Abstract
Ex vivo expansion and manipulation of human mesenchymal stem cells are important approaches to immunoregulatory and regenerative cell therapies. Although these cells show great potential for use, issues relating to their overall nature emerge as problems in the field. The need for extensive cell quantity amplification in vitro to obtain sufficient cell numbers for use, poses a risk of accumulating genetic and epigenetic abnormalities that could lead to sporadic malignant cell transformation. In this study, we have examined human mesenchymal stem cells derived from bone marrow, over extended culture time, using cytogenetic analyses, mixed lymphocyte reactions, proteomics and gene expression assays to determine whether the cultures would retain their potential for use in subsequent passages. Results indicate that in vitro cultures of these cells demonstrated chromosome variability after passage 4, but their immunomodulatory functions and differentiation capacity were maintained. At the molecular level, changes were observed from passage 5 on, indicating initiation of differentiation. Together, these results lead to the hypothesis that human mesenchymal stem cells cultures can be used successfully in cell therapy up to passage 4. However, use of cells from higher passages would have to be analysed case by case.
Introduction
Mesenchymal stem cells (MSCs) are multipotent cells that can differentiate in vitro into a variety of mesoderm cell lineages, including osteocytes, chondrocytes and adipocytes. Human MSCs (hMSCs) can be expanded and manipulated ex vivo and demonstrate immunomodulatory functions in vitro and in vivo; thus, they represent promising tools for use in immunoregulatory and regenerative cell therapies. Clinical use of hMSCs is an emerging field for treatment of several diseases, including non‐healing bone defects and fractures, inflammatory arthritis 1, inherited bone and cartilage disorders 2, cardiovascular disorders 3, graft‐versus‐host diseases 4, and for enhancement of haematopoietic stem‐cell transplantation 5. Although hMSCs have shown great potential for therapeutic application, little is known of their specific molecular mechanisms or impact of their culture expansion methods 6. Indeed, issues related to their nature have emerged as potential problems in this field.
Approximately 2 × 106 cells/kg are required for clinical application of hMSCs. Thus, extensive amplification in vitro without affecting cells' genomic characteristics and differentiation properties is necessary. However, such expansion could generate genetic and epigenetic instability, including chromosome alterations, and could also initiate cell senescence. Accumulation of changes during extended culture could lead to spontaneous malignant cell transformation 7, 8, 9.
Although it has been shown that hMSCs derived from bone marrow and adipose tissue can be continuously cultured in vitro for approximately 20 passages 10, it is unclear how many passages can be performed before the cells acquire chromosomal instability or lose multipotency; karyotypic aberrations have been observed in MSC cultures derived from mice and rats 11, 12. Some attempts have been made to examine such defects using different sources of hMSCs, different culture conditions and different passage densities; however, results of the studies are controversial. While some groups have shown retention of genetic stability up to passage three or four, others have shown that hMSC cultures maintain normal karyotype right up to passage 20 10, 13, 14, 15, 16.
In the study described here, we have analysed karyotype and multipotency of bone marrow‐derived hMSCs at passages 1–9. Our results identified in most cases, appearance of random chromosome alterations from passage 5 on, and also showed changes in cell proliferation rate, although the cells did not lose their differentiation capacity. Thus, we investigated whether the chromosome alterations would inhibit use of hMSCs cells in regenerative medicine.
Different approaches have been attempted for use of hMSCs in cell therapy. In some cases, cells are locally injected into the recipient organ, for example, in treatment of bone defects 17, 18; here, cells directly participate in the regeneration process. Furthermore, locally injected hMSCs secreted factors that stimulated cell population growth and differentiation of the stem cells, through paracrine effects or to counteract inflammation through immunomodulatory effects 19, 20. Alternatively, hMSCs can be intravenously infused to mediate repair of a microenvironment and facilitate treatment or prevention of graft‐versus‐host disease (GVHD) after haematopoietic stem‐cell transplantation 21. In this case, the immunomodulatory effect of hMSCs was exerted in peripheral blood, and the regenerative effect was expressed in the bone marrow. While intact hMSC genome is crucial for safety of cell therapy, injected cells do not normally survive over long periods of time in a recipient.
To further evaluate potential use of cultured hMSCs in cell therapy, we examined ability of hMSCs to inhibit in vitro lymphocyte proliferation. We showed that the capacity to inhibit this is maintained after a number of passages, even when hMSCs revealed chromosome instability. We used proteomic and transcriptomic approaches to verify molecular profiles and to examine potential modifications in multipotency of hMSCs from different culture passages (for example from passage 1 (P1) to passage 8 (P8)). Our results have indicated that cultured hMSCs develop molecular changes after passage 5.
Materials and methods
Isolation and culture of human bone marrow‐derived MSC
Bone marrow (BM)‐derived samples were obtained from healthy donors registered at the Bone Marrow Transplantation Unit, National Cancer Institute, Rio de Janeiro, Brazil, in accordance with guidelines of the local Ethics Committee and Helsinki Declaration. BM aspirates were obtained from 4 male donors (mean age 40.3, range 30–59) and 5 female donors (mean age 40.4, range 32–47). Mononuclear cells (MNC) were isolated from BM samples (5–10 ml) using density‐gradient centrifugation (Ficoll 1.077 g/ml; GE Healthcare Life Sciences, Piscataway, NJ, USA). Cells were plated in non‐coated 75‐cm2 polystyrene culture flasks (Techno Plastic Products AG, Trasadingen, Switzerland) at 500 000 cells/cm2 density, in low‐glucose Dulbecco's modified Eagle's medium (DMEM, Invitrogen, Carlsbad, CA, USA) supplemented with 15% foetal bovine calf serum (Hyclone, Thermo Scientific, Waltham, MA, USA), 100 IU/ml penicillin, 100 μg/ml streptomycin (Invitrogen) and 2 mM L‐glutamine (Invitrogen). After 48 h, non‐adherent cells were removed, and hMSCs were cultured until 80% confluence was obtained. Subsequently, cells were removed from plates using 0.05% trypsin (Invitrogen) treatment for 5 min at 37 °C, and replated at 2000 cells/cm2 density (passage 1) in a fresh culture flask. When 80% confluency was obtained (approximately 7 days culture), cells were trypsinized and replated in fresh culture flasks (passage 2). These processes were repeated up to passage 10, for all cultures. Population doubling of the cultured hMSCs was calculated at every passage, according to the equation log2 (number of harvested cells/number of seeded cells). For 10 passages, mean cumulative population doubling (CPD) was 24.8 and mean population doubling was 2.4 per passage.
Confirmation of multipotentiality
These experiments were performed in accordance with minimal criteria for defining multipotent mesenchymal stromal cells, as defined by the International Society for Cellular Therapy 22. The cultured plastic‐adherent cells, expressing CD73, CD90 and CD105, in the absence of lineage commitment markers such as CD14, CD19, CD34, CD45 and HLA‐DR (BD Biosciences, San Jose, CA, USA), were able to differentiate into adipocytes and osteoblasts. Ability of the hMSCs cultures to differentiate into adipocytes and osteoblasts was tested at passages 1, 5 and 8. To induce adipogenic differentiation, the cells were cultured with 10−8 M dexamethasone (Sigma‐Aldrich, St. Louis, MO, USA) and 5 μg/ml insulin (Sigma‐Aldrich); oil‐red‐O (Sigma‐Aldrich) staining was used to detect lipid accumulation. Osteogenic differentiation was induced using 10−8 M dexamethasone (Sigma‐Aldrich), 10 mM β‐glycerol‐phosphate (Sigma‐Aldrich) and 50 μg/ml ascorbic acid (Sigma‐Aldrich). Osteoblasts were identified using alizarine red S (Isofar, Brazil) staining.
