Abstract
Abstract. Objectives: Expression of the nuclear Ki‐67 protein (pKi‐67) is strongly associated with cell proliferation. For this reason, antibodies against this protein are widely used as prognostic tools for the assessment of cell proliferation in biopsies from cancer patients. Despite this broad application in histopathology, functional evidence for the physiological role of pKi‐67 is still missing. Recently, we proposed a function of pKi‐67 in the early steps of ribosomal RNA (rRNA) synthesis. Here, we have examined the involvement of pKi‐67 in this process by photochemical inhibition using chromophore‐assisted light inactivation (CALI). Materials and methods: Anti‐pKi‐67 antibodies were labelled with the fluorochrome fluorescein 5(6)‐isothiocyanate and were irradiated after binding to their target protein. Results: Performing CALI in vitro on cell lysates led to specific cross‐linking of pKi‐67. Moreover, the upstream binding factor (UBF) necessary for rRNA transcription was also partly subjected to cross‐link formation, indicating a close spatial proximity of UBF and pKi‐67. CALI in living cells, using micro‐injected antibody, caused a striking relocalization of UBF from foci within the nucleoli to spots located at the nucleolar rim or within the nucleoplasm. pKi‐67‐CALI resulted in dramatic inhibition of RNA polymerase I‐dependent nucleolar rRNA synthesis, whereas RNA polymerase II‐dependent nucleoplasmic RNA synthesis remained almost unaltered. Conclusions: Our data presented here argue for a crucial role of pKi‐67 in RNA polymerase I‐dependent nucleolar rRNA synthesis.
INTRODUCTION
Proliferation of all human cells is accompanied by prominent up‐regulation of pKi‐67 expression (Scholzen & Gerdes 2000). Until recently, it was thought that pKi‐67 was exclusively expressed during the active phases of the cell cycle (G1, S and G2, as well as mitosis) and was completely absent from G0 cells. However, new findings demonstrate that minor amounts of this protein can also be detected in quiescent cells (Bullwinkel et al. 2006). Due to the fact that in histopathology the fraction of positive tumour cell nuclei correlates well with the prognosis of various types of human neoplasms, antibodies against pKi‐67 are broadly used in routine tumour diagnostics (Scholzen & Gerdes 2000; Brown & Gatter 2002). Moreover, recent publications indicate that pKi‐67 is also used as a promising target for tumour therapy (2003, 2004). Despite these applications, the cellular function of the protein remains elusive. Immunolocalization studies in quiescent and proliferating cells indicate a possible role of pKi‐67 in ribosomal RNA (rRNA) transcription. This hypothesis is supported in that a fraction of nuclear pKi‐67 is physically associated with the rRNA genes (Bullwinkel et al. 2006).
In this study, we have utilized chromophore‐assisted light inactivation (CALI) to investigate the putative function of pKi‐67 for rRNA transcription in vivo. Methods commonly utilized for the elucidation of protein function are often based on inhibition of gene expression by the means of gene targeting, antisense or RNA interference strategies. In contrast, direct functional protein inactivation by light‐induced photochemical reactions offers the advantage to control the inactivation process spatially and temporally with high precision (Jay 1988; Liao et al. 1994; Surrey et al. 1998; Beck et al. 2002). CALI against pKi‐67 was performed, using fluorescein 5(6)‐isothiocyanate (FITC)‐labelled antibodies MIB‐1 and TuBB‐9 that are directed against different epitopes of pKi‐67 and exhibit different sub‐cellular staining patterns, probably caused by differential epitope masking (Bullwinkel et al. 2006). MIB‐1, recognizes an epitope located within the repetitive region of pKi‐67, encoded by exon 13, and stains mainly the outer rim of the nucleoli. In contrast, TuBB‐9 binds to an epitope located within the exon 9 encoded region, and exhibits a fine punctuate staining pattern colocalizing with components of the rRNA transcription machinery. Utilizing micro‐injection of antibodies followed by pulse labelling with RNA precursors, we showed that TuBB‐9‐CALI but not MIB‐1‐CALI lead to a profound inhibition of rRNA synthesis.
