Abstract
The cytoplasm seems to provide an environment that favors conversion of the prion protein (PrP) to a form with the physical characteristics of the PrPSc conformation, which is associated with transmissible spongiform encephalopathies. However, it is not clear whether PrP would ever exist in the cytoplasm under normal circumstances. We report that PrP accumulates in the cytoplasm when proteasome activity is compromised. The accumulated PrP seems to have been subjected to the normal proteolytic cleavage events associated with N- and C-terminal processing in the endoplasmic reticulum, suggesting that it arrives in the cytoplasm through retrograde transport. In the cytoplasm, PrP forms aggregates, often in association with Hsc70. With prolonged incubation, these aggregates accumulate in an “aggresome”-like state, surrounding the centrosome. A mutant (D177N), which is associated with a heritable and transmissible form of the spongiform encephalopathies, is less efficiently trafficked to the surface than wild-type PrP and accumulates in the cytoplasm even without proteasome inhibition. These results demonstrate that PrP can accumulate in the cytoplasm and is likely to enter this compartment through normal protein quality-control pathways. Its potential to accumulate in the cytoplasm has implications for pathogenesis.
The prion protein (PrP) is closely associated with a group of fatal neurodegenerative diseases. These take various forms that are manifested as sporadic, dominantly heritable, and transmissible disorders (1–3). The normal form of PrP, PrPC, is rich in α-helices, soluble in mild detergents, and highly sensitive to protease digestion (3, 4). During disease, it accumulates in different physical states (1, 5), the most well known of which is PrPSc, which is associated with infectious forms of prion diseases. PrPSc is rich in β-sheet and insoluble in mild detergents, and has a particular pattern of protease sensitivity and resistance (1, 3, 6).
Normally, PrP is a cell-surface N-linked glycoprotein that is widely expressed and particularly abundant in the brain, although its function is unknown (7). A 22-aa N-terminal signal peptide directs PrP into the endoplasmic reticulum (ER) cotranslationally. There, the signal peptide is removed, core N-linked oligosaccharides are added, and the C-terminal 23 aa are removed with the addition of a glycosyl-phosphatidylinositol (GPI) anchor. Properly folded PrPC then moves out of the ER, matures through the Golgi complex with additional carbohydrate modifications, and reaches the cell surface.
Despite decades of effort, how PrP causes fatal neurodegenerative disease remains unclear. However, misfolding and mistrafficking of PrP are hallmarks of various forms of diseases involving PrP, and they almost certainly play a role in pathogenesis. Mutations in the PrP hydrophobic region that enhance the formation of transmembrane forms of PrP cause neurodegeneration in transgenic mice (8). But studies with a series of mutants associated with transmissible spongiform encephalopathies indicated that most mutants that reside outside the hydrophobic region do not form detectable transmembrane forms (9). Instead, they are partially retained in the ER, which suggests misfolding and retention by the ER quality-control system (10). Recent understanding of protein folding and quality-control mechanisms has revealed that many misfolded secretory proteins are retained in the ER and subject to retrograde transport to the cytoplasm and degradation by the proteasome (11–13). Indeed, accumulation of PrP has been reported upon proteasome inhibition (14–16). However, detailed studies of the fate of two PrP mutants, Q217R and Y145stop, indicate that they accumulate in the ER and other membrane-bound compartments when proteasome activity is blocked (14, 15). Thus, it might be that, unlike other proteins, PrP is not subject to retrograde transport when triaged by the quality-control system and accumulates after proteasome inhibition as a secondary consequence of quality-control perturbations. These particular mutants, however, are associated with slowly progressing forms of disease that are not transmissible. They do not accumulate in the PrPSc conformation, even in diseased brains (5, 17). Thus, their behavior might not reflect that of wild-type PrP or that of mutants associated with transmissible forms of the disease.
Here, we show that wild-type PrP rapidly accumulates in the cytoplasm when the proteasome is inhibited. Moreover, one of the best-characterized mutants associated with transmissible forms of the disease accumulates in the cytoplasm even in the absence of proteasome inhibitors. These results indicate that PrP, a GPI-anchored plasma membrane protein, is subject to the same quality-control mechanism that many other proteins residing in the ER, or passing through it, are subjected to. Given the fact that reducing conditions in vitro (18) or exposure to the cytoplasmic environment in vivo favors the formation of a PrPSc-like conformation (19), these results have significant implications.
Materials and Methods
DNA Constructs.
The expression plasmid for mouse PrP carrying the 3F4 epitope was generated by cloning a BamHI and HindIII fragment of plasmid PrP(3F4)-pBC12/CMV into vector pCB6+ under a cytomegalovirus (CMV) promoter. The D177N mutant was generated with the QuickChange site-directed mutagenesis kit (Stratagene), using primers 5′-CAACTTCGTGCACAACTGCGTCAATATCACC-3′ and 5′-GGTGATATTGACGCAGTTGTGCACGAAGTTG-3′.
Cell Culture and Transfection.
