Abstract
Aetiology of intervertebral disc degeneration (IDD) is complex, with genetic, developmental, biochemical and biomechanical factors contributing to the disease process. It is becoming obvious that epigenetic processes influence evolution of IDD as strongly as the genetic background. Deregulated phenotypes of nucleus pulposus cells, including differentiation, migration, proliferation and apoptosis, are involved in all stages of progression of human IDD. Non‐coding RNAs, including microRNAs, have recently been recognized as important regulators of gene expression. Research into roles of microRNAs in IDD has been very active over the past 5 years. Our review summarizes current research enlightenment towards understanding roles of microRNAs in regulating nucleus pulposus cell functions in IDD. These exciting findings support the notion that specific modulation of microRNAs may represent an attractive approach for management of IDD.
Introduction
Intervertebral disc degeneration (IDD) is one of the major causes of low back pain, which compose a global burden with severe health care and socioeconomic costs 1, 2, 3. Even though IDD treatment has advanced remarkably over the last decade, many patients still do not achieve sustained remission, constituting a major and unmet clinical need 4. Aetiology of IDD is multifactorial, including genetic predisposition, lifestyles (for example, type of occupation, smoking, alcohol consumption) and aging 1, 5, 6, 7. Underlying IDD molecular and cellular mechanisms are still largely unknown. Thus, increasing numbers of studies support the observation that nucleus pulposus (NP) cells are important in maintaining integrity of intervertebral discs (IVD) by their roles in producing type II collagen, aggrecan and other components of the extracellularmatrix (ECM) 8, 9, 10. There is a growing body of evidence to support that microRNA (miRNA) can regulate many, if not all, aspects of cell activity, from differentiation and proliferation to apoptosis, to mediate their effects on a great range of physiological and pathological processes 11, 12, 13, 14. In this review, we summarize the current knowledge on roles of miRNAs in regulation of NP cell functions, including differentiation, proliferation, ECM synthesis, apoptosis, and the possible implications for IDD.
MicroRNA biogenesis and function
Over the last decade it has become better recognized that small ribonucleic acids (RNAs) are important components of gene regulatory networks 15, 16, 17. Among these, miRNAs are a class of small molecules and non‐coding single‐stranded RNAs of 18–22 nucleotides, that act as post‐transcriptional gene regulatory elements 18, 19, 20. Currently, miRNAs have been found in virtually all species of animals, plants and viruses examined; there are over 2042 mature human miRNA sequences listed in the miRNA registry 21, 22. It is estimated that there are approximately 1500 predicted miRNAs in the human genome that have the potential to regulate at least 20–30% of all human genes 23, 24.
MiRNAs are transcribed from their respective gene loci as primary miRNAs (pri‐miRNA) 25 and their biogenesis consists of a series of maturation steps 26. They can be derived from two sources: (i) Pri‐miRNAs can be transcribed from specific miRNA‐encoding regions of the genome (intergenic miRNA) by RNA polymerase II 27, 28. These pri‐miRNAs are then cleaved in the nucleus by a multiprotein complex, which contains an anchor protein DGCR8 (DiGeorge syndrome critical region gene 8), giving rise to 70‐nucleotide pre‐miRNAs 29, 30. (ii) Pri‐miRNAs can be derived from mRNA intronic sequences (mirtrons), whose maturation does not require Drosha/DGCR8 processing. These miRNAs are debranched and spliced by lariat debranching enzyme (Ldbr) and are co‐transcribed with host protein‐coding genes, forming pre‐miRNA hairpins as above 31, 32. Intriguingly, these intronic miRNAs are usually involved in the same biological pathways as host protein‐coding genes 33.
MiRNAs inhibit gene expression by selectively binding to complementary 3′untranslated regions (3′UTRs) of target mRNAs through complementary base pairing 34, 35. Most identified miRNAs are highly conserved among species 36. Expression of miRNAs has both spatial and temporal specificity, together with tissue and cell type specificity 37. They play crucial roles in diverse pathological conditions, such as in cancer, neurodegeneration and aging 38, 39, 40. Recent findings have revealed that miRNAs can be involved in cell proliferation, differentiation and apoptosis, and are thus involved in broader processes, such as animal development, homeostasis and bone metabolism 41, 42, 43, 44, 45. Recent progress in biology has shown that miRNAs are dysregulated in different cancer types, including gastric cancer, breast cancer, osteosarcoma, lung cancer and hepatocellular carcinoma 46, 47, 48, 49, 50. Thus, miRNAs have considerable potential to become a research focus for prevention and treatment of IDD, specially for targeting IDD‐related cell processes, such as NP cell proliferation and apoptosis.
