Abstract
Objectives
miRNAs play crucial roles in human tumourigenesis. This study was performed to measure expression and function of miR‐762 in breast cancer.
Materials and methods
Expression of miR‐762 in breast tissues and cell lines (SK‐BR‐3, DA‐MB‐435s, MCF‐7 and MDA‐MB‐231, HBL‐100) was measured by using real‐time RT‐PCR. We restored expression of miR‐762 in MCF‐7 cells to measure its functional roles. Luciferase assays were performed to reveal the target gene of miR‐762.
Results
Expression of miR‐762 was high in both breast cancer cell lines and specimens, and its overexpression increased breast cancer cell proliferation and invasion. Interferon regulatory factor 7 (IRF7) is a direct target of miR‐762 and overexpression of miR‐762 reduced expression of IRF7. Moreover, IRF7 was repressed, its levels inversely correlated to miR‐762 expression. IRF7 rescued miR‐762‐induced cell invasion and proliferation.
Conclusions
These results demonstrate that miR‐762 tumour effect was achieved by targeting IRF7 in human breast cancer specimens.
Introduction
Breast cancer is a leading malignant tumour in women of developed countries, and an alarming increase in its incidence has now been shown in developing countries too 1, 2, 3. Aggressive breast cancers have a high potential to become metastatic, a transition that makes clinical intervention difficult 4, 5. In recent years, although systemic treatments have improved outcomes, prognosis for patients with breast cancer still remains poor 6, 7, 8. Thus, it is important to find new therapeutic targets for this malignancy.
MicroRNAs (miRNAs) are a class of endogenous, typically 18–25 nucleotides long, non‐coding single RNAs, highly conserved and endogenously expressed, across many species 9, 10, 11. They regulate other types of gene expression at post‐transcriptional levels, and are aberrantly expressed in numerous human malignancies 12, 13, 14. Previous studies have shown that miRNAs play critical roles in biological events, such as cell proliferation, invasion, migration and apoptosis, glucose and lipid metabolism, and infection and immune responses 15, 16, 17, 18, 19. In addition, it has been well established that miRNAs participate in tumourigenesis, yet might also act as tumour suppressors or oncogenically 20, 21, 22.
The aim of our study was to investigate biological roles of miR‐762 in breast cancer. We also explored its potential application as a therapeutic target for breast cancer patients.
Materials and methods
Clinical breast cancer samples
Surgical tissues (both tumour and adjacent non‐tumour specimens) were collected from patients who underwent surgery between 2013 and 2014 at our department. All patients had provided written informed consent and the study was approved by the ethical board of the institute of Xijing Hospital, complied according to the Declaration of Helsinki. Tissues were frozen immediately in liquid nitrogen until use.
Cell lines and culture
Human breast cancer cell lines SK‐BR‐3, DA‐MB‐435s, MCF‐7 and MDA‐MB‐231 as well as the normal breast cell line, HBL‐100, were cultured in RPMI‐1640 medium. HBL‐100, MDA‐MB‐435s and SK‐BR‐3 were purchased from the Institute of Biochemistry and Cell Biology (Shanghai, China) and MDA‐MB‐231 and MCF‐7 were obtained from the Cell Center at the Xiangya School of Medicine (Changsha, China).
Oligonucleotide transfection
miR‐762 mimics, inhibitor and their controls were bought from GenePharma (Shanghai, China) and transfected with Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA), in MCF‐7 cells, following the manufacturer's instructions.
RNA isolation and quantitative real‐time PCR
Total RNA was extracted using TRIzol Reagent (Invitrogen) and miR‐762 quantification was measured using TaqMan miRNA Assay (Applied Biosystems, Foster City, CA, USA). Expression of IRF7 was measured by qRT‐PCR; U6 snRNA being used as control for miR‐762. Expression level of GAPDH was used as control for IRF7 expression (Table S1).
Luciferase reporter assay
The entire 3′‐UTR of IRF7 was amplified using PCR and PCR products were inserted into the p‐MIR‐reporter plasmid (Ambion, Austin, TX, USA). For the luciferase reporter assay, MCF‐7 cells were cultured in 24‐well plates and transfected with luciferase reporter plasmid, miR‐762 mimic or scrambled, using lipofectamine 2000 (Invitrogen). 24 h after transfection, cells were measured for relative luciferase activity using luciferase kits (Promega, Madison, WI, USA).
