitomycin C pre‐treatment reduced efficiency of hAMSC adipogenesis. On the other hand, the siDNER‐transfected group demonstrated greater adipogenic differentiation than was observed in the siControl group. These results indicated that inhibition of cell proliferation, specifically by DNER knockdown, enhanced hAMSC adipogenesis.
Figure 7.
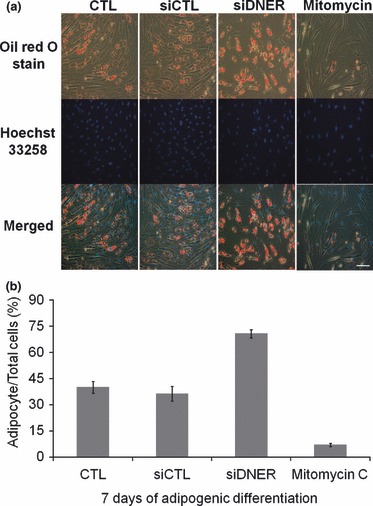
Growth arrest specifically because of DNER knockdown affects hAMSC adipogenesis. (a) Cells were transfected with siDNER or siControl or pre‐treated with Mitomycin C. The cells were then induced to undergo adipogenic differentiation for 7 days and were stained with Oil Red O and Hoechst 33258 (nuclei marker, blue). Bar represents 100 μm. (b) Percentage of numbers of adipocytes derived from hAMSC to total cells. The values are expressed as the mean ± SD for three independent experiments.
Discussion
In this study, the function of DNER during adipogenesis of hAMSC was evaluated. DNER, a neuron‐specific transmembrane protein, has been identified as a ligand of Notch during cell morphogenesis of Bergmann glial cells and is known to mediate functional communication via cell–cell interactions in mouse cerebellum (23, 29); however, the role of DNER in adipogenesis has not been studied previously. This is the first study demonstrates that DNER presence may contribute to adipogenesis in hAMSC.
That DNER was expressed in the cells and its expression decreased during adipogenic differentiation was observed here. Based on previous reports, we examined whether DNER was related to Notch signalling; however, we found that DNER might be regulated independently from Notch signalling in adipogenesis of hAMSC. We determined whether knockdown of DNER could modulate adipogenic differentiation in the cells using specific siRNA. From our results, we observed that cells ceased to proliferate and changed morphology to that of preadipocytes, which eventually led to increase in adipogenic differentiation. We also found that the proportion of cells in S phase decreased concurrently with diminution of CDK2, 4 and CCND1 expression levels.
C/EBPs appear to have diverse roles in regulation of preadipocyte differentiation. C/EBPβ and C/EBPδ are the first transcription factors induced after exposure of preadipocytes to differentiation medium and were thus postulated to be involved in directing the differentiation process. In accordance with this notion, it was found that expression of either C/EBPβ or C/EBPδ in preadipocytes accelerates the rate of C/EBPα induction and enhances adipogenesis (20) Subsequent to immortalization, lack of C/EBPδ promotes cell growth of mouse embryonic fibroblasts (19). When we looked at the expression pattern of C/EBPs in adipogenic differentiation process after DNER inhibition, we found that in early adipogenesis, C/EBPδ was significantly up‐regulated but C/EBPα increased late in adipogenesis.
As we observed cessation of cell population growth by DNER inhibition using siRNA, we confirmed that growth arrest by siDNER played a key role in adipogenesis. When we induced cell quiescence by using mitomycin C, we found no elevation of adipogenic differentiation thus confirming that inhibition of cell proliferation alone did not enhance adipogenesis.
These results may indicate that inhibition of DNER using siRNA during adipogenesis caused up‐regulation of C/EBPδ, which subsequently pushed the hAMSC into a quiescent state to bypass MCE and initiate the adipogenic machinery with more rapid expression of C/EBPα.
Taken together, our results show that a reduction of DNER was necessary but, perhaps, not sufficient for hAMSC adipogenesis. Inhibition of DNER enhanced adipocyte differentiation of the cells resulting in larger lipid droplets and higher numbers of adipocytes, caused by up‐regulation of C/EBPδ mRNA expression. This was a consequence of omission of MCE, which may accelerate lipid accumulation ability of the hAMSC. Down‐stream signalling of DNER in the context of hAMSC adipogenesis is very important and the mechanisms underlying cell population growth arrest and adipogenic enhancement are yet to be clearly elucidated.
In summary, we have assessed the effects of DNER on adipocyte differentiation of hAMSCs. These results revealed, for the first time to the best of our knowledge, that the function of DNER in these human cells was strikingly different from that observed in mouse cells, in which DNER functions as a Notch ligand. The functional differences in DNER between murine and human systems should be further evaluated by comparative research using human and murine models so that the role of DNER in hAMSC adipogenesis may be determined in more detail, which, it is hoped, will provide further insights into the phenomenon of obesity.