COS-1 cells and NT-2 human neuroblastoma cells were cultured in DMEM or OptiMEM (GIBCO/BRL) with 10% FBS. COS cells were transfected with Fugene 6 reagent (Roche Biochemicals) according to the manufacturer's directions. Amounts of plasmid DNA and Fugene 6 transfection reagents were chosen by preliminary experiments to ensure modest level of transfection in most experiments. High-level transfections were achieved by using high amounts of plasmid DNA and Fugene 6 reagents (see Fig. 3E), by using Lipofectamine transfection reagents (GIBCO/BRL), or by using GenePorter transfection reagents (Gene Therapy Systems, San Diego) (data not shown).
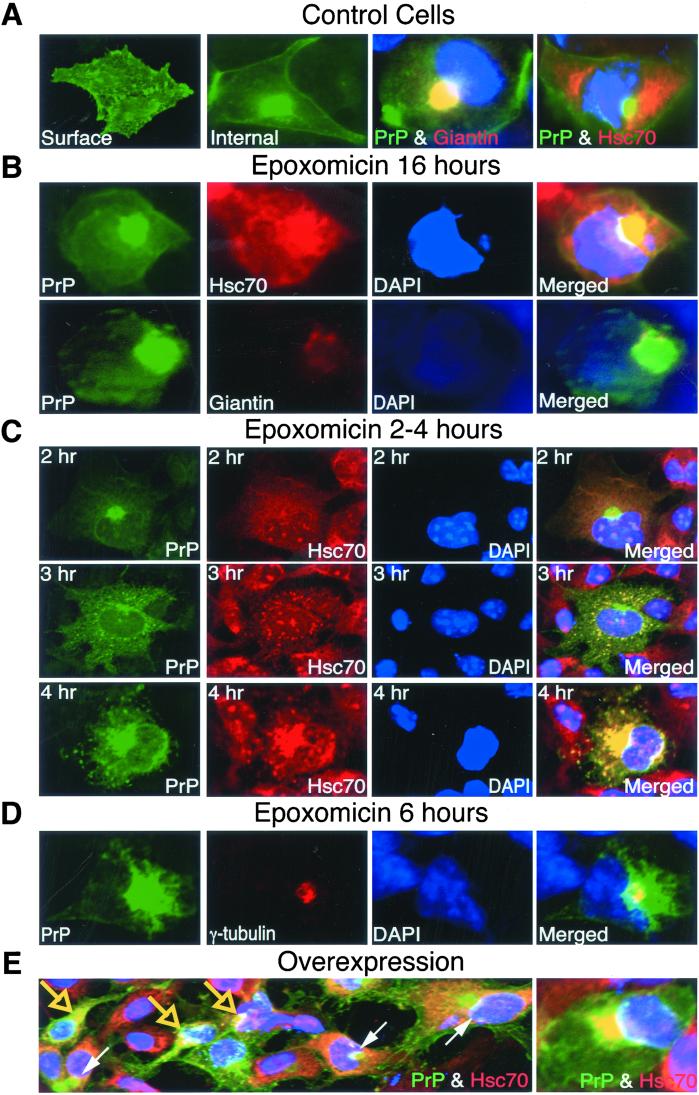
Figure 3.
Effect of proteasome inhibitor on PrP localization in COS cells. Cells expressing wild-type PrP were treated without (A) or with 5 μM epoxomicin for various lengths of time as indicated in B–D. (E) Cells without proteasome inhibition but overexpressing PrP because of high transfection levels. In all panels, PrP was stained with anti-PrP antibody (green). Giantin, hsc70, or γ-tubulin (red) staining with specific antibodies was indicated; nuclei were stained with DAPI (blue). All of the cells were fixed by methanol except for visualization of surface staining in cells fixed by paraformaldehyde. Large arrows in E, cells with cytoplasmic PrP colocalizing with Hsc70; small arrows in E, cells with PrP in the Golgi complex. (E, Right) Higher magnification of colocalization of PrP aggregates and hsc70.
Proteasome Inhibitor Treatment.
For NT-2 cells, MG132 (Calbiochem) was added when cells reached 80% confluence, and culture was continued at 37°C in a CO2 incubator for 12 hr. For transiently transfected cells, epoxomicin (Affinity, Nottingham, U.K.) at indicated concentrations was added to the culture media. Cells were cultured at 37°C with 5% CO2 for 16 hr unless indicated.
Analysis of PrP Aggregation and Immunoblot Analysis.
After transfection and treatment, cells growing on 6-well cell-culture plates were washed once with ice-cold PBS and lysed with 300 μl (per well) of lysis buffer [50 mM Tris⋅HCl, pH 7.5/150 mM NaCl/2 mM EDTA/0.5% (vol/vol) Triton X-100/0.5% (vol/vol) sodium deoxycholate] on ice. Cells were disrupted by sequential passages through 21- and 25-gauge needles 10 times on ice. Fifty microliters of lysates was sedimented at 16,000 × g for 30 min at 4°C. Proteins from supernatants were precipitated with 4 vol of 100% methanol (−20°C) and incubated at −20°C for at least 30 min. Pellet fraction and precipitated supernatant were sonicated in SDS/PAGE sample buffer containing 5% (wt/vol) SDS. For endoglycosidase H (Endo H) digestion, the pellet fraction was sonicated in the presence of Endo H buffer and incubated with 100 milliunits of Endo H at 37°C for 16 hr. Proteins were resolved on 14% polyacrylamide gels, transferred to poly(vinylidene difluoride) membrane, and reacted with 3F4 antibody at 1:5,000, R20 antibody at 1:1,000, R24 antibody at 1:500, or calnexin antibody (StressGen Biotechnologies, Victoria, BC, Canada) at 1:500.