Cytogenetic analysis
Nine hMSC cultures were cytogenetically defined at a number of passages. Prior to obtaining confluence, cells were incubated at 37 °C with colcemid (Sigma‐Aldrich) at 1 μg/ml for 2 h. Then they were trypsinized and processed using standard cytogenetic procedures. GTG‐banding was performed, and chromosomes were identified and analysed according to the International System of Human Cytogenetic Nomenclature (ISCN, 2009).
Quantitative PCR
Quantitative PCR (Q‐PCR) analyses were performed using one μg of mRNA, treated with DNAse Amplification Grade I (Invitrogen), which was reverse transcribed using Superscript II Reverse Transcriptase® (Invitrogen). Q‐PCR reactions contained 1× Power SYBR Green PCR Master Mix® (Applied Biosystems, Carlsbad, CA, USA), 2.5 μl of 100‐fold diluted cDNA and 0.1 μM of each primer. Quantitative determination of mRNA levels was performed using Rotor‐Gene 6000 Series Software (Corbett, Australia), and reactions were performed in a Rotor‐Gene 6000 thermocycler (Corbett, Australia) using the following program: 95 °C for 5 min and 45 cycles at 95 °C for 15 s followed by 60 °C for 1 min. Dissociation curve analysis was used to demonstrate equal amplification efficiency of a specific PCR product for all primers used in these studies. Determination of fold‐expression was calculated using the DDCt method according to Livak and Schmittgen 23. Data were analysed statistically using Prisma 4 and ANOVA.
Expression levels were estimated in triplicate, and β‐actin and GAPDH were used as reference genes for normalization. The following primers were used: CD44 Fw, 5′‐ AGACCCACTC AGCCAAATCT ‐3′/ Rev, 5′‐ CTTTCACAGGA GAGAACAGGTG‐3′; SCF Fw, 5′‐ ATGCAAATGGTTGCCTGGTCCC ‐3′/ Rev, 5′‐TGCCAATGTCCTTCTTGGCAGA ‐3′; c‐Kit Fw, 5′‐ATGGGAAAGAAGACAACG ACACGC ‐3′/ Rev, 5′‐ GCAGAC AGAGCCGATGGTAGG ‐3′; BMPR Fw, 5′‐ CGA AGTGCT GGACGAAAGCCT ‐3′/ Rev, 5′‐ TTGACACACACAACCTCACGCA –3′; β‐actin Fw, 5′‐ACCTGAGAACT CCACTACCCT‐3′/ Rev, 5′‐GGTCCCACCCATG TTCCAG ‐3′; GAPDH Fw, 5′‐ACCACAGTCCATGCCATCAC‐3′/ Rev, 5′‐CCACC ACCCTGTTGCTGTA ‐3′
Mixed lymphocyte reaction
Peripheral blood mononuclear cells (PBMCs) were isolated from heparinized samples of unrelated healthy volunteers, using density‐gradient centrifugation. For the mixed lymphocyte reaction (MLR), 5 × 105 carboxy fluorescein diacetate succinimidyl ester (CFSE)‐labelled responder PBMC (R) and 5 × 105 irradiated (2500 cGy) stimulator PBMC (S)/well were incubated in 24‐well flat‐bottom tissue‐culture plates containing RPMI‐1640 (Invitrogen) supplemented with 2 mM L‐glutamine (Invitrogen), 100 IU/ml penicillin, 100 μg/ml streptomycin (Invitrogen) and 10% FCS (Hyclone) in the absence or presence of 5 × 104 hMSCs. After seven days, cultures were analysed using flow cytometry in a FACScan instrument (Becton Dickinson, Franklin Lakes, NJ, USA) and data were analysed using CELLQuest software (Becton Dickinson). With Student's t‐test, P‐values < 0.05 were considered to be statistically significant. Statistical analyses and graphic representations were performed using GraphPad PrismTM software (GraphPad software, San Diego, CA, USA).
Expression chip array data analysis
Total RNA from hMSC cultures at passages 1, 5 and 8 were obtained using RNeasy Mini Kit (QIAGEN, Valencia, CA, USA) according to the manufacturer's instructions. One μg of total RNA was used to synthesize biotinylated cRNA according to GeneChip Whole‐Transcript (WT) Sense Target Labelling Assay (Affymetrix, Santa Clara, CA, USA). Biotinylated cRNA was subsequently hybridized to GeneChip human exon 1.0 ST array (Affymetrix), washed and stained according to the manufacturer's protocols. GeneChip arrays were scanned using GeneChip® Scanner 3000. Affymetrix Expression Console Software Version 1.0 was used to create summarized expression values (CHP‐files) and Robust Multichip Analysis (RMA) algorithm was applied. Data were analysed using Partek® software (http://www.partek.com) 24, with differentially expressed genes, ≥5‐fold change, as criteria to define overexpression or downregulation. Gene ontology analysis was conducted using Ingenuity Pathways Analysis (IPA) software, available at NCBI or http://www.ingenuity.com.
Two‐dimensional gel electrophoresis
To prepare protein extracts, 7 × 106 cells at passages 1, 5 and 8 were washed 3 times in phosphate‐buffered saline (PBS), centrifuged and resuspended in cold lysis buffer containing 50 mM Tris pH 7.5, 5 mM ethylene diamine tetraacetic acid (EDTA), 10 mM ethylene glycol tetraacetic acid (EGTA), 50 mM sodium fluoride, 20 mM β‐glycerol‐phosphate, 250 mM sodium chloride, 0.1% Triton X‐100, 20 mM sodium orthovanadate, and protease inhibitor mix (2.4 μg/ml AEBSF, 0.1 μg/ml Bestatin, 0.1 μg/ml Pepstatin, 0.1 μg/ml Leupeptin, and 1.8 μg/ml E‐64) (Calbiochem, Ardmore, PA, USA). After incubation for 30 minutes on ice, lysates were spun at 12 000 rpm at 4 °C, for 30 min. Total protein concentration obtained was determined using the Bradford assay 25. Five hundred μg of total cell protein extracts was precipitated using a 2D Cleanup Kit (GE Healthcare, Piscataway, NJ, USA) and resuspended in 200 μl of reswelling buffer containing 6 M urea, 2 M thiourea, 15 mM DTT, 2% (w/v) ASB14, 0.5% IPG buffer pH 3 (GE Healthcare, Sweden) and traces of bromophenol blue. Electrophoresis in the first dimension was performed using 11 cm of Immobiline DryStrip, pH 4‐7 (GE Healthcare), at 30 550 VhT.
IPG strips were run in ExcelGel SDS 8–18% according to the manufacturer's instructions (GE Healthcare) and ExcelGels were stained with colloidal Coomassie Blue G‐250 (Sigma‐Aldrich). All gels were scanned on an image scanner using LabScan software (GE Healthcare), analysed and compared using Image Master 2D Platinum 6.0 software (GE Healthcare), followed by additional visual analysis. Gels were analysed and averaged separately. Normalized spot volumes were compared and averaged for all visualized spots in each gel. After normalization of images, spot volume was calculated to quantify intensity of each spot. Comparison reports of qualitative and quantitative differences between samples were generated, and only spots that showed greater than 2‐fold difference between samples, were considered to be differentially expressed.
