Abstract
Abstract. Glyceraldehyde‐3‐phosphate dehydrogenase is a multifunctional protein possessing numerous cytoplasmic and nuclear functions associated with cellular proliferation. Despite the emerging role of glyceraldehyde‐3‐phosphate dehydrogenase in regulating the proliferative process, there is a paucity of data regarding its expression and intracellular distribution in non‐malignant proliferating hepatocytes. Thus the aim of the present study was to document the intracellular distribution of glyceraldehyde‐3‐phosphate dehydrogenase protein in proliferating hepatocytes derived from regenerating rat livers, and glyceraldehyde‐3‐phosphate dehydrogenase gene expression in fasted and re‐fed rats following partial hepatectomy (PHx). Glyceraldehyde‐3‐phosphate dehydrogenase mRNA and protein expression were documented by Northern and Western blot analyses, respectively, at various times following 70% PHx in adult Sprague–Dawley rats. At 24 h post‐surgery, glyceraldehyde‐3‐phosphate dehydrogenase mRNA expression was significantly increased in both PHx and sham operated rats (P < 0.001), respectively. Despite the increase in glyceraldehyde‐3‐phosphate dehydrogenase mRNA expression in both groups, only PHx rats had a significant increase in the nuclear fraction of glyceraldehyde‐3‐phosphate dehydrogenase protein (threefold increase compared to sham and baseline levels, P < 0.01), cytoplasmic levels of glyceraldehyde‐3‐phosphate dehydrogenase protein remained unaltered in both groups. In terms of the effects of feeding and fasting on rats there were no significant differences in glyceraldehyde‐3‐phosphate dehydrogenase mRNA levels, whether fasted or refed, in rats that had undergone PHx, 8 h earlier. On the other hand, glyceraldehyde‐3‐phosphate dehydrogenase mRNA levels were significantly increased in refed compared to fasted sham operated rats 8 h following surgery. Serum insulin concentrations were higher in the refed PHx and sham groups compared to their fasted counterparts. The results of this study indicate that although glyceraldehyde‐3‐phosphate dehydrogenase mRNA are altered to the same extent in PHx and sham‐operated rats following surgery, increases in the nuclear fraction of glyceraldehyde‐3‐phosphate dehydrogenase protein only occur in PHx rats. The results also indicate that glyceraldehyde‐3‐phosphate dehydrogenase expression is affected by the nutritional status of animals undergoing abdominal sham surgery.
Introduction
Glyceraldehyde‐3‐phosphate dehydrogenase (EC1.2.1.12) (GAPDH) is a key glycolytic enzyme which catalyses the reversible conversion of glyceraldehyde‐3‐phosphate to 1,3‐bisphosphoglycerate (Dandliker & Fox 1955; Fox & Dandliker 1956; Harris & Perham 1963). Traditionally, GAPDH was considered a highly conserved housekeeping protein solely involved in intermediary cellular metabolism, however, within recent years GAPDH has been implicated in a number of additional biological activities. Several independent studies employing cell culture systems have demonstrated that GAPDH is indeed a multifunctional protein possessing cytoplasmic and nuclear functions which include regulation of translation through interactions with RNA species (Singh & Green 1993; Nagy & Rigby 1995), DNA repair (Meyer‐Siegler et al. 1992a; Baxi & Vishwananatha 1995; McNulty & Tascanco 1995), regulation of DNA replication (Baxi & Vishwananatha 1995) and regulation of apoptosis (Ishitani & Chaung 1996; Ishitani et al. 1996a; Ishitani et al. 1996b).
Cell culture systems have also been employed to demonstrate that GAPDH expression is cell‐cycle dependent (Cool & Sirover 1989; Meyer‐Siegler et al. 1992b; Mansur et al. 1993). Most recently, interest in GAPDH has focused on the finding that GAPDH gene expression is markedly elevated in various malignant tissues: human lung (Tokunaga et al. 1987), cervical (Kim et al. 1998), pancreatic (Schek et al. 1988), colon (Schek et al. 1988) and hepatocellular carcinoma (HCC) (Gong et al. 1996); and several transformed cell lines (Bhatia et al. 1994).