MiRNAs in intervertebral disc degeneration
MiRNAs and NP cell apoptosis
NP cell death mediated by apoptosis is involved in various deleterious consequences of IDD, such as inflammation, degeneration and ECM degradation 51. NP cell apoptosis may have dual effects on IDD depending on the extent of the cell death 4, 52. On the one hand, NP cell apoptosis can offset effects of aberrant NP cell proliferation during IDD 53. On the other hand, apoptosis can be harmful as NP cells produce interstitial collagen fibres, which are critical for maintaining tensile strength of the fibrous cap 54. Thus, apoptosis of NP cells plays an important role in determining their stability.
MiR‐155 is one of the well‐documented miRNAs involved in regulation of apoptotic pathways and immunological responses 55. Its aberrant expression has been found to be associated with various diseases, such as lung, gastric, pancreatic and colon cancers 56, 57, 58, 59. Wang et al. have shown that 29 miRNAs exhibit significantly differential expression in degenerative NP cells, of which miR‐155 was found to be one of the most down‐regulated 60. Overexpression of miR‐155 can inhibit NP cell apoptosis by repressing FADD and caspase‐3 expression. In situ hybridization and immunohistochemistry have further revealed that miR‐155, when expressed in cytoplasm of human NP cells is inversely correlated with FADD and caspase‐3.
MiR‐27a is a further well‐studied miRNA expressed in diverse tissue types. Its aberrant expression is found to be associated with several diseases, including (once again) colorectal, bladder and gastric cancers 61, 62, 63. Moreover, previous studies have shown that miR‐27a is involved in cell proliferation, apoptosis and tumourigenesis 62, 64. It has been found that it affects apoptotic signalling pathways in initiation and progression of gliomas. Using real‐time RT‐PCR, Liu et al. 65 discovered that expression of miR‐27a is high in degenerative NP cells. Furthermore, its enforced expression inhibits phosphoinositide‐3 kinases (PI3K) expression by directly targeting its 3′‐UTR, and this inhibition is abolished by mutation of miR‐27a binding sites. In short, up‐regulation of miR‐27a initiates apoptosis of NP cells by targeting PI3K.
MiRNAs and NP cell proliferation
Increasing numbers of studies have demonstrated that formation of NP cell clusters and aberrant NP cell proliferation play a crucial role in IDD 8. More evidence has shown that miRNAs play an important role in control of NP cell proliferation by post‐transcriptional regulation of a number of genes 66.
Previous investigations revealed that miR‐10b is involved in regulation of cell proliferation in various cell types, especially in cancer cells, such as breast, liver, pancreatic and gastric cancers 67, 68, 69, 70. It is also deregulated in these cancers and its levels are closely associated with tumour progression and pathological grade 71, 72. In addition, our previous results have shown that miR‐10b is significantly down‐regulated in gastric cancer cell lines and tissues as demonstrated by quantitative real‐time PCR. Overexpression of miR‐10b in MGC‐803 and HGC‐27 cells dramatically suppressed cell proliferation, migration and invasion and induced apoptosis 11. Similar to its roles in cancer, miR‐10b is up‐regulated in degenerative NP tissues and is significantly associated with disc degeneration grade. Moreover, overexpression of miR‐10b significantly increased NP cell proliferation. In addition, miR‐10b promoted proliferation of NP cells by directly targeting HOXD10. MiR‐10b also induces Akt phosphorylation in a RhoC‐dependent manner 73.