Immunofluorescence.
Cells were cultured on glass coverslips in 24-well culture plates. For staining lysosomes, cells were incubated with 0.5 mg/ml Lucifer yellow CH in medium for 10 hr, followed by washing and continued culture in medium for 2 hr. To visualize surface staining, cells were fixed with 4% (vol/vol) paraformaldehyde (EM Laboratories, Elmsford, NY). For intracellular staining, cells were fixed with 100% cold methanol at −20°C for 20 min. After blocking with 5% normal goat serum in PBS for 45 min, cells were reacted with primary antibodies diluted in blocking buffer for 1 hr, washed four times with PBS, and incubated with secondary antibody (Alexa-labeled goat anti-mouse, rabbit, or rat IgG antibodies; Molecular Probes) diluted in blocking buffer for 1 hr. After washing twice with PBS, cells were incubated with 100 ng/ml 4′,6-diamidino-2-phenylindole (DAPI) (Sigma) in PBS for 10 min and washed three times with PBS. Coverslips were mounted on glass slides, and staining was visualized with a Zeiss Axioplan 2 fluorescence microscope (Zeiss). PrP was detected with 3F4 antibody at 1:500 dilution. Other antibodies were diluted as follows: rabbit anti-γ-tubulin antibody (Sigma), 1:2,500; rat anti-hsc70 antibody (StressGen Biotechnologies), 1:200; rabbit anti-BiP antibody (StressGen Biotechnologies), 1:200.
Results
Endogenous PrP from Human Neuroblastoma Cells Accumulates in the Cytoplasm When Proteasome Activity Is Blocked.
Misfolded or mistargeted proteins that appear in the cytoplasm are difficult to detect because the highly efficient proteasome prevents their accumulation. To determine whether endogenous wild-type PrP might be subject to proteasome degradation, we asked whether various proteasome inhibitors cause the protein to accumulate in the cytoplasm. In untreated neuroblastoma (NT-2) cells, endogenous PrP was primarily surface-localized, detected as bright staining over the whole surface when cells were fixed with paraformaldehyde (data not shown). When cells were fixed with methanol, surface staining was most readily detected by fluorescence at cell borders (Fig. 1A). After 12 hr of treatment with the proteasome inhibitor MG132, surface staining of PrP remained strong when cells were fixed with paraformaldehyde (data not shown), but was less apparent with methanol fixation because cell–cell contacts were reduced. Notably, methanol fixation revealed that substantial quantities of PrP had now accumulated internally in fine foci throughout the cells, partly concentrated around nuclei (Fig. 1B).
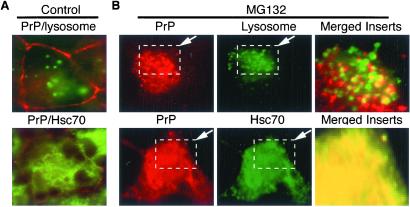
Figure 1.
Effect of proteasome inhibition on endogenous PrP in NT-2 cells. Cells were treated without (control) or with MG132 at indicated concentrations for 12 hr. (A) Merged images of cells costained with anti-PrP antibody (red) and Lucifer yellow CH (green, lysosome staining) (Upper) or with anti-PrP (red) and anti-Hsc70 antibodies (green) (Lower). (B, Upper) MG132-treated cells were stained with anti-PrP antibody (red) and Lucifer yellow CH (green). The regions outlined by dashed boxes were merged and magnified (Right). (Lower) MG132-treated cells were stained with anti-PrP antibody (red) and anti-Hsc70 antibody (green).
Costaining with PrP antibodies and the DNA-binding dye DAPI (data not shown) demonstrated that accumulating PrP foci were not inside nuclei but clustered outside. Staining with Lucifer yellow CH, which is taken up by endosomes and lysosomes (20, 21), indicated that very few PrP foci were coincident with endocytic compartments (Fig. 1B, Left and Center), even in regions where they were most concentrated and appeared to overlap at low magnification (Fig. 1B, merged and magnified Inset). Antibodies against BiP, a member of the Hsp70 chaperone family that is localized to the ER, revealed no colocalization with PrP foci (data not shown, but see Fig. 5B for similar analysis of a PrP mutant). In contrast, antibodies against a cytoplasmic form of Hsp70, Hsc70, showed strong colocalization with intracellular PrP aggregates in many cells treated with MG132, but never in untreated cells (compare lower panels in Fig. 1 A and B; yellow fluorescence in merged image indicates colocalization). These results indicate that when proteasome activity is compromised, a significant portion of endogenous wild-type PrP accumulates in the cytoplasm.