Results
Differentiation potential of hMSC cultures and maintenance of hMSC multipotency markers
To determine whether the expanded hMSC cultures maintained multipotency differentiation characteristics, we tested them at passages 1, 5 and 8 for differentiation into adipogenic and osteogenic cells. Our results showed that at all passages, hMSC cultures were able to differentiate into adipogenic and osteogenic cells that maintained their multipotency capacity (Fig. 1).
Figure 1.
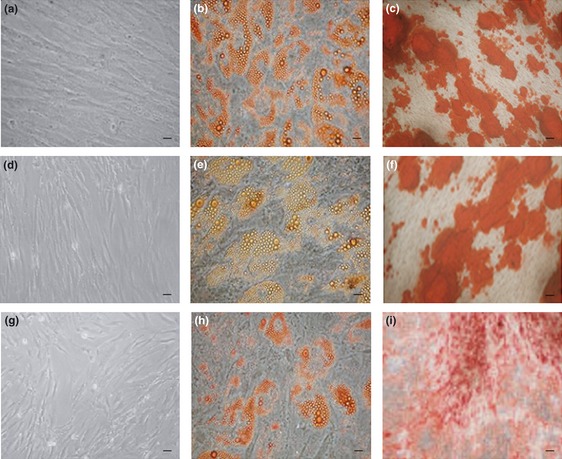
hMSC capacity for differentiation. (a), (d) and (g) – undifferentiated hMSCs at passages 1, 5 and 8. (b), (e) and (h) – adipogenic differentiation at passages 1, 5 and 8. Accumulation of neutral lipid vacuoles stained with oil red O indicate differentiation. (c), (f) and (i) – osteogenic differentiation at passages 1, 5 and 8. Calcium deposition stained with alizarin red indicate differentiation. Scale bars ‐ 26 μm (100× magnification).
As described in previous studies, a number of genes, including SCF, c‐Kit, BMPR and CD44, are considered to be hMSCs multipotency markers 26. Although they are not specific to hMSCs, they are expressed in progenitor cells 27, 28, 29. To further confirm multipotency during hMSC culture, we quantified expression of these markers during subsequent passages of all hMSC cultures. We performed Q‐PCR in nine different cell cultures from passage 1 to 9. Expression of SCF mRNA was reduced 2‐ to 5‐fold from passages 5 to 9 in all tested cultures (Fig. 2), although expression of c‐Kit, BMPR and CD44 was conserved throughout all passages, indicating profile maintenance. SCF is a gene related to cell proliferation and not to differentiation 30, 31, 32; thus, changes in SCF expression suggests that after passage 5, cells had reduced proliferation. These results could also indicate senescence during in vitro expansion. Under our culture conditions, typical morphological changes associated with senescence were observed from passage 8 (Fig. 3); cells were enlarged and had irregular shapes 33. These data could explain the delayed growth observed in our experiments. In early passages, cell cultures reached confluence at 7 days; however, after passage 6, confluence was obtained after 12 days here, under the same culture conditions.
Figure 2.
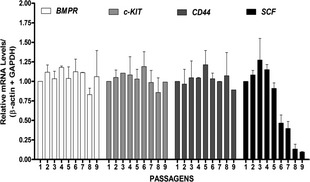
Maintenance of hMSC markers. Total RNA was isolated from hMSC cultures at different passages (1 to 9) and examined using Q‐PCR to determine changes in mRNA expression levels after normalisation to β‐actin and GAPDH expression. Analysis of fold changes in expression of SCF, CD44, c‐Kit and BMPR were performed at the different passages. Data are presented as fold induction relative to control group (passage 1). Mean ± SD (n = 9). *Statistical significance was considered at P < 0.001.
Figure 3.
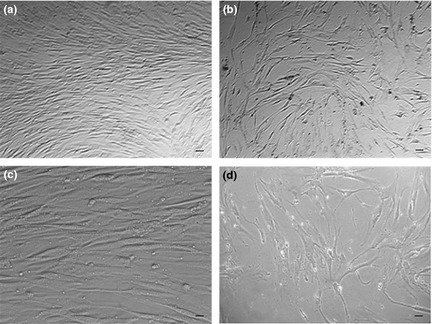
Morphologic changes indicating senescence. Using the same confluence stage, representative morphology of hMSCs is shown at early passage (P3) [(a) and (c)] and late passage (P8) [(b) and (d)]. (a) and (b): Scale bars ‐ 65 μm (40× magnification). (c) and (d): Scale bars ‐13 μm (200× magnification). Cells were seeded at the same density (2.000 cells/cm2), and at P3, the hMSC culture obtained confluence at 7 days, while at P8, confluence was obtained at 12 days.
hMSCs cytogenetic analysis
To verify whether the hMSCs maintained their normal karyotype in culture throughout all passages, the same nine cultures were tested for their genomic stability using conventional cytogenetic analysis. Our results showed that until passage 4, hMSCs from all donors were of normal diploid karyotype. In subsequent passages, seven of the nine cultures presented random aneuploidy, which is a characteristic of cytogenetic instability (Table 1). However, in most cases, these abnormalities were lost by the next passage. Nevertheless, in one culture, a clonal abnormality was identified and amplified from passage 6 to passage 8 (Table 1, Sample 1).
Table 1.
Cytogenetic analysis of hMSCs from different donors and passages
| Samples | Culture Passage | Karyotype |
|---|---|---|
| 1 | P1 | 46,XX[20] |
| P3 | 46,XX[21] | |
| P5 | 46,XX[20] | |
| P6 | 46,XX[17]/46,X,‐X,+16[20] a /36~42,XX,‐3,‐7,‐11,‐12,‐13,‐14,‐14,‐16,‐16,‐19[1]./5,‐8,‐11,‐14,‐20[1]/‐4,‐7,‐10,‐13[1] | |
| P8 | 46,XX[10]/46,X,‐X,+16[5] a/40~42,X,‐X,‐3,‐5,‐8,‐1‐,‐13[5] | |
| 2 | P1 | 46,XY[20] |
| P4 | 46,XY[22] | |
| P6 | 46,XY[20]43~45,XY,‐5,‐13,‐21[1]/‐6,‐9,‐20[1]/‐10,‐14,‐19[1] | |
| P8 | 46,XY[18]42~45,XY,‐4,‐8,‐13,‐21[1]/‐9,‐11,‐19[1]/‐5,‐20[1] | |
| 3 | P1 | 46,XX[21] |
| P4 | 46,XX[22] | |
| P6 | 46,XX[20] | |
| P8 | 46,XX[20] | |
| 4 | P1 | 46,XY[23] |
| P3 | 46,XY[25] | |
| P5 | 46,XY[20] | |
| P7 | 46,XY[20]/42~45,XY,‐4,‐6,‐13,‐20[1]/‐3,‐5,‐13,‐20[1]/‐6,‐10,‐14,‐19[1]/‐15,‐21[1]/‐8,‐15[1]/‐22[1] | |
| P9 | 46,XY[10]/42~45,XY,‐4,‐6,‐9,‐15[1]/‐8,‐13,‐18,‐21[1]/‐9,10,‐14,‐20[1]/‐5,‐8,‐22[1]/‐4,‐15[1]/‐22[1] | |
| 5 | P1 | 46,XY[20] |
| P3 | 46,XY[15] | |
| P5 | 46,XY[18] | |
| P6 | 45,XY,‐21[2]/46,XY[11] | |
| P8 | 44,XY,‐10,‐13[4]/46,XY[7] | |
| 6 | P1 | 46,XY[20] |
| P3 | 46,XY[22] | |
| P5 | 46,XY[13] | |
| P7 | 44,XY,‐6,‐15[2]/46,XY[8] | |
| 7 | P1 | 46,XX[20] |
| P2 | 46,XX[18] | |
| P5 | 47,XX,+mar[1]/46,XX[7] | |
| P7 | 46,XX[6]/45,XX,‐4[1] | |
| 8 | P1 | 46,XY[10] |
| P3 | 46,XY[15] | |
| P5 | 46,XY[8]/ 43~45,XY,‐10,‐13,‐19[1]/‐9,‐20,‐21[2]/‐10[1] | |
| 9 | P1 | 46,XX[21] |
| P4 | 46,XX[19] | |
| P6 | 46,XX[16] |
The bold text indicates the clonal abnormality.