MiR‐21, a well‐known miRNA, is most frequently dysregulated in different types of human cancer, such as those of the breast, lung, liver and stomach, and has been shown to be implicated in multiple cell processes, including migration, differentiation, proliferation, apoptosis and invasion 74, 75, 76, 77. Liu et al. reported that miR‐21 was up‐regulated in human degenerative NP tissues compared to normal NP 78. Moreover, enforced expression of miR‐21 promotes NP cell proliferation. In this regard, overexpression of miR‐21 led to increased phosphorylation of Akt by directly targeting PTEN. Furthermore, effects of miR‐21 on NP cell proliferation and cyclin D1 induction in human NP cells can be blocked by Ly294002, an AKT inhibitor.
miRNAs and ECM remodelling of NP cells
The disc is a complex structure highly specialized for mechanical functions such as spine connectivity, flexure, rotation and extension 79. Currently available evidence implicates loss of IVD ECM upon IDD, as a major cause of low back pain 80. During degeneration, IVD matrix undergoes structural, mechanical and molecular changes, which result in loss of demarcation between the outer annulus fibrosus and inner NP tissues 81, 82. ECM is constantly synthesized and degraded by disc cells, during which rates of the processes are normally in balance 83. However, this balance becomes shifted towards degradation in IDD, with alterations in collagen type and reduction in proteoglycan content, leading to loss of tissue integrity 84. Understanding which factors affect ECM changes is important to fully comprehend the possible implications for IDD.
Chen et al. found that ligamentum flavum (LF) thickness and expression of collagens I and III, as well as miR‐155, were higher in LF from lumbar spinal stenosis (LSS) patients than from lumbar disc herniation (LDH) patients 85. To this end, expression level of miR‐155 positively correlated with LF thickness and levels of types I and III collagen. Overexpression of miR‐155 increased mRNA and protein expression of collagens I and III in fibroblasts isolated from LF, while down‐regulated expression of miR‐155 had opposite effects.
Protein kinase C (PKC) signalling, a major regulator of chondrogenic differentiation, has also been implicated in pathological ECM remodelling. Tsirimonaki et al. demonstrated that PKCε activation induced up‐regulation of miR‐377, which was coupled to reductions in ADAMTS5 and cleaved aggrecan 86.
Conclusion
In this review, we summarized roles of miRNAs in function of NP cells and their contributions to IDD (Fig. 1 and Table 1); miRNAs critically affect them. As the aetiology of IDD is multifactorial, these data should improve our insights into pathogenesis of IDD and inform relationships between genetic predisposition and risk factor exposure. Taken together, miRNAs definitively represent an emerging area of research that will provide new insight into IDD pathogenesis with the hope of bringing about novel drug candidates and biomarkers.
Figure 1.
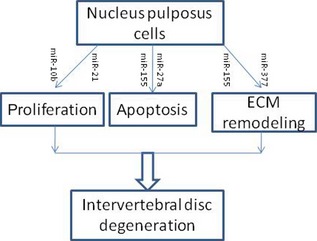
Mi RNA s in intervertebral disc degeneration. MiR‐10b and miR‐21 induce nucleus pulposus (NP) cell proliferation; miR‐155 and miR‐27a inhibit NP cell apoptosis; miR‐155 and miR‐377 are involved in NP cell ECM remodelling. ECM, extracellularmatrix.
Table 1.
The expression and function of miRNAs in intervertebral disc degeneration
| miRNA | Expression | Function | Target | References |
|---|---|---|---|---|
| miR‐155 | Decrease | Apoptosis | FADD, caspase‐3 | 60 |
| miR‐27a | Increase | Apoptosis | PI3K | 65 |
| miR‐10b | Increase | Proliferation | HOXD10 | 73 |
| miR‐21 | Increase | Proliferation | PTEN | 78 |
| miR‐155 | Decrease | ECM remodelling | Collagen I and III | 85 |
| miR‐377 | Increase | ECM remodelling | ADAMTS5 | 86 |
ECM, extracellularmatrix.
Acknowledgements
Funding: This work was supported by the National Natural Science Foundation of China (NSFC) (Grant Numbers: 81401847, 81272053 and 81330044).
Xin Yu and Zheng Li contributed equally to this work.