MATERIALS AND METHODS
Antibodies and labelling
The following mouse monoclonal antibodies were used: MIB‐1 (Key et al. 1993), TuBB‐9 (Bullwinkel et al. 2006), Mcm3 (Endl et al. 2001), ACT‐1 (Schwarting et al. 1987) and a rat monoclonal antibody against BrdUrd (Abcam). The human auto‐immune serum recognizing upstream binding factor (UBF), as well as the rabbit anti‐UBF serum were kindly provided by Manuel M. Valdivia, Cádiz, Spain.
Antibodies were conjugated to FITC (fluorescein 5(6)‐isothiocyanate, Sigma, St. Louis, MO, USA) by the method of Goding (1976). FITC was dissolved in dimethyl sulfoxide (1 mg/ml) and was mixed with the antibody in a ratio of 5 µg per 100 µg of antibody, which was dissolved in sodium carbonate buffer at a concentration of 1 mg/ml. Subsequently, the solution was incubated at room temperature on a shaker for 2 h and the labelled protein was purified with a NAP‐5‐Sephadex column (Amersham Biosciences, Little Chalfont, Buckinghamshire, UK). After elution with Tris‐buffered saline (TBS) (10 mm Tris/HCl, pH 8.2, 150 mm NaCl) the labelled antibodies were concentrated with Microcon tubes (Millipore, Schwalbach, Germany) and were re‐dissolved in TBS pH 7.5. Antibodies with comparable number of dye molecules per antibody were used for irradiation experiments. From the absorbance A(λ), the protein concentration (cprot) and the average number of fluorochromes per antibody (η) were calculated, using the following formulae for the FITC‐conjugates.
Cell culture, microinjection and preparation of cell lysate
Cells of the HeLa subline HEp‐2 (ATCC CCL‐23) were grown in Dulbecco's modified Eagle's medium, supplemented with 10% foetal calf serum, 2%l‐glutamine, 0.95% glucose, 50 µg/ml streptomycin and 50 U/ml penicillin. To assess ongoing RNA synthesis, the medium was supplemented with 2 mm 5‐fluorouridine (FU) (Boisvert et al. 2000) and cells were incubated for 10 min at 37 °C before being processed for immunostaining as described below. Fluorescence‐labelled antibodies were micro‐injected into the nucleus of HEp‐2 cells with an Eppendorf Micromanipulator 5171 and Transjector 5246 using Eppendorf glass Femtotips. For microinjection, 3–4 × 105 cells were plated on 35‐mm glass‐bottom dishes (MatTek, Ashland, MA, USA) and were incubated overnight.
For preparation of cell lysates, 2 × 107 HEp‐2 cells were centrifuged at 350 g and 4 °C for 10 min. Pellets were dissolved in 1 ml lysis buffer [100 mm Na‐citrate, pH 7.5, 1% (v/v) NP‐40, 1 mm dithiothreitol, protease inhibitors (Complete Mini, EDTA‐free, Roche Diagnostics, Penzberg, Germany)]. The cells were incubated on ice for 20 min and were briefly vortexed every 5 min. Subsequently, the lysate was cleared by centrifugating twice for 10 min at 16 000 g at 4 °C.
CALI on cell lysates
Fluorochrome‐labelled antibodies were added to 20 µl of cell lysate at 40 µg/ml end concentration and were incubated for 30 min at 4 °C. A 488 nm cw argon laser (270 Exciter, Spectra‐Physics, Mountain View, CA, USA) was used for irradiation of the samples. Immediately after irradiation samples were diluted 1 : 10 with sodium dodecyl sulfate (SDS) sample buffer (25 mm Tris/HCl, 50 mm dithiothreitol, 10% (v/v) glycerol, 2% (w/v) SDS, 0.02% (w/v) bromphenol blue, pH 6.8) and SDS/PAGE was performed essentially as described earlier (Endl & Gerdes 2000). Blots were blocked with 4% cold water fish skin gelatine (Sigma) in TBS‐T (0.1 m Tris/HCl, pH 8, 150 mm NaCl, 0.05% (v/v) Tween 20 for detection with anti‐pKi‐67 and anti‐Mcm3 antibodies or with 5% milk powder in TBS‐T for detection with human autoimmune serum recognizing UBF.
CALI on microinjected cells
Cells were irradiated 1–2 h after microinjection on a Nikon‐Eclipse TE2000‐U inverted microscope with a mercury‐vapour lamp and an excitation filter range from 465 nm to 495 nm. The beam of the 20× objective lens (Plan Fluor) with a numerical aperture of 0.45 had an irradiance of 570 mW/cm2 in the focal plane. The efficacy of the polychromatic excitation of FITC was calculated to be 90% when compared to excitation of a laser at 488 nm. After irradiation, cells were incubated for 20 min at 37 °C before ongoing RNA synthesis was pulse labelled with 5‐fluorouridine.