Cell proliferation and invasion assay
Cell Counting Kit‐8 (CCK‐8, Dojindo) was used to measure cell viability, following the manufacturer's protocol. For invasion, Matrigel (1 mg/ml) was added to media without serum, and coated on membranes (BD Biosciences, San Jose, CA, USA). Cells were plated in medium without serum or growth factors, supplemented with serum as chemoattractant in lower chambers. Cells from lower surfaces of membranes were stained with crystal violet and counted.
Western blotting
Total Protein Extraction Kit (KeyGen, Nanjing, China) was used to extract total protein and western blotting was performed as previously described 23. Primary antibodies were rabbit polyclonal anti‐IRF7 (Santa Cruz Biotechnology, Santa Cruz, CA, USA) and mouse monoclonal anti‐GAPDH (Cell Signaling Technology, Beverly, MA, USA).
Statistical analysis
Data are displayed as mean ± SD. Correlation between miR‐762 and IRF7 expression was compared using Pearson's correlation method. Significant differences were assessed using Student's t‐test. P < 0.05 was considered significant.
Results
miR‐762 expression was up‐regulated in the breast cancer cell lines and tissues
qRT‐PCR analysis indicated that miR‐762 expression was up‐regulated in the human breast cancer tissues compared to their normal tissue counterparts (Fig. 1a–c). Moreover, we also found that expression of miR‐762 was higher in human breast cancer cell lines (SK‐BR‐3, DA‐MB‐435s, MCF‐7 and MDA‐MB‐231) compared to the normal breast cell line (HBL‐100) (Fig. 1d).
Figure 1.
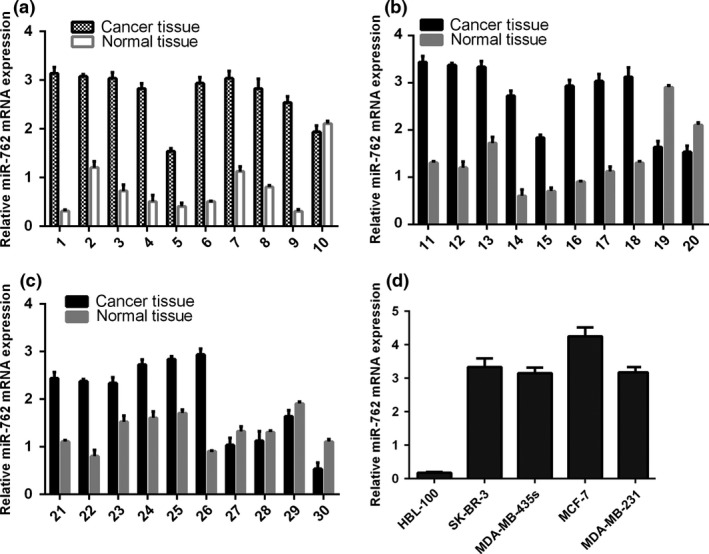
miR‐762 expression up‐regulated in both human breast cancer tissues and cell lines. (a) qRT‐PCR analysis of miR‐762 expression in human breast cancer tissues and distant non‐tumour tissues. (b) Expression of miR‐762 was measured using qRT‐PCR. (c) Expression of miR‐762 was measured using qRT‐PCR. (d) qRT‐PCR analysis of miR‐762 expression in four human breast cancer cell lines and a normal human breast epithelial cell line. Level of miR‐762 expression was normalized to U6.
Overexpression of miR‐762 promoted breast cancer cell proliferation and invasion
miR‐762 expression increased in transfected MCF‐7 cells using miR‐762 mimics (Fig. 2a), and decreased in transfected MCF‐7 cells, by using the miR‐762 inhibitor (Fig. 2b). CCK‐8 viability assay showed that population growth was higher in MCF‐7 cells transfected with miR‐762 mimics compared to cells transfected with scramble mimics or were untreated (Fig. 2c). Meanwhile, miR‐762 inhibitor reduced proliferation of the MCF‐7 cells (Fig. 2d). Invasiveness of cells transfected with miR‐762 mimics was dramatically increased compared to the scramble and control groups of cells and invasiveness of cells transfected with miR‐762 inhibitor was reduced compared to scramble and control group cells (Fig. 2e).
Figure 2.