References
- 1. Masuda K, Oegema TR Jr, An HS (2004) Growth factors and treatment of intervertebral disc degeneration. Spine (Phila Pa 1976) 29, 2757–2769. [DOI] [PubMed] [Google Scholar]
- 2. Millecamps M, Tajerian M, Naso L, Sage EH, Stone LS (2012) Lumbar intervertebral disc degeneration associated with axial and radiating low back pain in ageing SPARC‐null mice. Pain 153, 1167–1179. [DOI] [PubMed] [Google Scholar]
- 3. Raj PP (2008) Intervertebral disc: anatomy‐physiology‐pathophysiology‐treatment. Pain Pract. 8, 18–44. [DOI] [PubMed] [Google Scholar]
- 4. Loreto C, Musumeci G, Castorina A, Martinez G (2011) Degenerative disc disease of herniated intervertebral discs is associated with extracellular matrix remodeling, vimentin‐positive cells and cell death. Ann. Anat. 193, 156–162. [DOI] [PubMed] [Google Scholar]
- 5. Adams MA, Roughley PJ (2006) What is intervertebral disc degeneration, and what causes it? Spine (Phila Pa 1976) 31, 2151–2161. [DOI] [PubMed] [Google Scholar]
- 6. Furukawa T, Ito K, Nuka S, Hashimoto J, Takei H, Takahara M et al (2009) Absence of biglycan accelerates the degenerative process in mouse intervertebral disc. Spine (Phila Pa 1976) 34, 911–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mayer JE, Iatridis JC, Chan D, Qureshi SA, Gottesman O, Hecht AC (2013) Genetic polymorphisms associated with intervertebral disc degeneration. Spine J. 13, 299–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Li Z, Shen J, Wu WK, Yu X, Liang J, Qiu G et al (2012) Leptin induces cyclin D1 expression and proliferation of human nucleus pulposus cells via JAK/STAT, PI3K/Akt and MEK/ERK pathways. PLoS ONE 7, e53176. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 9. Li Z, Liang J, Wu WK, Yu X, Yu J, Weng X et al (2014) Leptin activates RhoA/ROCK pathway to induce cytoskeleton remodeling in nucleus pulposus cells. Int. J. Mol. Sci. 15, 1176–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Li Z, Shen J, Wu WK, Yu X, Liang J, Qiu G et al (2013) The role of leptin on the organization and expression of cytoskeleton elements in nucleus pulposus cells. J. Orthop. Res. 31, 847–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Li Z, Lei H, Luo M, Wang Y, Dong L, Ma Y et al (2015) DNA methylation downregulated mir‐10b acts as a tumor suppressor in gastric cancer. Gastric Cancer 18, 43–54. [DOI] [PubMed] [Google Scholar]
- 12. Brenner B, Hoshen MB, Purim O, David MB, Ashkenazi K, Marshak G et al (2011) MicroRNAs as a potential prognostic factor in gastric cancer. World J. Gastroenterol. 17, 3976–3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zheng F, Liao YJ, Cai MY, Liu YH, Liu TH, Chen SP et al (2012) The putative tumour suppressor microRNA‐124 modulates hepatocellular carcinoma cell aggressiveness by repressing ROCK2 and EZH2. Gut 61, 278–289. [DOI] [PubMed] [Google Scholar]
- 14. Zhao H, Guo M, Zhao G, Ma Q, Ma B, Qiu X et al (2012) miR‐183 inhibits the metastasis of osteosarcoma via downregulation of the expression of Ezrin in F5M2 cells. Int. J. Mol. Med. 30, 1013–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Malumbres M (2012) miRNAs and cancer: an epigenetics view. Mol. Aspects Med. 34, 863–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Liang W, Gao B, Fu P, Xu S, Qian Y, Fu Q. (2013) The miRNAs in the pathgenesis of osteosarcoma. Front. Biosci. (Landmark Ed.) 18, 788–794. [DOI] [PubMed] [Google Scholar]
- 17. Farazi TA, Spitzer JI, Morozov P, Tuschl T (2011) miRNAs in human cancer. J. Pathol. 223, 102–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fabbri M, Calin GA (2010) Epigenetics and miRNAs in human cancer. Adv. Genet. 70, 87–99. [DOI] [PubMed] [Google Scholar]