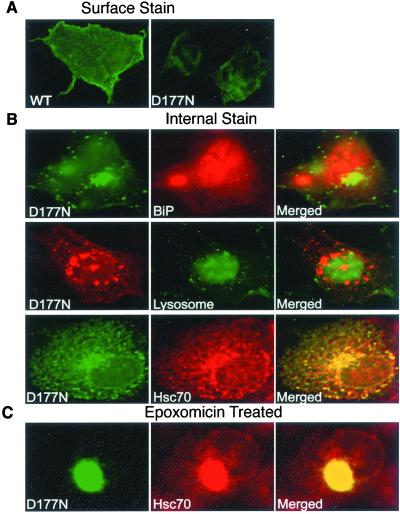
Figure 5.
Effect of the D177N mutation on PrP localization. COS cells were transfected with plasmids encoding either wild-type PrP or the D177N mutant. (A) Surface staining of paraformaldehyde-fixed cells expressing wild-type PrP (WT) or PrPD177N (D177N) by using 3F4 antibody. (B) Intracellular staining of PrPD177N. Cells were costained with 3F4 antibody and anti-BiP antibody (Top), Lucifer yellow CH (endocytic compartments) (Middle), or anti-Hsc70 antibody (Bottom). (C) Cells expressing PrPD177N were treated with 5 μM epoxomicin for 16 hr and costained with 3F4 and anti-Hsc70 antibodies.
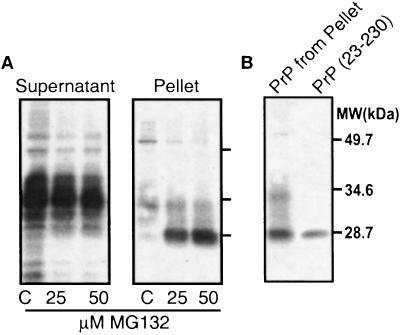
To further investigate the protein's physical state, cells were lysed with mild detergents and subjected to centrifugation. PrP from control cells was soluble and migrated heterogeneously during electrophoresis (Fig. 2A, lanes C), as expected from its normal, diverse glycosylation state (22, 23). Some PrP from proteasome-treated cells remained soluble, but a substantial fraction became insoluble. Similar results were obtained with another proteasome inhibitor, epoxomicin (data not shown).
Figure 2.
Physical state of PrP in NT-2 cells with or without proteasome inhibition. (A) Detergent cell lysates were sedimented, and PrP in the supernatant and pellet fractions was detected by immunoblot analysis with anti-PrP 3F4 monoclonal antibody. (B) Comigration of PrP from the pellet fraction with the recombinant mature fragment of PrP-(23–230), which lacks both N- and C-terminal signal sequences.
This aggregated, detergent-insoluble form of PrP (Fig. 2A, pellet) seemed to be derived from protein that had fully entered the ER for processing and had then undergone retrograde transport. That is, it did not migrate at the position of full-length PrP but rather at the position of recombinant, unglycosylated PrP lacking both the N-terminal ER translocation signal peptide and the 23-aa C-terminal GPI signal sequence (Fig. 2B). N- and C-terminal cleavage of protein accumulating after proteasome inhibition was confirmed in other cell types, in which PrP accumulated at sufficient levels for additional analysis (see below).
PrP Also Accumulates in the Cytoplasm of Other Cell Types When Proteasome Activity Is Blocked.
Within 12 hr of proteasome inhibition, many NT-2 cells detached from the culture dish and began to die. (Floating cells were discarded before harvesting samples for the above analyses.) Longer treatments produced greater cell loss. The selective toxicity of cytoplasmic forms of PrP in neuronal cells will be the subject of another article. Here, we examine other cell types that retained much higher levels of viability with proteasome inhibition. This allowed us to follow the fate of cytoplasmic PrP for longer periods, with interpretations less complicated by toxicity, and to take advantage of transient transfections to examine the trafficking of a PrP mutant.
As expected, in transfected COS (monkey kidney) cells expressing wild-type PrP, most of the protein localized to the cell surface. Paraformaldehyde fixation produced bright staining over the entire surface (Fig. 3A). This was lost when cells were treated with mild detergents (data not shown) or were fixed with methanol (Fig. 3A, Internal). Some PrP could then be detected internally, concentrated in perinuclear regions. Note that in this case, internal PrP did not colocalize with cytoplasmic Hsc70. Rather, it colocalized with the Golgi marker giantin (Fig. 3A). Thus, even internally localized protein was not cytoplasmic but was in the process of being transported through the secretory pathway.
When COS cells were treated overnight with epoxomicin (24), they retained high levels of viability. Surface staining remained strong (paraformaldehyde fixation; data not shown). In these cells, PrP also accumulated in large perinuclear aggregates. However, in striking contrast with untreated cells, the perinuclear PrP aggregates of cells treated with the proteasome inhibitor colocalized with cytoplasmic Hsc70, not with giantin (Fig. 3B). Note that the pattern of giantin staining itself changed, in a manner typical of cells that contain cytoplasmic aggresomes or inclusion bodies (25–27).