Despite the emerging consensus that GAPDH plays an important role in cell proliferation, a number of important issues regarding GAPDH remain unresolved. For example, it has yet to be determined whether the nuclear localization of GAPDH in proliferating cells in vitro (Cool & Sirover 1989) extends to proliferating cells in vivo where hormonal influences such as circulating insulin levels, which are known to effect GAPDH expression, are likely to be relevant. In addition, because GAPDH mRNA expression continues to be used as a marker for mRNA loading in Northern blot analysis, it is important to document whether its expression is altered in rapidly proliferating cells, such as hepatocytes, following partial hepatectomy (PHx) and whether the fasting versus fed state of the animal effects GAPDH expression. Finally, whether the association of increased GAPDH expression and carcinoma can be utilized for diagnostic purposes in patients with malignancies has yet to be determined.
Thus, in the present study, we documented the intracellular distribution (nuclear versus cytosolic) of GAPDH in proliferating hepatocytes derived from regenerating rat livers in situ, and GAPDH mRNA expression at various times post‐PHx in fasted and re‐fed rats. We also documented serum GAPDH enzyme activity and protein levels in cirrhotic patients with and without HCC.
MATERIALS AND METHODS
Human study
Patient population
Blood samples were collected from five untreated, histologically confirmed, cirrhotic patients with radiological and/or histological evidence of HCC, five cirrhotic patients with no evidence of HCC and five healthy controls. There were no significant differences in age, gender or underlying causes of liver disease in the two cirrhotic groups. Sera were separated from whole blood at the time of collection and stored at –40 °C until used for further analyses.
Enzymatic analyses
GAPDH enzyme activity was determined spectrophotometrically by monitoring the appearance of NADH (Velick 1955) in a 3‐ml reaction mixture containing sodium pyrophosphate, sodium arsenate, nicotinamide adenine dinucleotide (NAD+), dithiothreitol (DTT) and dl‐glyceraldehyde‐3‐phosphate. Changes in the absorbance at 340 nm were documented during the first 5 min of the assay and the initial linear portion of the curve was used to quantify GAPDH activity. The lower limit of detection of the assay was 10 µg/ml. Commercial GAPDH from human erythrocytes (Sigma, St Louis, MO, USA) served as a positive control.
Western blot analyses
One hundred micrograms of serum protein was separated on a 10% polyacrylamide gel in the presence of sodium dodecyl sulphate (SDS) and subsequently transferred to nitrocellulose. Membranes were then exposed to a 1 : 400 dilution of mouse monoclonal antihuman GAPDH antibodies (kindly provided by Dr M. Sirover, Fels Institute, Philadelphia, PA, USA). Immunostained bands were visualized using horseradish peroxidase‐conjugated rabbit anti‐mouse immunoglobulin G (IgG) and an enhancing chemiluminescent solution (ECL) (ECL Kit, Amersham, Amersham, UK). The immunostaining procedure was able to detect as little as 10 pg of protein. Relative densities were assessed using an HP scanner and a Macintosh computer with National Institutes of Health (NIH), Bethesda, MD, USA image software. Commercial GAPDH from human erythrocytes (Sigma) served as a positive control.
Animal study
Animals and surgery
Ten adult male Sprague–Dawley rats (250–300 g body weight) were maintained on rat chow and water ad libitum until the day before surgery, when food but not water was withdrawn. All animals were kept in identical housing on a cycle of 12 h of light and 12 h of dark. 70% PHx were performed between 9 a.m. and noon each day while the animals were under light ether anaesthesia according to the method described by Higgins and Anderson (Higgins & Anderson 1931). Resected liver tissue was collected and served as a baseline control. Sham operations, in which the appropriate portions of the liver were exteriorized for the same length of time as rats undergoing PHx, were also carried out. All animals were provided 20% sucrose solutions for nutritional support during the immediate 24 h following surgery.
Rats were sacrificed 24 h after surgery by exanguination under ether anaesthesia, following which livers were excised, weighed, frozen in liquid nitrogen and stored at –70 °C until further analyses were performed.
Northern blot analyses
Total RNA was extracted from tissue samples by the previously described LiCl–urea method (Assy et al. 1998). The RNA was transferred to GT membranes (Bio‐Rad, Hercules, CA, USA), and baked at 80 °C for 2 h. Membranes were then prehybridized in hybridization solution (50% formamide, 0.25 mol/l NaCl, 0.2 mol/l NaCl and 7% SDS for 2 h at 42 °C. Following prehybridization, a radiolabelled GAPDH cDNA probe (Edwards et al. 1985) was added to the hybridization mixture over night at 42 °C. Membranes were then washed twice with 2× sodium chloride/sodium citrate (SSC)/0.1% SDS at room temperature for 15 min and 0.2× SSC/0.1% SDS at 65 °C for 15 min Following exposure to X‐ray film (X‐OMAT‐AR, Kodak, Rochester, NY, USA), the membranes were stripped in 0.1× SSC/0.5% SDS at 95 °C for 40 min and reprobed with radio‐labelled 28S‐rRNS cDNA under the conditions described above. Optical densities were determined by an NIH Image Program. GAPDH gene expression was normalized to 28S‐rRNA to permit quantitative comparisons throughout the study.