Supporting information
Figure S1. Cell proliferation was measured via an MTT assay in hAMSC transfected with siDNER or siControl. The MTT assays were conducted after adipogenic differentiation at the indicated time points. The values indicated are expressed as the means ± SD for three independent experiments. Asterisks indicate statistically significant differences (*P < 0.05).
Figure S2. hAMSCs were transfected with siDNER or siControl or pretreated with Mitomycin C. Cell cycle distribution of hAMSC was analysed by flow cytometry as described in Materials and methods. The percentage of cells in each phase of the cell cycle was indicated as a bar graph.
Table S1. Primers used for PCRs
Table S2. Primers used for real‐time quantitative PCRs
Please note: Wiley‐Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Supporting info item
Supporting info item
Supporting info item
Acknowledgements
This study was supported by a grant from the Korean Science & Engineering Foundation (M10641450002‐06N4145‐00200). This study was also further supported by the BK21Program for Veterinary Science and a Korean Research Foundation Grant (KRF‐2008‐005‐J02903).
References
- 1. Sciaudone M, Gazzerro E, Priest L, Delany AM, Canalis E (2003) Notch 1 impairs osteoblastic cell differentiation. Endocrinology 144, 5631–5639. [DOI] [PubMed] [Google Scholar]
- 2. Garces C, Ruiz‐Hidalgo MJ, Font de Mora J, Park C, Miele L, Goldstein J et al. (1997) Notch‐1 controls the expression of fatty acid‐activated transcription factors and is required for adipogenesis. J. Biol. Chem. 272, 29729–29734. [DOI] [PubMed] [Google Scholar]
- 3. Ross DA, Rao PK, Kadesch T (2004) Dual roles for the Notch target gene Hes‐1 in the differentiation of 3T3‐L1 preadipocytes. Mol. Cell. Biol. 24, 3505–3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hauner H, Entenmann G (1991) Regional variation of adipose differentiation in cultured stromal‐vascular cells from the abdominal and femoral adipose tissue of obese women. Int. J. Obes. 15, 121–126. [PubMed] [Google Scholar]
- 5. De Girolamo L, Sartori MF, Arrigoni E, Rimondini L, Albisetti W, Weinstein RL et al. (2008) Human adipose‐derived stem cells as future tools in tissue regeneration: osteogenic differentiation and cell‐scaffold interaction. Int. J. Artif. Organs 31, 467–479. [DOI] [PubMed] [Google Scholar]
- 6. Yu JM, Jun ES, Bae YC, Jung JS (2008) Mesenchymal stem cells derived from human adipose tissues favor tumor cell growth in vivo. Stem Cells Dev. 17, 463–473. [DOI] [PubMed] [Google Scholar]
- 7. Yeh WC, Bierer BE, McKnight SL (1995a) Rapamycin inhibits clonal expansion and adipogenic differentiation of 3T3‐L1 cells. Proc. Natl. Acad. Sci. USA 92, 11086–11090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tang QQ, Otto TC, Lane MD (2003) Mitotic clonal expansion: a synchronous process required for adipogenesis. Proc. Natl. Acad. Sci. USA 100, 44–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Entenmann G, Hauner H (1996) Relationship between replication and differentiation in cultured human adipocyte precursor cells. Am. J. Physiol. 270, C1011–C1016. [DOI] [PubMed] [Google Scholar]
- 10. Bell A, Grunder L, Sorisky A (2000) Rapamycin inhibits human adipocyte differentiation in primary culture. Obes. Res. 8, 249–254. [DOI] [PubMed] [Google Scholar]
- 11. Farmer SR (2006) Transcriptional control of adipocyte formation. Cell Metab. 4, 263–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Takiguchi M (1998) The C/EBP family of transcription factors in the liver and other organs. Int. J. Exp. Pathol. 79, 369–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ramji DP, Foka P (2002) CCAAT/enhancer‐binding proteins: structure, function and regulation. Biochem. J. 365, 561–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Billiard J, Grewal SS, Lukaesko L, Stork PJ, Rotwein P (2001) Hormonal control of insulin‐like growth factor I gene transcription in human osteoblasts: dual actions of cAMP‐dependent protein kinase on CCAAT/enhancer‐binding protein delta. J. Biol. Chem. 276, 31238–31246. [DOI] [PubMed] [Google Scholar]
- 15. Breed DR, Margraf LR, Alcorn JL, Mendelson CR (1997) Transcription factor C/EBPdelta in fetal lung: developmental regulation and effects of cyclic adenosine 3′,5′‐monophosphate and glucocorticoids. Endocrinology 138, 5527–5534. [DOI] [PubMed] [Google Scholar]