A time course of PrP localization after proteasome inhibition revealed that cytoplasmic accumulation of PrP occurred very rapidly. Initially, the protein was present in small foci (Fig. 3C) similar to those observed in neuroblastoma cells (Fig. 1B). As incubation continued, foci in a greater number of cells gradually became intensely positive for Hsc70 staining. With longer treatments, PrP began to coalesce into a single large aggregate (after about 6 hr). This coalescence localized around the centrosome and costained with γ-tubulin antibody (Fig. 3D), as do the aggresomes or inclusion bodies formed by certain other misfolded proteins that accumulate cytoplasmically (25, 27).
Similar results were obtained with mouse fibroblast NIH 3T3 cells and with two chemically distinct proteasome inhibitors, MG132 and lactacystin (data not shown). Thus, accumulation of PrP in the cytoplasm was a general consequence of proteasome inhibition rather than an artifact of a particular cell line or an aberrant effect of a particular drug. Finally, cytoplasmic accumulation was not an artifact of proteasome inhibition per se. We used very modest levels of transfection in these experiments. However, in separate experiments employing higher levels of transfection with the same PrP expression plasmid (see Materials and Methods), PrP accumulated in the cytoplasm even when cells were cultured in the absence of proteasome inhibitors (Fig. 3E). Presumably, in this case, the ability of the normal cytoplasmic degradation machinery to clear misfolded PrP was compromised by excess substrate.
PrP Accumulates in the Cytoplasm by Retrograde Transport.
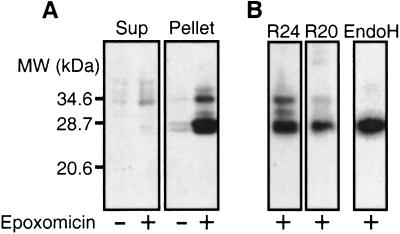
Western blotting demonstrated that after epoxomicin treatment, PrP accumulated to much higher levels. Most of this protein was insoluble in mild detergents (compare Fig. 4A supernatant and pellet), and it was enriched in a lower molecular weight species. This band comigrated with both a recombinant unglycosylated PrP lacking the N- and C-terminal signal sequences (as did the accumulated PrP of neuroblastoma cells; Fig. 2B) and with the unglycosylated isoform of mature, ER-processed PrP from untreated neuroblastoma cells (data not shown). The size of this species, together with its reaction (Fig. 4B) with antibodies specific for residues 23–37 (R24) and residues 218–232 (R20), located at either end of mature PrP (28), indicated that it must be derived from the central portion of PrP. That is, the protein accumulated in the cytoplasm after it had fully entered the ER and been subjected to N- and C-terminal proteolytic processing.
Figure 4.
The physical state of PrP in transfected cells. (A) COS cells transfected with wild-type PrP were incubated with or without 1 μM epoxomicin for 16 hr. Detergent lysates were fractionated by centrifugation, and PrP in the supernatant (Sup) or pellet fractions was detected by immunoblot analysis using 3F4 antibody. (B) PrP in the pellet fraction was detected by immunoblot analysis with antibodies specific for the N-terminal (R24) or C-terminal (R20) regions of mature PrP as indicated. EndoH, PrP in the Endo H-digested pellet fraction was detected by 3F4 antibody.
The higher molecular weight protein that accumulated after proteasome inhibition and fractionated in the pellet was sensitive to Endo H digestion (Fig. 4B). Thus, it also derived from protein that had entered the ER for processing. Although glycosylated, this species had not passed the quality-control system for passage to the Golgi complex, where further carbohydrate modifications would have rendered it Endo H resistant.
Mutant PrP Accumulates in the Cytoplasm Even Without Proteasome Inhibition.
If the appearance of PrP in the cytoplasm is relevant to disease, it should more readily occur in cells expressing mutant PrPs associated with transmissible, familial forms of transmissible spongiform encephalopathies. To test the above prediction, we created a point mutation in mouse PrP that changes Asp to Asn at residue 177 (D177N). This mutation (D178N in human PrP) is one of the more subtle genetic changes associated with transmissible spongiform encephalopathies and one of the most common.
Wild-type PrP and PrPD177N were examined at equivalent transfection efficiencies as determined by analysis of β-galactosidase expression from a cotransfected plasmid (data not shown). However, cells producing PrPD177N exhibited much less cell-surface staining than cells producing wild-type PrP (paraformaldehyde fixation, Fig. 5A). They also exhibited a different pattern of internal accumulation (compare methanol fixation, Fig. 5B with Fig. 3A). Specifically, many cytoplasmic aggregates of PrPD177N were visible even in the absence of the proteasome inhibitors. These internal foci did not colocalize with BiP or with Lucifer yellow CH, indicating they were not in the ER or in the endocytic compartment (Fig. 5B, Top and Middle). However, they strongly colocalized with the cytoplasmic chaperone Hsc70, especially in cells with higher levels of PrP accumulation (Fig. 5B, Bottom). Thus, even in the absence of the proteasome inhibitor, a substantial fraction of the mutant protein accumulated in the cytoplasmic compartment.