- 19. Kozlowska E, Krzyzosiak WJ, Koscianska E (2013) Regulation of huntingtin gene expression by miRNA‐137, ‐214, ‐148a, and their respective isomiRs. Int. J. Mol. Sci. 14, 16999–17016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ishikawa K, Ishikawa A, Shoji Y, Imai T (2014) A genotoxic stress‐responsive miRNA, miR‐574‐3p, delays cell growth by suppressing the enhancer of rudimentary homolog gene in vitro. Int. J. Mol. Sci. 15, 2971–2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fayyad‐Kazan H, Bitar N, Najar M, Lewalle P, Fayyad‐Kazan M, Badran R et al (2013) Circulating miR‐150 and miR‐342 in plasma are novel potential biomarkers for acute myeloid leukemia. J. Transl. Med. 11, 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bianchi N, Zuccato C, Finotti A, Lampronti I, Borgatti M, Gambari R. (2012) Involvement of miRNA in erythroid differentiation. Epigenomics 4, 51–65. [DOI] [PubMed] [Google Scholar]
- 23. Dassow H, Aigner A (2013) MicroRNAs (miRNAs) in colorectal cancer: from aberrant expression towards therapy. Curr. Pharm. Des. 19, 1242–1252. [DOI] [PubMed] [Google Scholar]
- 24. Marcinkowska M, Szymanski M, Krzyzosiak WJ, Kozlowski P (2011) Copy number variation of microRNA genes in the human genome. BMC Genom. 12, 183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Papagiannakopoulos T, Kosik KS (2008) MicroRNAs: regulators of oncogenesis and stemness. BMC Med. 6, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sato F, Tsuchiya S, Meltzer SJ, Shimizu K (2011) MicroRNAs and epigenetics. FEBS J. 278, 1598–1609. [DOI] [PubMed] [Google Scholar]
- 27. Nelson KM, Weiss GJ (2008) MicroRNAs and cancer: past, present, and potential future. Mol. Cancer Ther. 7, 3655–3660. [DOI] [PubMed] [Google Scholar]
- 28. Han L, Witmer PD, Casey E, Valle D, Sukumar S (2007) DNA methylation regulates MicroRNA expression. Cancer Biol. Ther. 6, 1284–1288. [DOI] [PubMed] [Google Scholar]
- 29. Chen Z, Wu J, Yang C, Fan P, Balazs L, Jiao Y et al (2012) DiGeorge syndrome critical region 8 (DGCR8) protein‐mediated microRNA biogenesis is essential for vascular smooth muscle cell development in mice. J. Biol. Chem. 287, 19018–19028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fan P, Chen Z, Tian P, Liu W, Jiao Y, Xue Y et al (2013) miRNA biogenesis enzyme Drosha is required for vascular smooth muscle cell survival. PLoS ONE 8, e60888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fernandez‐Hernando C, Ramirez CM, Goedeke L, Suarez Y (2013) MicroRNAs in metabolic disease. Arterioscler. Thromb. Vasc. Biol. 33, 178–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Grant JS, White K, Maclean MR, Baker AH (2013) MicroRNAs in pulmonary arterial remodeling. Cell. Mol. Life Sci. 70, 4479–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rottiers V, Naar AM (2012) MicroRNAs in metabolism and metabolic disorders. Nat. Rev. Mol. Cell Biol. 13, 239–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Guancial EA, Bellmunt J, Yeh S, Rosenberg JE, Berman DM (2014) The evolving understanding of microRNA in bladder cancer. Urol. Oncol. 32(41), e31–e40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yoshino H, Seki N, Itesako T, Chiyomaru T, Nakagawa M, Enokida H. (2013) Aberrant expression of microRNAs in bladder cancer. Nat. Rev. Urol. 10, 396–404. [DOI] [PubMed] [Google Scholar]