P‐ Passage.
Chromosomes were analysed according to the International System of Human Cytogenetic Nomenclature (ISCN, 2009).
In vitro hMSC‐mediated T‐cell inhibition
To further determine whether our hMSC cultures maintain their ability to inhibit proliferation of lymphocytes through alloantigen stimulation at different passages, we performed mixed lymphocyte reaction assays (MLR) in the absence or presence of hMSCs. hMSCs from different passages were added to MLR on day 0, and cells were harvested on day 7. Data collected showed that hMSC cultures from the same 9 different donors previously tested for cytogenetics, were capable of inhibiting lymphocyte proliferation by 70%, in response to alloantigen stimulation at different passages (P1 to P9) (Fig. 4). The low peak of data shown in Fig. 4b in early and late passages represents reduced proliferation of the cells as a consequence of hMSC‐induced immunosuppression of lymphocyte proliferation. These results confirmed that hMSC cultures, even at subsequent passages, had in vitro immunomodulatory properties.
Figure 4.
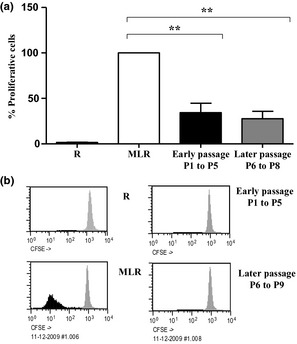
hMSC‐mediated T cell inhibition. (a) hMSC inhibition of MLR. Lymphocyte proliferation was assessed using MLR in absence (MLR) or presence of 10% hMSCs/well, administered at early (P1 to P5) or late (P6 to P10) passage. Results expressed as mean ± sd for independent experiments using 9 hMSC cultures (P < 0.01); (b) Representative assay showing MLR in absence (MLR) or presence of 10% MSC/well at early (P1 to P5) or late (P6 to P9) passages. Black represents proliferative cells and grey represents non‐proliferative cells. R ‐ responders without stimulators.
Proteomic and transcriptomic profile similarity of hMSCs at different passages
Although hMSCs, even with chromosomal instability, are able to differentiate into various lineages and inhibit lymphocyte proliferation, it is possible that multipotency would not be maintained. Therefore, we tested whether molecular profiles of these cells would be conserved during passaging. For this purpose, we performed comparative expression analysis, using proteomic and transcriptome approaches, using three different hMSC cultures from different donors that showed chromosomal instability at passages 1, 5 and 8. Samples 1, 2 and 5 shown in Table 1 were used in these analyses.
Using proteomic approaches, we compared and analysed the cultures using 2D electrophoresis. One representative gel from one donor culture at passages 1, 5 and 8 is shown in Fig. 5. After image analysis, all gels demonstrated 195 ± 3 spots, a 95% match.
Figure 5.
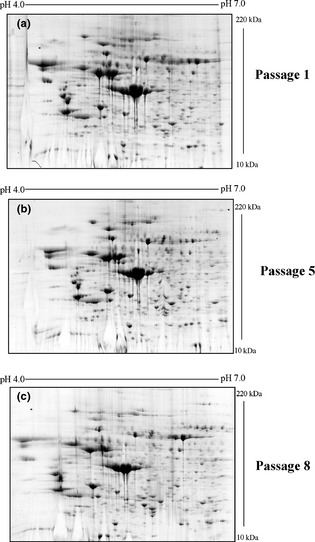
hMSC proteomic profile obtained using cells at passages 1, 5 and 8 from the same donor. (a) 2‐DE gel ‐ passage 1. (b) 2‐DE gel ‐ passage 5. (c) 2‐DE gel ‐ passage 8. Values on horizontal axes represent pI, and values on vertical axes represent molecular weight.
In our previous work, proteins spots were identified, and results demonstrated that hMSCs would have similar protein expression patterns at the first passage 34. Here we observed similar results in all cultures at all passages analysed, suggesting high similarity among different ones, as shown in the scatter plot in Fig. 6. These results suggest that protein profile of hMSC cultures derived from different passages and different donors is equivalent, at least based on the results from this approach.
Figure 6.
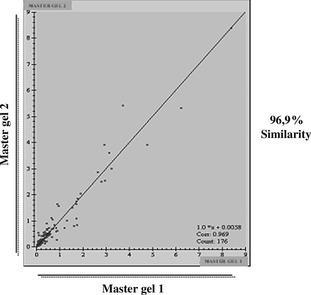
Bidimensional gel analysis using Image Master 2D Platinum 6.0. Values on horizontal axes represent master gel 1 products from pool of 2‐DE gels from passages 1, 5 and 8 from the same donor. Values on vertical axes represent master gel 2 products from pool of 2‐DE gels from passages 1, 5 and 8 from the same donor, which were different from those of master gel 1. Scatter plot calculated using results from analysed gels and shows relationship between spot values (% volume), which show 96.9% similarity.
To determine the global pattern of gene expression in hMSC cultures, we performed a transcriptome comparison analysis using an expression chip array assay. For this study, we used the same donors analysed in the proteomic assay at passages 1, 5 and 8. mRNA in hMSCs from these passages was amplified, followed by labelling and hybridization to the Affymetrix GeneChip human exon 1.0 ST array. After normalization and analysis of the microarray data using Partek® software (http://www.partek.com), we observed alterations in some genes at passage 5. Using a 5‐fold change as a cut‐off in expression level, we observed 21 and 91 differentially expressed genes at passages 5 and 8, respectively, compared to passage 1 (Tables 2 and 3).
Table 2.