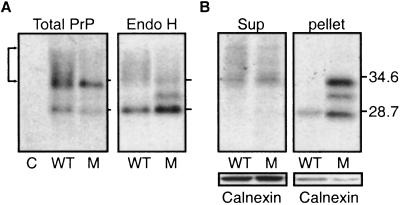
When the proteins of cells cultured in the absence of proteasome inhibitors were analyzed on SDS gels, a substantial fraction of wild-type PrP exhibited the diffuse, slow migration (Fig. 6A, total PrP, bracket) that characterizes proteins that have passed from the ER to the Golgi complex for oligosaccharide modification (29). A smaller fraction of PrPD177N exhibited this migration pattern. Furthermore, Endo H had a greater effect on the migration of PrPD177N than on the migration of wild-type PrP (Fig. 6A, Endo H). Taken together with the differences in intracellular localization, the differences in electrophoretic migration and Endo H sensitivity indicate that, in cells that had been carefully matched for similar transfection levels, a smaller fraction of PrPD177N than of wild-type PrP passed through the ER quality-control system.
Figure 6.
Physical state of PrPD177. COS cells were transfected as in Fig. 4. The activity of a cotransfected β-galactosidase construct (data not shown) demonstrated equivalent transfection efficiency. PrP was detected by 3F4 antibody. C, vector control; WT, PrP; M, PrPD177. (A, Left) Total PrP in lysates. (Right) PrP in Endo H-digested lysates. (B) PrP in supernatant and pellet fractions of cell lysates. The same blots were also probed with calnexin antibody.
The two proteins also showed different propensities to accumulate in detergent-insoluble aggregates in the absence of proteasome inhibitors. Wild-type PrP was mostly soluble, whereas the majority of PrPD177N was insoluble (Fig. 6B). Staining the same blots with calnexin (Fig. 6B) and several other proteins (data not shown) demonstrated that accumulation was highly selective.
After proteasome inhibition, both wild-type PrP and PrPD177N accumulated in an insoluble form, but PrPD177N accumulated to a higher level (data not shown). In this case, colocalization with Hsc70 was observed in every transfected cell, and aggregates coalesced into large perinuclear inclusion bodies (Fig. 5C), as described earlier for wild-type PrP (Fig. 3).
Discussion
Our work demonstrates that a significant fraction of PrP, a cell-surface GPI-anchored protein, accumulates in the cytoplasm when the activity of the primary quality-control system for cytoplasmic protein degradation, the proteasome, is compromised. In the cytoplasmic compartment PrP aggregates, often, but not always, associating with Hsc70. With prolonged accumulation, the aggregates coalesce and associate with the centrosome, as has been reported for several other proteins that accumulate in the cytoplasm when their degradation is blocked.
Cytoplasmic accumulation of PrP is not specific to a particular cell line, transfection procedure, or proteasome inhibitor. It is observed with the endogenous PrP protein of neuroblastoma cells as well as with PrP protein in transfected COS cells (monkey kidney cells) and NIH 3T3 cells (mouse fibroblasts). It occurs also with diverse proteasome inhibitors (MG132, lactacystin, epoxomicin), which have different chemical structures and mechanisms of action. Furthermore, cytoplasmic accumulation of PrP is not an artifact of proteasome inhibition per se. A substantial fraction of a PrP mutant that is associated with transmissible spongiform encephalopathy diseases, D177N, accumulates in the cytoplasm in the absence of proteasome inhibition. Moreover, when high levels of DNA are used for transfection, even wild-type PrP accumulates in the cytoplasm. Presumably, in this case the quality-control machinery is simply overwhelmed by excess substrate.
The PrP protein that accumulates in the cytoplasm seems to have completed entrance into the ER and to have been delivered to the cytoplasmic compartment by retrograde transport. First, the size of the most prominent species is that expected for mature PrP, and it comigrates on SDS gels with a recombinant protein [PrP-(23–230)] equivalent to the mature fragment. Proteins that retained either the N or C termini would have readily been distinguished from the mature species on such gels (14, 15). Second, it reacts with two antibodies that recognize sequences near either end of mature PrP. Given the protein's size, it could react with both antibodies only if it represented the middle section of PrP, with sequence removed from either end. The enzymes for removing the N-terminal signal sequence that directs transport into the secretory compartment and the C-terminal signal for proteolytic cleavage and attachment of GPI are located in the ER. It is conceivable that PrP accumulates in the cytoplasm through indirect effects of proteasome inhibition that cause mistargeting, rather than through normal delivery to the ER, recognition by the quality-control system, and retrograde transport. However, this would require that both N-terminal and C-terminal sequences are removed in the cytoplasm by some unknown proteolytic activity and that both cleavages fortuitously occur close enough to the bona fide cleavage sites to generate the right-sized fragment, a scenario that seems extremely unlikely.