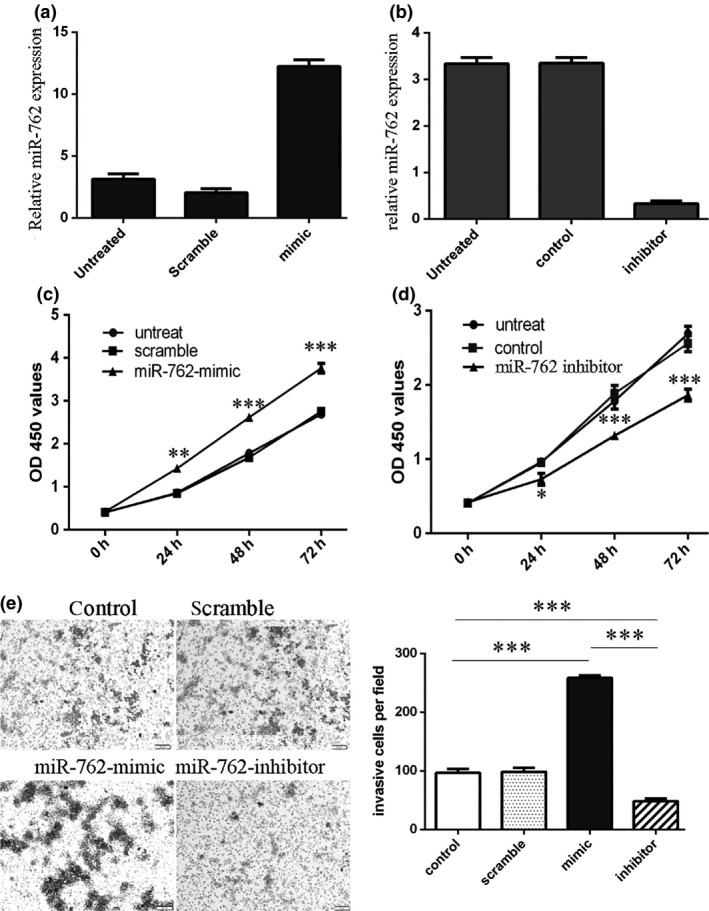
Overexpression of miR‐762 promoted breast cancer cell proliferation and invasion. (a) qRT‐PCR analysis of miR‐762 in MCF‐7 cells on transfection of miR‐762. Expression of miR‐762 in MCF‐7 cells transfected with miR‐762 mimics was up‐regulated. U6 snRNA was used as internal control. (b) Expression of miR‐762 in MCF‐7 cells transfected with miR‐762 inhibitor was down‐regulated. U6 snRNA was used as internal control. (c) CCK‐8 cell viability assay showed that miR‐762 mimics promoted proliferation of MCF‐7 cells. (d) miR‐762 inhibitor reduced proliferation of MCF‐7 cells. (e) Invasion analysis of MCF‐7 cells after treatment with miR‐762 mimics, inhibitors or scramble or control; relative ratio of invasive cells per field is shown below, *P < 0.05, **P < 0.01, and ***P < 0.001.
IRF7 was a direct target of miR‐762 in the breast cancer cells
IRF7 was one of the predicted miR‐762 targets whose 3′UTR contained putative miR‐762 target sites (Fig. 3a). The luciferase reporter assay result showed that relative luciferase activity of the reporter (which contains wildtype 3′UTR of IRF7) was reduced in the miR‐762 group compared to the scramble group. However, relative luciferase activity of mutant IRF7 3′UTR reporter showed no significant difference from the control group (Fig. 3b). In MCF‐7 cells transfected by miR‐762 mimics, IRF7 was reduced at both mRNA (Fig. 3c) and protein levels (Fig. 3d) compared to control cells.
Figure 3.
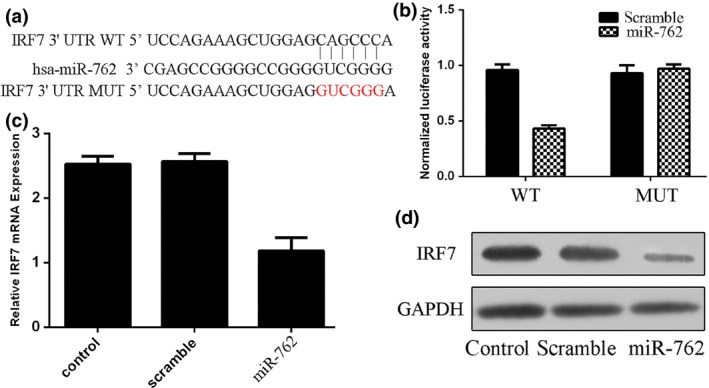
IRF 7 was a direct target of miR‐762 in the breast cancer cells. (a) Targetscan prediction of miR‐762 targeting of IRF7 at the IRF7 mRNA 3′‐UTR. (b) miR‐762‐mediated directly inhibitory effects of IRF7 by luciferase reporter assay, in which expression of the reporter containing IRF7 3′UTR was suppressed by miR‐762, but not in the mutated construct. (c) miR‐762‐mediated suppression of IRF7 mRNA levels in MCF‐7 cells and analysed by qRT‐PCR. (d) Western blot analysis showing that miR‐762 inhibited protein expression of IRF7 in MCF‐7 cells.