Genes upregulated in hMSC culture (fold change > 5)
| Gene Symbol | Description | Fold change‐P5 | Fold change‐P8 | |
|---|---|---|---|---|
| 1 | PRUNE2 | Prune homolog 2 (Drosophila) | 5.34 up | 12.05 up |
| 2 | EPGN | Epithelial mitogen homolog (mouse) | — | 12.47 up |
| 3 | C4orf3 | Chromosome 4 open reading frame 3 | 5.91 up | 12.50 up |
| 4 | SLC7A11 | Solute carrier family 7 | 5.16 up | 12.72 up |
| 5 | TUBA1B | Tubulin, alpha 1b | 49.14 up | 17.39 up |
| 6 | ACTC1 | Actin, alpha, cardiac muscle 1 | — | 18.17 up |
| 7 | CHORDC1 | Cysteine and histidine‐rich domain (CHORD)‐containing 1 | 17.76 up | 28.48 up |
| 8 | FAM36A | Family with sequence similarity 36, member A | — | 5.00 up |
| 9 | MAPRE3 | Microtubule‐associated protein, RP/EB family, member 3 | — | 5.00 up |
| 10 | UAP1 | UDP‐N‐acetylglucosamine pyrophosphorylase 1 | — | 5.01 up |
| 11 | PREPL | Prolyl endopeptidase‐like | — | 5.02 up |
| 12 | VGLL3 | Vestigial‐like 3 (Drosophila) | — | 5.05 up |
| 13 | PGRMC2 | Progesterone receptor membrane component 2 | — | 5.07 up |
| 14 | PLD5 | Phospholipase D family, member 5 | — | 5.08 up |
| 15 | AMIGO2 | Adhesion molecule with IgG‐like domain 2 | — | 5.10 up |
| 16 | ANKRD1 | Ankyrin repeat domain 1 (cardiac muscle) | — | 5.10 up |
| 17 | SC4MOL | Sterol‐C4‐methyl oxidase‐like | — | 5.10 up |
| 18 | SERPINB2 | Serpin peptidase inhibitor, clade B (ovalbumin), member | 9.07 up | 5.10 up |
| 19 | FAM198B | Family with sequence similarity 198, member B | — | 5.12 up |
| 20 | C4orf18 | Chromosome 4 open reading frame 18 | — | 5.16 up |
| 21 | BCAP29 | B‐cell receptor‐associated protein 29 | — | 5.18 up |
| 22 | HAPLN1 | Hyaluronan and proteoglycan link protein 1 | — | 5.18 up |
| 23 | LYPD6B | LY6/PLAUR domain containing 6B | — | 5.26 up |
| 24 | MANBA | Mannosidase, beta A, lysosomal | — | 5.27 up |
| 25 | LMBR1 | Limb region 1 homolog (mouse) | — | 5.33 up |
| 26 | COL8A1 | Collagen, type VIII, alpha 1 | — | 5.34 up |
| 27 | ATP11B | ATPase, class VI, type 11B | — | 5.35 up |
| 28 | FMN2 | Formin 2 | — | 5.39 up |
| 29 | PLOD2 | Procollagen‐lysine, 2‐oxoglutarate 5‐dioxygenase 2 | — | 5.42 up |
| 30 | NEK7 | NIMA (never in mitosis gene a)‐related kinase 7 | — | 5.45 up |
| 31 | TOMM20 | Translocase of outer mitochondrial membrane 20 homolog | — | 5.50 up |
| 32 | SEPT7 | Septin 7 | — | 5.53 up |
| 33 | IVNS1ABP | Influenza virus NS1A binding protein | — | 5.56 up |
| 34 | ITGA6 | Integrin, alpha 6 | 5.05 up | 5.58 up |
| 35 | ELTD1 | Latrophilin and seven transmembrane domain containing | 6.08 up | 5.63 up |
| 36 | HINT3 | Histidine triad nucleotide binding protein 3 | — | 5.64 up |
| 37 | OGFRL1 | Opioid growth factor receptor‐like 1 | — | 5.64 up |
| 38 | SERPINI1 | Serpin peptidase inhibitor, clade I (neuroserpin) | — | 5.64 up |
| 39 | TOR1AIP2 | Torsin A interacting protein 2 | — | 5.66 up |
| 40 | PDE11A | Phosphodiesterase 11A | — | 5.69 up |
| 41 | RAP1B | RAP1B, member of RAS oncogene family | 5.23 up | 5.71 up |
| 42 | FAM174A | Family with sequence similarity 174, member A | — | 5.73 up |
| 43 | XRCC4 | X‐ray repair complementing defective repair in Chinese ham | — | 5.73 up |
| 44 | SYT14 | Synaptotagmin XIV | — | 5.78 up |
| 45 | EVI5 | Ecotropic viral integration site 5 | — | 5.81 up |
| 46 | STC1 | Stanniocalcin 1 | — | 5.87 up |
| 47 | ME1 | Malic enzyme 1, NADP(+)‐dependent, cytosolic | — | 6.08 up |
| 48 | DLG1 | Discs, large homolog 1 (Drosophila) | 6.23 up | 6.11 up |
| 49 | SLITRK1 | SLIT and NTRK‐like family, member 1 | — | 6.13 up |
| 50 | FAM38B | Family with sequence similarity 38, member B | — | 6.32 up |
| 51 | MCART1 | Mitochondrial carrier triple repeat 1 | — | 6.32 up |
| 52 | PCDH10 | Protocadherin 10 | — | 6.44 up |
| 53 | PLCB4 | Phospholipase C, beta 4 | — | 6.46 up |
| 54 | FLRT2 | Fibronectin leucine‐rich transmembrane protein 2 | — | 6.62 up |
| 55 | SCN9A | Sodium channel, voltage‐gated, type IX, alpha subunit | — | 6.66 up |
| 56 | ERV3 | Endogenous retroviral sequence 3 | — | 6.74 up |
| 57 | SCIN | Scinderin | — | 6.99 up |
| 58 | CD36 | CD36 molecule (thrombospondin receptor) | — | 7.07 up |
| 59 | FGF7 | Fibroblast growth factor 7 (keratinocyte growth factor) | — | 7.17 up |
| 60 | CPE | Carboxypeptidase E | — | 7.19 up |
| 61 | VANGL1 | Vang‐like 1 (van Gogh, Drosophila) | 7.50 up | 7.20 up |
| 62 | PVRL3 | Poliovirus receptor‐related 3 | — | 7.27 up |
| 63 | EPHA5 | EPH receptor A5 | — | 7.39 up |
| 64 | HIST1H2BK | Histone cluster 1, H2bk | 5.60 up | 7.72 up |
| 65 | MAMDC2 | MAM domain containing 2 | — | 7.78 up |
| 66 | ATRNL1 | Attractin‐like 1 | — | 7.89 up |
| 67 | COL11A1 | Collagen, type XI, alpha 1 | — | 7.90 up |
| 68 | ARHGAP29 | Rho GTPase activating protein 29 | 9.08 up | 8.62 up |
| 69 | DPP4 | Dipeptidyl‐peptidase 4 | 5.14 up | 9.62 up |
| 70 | ANKRD36B | Ankyrin repeat domain 36B | 7.58 | — |
| 71 | CCDC99 | Coiled‐coil domain containing 99 | 5.03 up | — |
| 72 | USP53 | Ubiquitin‐specific peptidase 53 | 5.19 up | — |
The fold change is related to passage 1.
(—) Not differentially expressed.
Table 3.