Immunofluorescence analyses
Cells were fixed for 10 min in phosphate‐buffed saline (PBS), 2% formaldehyde and then were permeabilized for 10 min with PBS, 0.25% Triton X‐100. After washing in TBS, pH 7.5, they were incubated for 30 min with the primary antibody diluted in TBS, 10% bovine serum albumin. Monoclonal rat anti‐BrdUrd antibody (Abcam) was used because of its cross‐reactivity for detection of 5‐fluorouridine at a concentration of 5 µg/ml. Rabbit anti‐UBF serum was diluted 1 : 600. Subsequently, the dishes were washed with TBS and cells were incubated for 30 min with the secondary antibodies, goat antimouse immunoglobulin G (IgG) (H + L), goat antirabbit IgG (H + L) and goat antirat IgG (H + L) conjugated to Alexa Fluor 488, Alexa Fluor 546 or Alexa Flour 647 (Molecular Probes) at a concentration of 7 µg/ml. After washing with TBS, the cells were treated for 10 min with PBS and 2% formaldehyde. Specimens were analysed with a Leica TCS‐SP confocal scanning microscope equipped with an acousto‐optical tunable filter and a 63× numerical aperture 1.32 oil‐immersion objective lens (HCX PL APO, Leica microsystems). Images were acquired with the Leica TCSNT software and were further processed and assembled using Adobe Photoshop 6.0.
RESULTS
To test the antibody conjugates, cell lysates from proliferating pKi‐67 expressing HEp‐2 cells were incubated with FITC‐labelled MIB‐1 or TuBB‐9. After irradiation, they were analysed by Western blotting (Fig. 1a). In non‐irradiated samples (lanes 1–3), the characteristic pKi‐67 double band is visible at approximately 315 and 350 kDa (Gerlach et al. 1998), whereas in irradiated samples it was barely detectable (lanes 4 and 5). Instead we observed reactivity of higher apparent molecular mass, which most likely resulted from covalent cross‐linking of the target protein, as reported previously after FITC irradiation (Lepock et al. 1978).
Figure 1.

CALI in HEp‐2 cell lysates. Samples were incubated as indicated. Immunoblot in (a) shows pKi‐67 detected with the MIB‐1 antibody. In non‐irradiated samples (lanes 1–3) the pKi‐67 main bands are visible at approximately 315 and 350 kDa. After irradiation with a 488 nm cw argon laser at 500 mW/cm2 for 15 min these bands are absent and a reactivity of higher molecular weight appears (lanes 4–5). In samples from the same experiment Mcm3 (b) and UBF (c) were detected. The main bands of Mcm3 and UBF are clearly visible after CALI against pKi‐67 (compare lanes 4–5 in a with b, c). Double arrows show the characteristic pKi‐67 and the UBF double band. Numbers on left hand side indicate the size in kDa.
To investigate the specificity of this effect, samples from the same experiment were analysed for nuclear mini‐chromosome maintenance protein 3 (Mcm3) and the nucleolar UBF (Fig. 1b,c). Mcm proteins play a crucial role during initiation and elongation of DNA replication (Alexandrow et al. 2002) and are presumably functionally unrelated to pKi‐67. As shown (Fig. 1b), the Mcm3 main band at approximately 100 kDa is barely affected by irradiation, indicating that the effect of CALI‐conjugates after irradiation is specific for pKi‐67 (compare Fig. 1a to Fig. 1b). However, the 50 kDa band originating from the heavy chains of the fluorochrome‐labelled anti‐pKi‐67 antibodies became weaker after irradiation (Fig. 1b, lanes 4 and 5), indicating that the labelled antibodies were also subjected to cross‐link formation. UBF, a transcription factor for RNA polymerase I (RNA Pol I), has recently been demonstrated to colocalize with pKi‐67 (Bullwinkel et al. 2006). The UBF doublet band at approximately 100 kDa was only slightly affected by CALI against pKi‐67 (Fig. 1c). However, irradiated samples exhibited an additional reactivity above 160 kDa, indicating participation of an UBF sub‐fraction in cross‐link formation.