Taken together, our results strongly suggest that PrP is triaged by the ER quality-control system that shunts unfolded or misfolded proteins to the cytoplasm for degradation by the proteasome. If so, it is not surprising that PrP is not normally detectable in the cytoplasm because this system is usually so efficient. The rapidity with which PrP accumulates in the cytoplasm after proteasome inhibition suggests there is a constant flux of unfolded or misfolded PrP to this compartment, which is detected only when the activity of the quality-control system is compromised. The observation that the N-terminal region of PrP is not well folded (4, 30) would further suggest that a fraction of the protein is constantly subject to this control pathway. It is also possible that PrP appears in the cytoplasm only when the folding capacity of the system is compromised. Even if this is the case, PrP would still be likely to appear in the cytoplasm under normal circumstances when aging, natural biological traumas, or environmental stresses compromise the system.
The PrP that accumulates cytoplasmically is enriched in unglycosylated species. There are two, not mutually exclusive, explanations. First, cytoplasmic deglycosidases can remove oligosaccharides from ER proteins after they are transported to the cytoplasm (31). Second, unglycosylated proteins have a greater tendency to misfold and are therefore more likely to be subject to retrograde transport (32). The accumulation of unglycosylated species is of interest because previous studies from several laboratories suggest they are more likely to convert to a PrPSc-like form (18, 19, 33).
Indeed, in separate experiments, we have found that cytoplasmic PrP is selectively toxic to neuronal cells and that a fraction of the protein converts to a PrPSc-like conformation in a manner that critically depends on the rate at which the protein appears in the cytoplasm (unpublished results). Together, these data support our previous suggestion that PrPSc might arise de novo in sporadic and familial forms of the prion diseases on the rare occasions when unfolded or misfolded PrP is retrograde transported to the cytoplasm but degradation fails. This would also be consistent with previous observation of cytoplasmic PrP during disease progression (34). In any case, our data strongly suggest that PrP does appear in the cytoplasmic compartment under some normal biological circumstances, which may contribute to the pathogenesis of prion disease.
Acknowledgments
We thank Byron Caughey, Sue Priola, Jim Mastrianni, David Harris, Adam Linstedt, and Elaine Fuchs for reagents. This work was supported by the National Institutes of Health and by the Howard Hughes Medical Institute.
Abbreviations
- PrP
prion protein
- PrPSc
PrP in the conformation associated with transmissible spongiform encephalopathies
- ER
endoplasmic reticulum
- Endo H
endoglycosidase H
- GPI
glycosyl-phosphatidylinositol
- DAPI
4′,6-diamidino-2-phenylindole
Note Added in Proof.
After our studies were completed, Yedida et al. (35) reported evidence that wild-type PrP is subject to ubiquitination and degradation by the proteasome. This work had a somewhat different focus, but partially complements the work in this study.
References
- 1.Prusiner S B. Proc Natl Acad Sci USA. 1998;95:13363–13383. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weissmann C. Trends Cell Biol. 1994;4:10. doi: 10.1016/0962-8924(94)90032-9. [DOI] [PubMed] [Google Scholar]
- 3.Caughey B, Chesebro B. Trends Cell Biol. 1997;7:56–62. doi: 10.1016/S0962-8924(96)10054-4. [DOI] [PubMed] [Google Scholar]
- 4.Riek R, Hornemann S, Wider G, Glockshuber R, Wüthrich K. FEBS Lett. 1997;413:282–288. doi: 10.1016/s0014-5793(97)00920-4. [DOI] [PubMed] [Google Scholar]
- 5.Tagliavini F, Prelli F, Porro M, Rossi G, Giaccone G, Farlow M R, Dlouhy S R, Ghetti B, Bugiani O, Frangione B. Cell. 1994;79:695–703. doi: 10.1016/0092-8674(94)90554-1. [DOI] [PubMed] [Google Scholar]
- 6.Parchi P, Zou W, Wang W, Brown P, Capellari S, Ghetti B, Kopp N, Schulz-Schaeffer W J, Kretzschmar H A, Head M W, et al. Proc Natl Acad Sci USA. 2000;97:10168–10172. doi: 10.1073/pnas.97.18.10168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harris D A, Gorodinsky A, Lehmann S, Moulder K, Shyng S L. Curr Top Microbiol Immunol. 1996;207:77–93. doi: 10.1007/978-3-642-60983-1_7. [DOI] [PubMed] [Google Scholar]