Genes downregulated in hMSC culture (fold change > 5)
| Gene Symbol | Description | Fold change‐P5 | Fold change‐P8 | |
|---|---|---|---|---|
| 1 | NRN1 | Neuritin 1 | — | 7.67 down |
| 2 | IFNA21 | Interferon, alpha 21 | — | 7.23 down |
| 3 | KRTAP13‐3 | Keratin‐associated protein 13‐3 | — | 6.64 down |
| 4 | HIST1H4L | Histone cluster 1, H4l | — | 6.58 down |
| 5 | TRHDE | Thyrotropin‐releasing hormone degrading enzyme | — | 6.32 down |
| 6 | MT1IP | Metallothionein 1I (pseudogene) | — | 6.23 down |
| 7 | PTGER2 | Prostaglandin E receptor 2 (subtype EP2), 53 kDa | — | 6.13 down |
| 8 | PCDH11Y | Protocadherin 11 Y‐linked | 6.55 down | 6.05 down |
| 9 | HEPH | Hephaestin | — | 5.99 down |
| 10 | OSTBETA | Organic solute transporter beta | — | 5.90 down |
| 11 | FBLN1 | Fibulin 1 | — | 5.83 down |
| 12 | OSTbeta | Organic solute transporter beta | — | 5.69 down |
| 13 | LCE2D | Late cornified envelope 2D | — | 5.36 down |
| 14 | HGF | Hepatocyte growth factor | — | 5.11 down |
| 15 | DPPA3 | Developmental pluripotency associated 3 | — | 5.07 down |
| 16 | OR5M3 | Olfactory receptor, family 5, subfamily M, member 3 | — | 5.03 down |
| 17 | EIF2S2 | Eukaryotic translation initiation factor 2, subunit 2 bet | 7.36 down | — |
| 18 | IFNA17 | Interferon, alpha 17 | 6.44 down | — |
| 19 | MS4A4A | Membrane‐spanning 4‐domains, subfamily A | 6.33 down | — |
The fold change is related to passage 1.
(—) Not differentially expressed.
In silico analysis of these results using Ingenuity software (NCBI) showed that overexpressed genes at P5 and P8 were associated with the cell cycle, cell expansion and proliferation, cell‐to‐cell signalling and their interactions, and cell differentiation, among other functions. Tables 4 and 5 describe biological processes and functional interaction network classes, respectively, which are associated with the genes that were over expressed at P5 and P8. Networks associated with genes upregulated and downregulated at both P5 and P8 are described in Supplementary Figs 1 and 2.
Table 4.
Biological processesa including molecular and cellular classes associated with genes overexpressed in P5 and P8
| Cell‐to cell signalling and interaction |
| Cell cycle |
| Cell morphology |
| Cellular organization |
| Cellular development |
| Molecular transport |
| Carbohydrate metabolism |
| Lipid metabolism |
Gene Ontology (GO) terms were used to define the biological processes using the Ingenuity Pathways Analysis (Ingenuity® Systems). Processes listed in the table are those statistically most relevant (with the smallest P values).
Table 5.
Functional interaction networks classesa associated with genes overexpressed in P5 and P8
| Cell cycle, cell death |
| Cellular growth and proliferation |
| Cell‐to‐cell signalling and interaction |
| Gene expression |
| Cellular function and maintenance |
| Cellular compromise |
| Cellular development |
| Molecular transport |
aGene Ontology (GO) terms were used to define the biological processes using the Ingenuity Pathways Analysis (Ingenuity® Systems). The processes listed in the table are the most statistically relevant (with the smallest P values).
Interestingly, at passage 5, we observed upregulation of genes such as PRUNE2, DLG1, and VANGL1, which are involved in differentiation in Drosophila melanogaster. Indeed, at passage 8, we observed over expression of genes, such as FGF7 and COL11A1, indicative of cell differentiation. We also observed upregulation of ARHGAP29, which is associated with cell senescence, at both passages 5 and 8.
Together, these findings show that although results of the proteomic analysis indicated that the same protein profile would be maintained through all passages, levels of gene expression revealed that small but important changes occur from passage 5 on.
Discussion
Clinical application of hMSCs requires use of a significant number of cells and, considering low numbers of hMSCs in vivo 35, their extensive ex vivo expansion is required. However, such expansion increases the risk of genetic and epigenetic changes. Cell proliferation in vitro could result in occurrence of mutations and chromosomal aberrations, eventually leading to malignant transformation. Thus, cytogenetic and molecular studies must be used to determine appropriate hMSCs to use for cell therapy 36, 37.
Although it has been shown that hMSCs can be continuously cultured in vitro, it is unclear how many passages can be performed before they acquire chromosomal instability or lose their multipotency. Using different sources, culture conditions and passage densities, some groups have attempted studies concerning chromosome stability of human MSCs in culture. Zhang and co‐workers, using 106 cells/cm2 cultured in DMEM/15% FBS (foetal bovine serum) medium, showed that in vitro cultured bone marrow MSCs retained normal cytogenetics until passage three, indicating that genetic stability was retained until this passage 12. In addition, Bochkov and co‐workers evaluated chromosome variability of hMSCs derived from adipose tissue, cultured in DMEM/F12 10% FCS (foetal calf serum) at 1.5 × 103 cells/cm2 density. This group verified that at early passages (P1 to P5), the cells had normal karyotypes, while at later passage cultures (P6 to P14) they began to show chromosomal aberrations 13. However, Bernado et al. revealed that MSCs derived from bone marrow and cultured in Mesencult/10% FCS medium at 1.6 × 104 cells/cm2 density did not undergo spontaneous transformation and thus that these MSCs could be safely expanded in vitro without developing chromosomal abnormalities. Moreover, Izadpanah and colleagues verified that hMSC cultures derived from bone marrow and adipose tissue and cultured in α‐MEM /20% FBS medium displayed normal karyotypes up to passage 20 14, 15. Furthermore, Tarte and colleagues showed that occurrence of aneuploidy in bone marrow‐derived hMSCs did not persist in culture and was not associated with transformation events 16. Thus, the determinants that define genetic stability remain unknown.
In this study, karyotypic analysis of each culture was performed which showed that different cultures underwent genetic instability after passage 4. Interestingly, most of abnormalities observed using conventional cytogenetic analyses were not clonal, which suggests that the alterations could be lost over subsequent passages. Moreover, during cell therapy, such hMSCs would not survive for long in tissues due to their inability to stabilize in this niche; they would,most likely, undergo apoptosis. However, results from our assays showed one culture that acquired clonal abnormalities at passage 6.
To better understand the consequences of this alteration, we decided to further characterize these cells. We analysed their immunomodulatory and molecular profiles, with and without clonal abnormalities, from different passages. Our results showed that hMSC cultures could be cultured long‐term in vitro without losing their immunomodulatory capacity and multipotency, even when the analysis included samples with karyotypic abnormalities. However, molecular analysis revealed that some changes could be observed along the passages. Such changes were the same in all analysed samples, but were not related to presence of the abnormality.
The proteomes and transcriptomes of mesenchymal stem cells in undifferentiated and differentiated states have previously been characterized, but these studies were not focused on variations observed in extensively expanded cultures 26. Analysis of molecular profiles of our hMSC cultures revealed that they underwent molecular changes from passage 5 on. Although we did not observe differences in their proteomic profiles using 2D analysis, the expression chip array assay (which had 5‐fold expression level as cut‐off), revealed that 21 genes underwent alterations after passage 5, and numbers of genes differentially expressed increased by passage 8. Analysis using Ingenuity software (IPA) showed that these overexpressed genes were associated with cell growth and proliferation, cell‐cell signalling interactions, and cell differentiation, suggesting that they had begun differentiation from passage 5.
Notably, some of the altered genes present in passage 5 are homologous to Drosophila melanogaster development genes, such as PRUNE2, which plays an important role in regulating differentiation 38, DLG1, which is associated with cell proliferation 39, and VANGL1, which is associated with neural differentiation 40. Homology of these genes between insect and mammal suggests that important changes are taking place in these cells.