Up‐regulation of miR‐762 highly correlated with down‐regulation of IRF7 expression
Compared to their non‐tumour counterparts, expression of IRF7 protein was higher in tumour tissues from four different patients (Fig. 4a,b). mRNA and protein expression of IRF7 was lower in the malignant cell lines (SK‐BR‐3, DA‐MB‐435s, MCF‐7 and MDA‐MB‐231) compared to normal breast cell lines (HBL‐100) (Fig. 4c,d). qRT‐PCR analysis showed that IRF7 expression was up‐regulated in all breast cancer cases compared to normal tissues (Fig. 4e). Furthermore, between pairs of breast cancer tissues, there was statistically significant inverse correlation between IRF7 and miR‐762 expression (Fig. 4f).
Figure 4.
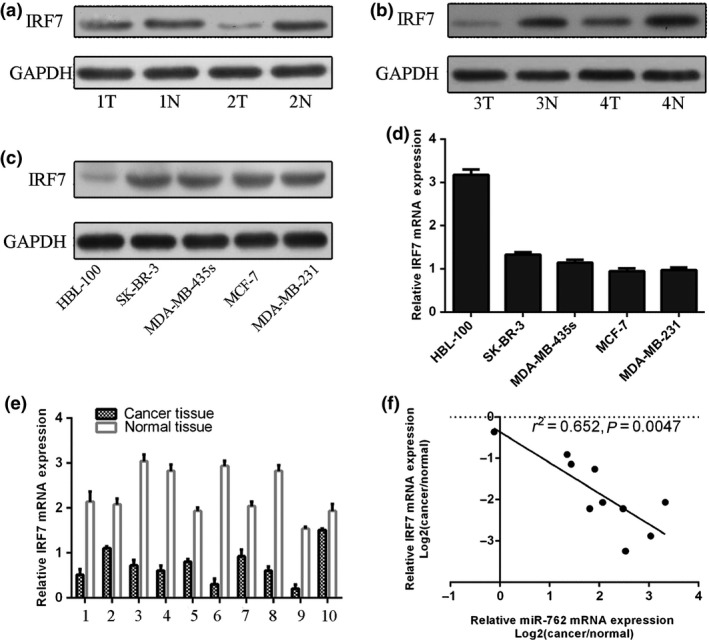
Up‐regulation of miR‐762 highly correlated with down‐regulation of IRF 7 expression in human breast cancer tissues. (a) and (b) IRF7 protein was analysed by western blotting in four pairs of breast cancer tissues. (c) Western blot analysis of IRF7 protein expression in four human breast cancer cell lines and a normal human breast epithelial cell line. GAPDH was also detected as a loading control. (d) qRT‐PCR analysis of IRF7 mRNA expression in four human breast cancer cell lines and a normal human breast epithelial cell line. (e) qRT‐PCR analysis of IRF7 expression in 10 human breast cancer tissues and distant non‐tumour tissues. (f) Inverse correlation in breast cancer tissues was analysed by Pearson's correlation method.
miR‐762‐mediated inhibition of IRF7 was involved in tumour invasion of the breast cancer cells
Ectopic expression of IRF7 enhanced IRF7 protein expression and miR‐762 mimic inhibited part of the function of IRF7 cDNA plasmid (Fig. 5a). The CCK‐8 cell viability assay and the invasion assay showed that miR‐762 mimics promoted cell proliferation and invasion. When miR‐762 and pcDNA‐IRF7 were co‐transfected into MCF‐7 cells, miR‐762 expression reduced IRF7‐induced breast cell proliferation and invasion (Fig. 5b,c).
Figure 5.