After demonstrating the specificity of anti‐pKi‐67‐CALI in vitro, we next adopted the method for the analysis of pKi‐67 function in living cells. Recently, a role for pKi‐67 in rRNA transcription or in the early stages of rRNA processing has been proposed (Bullwinkel et al. 2006), but functional in vivo evidence was still missing. Therefore, we investigated the consequences of pKi‐67 inactivation by CALI on RNA synthesis in living cells. The labelled MIB‐1 and TuBB‐9 antibodies were micro‐injected into nuclei of HEp‐2 cells and were irradiated with a fluorescence microscopy. As depicted in Fig. 2, TuBB‐9‐CALI with a radiant exposure of 30 J/cm2– corresponding to an irradiation time of 1 min – was sufficient to specifically inhibit nucleolar rRNA synthesis in approximately 70% of the cells. In contrast, similar experiments with MIB‐1 did not evoke specific effects on rRNA synthesis. However, further elongation of the irradiation time led to inhibition of RNA synthesis in the entire nucleus, irrespective of the antibody conjugate used and was also apparent with fluorochrome conjugates of the ACT‐1 control antibody that does not recognize any epitope in HEp‐2 cells (data not shown).
Figure 2.

Inhibition of nucleolar RNA synthesis by CALI. Cells were injected with antibody conjugates as indicated and analysed for ongoing RNA synthesis as described in Figure 3. The fractions of cells with inhibition of nucleolar RNA synthesis as well as complete inhibition of RNA synthesis were determined by counting 30–60 microinjected cells. Radiant exposure was 570 mW/cm2.
Figure 3 illustrates in detail CALI effects at low radiant exposure (45 J/cm2). In non‐irradiated, TuBB‐9‐FITC micro‐injected cells, numerous spots were visible inside the nucleoli (Fig. 3a), which exhibited colocalization with UBF (Fig. 3b). As depicted in Fig. 3c, newly synthesized RNA, visualized by pulse‐labelling with 5‐fluorouridine (FU), was apparent in the nucleoplasm (mRNA), as well as in nucleoli (rRNA). After irradiation (Fig. 3d–f), Pol I‐dependent rRNA synthesis in nucleoli was absent in most of the injected cells, while the Pol II‐dependent mRNA synthesis in the nucleoplasm was only slightly affected (Fig. 3f). This was most clearly seen in the two micro‐injected cells at the bottom of Fig. 3d–f. For comparison, Pol I transcription was not affected in the upper cell of that panel, which was not microinjected. In addition to inhibition of rRNA synthesis, notable redistribution of the TuBB‐9 antibody and UBF could be observed (Fig. 3d,e). At higher magnification, it was apparent that both TuBB‐9 and UBF redistributed to spots, which were located either within the nucleoplasm (Fig. 3g–i, arrowheads) or at the rim of nucleoli (Fig. 3g–i, arrows). In some cases, very large clusters formed at rims of the nucleoli (Fig. 3j–l). In contrast to results obtained with the TuBB‐9 antibody, performing CALI experiments with MIB‐1 did not lead to specific effects on rRNA synthesis or protein localization (Fig. 3m–r).
Figure 3.