Development genes play fundamental roles in a range of processes, specially those involving cell differentiation, proliferation and apoptosis. The balance of these processes maintains cell homeostasis and preserves genomic stability. Thus, changes in expression of developmental genes after passage 5 are indicative of alterations in balance of cells of the culture, which could be associated with cell instability. This results from chromosomal anomalies or epigenetic changes that alter gene expression.
While these genes are not classified as oncogenes, their expression could lead to malignant transformation. Changes in expression of PRUNE2, DGL1 and VANGL1 have previously been associated with specific types of cancer 41, 42, 43. Moreover, the hMSC cultures might begin differentiation processes at passage 8, as we observed expression of genes, such as FGF7, which play regulatory roles in differentiation of adipocytes 44, and COL11A1I, a chondrogenic marker gene 45.
Interestingly, ARHGAP29 was upregulated in both passages 5 and 8. This gene has recently been associated with senescence as a part of the senescence‐associated gene expression signature 37. This finding is consistent with changes in SCF expression after passage 5 in hMSC culture that we observed in this study, suggesting reduction in proliferation and potential induction of senescence.
Downregulation of IFNA21 and IFNA17 after passage 5 could be associated with secretion. It is well known that hMSCs have immunomodulatory properties. However, this function must be activated by a stimulus, mediated by secreted factors. We used hMSCs from different passages in the chip array assay, but all cells were in an inactive state. Thus, our results do not provide evidence for immunomodulatory capacity of hMSCs. Indeed, altered expression of genes, such as IFNA21 and IFNA17, at passages 5 and 8 could not be associated with immunomodulatory properties, or in presence of stimulus‐induced changes in expression of all the genes.
Taken together, the results of this work suggest that hMSC cultures were genetically stable up to passage 4. Until then, not only was it possible to obtain clinical grade expansion for clinical applications, but use of the cells would appear to be a safe and effective tool for cell therapy. However, use of cells from more advanced culture passages would seem to have to be analysed on a case‐by‐case basis.
Supporting information
Fig. S1. (a) Networks related to genes downregulated in hMSC culture passage 5 (P5), (b) Networks related to genes overexpressed in hMSC culture passage 5 (P5)
Fig. S2. (a) Networks related to genes downregulated in hMSC culture passage 8 (P8) (b) Networks related to genes overexpressed in hMSC culture passage 8 (P8)
Acknowledgements
This work was supported by grants from CNPq, FINEP and FAPERJ.
References
- 1. Kassem M, Abdallah BM (2008) Human bone‐marrow‐derived mesenchymal stem cells: biological characteristics and potential role in therapy of degenerative diseases. Cell Tissue Res. 331, 157–163. [DOI] [PubMed] [Google Scholar]
- 2. Bolland BJ, Tilley S, New AM, Dunlop DG, Oreffo RO (2007) Adult mesenchymal stem cells and impaction grafting: a new clinical paradigm shift. Expert Rev. Med. Devices 4, 393–404. [DOI] [PubMed] [Google Scholar]
- 3. Abdel‐Latif A, Bolli R, Tleyjeh IM, Montori VM, Perin EC, Hornung CA et al (2007) Adult bone marrow‐derived cells for cardiac repair: a systematic review and meta‐analysis. Arch. Intern. Med. 167, 989–997. [DOI] [PubMed] [Google Scholar]
- 4. Ringdén O, Uzunel M, Rasmusson I, Remberger M, Sundberg B, Lönnies H et al (2006) Mesenchymal stem cells for treatment of therapy‐resistant graft‐versus‐host disease. Transplantation 81, 1390–1397. [DOI] [PubMed] [Google Scholar]
- 5. Giordano A, Galderisi U, Marino IR (2007) From the laboratory bench to the patient's bedside: an update on clinical trials with mesenchymal stem cells. J. Cell. Physiol. 211, 27–35. [DOI] [PubMed] [Google Scholar]
- 6. Wagner W, Wein F, Roderburg C, Saffrich R, Diehlmann A, Eckstein V et al (2008) Adhesion of human hematopoietic progenitor cells to mesenchymal stromal cells involves CD44. Cells Tissues Organs 188, 160–169. [DOI] [PubMed] [Google Scholar]
- 7. Rubio D, Garcia‐Castro J, Martín MC, de la Fuente R, Cigudosa JC, Lloyd AC et al (2005) Spontaneous human adult stem cell transformation. Cancer Res. 65, 3035–3039. [DOI] [PubMed] [Google Scholar]
- 8. Lepperdinger G, Brunauer R, Jamnig A, Laschober G, Kassem M (2008) Controversial issue: is it safe to employ mesenchymal stem cells in cell‐based therapies? Exp. Gerontol. 43, 1018–1023. [DOI] [PubMed] [Google Scholar]
- 9. Sensebé L, Bourin P (2009) Mesenchymal stem cells for therapeutic purposes. Transplantation 15(9 Suppl), S49–S53. Review. [DOI] [PubMed] [Google Scholar]
- 10. Izadpanah R, Kaushal D, Kriedt C, Tsien F, Patel B, Dufour J et al (2008) Long‐term in vitro expansion alters the biology of adult mesenchymal stem cells. Cancer Res. 68, 4229–4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Foudah D, Redaelli S, Donzelli E, Bentivegna A, Miloso M, Dalprà L et al (2009) Monitoring the genomic stability of in vitro cultured rat boné‐marrow‐derived mesenchymal stem cells. Chromosome Res. 17, 1025–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Furlani D, Li W, Pittermann E, Klopsch C, Wang L, Knopp A et al (2009) A transformed cell population derived from cultured mesenchymal stem cells has no function effect after transplantation into the injured heart. Cell Transplant. 18, 319–331. [DOI] [PubMed] [Google Scholar]
- 13. Zhang ZX, Guan LX, Zhang K, Wang S, Cao PC, Wang YH et al (2007) Cytogenetic analysis of human bone marrow‐derived mesenchymal stem cells passaged in vitro . Cell Biol. Int. 31, 645–648. [DOI] [PubMed] [Google Scholar]
- 14. Bochkov NP, Voronina ES, Kosyakova NV, Liehr T, Rzhaninova AA, Katosova LD et al (2007) Chromosome variability of human multipotent mesenchymal stromal cells. Bull. Exp. Biol. Med. 143, 122–126. [DOI] [PubMed] [Google Scholar]
- 15. Bernardo ME, Zaffaroni N, Novara F, Cometa AM, Avanzini MA, Moretta A et al (2007) Human bone marrow derived mesenchymal stem cells do not undergo transformation after long‐term in vitro culture and do not exhibit telomere maintenance mechanisms. Cancer Res. 67, 9142–9149. [DOI] [PubMed] [Google Scholar]
- 16. Tarte K, Gaillard J, Lataillade JJ, Fouillard L, Becker M, Mossafa H et al (2010) Clinical‐grade production of human mesenchymal stromal cells: occurrence of aneuploidy without transformation. Blood 25, 1549–1553. [DOI] [PubMed] [Google Scholar]