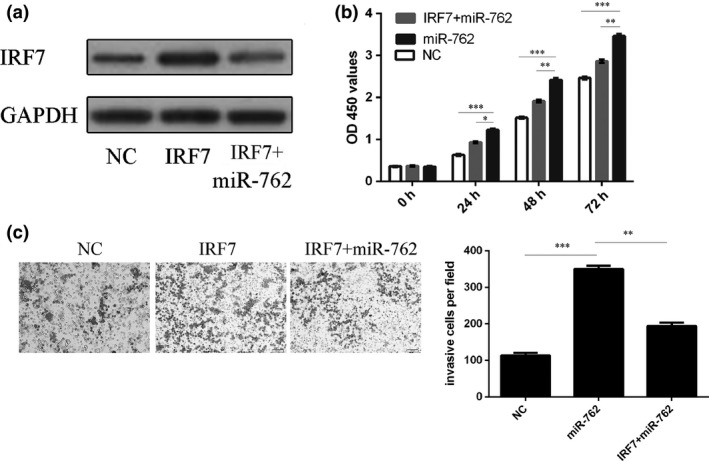
miR‐762‐mediated inhibition of IRF 7 was involved in tumour invasion of breast cancer cells. (a) Western blotting was performed to examine effects of pCDNA‐IRF7 on expression of IRF7. GAPDH was also detected as loading control. (b) Cell growth of MCF‐7 cells co‐transfected with either miR‐762 mimic and 2.0 μg pCDNA‐IRF7 or pCDNA empty vector using CCK‐8 viability assay. (c) Invasion of MCF‐7 cells co‐transfected with either miR‐762 mimic and 2.0 μg pCDNA‐IRF7 or pCDNA empty vector using invasion assay. *P < 0.05, **P < 0.01, and ***P < 0.001.
Discussion
miRNAs are a group of endogenous, small (18–25‐nucleotide long) and non‐coding RNA molecules that regulate the expression of specific mRNAs by either translational inhibition or mRNA degradation 24, 25, 26, 27. More than 50% miRNAs are reported to be located in cancer‐associated genomic break points and function as tumour suppressors or oncogenically 28, 29, 30, 31. Our current study provides the first evidence that miR‐762 promotes proliferation and invasion of breast cancer cells. Expression of miR‐762 was significantly higher in breast cancer cell lines and clinical breast cancer tissues, indicating its possible involvement in cancer development. We subsequently confirmed that miR‐762 significantly promoted breast cancer cell proliferation and invasion in vitro. Our research to unravel biological roles of miR‐762 in breast cancer development identified IRF7 as a downstream target. We revealed that IRF7 was down‐regulated in breast cancer cell lines, and exogenous miR‐762 down‐regulated IRF7 protein and mRNA levels. Furthermore, luciferase reporter assays revealed that miR‐762 directly targetted the 3′‐UTR of IRF7 mRNA. Our findings suggest that miR‐762 has a fundamental role in breast cancer tumourigenesis through affecting cancer cell proliferation and invasion.
Previous study has shown that miR‐762 is up‐regulated in human corneal epithelial cells in response to tear fluid and Pseudomonas aeruginosa antigens, and genes coding for RNase7 and ST2 have been identified as target genes of miR‐762 28. Moreover, miR‐762 suppresses expression of IFITM5 at the translational level in Saos‐2 cells, and probably contributes to progression of bone mineralization 32. However, the role of miR‐762 in breast cancer has remained unclear. In our study, miR‐762 was frequently up‐regulated in breast tumour tissues and cell lines; its overexpression enhanced breast cancer cell proliferation and invasion. Ability of miR‐762 to promote cell proliferation and invasion was confirmed by both overexpression and down‐regulation experiments. Thus, our results suggested miR‐762 to be a novel oncogenic RNA in breast cancer.
To explore the molecular mechanisms by which miR‐762 enhances breast cancer cell line growth and invasion, IRF7 was identified as a direct target of miR‐762 in breast cancer cells. First, the complementary sequence of miR‐762 was identified in the 3′UTR of IRF7 mRNA. Second, miR‐762 reduced IRF7 mRNA and protein levels in the cells. Third, up‐regulation of miR‐762 reduced activity of a luciferase reporter containing the 3′UTR sequence of IRF7. More importantly, mRNA levels of miR‐762 inversely correlated with IRF7 levels in the breast cancer tissues and IRF7 rescued miR‐329‐induced promotion of cell invasion and proliferation. These data indicate that miR‐762, by down‐regulating IRF7, promoted breast cancer cell proliferation and invasion in our experiments.