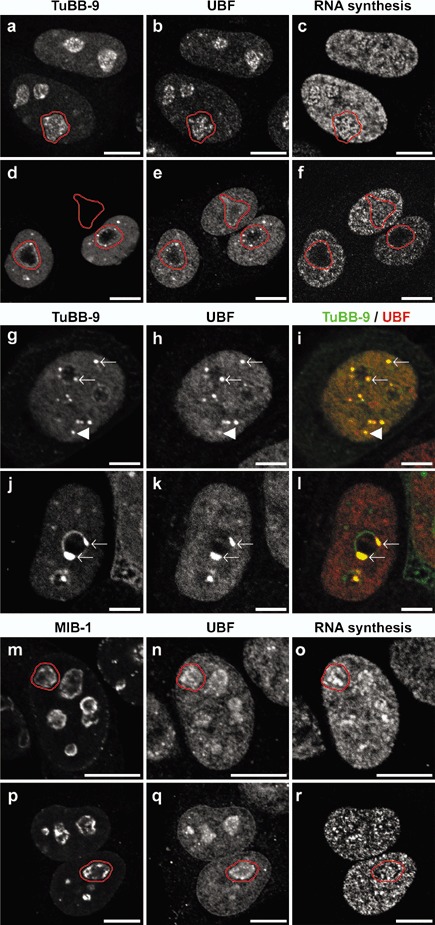
Inhibition of nucleolar RNA transcription and protein relocalisation induced by CALI in living cells. Cells were either microinjected with TuBB‐9‐FITC (a–l) or MIB1‐FITC (m–r). In non‐irradiated control cells (a–c) the microinjected TuBB‐9‐FITC antibody (a) accumulates in small foci within the nucleoli (one nucleolus outlined in red) co‐localizing with UBF (b). In contrast, irradiation of cells with a fluorescence microscope for 90 s at 570 mW/cm2 (d–l) leads to a prominent redistribution of the microinjected antibody to distinct intranuclear spots (d, g, j). These spots are frequently located at the rim of the nucleoli (arrows), often forming large complexes (j) as well as to smaller spots within the nucleoplasm (arrowheads). Interestingly, these spots completely co‐localize with UBF staining (e, h, k) which is also apparent by the yellow colour in the merged images (i, l) where TuBB‐9 staining is shown in green and UBF staining in red. While control cells exhibit a distinctive RNA synthesis within nucleoli and nucleoplasm (c), irradiation of microinjected cells leads to a dramatic inhibition of the nucleolar rRNA synthesis whereas the nucleoplasmic RNA synthesis is almost unaffected (f). The microinjected MIB‐1‐FITC antibody accumulates at the outer rim of the nucleoli (m) in non‐irradiated control cells (m–o, one nucleolus outlined in red). In contrast to cells microinjected with TuBB‐9‐FITC, after irradiation (p–r) no effect on protein localisation or RNA synthesis could be observed. Bars: 10 µm; g–l: 5 µm.
Specificity of TuBB‐9‐mediated CALI was further examined by fibrillarin staining. Fibrillarin is a nucleolar protein involved in early steps of rRNA processing and localizes in close vicinity to the rRNA transcription machinery (Ochs et al. 1985; Biggiogera et al. 2001; Hernandez‐Verdun & Roussel 2003). As shown in Fig. 4, TuBB‐9‐CALI had no influence on fibrillarin localization, indicating that TuBB‐9‐CALI did not lead to pleiotropic effects within the nucleolus.
Figure 4.
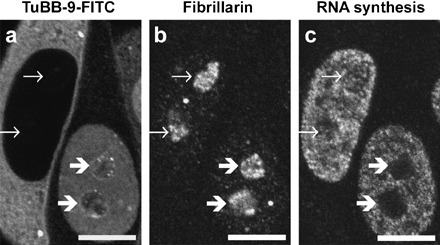
CALI with TuBB‐9‐FITC does not alter fibrillarin localisation inside the nucleoli. CALI after nuclear microinjection of TuBB‐9‐FITC (right hand cell) does not affect fibrillarin localisation (b) whereas nucleolar RNA synthesis (c) is inhibited (thick arrows). Microinjection into the cytoplasm which is shown as a control, does affect neither fibrillarin localisation nor RNA synthesis (left hand cell, thin arrows). Irradiation 15 min at 570 mJ/cm2. Bars 10 µm.
DISCUSSION
The CALI experiments performed on cell lysates (Fig. 1) vividly demonstrated the usefulness of the method to affect pKi‐67 with high specificity. While the target protein was effectively converted to molecular species with higher apparent molecular masses, Mcm3 control protein was virtually unaffected. The shift of molecular mass was presumably due to covalent cross‐linking induced by a FITC free radical intermediate (Lepock et al. 1978). Interestingly, Pol I transcription factor UBF was also partly affected by CALI against pKi‐67. Although the staining intensity of the UBF doublet band at approximately 100 kDa was almost unaltered, several additional distinct bands above 160 kDa emerged after CALI treatment (Fig. 1c). UBF, which is required for transcription of the rRNA genes (Bell et al. 1988), has recently been demonstrated to colocalize with pKi‐67 in vivo (Bullwinkel et al. 2006). The range of FITC‐CALI effects is rather small and was initially estimated to have a maximum of 30 nm (Surrey et al. 1998); this was later defined to be 4 nm for half‐maximal damage (Beck et al. 2002). Thus, occurrence of pKi‐67‐CALI‐induced UBF cross‐links indicates close proximity of pKi‐67 and a fraction of UBF, arguing for an interaction between the molecules in vitro. CALI‐induced cross‐linking has recently also been shown in calmodulin and a variety of binding partners (Yan et al. 2006).