- 17. Ohgushi H, Goldberg VM, Caplan AI. (1989) Repair of bone defects with marrow cells and porous ceramic. Experiments in rats. Acta Orthop Scand. 60, 334–339. [DOI] [PubMed] [Google Scholar]
- 18. Bruder SP, Fink DJ, Caplan AI. (1994) Mesenchymal stem cells in bone development, bone repair, and skeletal regeneration therapy. J. Cell. Biochem. 56, 283–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Caplan AI. (2009) Why are MSCs therapeutic? New data: new insight J Pathol. 217, 318–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Meirelles Lda S, Fontes AM, Covas DT, Caplan AI. (2009) Mechanisms involved in the therapeutic properties of mesenchymal stem cells. Cytokine Growth Factor Rev. 20, 419–427. [DOI] [PubMed] [Google Scholar]
- 21. Battiwalla M, Hematti P (2009) Mesenchymal stem cells in hematopoietic stem cell transplantation. Cytotherapy 11, 503–515. Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dominici M, Le Blanc K, Mueller I, Slaper‐Cortenbach I, Marini F, Krause D et al (2006) E. Minimal criteria for defining multipotent mesenchymal stromal cells: The International Society for Cellular Therapy position statement. Cytotherapy 8, 315–317. [DOI] [PubMed] [Google Scholar]
- 23. Livak KJ, Schmittgen TD. (2001) Analysis of relative gene expression data using real‐time quantitative PCR and the 2(‐Delta Delta C(T)) Method. Methods 25, 402–408. [DOI] [PubMed] [Google Scholar]
- 24. Partek Inc . Partek® Discovery SuiteTM 2008. Version: revision 6.3. St. Louis. [Google Scholar]
- 25. Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein‐dye binding. Anal Biochem. 72, 248–254. [DOI] [PubMed] [Google Scholar]
- 26. Park HW, Shin JS, Kim CW (2007) Proteome of mesenchymal stem cells. Proteomics 7, 2881–2894. [DOI] [PubMed] [Google Scholar]
- 27. Wagner W, Feldmann RE Jr, Seckinger A, Maurer MH, Wein F, Blake J et al (2006) The heterogeneity of human mesenchymal stem cell preparations–evidence from simultaneous analysis of proteomes and transcriptomes. Exp. Hematol. 34, 536–548. [DOI] [PubMed] [Google Scholar]
- 28. Feldmann RE Jr, Bieback K, Maurer MH, Kalenka A, Bürgers HF, Gross B et al (2005) Stem cell proteomes: a profile of human mesenchymal stem cells derived from umbilical cord blood. Electrophoresis 26, 2749–2758. [DOI] [PubMed] [Google Scholar]
- 29. Van Hoof D, Passier R, Ward‐Van Oostwaard D, Pinkse MW, Heck AJ, Mummery CL et al (2006) A quest for human and mouse embryonic stem cell‐specific proteins. Mol. Cell. Proteomics 5, 1261–1273. [DOI] [PubMed] [Google Scholar]
- 30. Muta K, Krantz SB, Bondurant MC, Wickrema A (1994) Distinct roles of erythropoietin, insulin‐like growth factor I, and stem cell factor in the development of erythroid progenitor cells. J. Clinical Investigation 94, 34–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hirano K, Shishido‐Hara Y, Kitazawa A, Kojima K, Sumiishi A, Umino M et al (2008) Expression of stem cell factor (SCF), a KIT ligand, in gastrointestinal stromal tumors (GISTs): A potential marker for tumor proliferation. Pathology – Research and Practice 204, 799–807. [DOI] [PubMed] [Google Scholar]
- 32. Levina V, Marrangoni A, Wang T, Parikh S, Su Y, Herberman R et al (2010) Elimination of human lung cancer stem cells through targeting of the stem cell factor‐c‐kit autocrine signaling loop. Cancer Res. 70, 338–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wagner W, Horn P, Castoldi M, Diehlmann A, Bork S, Saffrich R et al (2008) Replicative senescence of mesenchymal stem cells: a continuous and organized process. PLoS One 21, e2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lazzarotto‐Silva C, Binato R, Rocher BD, Costa JA, Pizzatti L, Bouzas LF et al (2009) Similar proteomic profiles of human mesenchymal stromal cells from different donors. Cytotherapy 11, 268–277. [DOI] [PubMed] [Google Scholar]
- 35. Le Blanc K, Samuelsson H, Gustafsson B, Remberger M, Sundberg B, Arvidson J et al (2007) Transplantation of mesenchymal stem cells to enhance engraftment of hematopoietic stem cells. Leukemia 21, 1733–1738. [DOI] [PubMed] [Google Scholar]
- 36. Caplan A.I (2007) Adult mesenchymal stem cells for tissue engineering versus regenerative medicine. J. Cell. Physiol. 213, 341–347. [DOI] [PubMed] [Google Scholar]
- 37. Wagner W, Bork S, Lepperdinger G, Joussen S, Ma N, Strunk D et al (2010) How to track cellular aging of mesenchymal stem cells? Aging 2, 224–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ishidate T, Matsumine A, Toyoshima K, Akiyama T. (2000) The APC‐hDLG complex negatively regulates cell cycle progression from the G0/G1 to S phase. Oncogene 19, 365–372. [DOI] [PubMed] [Google Scholar]
- 39. Machida T, Fujita T, Ooo ML, Ohira M, Isogai E, Mihara M et al (2006) Increased expression of proapoptotic BMCC1, a novel gene with the BNIP2 and Cdc42GAP homology (BCH) domain, is associated with favorable prognosis in human neuroblastomas. Oncogene 25, 1931–1942. [DOI] [PubMed] [Google Scholar]
- 40. Kibar Z, Torban E, McDearmid JR, Reynolds A, Berghout J, Mathieu M et al (2007) Mutations in VANGL1 associated with neural‐tube defects. N Engl J Méd. 356, 1432–1437. [DOI] [PubMed] [Google Scholar]
- 41. Salagierski M, Verhaegh GW, Jannink SA, Smit FP, Hessels D, Schalken JA. (2010) Differential expression of PCA3 and its overlapping PRUNE2 transcript in prostate cancer. Prostate 70, 70–78. [DOI] [PubMed] [Google Scholar]
- 42. Vázquez‐Ulloa E, Lizano M, Avilés‐Salas A, Alfaro‐Moreno E, Contreras‐Paredes A. (2011) Abnormal distribution of hDlg and PTEN in premalignant lesions and invasive cervical cancer. Gynecol. Oncol. 122, 663–668. [DOI] [PubMed] [Google Scholar]
- 43. Anastas JN, Biechele TL, Robitaille M, Muster J, Allison KH, Angers S et al (2011) A protein complex of SCRIB, NOS1AP and VANGL1 regulates cell polarity and migration, and is associated with breast cancer progression. Oncogene 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mejhert N, Galitzky J, Pettersson AT, Bambace C, Blomqvist L, Bouloumié A et al (2010) Mapping of the fibroblast growth factors in human white adipose tissue. J. Clin. Endocrinol. Metab. 95, 2451–2457. [DOI] [PubMed] [Google Scholar]
- 45. Yang B, Guo H, Zhang Y, Chen L, Ying D, Dong S. (2011) MicroRNA‐145 regulates chondrogenic differentiation of mesenchymal stem cells by targeting Sox9. PLoS ONE 6, e21679. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. (a) Networks related to genes downregulated in hMSC culture passage 5 (P5), (b) Networks related to genes overexpressed in hMSC culture passage 5 (P5)
Fig. S2. (a) Networks related to genes downregulated in hMSC culture passage 8 (P8) (b) Networks related to genes overexpressed in hMSC culture passage 8 (P8)