In conclusion, although further work is required to fully understand the mechanisms by which miR‐762 influences tumour metastasis, identification of miR‐762 as a regulator of tumour cell proliferation and invasion suggests it may play an important role in mediating oncogenesis of breast cancer. In this way, miR‐762 may become a favoured therapeutic target for therapy of breast cancer.
Supporting information
Table S1 Primer sequence
Yongping Li and Ruixue Huang are co‐first authors.
References
- 1. Wang Z, Wang N, Liu P, Chen Q, Situ H, Xie T et al (2014) MicroRNA‐25 regulates chemoresistance‐associated autophagy in breast cancer cells, a process modulated by the natural autophagy inducer isoliquiritigenin. Oncotarget 5, 7013–7026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wang PY, Gong HT, Li BF, Lv CL, Wang HT, Zhou HH et al (2013) Higher expression of circulating miR‐182 as a novel biomarker for breast cancer. Oncol. Lett. 6, 1681–1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zhao Y, Li Y, Lou G, Zhao L, Xu Z, Zhang Y et al (2012) MiR‐137 targets estrogen‐related receptor alpha and impairs the proliferative and migratory capacity of breast cancer cells. PLoS ONE 7, e39102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kulkarni S, Augoff K, Rivera L, McCue B, Khoury T, Groman A et al (2012) Increased expression levels of WAVE3 are associated with the progression and metastasis of triple negative breast cancer. PLoS ONE 7, e42895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Soria‐Valles C, Gutierrez‐Fernandez A, Guiu M, Mari B, Fueyo A, Gomis RR et al (2013) The anti‐metastatic activity of collagenase‐2 in breast cancer cells is mediated by a signaling pathway involving decorin and miR‐21. Oncogene 33, 3054–3063. [DOI] [PubMed] [Google Scholar]
- 6. Ma L, Teruya‐Feldstein J, Weinberg RA (2007) Tumour invasion and metastasis initiated by microRNA‐10b in breast cancer. Nature 449, 682–688. [DOI] [PubMed] [Google Scholar]
- 7. Jovanovic J, Ronneberg JA, Tost J, Kristensen V (2010) The epigenetics of breast cancer. Mol. Oncol. 4, 242–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Liu Y, Zhao J, Zhang PY, Zhang Y, Sun SY, Yu SY et al (2012) MicroRNA‐10b targets E‐cadherin and modulates breast cancer metastasis. Med. Sci. Monit. 18: BR299–BR308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Schirmer U, Doberstein K, Rupp AK, Bretz NP, Wuttig D, Kiefel H et al (2014) Role of miR‐34a as a suppressor of L1CAM in endometrial carcinoma. Oncotarget 5, 462–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bier A, Giladi N, Kronfeld N, Lee HK, Cazacu S, Finniss S et al (2013) MicroRNA‐137 is downregulated in glioblastoma and inhibits the stemness of glioma stem cells by targeting RTVP‐1. Oncotarget 4, 665–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ohno M, Otsuka M, Kishikawa T, Shibata C, Yoshikawa T, Takata A et al (2014) Specific delivery of microRNA93 into HBV‐replicating hepatocytes downregulates protein expression of liver cancer susceptible gene MICA. Oncotarget 5, 5581–5590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhang C, Liu J, Wang X, Wu R, Lin M, Laddha SV et al (2014) MicroRNA‐339‐5p inhibits colorectal tumorigenesis through regulation of the MDM2/p53 signaling. Oncotarget 5, 9106–9117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wu W, He X, Kong J, Ye B (2012) Mir‐373 affects human lung cancer cells' growth and its E‐cadherin expression. Oncol. Res. 20, 163–170. [DOI] [PubMed] [Google Scholar]
- 14. Xiong X, Ren HZ, Li MH, Mei JH, Wen JF, Zheng CL (2011) Down‐regulated miRNA‐214 induces a cell cycle G1 arrest in gastric cancer cells by up‐regulating the PTEN protein. Pathol. Oncol. Res. 17, 931–937. [DOI] [PubMed] [Google Scholar]
- 15. Fei B, Wu H (2013) MiR‐378 inhibits progression of human gastric cancer MGC‐803 cells by targeting MAPK1 in vitro. Oncol. Res. 20, 557–564. [DOI] [PubMed] [Google Scholar]