- 36. Kobayashi E, Hornicek FJ, Duan Z (2012) MicroRNA Involvement in Osteosarcoma. Sarcoma 2012, 359739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gillen AE, Gosalia N, Leir SH, Harris A (2011) MicroRNA regulation of expression of the cystic fibrosis transmembrane conductance regulator gene. Biochem. J. 438, 25–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bou Kheir T, Futoma‐Kazmierczak E, Jacobsen A, Krogh A, Bardram L, Hother C et al (2011) miR‐449 inhibits cell proliferation and is down‐regulated in gastric cancer. Mol. Cancer. 10, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Montag J, Hitt R, Opitz L, Schulz‐Schaeffer WJ, Hunsmann G, Motzkus D. (2009) Upregulation of miRNA hsa‐miR‐342‐3p in experimental and idiopathic prion disease. Mol. Neurodegener. 4, 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wilfred BR, Wang WX, Nelson PT (2007) Energizing miRNA research: a review of the role of miRNAs in lipid metabolism, with a prediction that miR‐103/107 regulates human metabolic pathways. Mol. Genet. Metab. 91, 209–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cao H, Hu X, Zhang Q, Wang J, Li J, Liu B et al (2014) Upregulation of let‐7a inhibits vascular smooth muscle cell proliferation in vitro and in vein graft intimal hyperplasia in rats. J. Surg. Res. 192, 223–33. [DOI] [PubMed] [Google Scholar]
- 42. Majid S, Dar AA, Saini S, Shahryari V, Arora S, Zaman MS et al (2012) MicroRNA‐1280 inhibits invasion and metastasis by targeting ROCK1 in bladder cancer. PLoS ONE 7, e46743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Shi Z, Wei Q, Zhang M, She J (2014) MicroRNAs in bladder cancer: expression profiles, biological functions, regulation, and clinical implications. Crit. Rev. Eukaryot. Gene Expr. 24, 55–75. [DOI] [PubMed] [Google Scholar]
- 44. Bae Y, Yang T, Zeng HC, Campeau PM, Chen Y, Bertin T et al (2012) miRNA‐34c regulates Notch signaling during bone development. Hum. Mol. Genet. 21, 2991–3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Xie W, Li Z, Li M, Xu N, Zhang Y (2013) miR‐181a and inflammation: miRNA homeostasis response to inflammatory stimuli in vivo. Biochem. Biophys. Res. Commun. 430, 647–652. [DOI] [PubMed] [Google Scholar]
- 46. Bandres E, Bitarte N, Arias F, Agorreta J, Fortes P, Agirre X et al (2009) microRNA‐451 regulates macrophage migration inhibitory factor production and proliferation of gastrointestinal cancer cells. Clin. Cancer Res. 15, 2281–2290. [DOI] [PubMed] [Google Scholar]
- 47. Pichiorri F, Palmieri D, De Luca L, Consiglio J, You J, Rocci A et al (2013) In vivo NCL targeting affects breast cancer aggressiveness through miRNA regulation. J. Exp. Med. 210, 951–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Duan Z, Choy E, Harmon D, Liu X, Susa M, Mankin H et al (2011) MicroRNA‐199a‐3p is downregulated in human osteosarcoma and regulates cell proliferation and migration. Mol. Cancer Ther. 10, 1337–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Furuta M, Kozaki KI, Tanaka S, Arii S, Imoto I, Inazawa J. (2010) miR‐124 and miR‐203 are epigenetically silenced tumor‐suppressive microRNAs in hepatocellular carcinoma. Carcinogenesis 31, 766–776. [DOI] [PubMed] [Google Scholar]
- 50. Kumar MS, Armenteros‐Monterroso E, East P, Chakravorty P, Matthews N, Winslow MM et al (2014) HMGA2 functions as a competing endogenous RNA to promote lung cancer progression. Nature 505, 212–217. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 51. Wei A, Brisby H, Chung SA, Diwan AD (2008) Bone morphogenetic protein‐7 protects human intervertebral disc cells in vitro from apoptosis. Spine J. 8, 466–474. [DOI] [PubMed] [Google Scholar]
- 52. Jiang L, Zhang X, Zheng X, Ru A, Ni X, Wu Y et al (2013) Apoptosis, senescence, and autophagy in rat nucleus pulposus cells: implications for diabetic intervertebral disc degeneration. J. Orthop. Res. 31, 692–702. [DOI] [PubMed] [Google Scholar]
- 53. Ha KY, Kim BG, Kim KW, Oh IS, Seo JY (2011) Apoptosis in the sequestrated nucleus pulposus compared to the remaining nucleus pulposus in the same patient. Spine (Phila Pa 1976) 36: 683–689. [DOI] [PubMed] [Google Scholar]
- 54. Murata Y, Nannmark U, Rydevik B, Takahashi K, Olmarker K (2008) The role of tumor necrosis factor‐alpha in apoptosis of dorsal root ganglion cells induced by herniated nucleus pulposus in rats. Spine (Phila Pa 1976) 33: 155–162. [DOI] [PubMed] [Google Scholar]