- 8.Hegde R S, Mastrianni J A, Scott M R, DeFea K A, Tremblay P, Torchia M, DeArmond S J, Prusiner S B, Lingappa V R. Science. 1998;279:827–834. doi: 10.1126/science.279.5352.827. [DOI] [PubMed] [Google Scholar]
- 9.Stewart R S, Harris D A. J Biol Chem. 2001;276:2212–2220. doi: 10.1074/jbc.M006763200. [DOI] [PubMed] [Google Scholar]
- 10.Ivanova L, Barmada S, Kummer T, Harris D A. J Biol Chem. 2001;276:42409–42421. doi: 10.1074/jbc.M106928200. [DOI] [PubMed] [Google Scholar]
- 11.Kopito R R. Cell. 1997;88:427–430. doi: 10.1016/s0092-8674(00)81881-4. [DOI] [PubMed] [Google Scholar]
- 12.Bonifacino J S, Weissman A M. Annu Rev Cell Dev Biol. 1998;14:19–57. doi: 10.1146/annurev.cellbio.14.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Plemper R K, Wolf D H. Trends Biochem Sci. 1999;24:266–270. doi: 10.1016/s0968-0004(99)01420-6. [DOI] [PubMed] [Google Scholar]
- 14.Zanusso G, Petersen R B, Jin T, Jing Y, Kanoush R, Ferrari S, Gambetti P, Singh N. J Biol Chem. 1999;274:23396–23404. doi: 10.1074/jbc.274.33.23396. [DOI] [PubMed] [Google Scholar]
- 15.Jin T, Gu Y, Zanusso G, Sy M, Kumar A, Cohen M, Gambetti P, Singh N. J Biol Chem. 2000;275:38699–38704. doi: 10.1074/jbc.M005543200. [DOI] [PubMed] [Google Scholar]
- 16.Stewart R S, Drisaldi B, Harris D A. Mol Biol Cell. 2001;12:881–889. doi: 10.1091/mbc.12.4.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kitamoto T, Iizuka R, Tateishi J. Biochem Biophys Res Commun. 1993;192:525–531. doi: 10.1006/bbrc.1993.1447. [DOI] [PubMed] [Google Scholar]
- 18.Jackson G S, Hosszu L L, Power A, Hill A F, Kenney J, Saibil H, Craven C J, Waltho J P, Clarke A R, Collinge J. Science. 1999;283:1935–1937. doi: 10.1126/science.283.5409.1935. [DOI] [PubMed] [Google Scholar]
- 19.Ma J, Lindquist S. Nat Cell Biol. 1999;1:358–361. doi: 10.1038/14053. [DOI] [PubMed] [Google Scholar]
- 20.Geze M, Morliere P, Maziere J C, Smith K M, Santus R. J Photochem Photobiol B. 1993;20:23–35. doi: 10.1016/1011-1344(93)80128-v. [DOI] [PubMed] [Google Scholar]
- 21.Swanson J, Bushnell A, Silverstein S C. Proc Natl Acad Sci USA. 1987;84:1921–1925. doi: 10.1073/pnas.84.7.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Collinge J, Sidle K C, Meads J, Ironside J, Hill A F. Nature (London) 1996;383:685–690. doi: 10.1038/383685a0. [DOI] [PubMed] [Google Scholar]
- 23.Stimson E, Hope J, Chong A, Burlingame A L. Biochemistry. 1999;38:4885–4895. doi: 10.1021/bi982330q. [DOI] [PubMed] [Google Scholar]
- 24.Meng L, Mohan R, Kwok B H, Elofsson M, Sin N, Crews C M. Proc Natl Acad Sci USA. 1999;96:10403–10408. doi: 10.1073/pnas.96.18.10403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnston J A, Ward C L, Kopito R R. J Cell Biol. 1998;143:1883–1898. doi: 10.1083/jcb.143.7.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garcia-Mata R, Bebok Z, Sorscher E J, Sztul E S. J Cell Biol. 1999;146:1239–1254. doi: 10.1083/jcb.146.6.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wigley W C, Fabunmi R P, Lee M G, Marino C R, Muallem S, DeMartino G N, Thomas P J. J Cell Biol. 1999;145:481–490. doi: 10.1083/jcb.145.3.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Caughey B, Kocisko D A, Raymond G J, Lansbury P T., Jr Chem Biol. 1995;2:807–817. doi: 10.1016/1074-5521(95)90087-x. [DOI] [PubMed] [Google Scholar]
- 29.Close B E, Colley K J. J Biol Chem. 1998;273:34586–34593. doi: 10.1074/jbc.273.51.34586. [DOI] [PubMed] [Google Scholar]
- 30.Donne D G, Viles J H, Groth D, Mehlhorn I, James T L, Cohen F E, Prusiner S B, Wright P E, Dyson H J. Proc Natl Acad Sci USA. 1997;94:13452–13457. doi: 10.1073/pnas.94.25.13452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suzuki T, Park H, Hollingsworth N M, Sternglanz R, Lennarz W J. J Cell Biol. 2000;149:1039–1052. doi: 10.1083/jcb.149.5.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parodi A J. Annu Rev Biochem. 2000;69:69–93. doi: 10.1146/annurev.biochem.69.1.69. [DOI] [PubMed] [Google Scholar]
- 33.Lehmann S, Harris D A. J Biol Chem. 1997;272:21479–21487. doi: 10.1074/jbc.272.34.21479. [DOI] [PubMed] [Google Scholar]
- 34.Tanaka S, Saito M, Morimatsu M, Ohama E. Neuropathology. 2000;20:124–133. doi: 10.1046/j.1440-1789.2000.00285.x. [DOI] [PubMed] [Google Scholar]
- 35.Yedidda Y, Horonchik L, Tazban S, Yanal A, Taraboulour A. EMBO J. 2001;20:5383–5391. doi: 10.1093/emboj/20.19.5383. [DOI] [PMC free article] [PubMed] [Google Scholar]