The interaction between pKi‐67 and UBF might also be of relevance in vivo, as can be deduced from CALI experiments of TuBB‐9‐FITC micro‐injected cells (Fig. 3a–l). After irradiation, TuBB‐9 redistributed to spots located within the nucleoplasm or at the rim of the nucleoli, and surprisingly, UBF was co‐recruited to the same sites. While the underlying mechanism for formation of these spots is obscure, it clearly correlates with inhibition of the nucleolar RNA synthesis.
In contrast, after CALI with MIB‐1‐FITC, neither the antibody nor UBF showed altered intranuclear distribution, and nucleolar RNA synthesis was unaffected (Fig. 3m–r). As mentioned above, anti‐pKi‐67 antibodies TuBB‐9 and MIB‐1 differ in their nuclear staining patterns probably resulting from epitope masking caused by interacting molecules. In this context, it has been proposed that only the pKi‐67 fraction recognized by TuBB‐9 and not the fraction recognized by MIB‐1 may play a role in rRNA synthesis (Bullwinkel et al. 2006). Interestingly, CALI experiments in vitro did not reveal any difference between TuBB‐9 and MIB‐1 with respect to their ability to induce UBF cross‐link formation (Fig. 1, lanes 4 and 5). This indicates that differential epitope masking may not take place in cell lysates.
Data presented here provide the first functional evidence that a fraction of pKi‐67 is involved in rRNA synthesis in vivo. However, the underlying mechanism of this inhibition is not yet clear. The effect may either be caused by direct functional inactivation of pKi‐67 or a member of the Pol I transcription machinery that is found in very close proximity. A good candidate for the latter would be UBF, which was identified as a putative binding partner of pKi‐67. Availability of rRNA molecules is a key regulator for ribosome production and thus determines the potential for cell growth and proliferation (for review see Grummt 2003). Our findings that pKi‐67 is a potential UBF binding partner and participates in rRNA synthesis indicates a functional role in adjusting ribosome biosynthesis with respect to cell proliferation.
ACKNOWLEDGEMENTS
We would like to thank Margrit Kernbach and Bettina Baron‐Lühr for excellent technical assistance. This work was funded by the German Bundesministerium für Bildung und Forschung (grant no. 13 N8459).
REFERENCES
- Alexandrow MG, Ritzi M, Pemov A, Hamlin JL (2002) A potential role for mini‐chromosome maintenance (MCM) proteins in initiation at the dihydrofolate reductase replication origin. J. Biol. Chem. 277, 2702–2708. [DOI] [PubMed] [Google Scholar]
- Beck S, Sakurai T, Eustace BK, Beste G, Schier R, Rudert F, Jay DG (2002) Fluorophore‐assisted light inactivation: a high‐throughput tool for direct target validation of proteins. Proteomics 2, 247–255. [DOI] [PubMed] [Google Scholar]
- Bell SP, Learned RM, Jantzen HM, Tjian R (1988) Functional cooperativity between transcription factors UBF1 and SL1 mediates human ribosomal RNA synthesis. Science 241, 1192–1197. [DOI] [PubMed] [Google Scholar]
- Biggiogera M, Malatesta M, Abolhassani‐Dadras S, Amalric F, Rothblum LI, Fakan S (2001) Revealing the unseen: the organizer region of the nucleolus. J. Cell Sci. 114, 3199–3205. [DOI] [PubMed] [Google Scholar]
- Boisvert FM, Hendzel MJ, Bazett‐Jones DP (2000) Promyelocytic leukemia (PML) nuclear bodies are protein structures that do not accumulate RNA. J. Cell Biol. 148, 283–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DC, Gatter KC (2002) Ki67 protein: the immaculate deception? Histopathology 40, 2–11. [DOI] [PubMed] [Google Scholar]
- Bullwinkel J, Baron‐Lühr B, Lüdemann A, Wohlenberg C, Gerdes J, Scholzen T (2006) Ki‐67 protein is associated with ribosomal RNA transcription in quiescent and proliferating cells. J. Cell. Physiol. 206, 624–635. [DOI] [PubMed] [Google Scholar]