- 55. Wei Y, Nazari‐Jahantigh M, Neth P, Weber C, Schober A (2013) MicroRNA‐126, ‐145, and ‐155: a therapeutic triad in atherosclerosis? Arterioscler. Thromb. Vasc. Biol. 33, 449–454. [DOI] [PubMed] [Google Scholar]
- 56. Wang Y, Li J, Tong L, Zhang J, Zhai A, Xu K et al (2013) The prognostic value of miR‐21 and miR‐155 in non‐small‐cell lung cancer: a meta‐analysis. Jpn. J. Clin. Oncol. 43, 813–820. [DOI] [PubMed] [Google Scholar]
- 57. Li CL, Nie H, Wang M, Su LP, Li JF, Yu YY et al (2012) microRNA‐155 is downregulated in gastric cancer cells and involved in cell metastasis. Oncol. Rep. 27, 1960–1966. [DOI] [PubMed] [Google Scholar]
- 58. Greither T, Grochola LF, Udelnow A, Lautenschlager C, Wurl P, Taubert H. (2010) Elevated expression of microRNAs 155, 203, 210 and 222 in pancreatic tumors is associated with poorer survival. Int. J. Cancer 126, 73–80. [DOI] [PubMed] [Google Scholar]
- 59. Pu J, Bai D, Yang X, Lu X, Xu L, Lu J. (2012) Adrenaline promotes cell proliferation and increases chemoresistance in colon cancer HT29 cells through induction of miR‐155. Biochem. Biophys. Res. Commun. 428, 210–215. [DOI] [PubMed] [Google Scholar]
- 60. Wang HQ, Yu XD, Liu ZH, Cheng X, Samartzis D, Jia LT et al (2011) Deregulated miR‐155 promotes Fas‐mediated apoptosis in human intervertebral disc degeneration by targeting FADD and caspase‐3. J. Pathol. 225, 232–242. [DOI] [PubMed] [Google Scholar]
- 61. Zhao X, Yang L, Hu J (2011) Down‐regulation of miR‐27a might inhibit proliferation and drug resistance of gastric cancer cells. J. Exp. Clin. Cancer Res. 30, 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Drayton RM, Dudziec E, Peter S, Bertz S, Hartmann A, Bryant HE et al (2014) Reduced expression of miRNA‐27a modulates cisplatin resistance in bladder cancer by targeting the cystine/glutamate exchanger SLC7A11. Clin. Cancer Res. 20, 1990–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Hezova R, Kovarikova A, Bienertova‐Vasku J, Sachlova M, Redova M, Vasku A et al (2012) Evaluation of SNPs in miR‐196‐a2, miR‐27a and miR‐146a as risk factors of colorectal cancer. World J. Gastroenterol. 18, 2827–2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Liu T, Tang H, Lang Y, Liu M, Li X (2009) MicroRNA‐27a functions as an oncogene in gastric adenocarcinoma by targeting prohibitin. Cancer Lett. 273, 233–242. [DOI] [PubMed] [Google Scholar]
- 65. Liu G, Cao P, Chen H, Yuan W, Wang J, Tang X. (2013) MiR‐27a regulates apoptosis in nucleus pulposus cells by targeting PI3K. PLoS ONE 8, e75251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Wang W, Zhao LJ, Tan YX, Ren H, Qi ZT (2012) MiR‐138 induces cell cycle arrest by targeting cyclin D3 in hepatocellular carcinoma. Carcinogenesis 33, 1113–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Ma L (2010) Role of miR‐10b in breast cancer metastasis. Breast Cancer Res. 12, 210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Wang YY, Ye ZY, Zhao ZS, Li L, Wang YX, Tao HQ et al (2013) Clinicopathologic significance of miR‐10b expression in gastric carcinoma. Hum. Pathol. 44, 1278–85. [DOI] [PubMed] [Google Scholar]
- 69. Li QJ, Zhou L, Yang F, Wang GX, Zheng H, Wang DS et al (2012) MicroRNA‐10b promotes migration and invasion through CADM1 in human hepatocellular carcinoma cells. Tumour Biol. 33, 1455–1465. [DOI] [PubMed] [Google Scholar]
- 70. Frampton AE, Krell J, Zhang Y, Stebbing J, Castellano L, Jiao LR. (2012) The role of miR‐10b in metastatic pancreatic ductal adenocarcinoma. Surgery 152, 936–8. [DOI] [PubMed] [Google Scholar]
- 71. Ma L, Teruya‐Feldstein J, Weinberg RA (2007) Tumour invasion and metastasis initiated by microRNA‐10b in breast cancer. Nature 449, 682–688. [DOI] [PubMed] [Google Scholar]