- 16. Wang Z, Yin B, Wang B, Ma Z, Liu W, Lv G (2014) MicroRNA‐210 promotes proliferation and invasion of peripheral nerve sheath tumor cells targeting EFNA3. Oncol. Res. 21, 145–154. [DOI] [PubMed] [Google Scholar]
- 17. Zhang WH, Gui JH, Wang CZ, Chang Q, Xu SP, Cai CH (2012) The identification of miR‐375 as a potential biomarker in distal gastric adenocarcinoma. Oncol. Res. 20, 139–147. [DOI] [PubMed] [Google Scholar]
- 18. Misawa A, Katayama R, Koike S, Tomida A, Watanabe T, Fujita N (2010) AP‐1‐Dependent miR‐21 expression contributes to chemoresistance in cancer stem cell‐like SP cells. Oncol. Res. 19, 23–33. [DOI] [PubMed] [Google Scholar]
- 19. Ohdaira H, Sekiguchi M, Miyata K, Yoshida K (2012) MicroRNA‐494 suppresses cell proliferation and induces senescence in A549 lung cancer cells. Cell Prolif. 45, 32–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li J, You T, Jing J (2014) MiR‐125b inhibits cell biological progression of Ewing's sarcoma by suppressing the PI3K/Akt signalling pathway. Cell Prolif. 47, 152–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li M, Yu M, Liu C, Zhu H, He X, Peng S (2013) miR‐34c works downstream of p53 leading to dairy goat male germline stem‐cell (mGSCs) apoptosis. Cell Prolif. 46, 223–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Huang J, Zhang SY, Gao YM, Liu YF, Liu YB, Zhao ZG et al (2014) MicroRNAs as oncogenes or tumour suppressors in oesophageal cancer: potential biomarkers and therapeutic targets. Cell Prolif. 47, 277–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li Z, Shen J, Wu WK, Yu X, Liang J, Qiu G et al (2012) Leptin induces cyclin D1 expression and proliferation of human nucleus pulposus cells via JAK/STAT, PI3K/Akt and MEK/ERK pathways. PLoS ONE 7, e53176. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 24. Zhou J, Wang W, Gao Z, Peng X, Chen X, Chen W et al (2013) MicroRNA‐155 promotes glioma cell proliferation via the regulation of MXI1. PLoS ONE 8, e83055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hui W, Yuntao L, Lun L, WenSheng L, ChaoFeng L, HaiYong H et al (2013) MicroRNA‐195 inhibits the proliferation of human glioma cells by directly targeting cyclin D1 and cyclin E1. PLoS ONE 8, e54932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rao SA, Arimappamagan A, Pandey P, Santosh V, Hegde AS, Chandramouli BA et al (2013) miR‐219‐5p inhibits receptor tyrosine kinase pathway by targeting EGFR in glioblastoma. PLoS ONE 8, e63164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chiang CH, Hou MF, Hung WC (2013) Up‐regulation of miR‐182 by beta‐catenin in breast cancer increases tumorigenicity and invasiveness by targeting the matrix metalloproteinase inhibitor RECK. Biochim. Biophys. Acta 1830, 3067–3076. [DOI] [PubMed] [Google Scholar]
- 28. Mun J, Tam C, Chan G, Kim JH, Evans D, Fleiszig S (2013) MicroRNA‐762 is upregulated in human corneal epithelial cells in response to tear fluid and Pseudomonas aeruginosa antigens and negatively regulates the expression of host defense genes encoding RNase7 and ST2. PLoS ONE 8, e57850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Morishita A, Masaki T (2014) miRNA in hepatocellular carcinoma. Hepatol. Res. 45, 128–141. [DOI] [PubMed] [Google Scholar]
- 30. Volinia S, Galasso M, Costinean S, Tagliavini L, Gamberoni G, Drusco A et al (2010) Reprogramming of miRNA networks in cancer and leukemia. Genome Res. 20, 589–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lee YM, Lee JY, Ho CC, Hong QS, Yu SL, Tzeng CR et al (2011) miRNA‐34b as a tumor suppressor in estrogen‐dependent growth of breast cancer cells. Breast Cancer Res. 13, R116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mo X, Lu Y, Han J (2014) Effects of targeted modulation of miR‐762 on expression of the IFITM5 gene in Saos‐2 cells. Intractable Rare Dis. Res. 3, 12–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 Primer sequence