- Endl E, Gerdes J (2000) Posttranslational modifications of the KI‐67 protein coincide with two major checkpoints during mitosis. J. Cell. Physiol. 182, 371–380. [DOI] [PubMed] [Google Scholar]
- Endl E, Kausch I, Baack M, Knippers R, Gerdes J, Scholzen T (2001) The expression of Ki‐67, MCM3, and p27 defines distinct subsets of proliferating, resting, and differentiated cells. J. Pathol. 195, 457–462. [DOI] [PubMed] [Google Scholar]
- Gerlach C, Kubbutat M, Schwab U, Key G, Flad HD, Gerdes J (1998) Proliferation‐associated Ki‐67 protein is a target for autoantibodies in the human autoimmune disease systemic lupus erythematosus. Lab. Invest. 78, 129–130. [PubMed] [Google Scholar]
- Goding JW (1976) Conjugation of antibodies with fluorochromes: modifications to the standard methods. J. Immunol. Methods 13, 215–226. [DOI] [PubMed] [Google Scholar]
- Grummt I (2003) Life on a planet of its own: regulation of RNA polymerase I transcription in the nucleolus. Genes Dev. 17, 1691–1702. [DOI] [PubMed] [Google Scholar]
- Hernandez‐Verdun D, Roussel P (2003) Regulators of nucleolar functions. Prog. Cell Cycle Res. 5, 301–308. [PubMed] [Google Scholar]
- Jay DG (1988) Selective destruction of protein function by chromophore‐assisted laser inactivation. Proc. Natl. Acad. Sci. USA 85, 5454–5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kausch I, Lingnau A, Endl E, Sellmann K, Deinert I, Ratliff TL, Jocham D, Sczakiel G, Gerdes J, Böhle A (2003) Antisense treatment against Ki‐67 mRNA inhibits proliferation and tumor growth in vitro and in vivo . Int. J. Cancer 105, 710–716. [DOI] [PubMed] [Google Scholar]
- Kausch I, Jiang H, Brocks C, Bruderek K, Krüger S, Sczakiel G, Jocham D, Böhle A (2004) Ki‐67‐directed antisense therapy in an orthotopic renal cell carcinoma model. Eur. Urol. 46, 118–124. [DOI] [PubMed] [Google Scholar]
- Key G, Becker MH, Baron B, Duchrow M, Schlüter C, Flad HD, Gerdes J (1993) New Ki‐67‐equivalent murine monoclonal antibodies (MIB 1–3) generated against bacterially expressed parts of the Ki‐67 cDNA containing three 62 base pair repetitive elements encoding for the Ki‐67 epitope. Lab. Invest. 68, 629–636. [PubMed] [Google Scholar]
- Lepock JR, Thompson JE, Kruuv J (1978) Photoinduced cross‐linking of membrane proteins by fluorescein isothiocyanate. Biochem. Biophys. Res. Commun. 85, 344–350. [DOI] [PubMed] [Google Scholar]
- Liao JC, Roider J, Jay DG (1994) Chromophore‐assisted laser inactivation of proteins is mediated by the photogeneration of free radicals. Proc. Natl. Acad. Sci. USA 91, 2659–2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochs RL, Lischwe MA, Shen E, Carroll RE, Busch H (1985) Nucleologenesis: composition and fate of prenucleolar bodies. Chromosoma 92, 330–336. [DOI] [PubMed] [Google Scholar]
- Scholzen T, Gerdes J (2000) The Ki‐67 protein: from the known and the unknown. J. Cell. Physiol. 182, 311–322. [DOI] [PubMed] [Google Scholar]
- Schwarting R, Gerdes J, Ziegler A, Stein H (1987) Immunoprecipitation of the interleukin‐2 receptor from Hodgkin's disease derived cell lines by monoclonal antibodies. Hematol. Oncol. 5, 57–64. [DOI] [PubMed] [Google Scholar]
- Surrey T, Elowitz MB, Wolf PE, Yang F, Nédélec F, Shokat K, Leibler S (1998) Chromophore‐assisted light inactivation and self‐organization of microtubules and motors. Proc. Natl. Acad. Sci. USA 95, 4293–4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan P, Xiong Y, Chen B, Negash S, Squier TC, Mayer MU (2006) Fluorophore‐assisted light inactivation of calmodulin involves singlet‐oxygen mediated cross‐linking and methionine oxidation. Biochemistry 45, 4736–4748. [DOI] [PubMed] [Google Scholar]