- 72. Nakata K, Ohuchida K, Mizumoto K, Kayashima T, Ikenaga N, Sakai H. (2011) MicroRNA‐10b is overexpressed in pancreatic cancer, promotes its invasiveness, and correlates with a poor prognosis. Surgery 150, 916–922. [DOI] [PubMed] [Google Scholar]
- 73. Yu X, Li Z, Shen J, Wu WK, Liang J, Weng X et al (2013) MicroRNA‐10b promotes nucleus pulposus cell proliferation through RhoC‐Akt pathway by targeting HOXD10 in intervetebral disc degeneration. PLoS ONE 8, e83080. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 74. Cao Z, Yoon JH, Nam SW, Lee JY, Park WS (2012) PDCD4 expression inversely correlated with miR‐21 levels in gastric cancers. J. Cancer Res. Clin. Oncol. 138, 611–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Iyevleva AG, Kuligina E, Mitiushkina NV, Togo AV, Miki Y, Imyanitov EN. (2012) High level of miR‐21, miR‐10b, and miR‐31 expression in bilateral vs. unilateral breast carcinomas. Breast Cancer Res. Treat. 131, 1049–1059. [DOI] [PubMed] [Google Scholar]
- 76. Haigl B, Vanas V, Setinek U, Hegedus B, Gsur A, Sutterlüty‐Fall H. (2014) Expression of microRNA‐21 in non‐small cell lung cancer tissue increases with disease progression and is likely caused by growth conditional changes during malignant transformation. Int. J. Oncol. 44, 1325–1334. [DOI] [PubMed] [Google Scholar]
- 77. Huang YH, Lin YH, Chi HC, Liao CH, Liao CJ, Wu SM et al (2013) Thyroid hormone regulation of miR‐21 enhances migration and invasion of hepatoma. Cancer Res. 73, 2505–2517. [DOI] [PubMed] [Google Scholar]
- 78. Liu H, Huang X, Liu X, Xiao S, Zhang Y, Xiang T et al (2014) miR‐21 promotes human nucleus pulposus cell proliferation through PTEN/AKT signaling. Int. J. Mol. Sci. 15, 4007–4018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Tian Y, Yuan W, Fujita N, Wang J, Wang H, Shapiro IM et al (2013) Inflammatory cytokines associated with degenerative disc disease control aggrecanase‐1 (ADAMTS‐4) expression in nucleus pulposus cells through MAPK and NF‐kappaB. Am. J. Pathol. 182, 2310–2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Roughley PJ (2004) Biology of intervertebral disc aging and degeneration: involvement of the extracellular matrix. Spine (Phila Pa 1976) 29: 2691–2699. [DOI] [PubMed] [Google Scholar]
- 81. Huang M, Wang HQ, Zhang Q, Yan XD, Hao M, Luo ZJ. (2012) Alterations of ADAMTSs and TIMP‐3 in human nucleus pulposus cells subjected to compressive load: implications in the pathogenesis of human intervertebral disc degeneration. J. Orthop. Res. 30, 267–273. [DOI] [PubMed] [Google Scholar]
- 82. Hayes AJ, Benjamin M, Ralphs JR (2001) Extracellular matrix in development of the intervertebral disc. Matrix Biol. 20, 107–121. [DOI] [PubMed] [Google Scholar]
- 83. Clouet J, Pot‐Vaucel M, Grimandi G, Masson M, Lesoeur J, Fellah BH et al (2011) Characterization of the age‐dependent intervertebral disc changes in rabbit by correlation between MRI, histology and gene expression. BMC Musculoskelet. Disord. 12, 147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Chen WH, Lo WC, Lee JJ, Su CH, Lin CT, Liu HY et al (2006) Tissue‐engineered intervertebral disc and chondrogenesis using human nucleus pulposus regulated through TGF‐beta1 in platelet‐rich plasma. J. Cell. Physiol. 209, 744–754. [DOI] [PubMed] [Google Scholar]
- 85. Chen J, Liu Z, Zhong G, Qian L, Li Z, Qiao Z et al (2014) Hypertrophy of ligamentum flavum in lumbar spine stenosis is associated with increased miR‐155 level. Dis. Markers 2014, 786543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Tsirimonaki E, Fedonidis C, Pneumaticos SG, Tragas AA, Michalopoulos I, Mangoura D. (2013) PKCepsilon signalling activates ERK1/2, and regulates aggrecan, ADAMTS5, and miR377 gene expression in human nucleus pulposus cells. PLoS ONE 8, e82045. [DOI] [PMC free article] [PubMed] [Google Scholar]