- 16. Cassel TN, Nordlund‐Moller L, Andersson O, Gustafsson JA, Nord M (2000) C/EBPalpha and C/EBPdelta activate the clara cell secretory protein gene through interaction with two adjacent C/EBP‐binding sites. Am. J. Respir. Cell Mol. Biol. 22, 469–480. [DOI] [PubMed] [Google Scholar]
- 17. Gigliotti AP, DeWille JW (1998) Lactation status influences expression of CCAAT/enhancer binding protein isoform mRNA in the mouse mammary gland. J. Cell. Physiol. 174, 232–239. [DOI] [PubMed] [Google Scholar]
- 18. Gigliotti AP, DeWille JW (1999) Local signals induce CCAAT/enhancer binding protein‐delta (C/EBP‐delta) and C/EBP‐beta mRNA expression in the involuting mouse mammary gland. Breast Cancer Res. Treat. 58, 57–63. [DOI] [PubMed] [Google Scholar]
- 19. Huang AM, Montagna C, Sharan S, Ni Y, Ried T, Sterneck E (2004) Loss of CCAAT/enhancer binding protein delta promotes chromosomal instability. Oncogene 23, 1549–1557. [DOI] [PubMed] [Google Scholar]
- 20. Yeh WC, Cao Z, Classon M, McKnight SL (1995b) Cascade regulation of terminal adipocyte differentiation by three members of the C/EBP family of leucine zipper proteins. Genes Dev. 9, 168–181. [DOI] [PubMed] [Google Scholar]
- 21. Eiraku M, Hirata Y, Takeshima H, Hirano T, Kengaku M (2002) Delta/notch‐like epidermal growth factor (EGF)‐related receptor, a novel EGF‐like repeat‐containing protein targeted to dendrites of developing and adult central nervous system neurons. J. Biol. Chem. 277, 25400–25407. [DOI] [PubMed] [Google Scholar]
- 22. Nishizumi H, Komiyama T, Miyabayashi T, Sakano S, Sakano H (2002) BET, a novel neuronal transmembrane protein with multiple EGF‐like motifs. Neuroreport 13, 909–915. [DOI] [PubMed] [Google Scholar]
- 23. Eiraku M, Tohgo A, Ono K, Kaneko M, Fujishima K, Hirano T et al. (2005) DNER acts as a neuron‐specific Notch ligand during Bergmann glial development. Nat. Neurosci. 8, 873–880. [DOI] [PubMed] [Google Scholar]
- 24. Park JR, Jung JW, Lee YS, Kang KS (2008) The roles of Wnt antagonists Dkk1 and sFRP4 during adipogenesis of human adipose tissue‐derived mesenchymal stem cells. Cell Prolif. 41, 859–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Park JR, Park JS, Jo EH, Hwang JW, Kim SJ, Ra JC et al. (2006) Reversal of the TPA‐induced inhibition of gap junctional intercellular communication by Chaga mushroom (Inonotus obliquus) extracts: effects on MAP kinases. Biofactors 27, 147–155. [DOI] [PubMed] [Google Scholar]
- 26. Jo EH, Kim SH, Ra JC, Kim SR, Cho SD, Jung JW et al. (2005) Chemopreventive properties of the ethanol extract of chinese licorice (Glycyrrhiza uralensis) root: induction of apoptosis and G1 cell cycle arrest in MCF‐7 human breast cancer cells. Cancer Lett. 230, 239–247. [DOI] [PubMed] [Google Scholar]
- 27. Otto TC, Lane MD (2005) Adipose development: from stem cell to adipocyte. Crit. Rev. Biochem. Mol. Biol. 40, 229–242. [DOI] [PubMed] [Google Scholar]
- 28. Fang L, Igarashi M, Leung J, Sugrue MM, Lee SW, Aaronson SA (1999) p21Waf1/Cip1/Sdi1 induces permanent growth arrest with markers of replicative senescence in human tumor cells lacking functional p53. Oncogene 18, 2789–2797. [DOI] [PubMed] [Google Scholar]
- 29. Tohgo A, Eiraku M, Miyazaki T, Miura E, Kawaguchi SY, Nishi M et al. (2006) Impaired cerebellar functions in mutant mice lacking DNER. Mol. Cell. Neurosci. 31, 326–333. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Cell proliferation was measured via an MTT assay in hAMSC transfected with siDNER or siControl. The MTT assays were conducted after adipogenic differentiation at the indicated time points. The values indicated are expressed as the means ± SD for three independent experiments. Asterisks indicate statistically significant differences (*P < 0.05).
Figure S2. hAMSCs were transfected with siDNER or siControl or pretreated with Mitomycin C. Cell cycle distribution of hAMSC was analysed by flow cytometry as described in Materials and methods. The percentage of cells in each phase of the cell cycle was indicated as a bar graph.
Table S1. Primers used for PCRs
Table S2. Primers used for real‐time quantitative PCRs
Please note: Wiley‐Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Supporting info item
Supporting info item
Supporting info item