Protein isolation and subcellular fractionation
The frozen liver tissue was suspended in four volumes of buffer A [50 mm Tris‐HCl (pH 8.0), 25 mm KCl, 5 mm MgCl2, 1 mm DTT, 1 mm ethylenediaminetetraacetic acid (EDTA), 250 mm surose, 0.7 g/ml pepstatin A, 1 mm phenylmethylsulphonyl fluoride, 0.5 g/ml leupeptin, 1 g/ml aprotinin], cut into pieces and homogenized with 15 strokes (700 r.p.m.) using a Potter‐Elvejhem apparatus of 90–120 µm clearance. The homogenate was filtered through four layers of cheesecloth, then centrifuged at 800 g for 10 min. The supernatant was collected and the sediment of crude nuclei was resuspended in an equal volume of buffer A. Homgenization, centrifugation and resuspension was repeated twice, with the supernatant being collected and pooled each time (pooled crude cytosolic fraction). The final nuclear pellet was resupended in 5 ml/g of buffer A.
Cytosolic preparation
Ammonium sulphate fractionation was performed following the method of Scheek & Slater (1982). Following the final precipitation the protein was removed by centrifugation (10 000 g for 15 min), and the pellet resuspended in 2 ml of buffer B [20 mm Tris–HCl (pH 7.5), 1 mm DTT, 1 mm EDTA, 10% glycerol] and dialysed overnight against buffer B to remove ammonium sulphate. Dialysed samples were stored frozen at –70 °C until diethylaminoethyl (DEAE) chromatography was performed.
Nuclear preparation
Nuclei purification and protein precipitation were performed according to the method described by Domena & Mosbaugh (1985). Following precipitation the protein was removed by centrifugation (15 000 g for 15 min), and the precipitate resuspended in 1–2 ml of buffer B and dialysed against the same buffer. Precipitate formed during dialysis was removed by centrifugation (15 000 g for 10 min). The supernatant was stored at –70 °C until DEAE chromatography was performed.
DEAE–Sepharose chromatography
A 10‐ml column containing 3 ml of DEAE Sepharose CL‐6B resin (Pharmacia, Quebec, Canada) was equilibrated with buffer B. Cytosolic and nuclear fractions were loaded and the column washed with three bed volumes of buffer B. Eluted fractions were stored at –70 °C until Western blot analysis were performed.
Western blot analyses
Protein was quantified in the eluted fractions using the Bio‐Rad protein assay. Those containing the highest concentration of protein were used for subsequent analyses. Equal amounts of protein (40 µg per lane) were separated, transferred, and quantified as described above, other than immunoblotting which was performed with a 1 : 200 dilution of monoclonal anti‐rabbit GAPDH (Biodesign International. Kennebunk, ME, USA).
Refed/fasting study
Sixteen male Sprague–Dawley rats were fasted overnight, prior to surgery. At surgery, eight rats underwent PHx while the remaining eight had sham operations (as described above). Post‐operative nutritional support, the 20% sucrose solution, was provided to four of the animals in each group, while the remaining four remained fasted during the post‐operative period. All animals were sacrificed at 8 h post‐operatively, when blood samples were collected via cardiac puncture, and the remnant livers were excised and frozen in liquid nitrogen. GAPDH expression was determined in these liver samples by Northern blot analyses and serum insulin concentrations by a commercial immunoassay kit (Mercodia Rat Insulin ELISA, ALPCO Ltd. Windham, NH, USA).
RESULTS
Human study
GAPDH enzymatic activity could not be detected in the sera from HCC patients, non‐HCC cirrhotic or healthy controls. Similarly, the results of Western blot analyses revealed that the 37 kDa subunit of GAPDH was not present in the sera from these groups (data not shown).
Animal study
GAPDH expression in regenerating hepatocytes
Figure 1 provides the results of GAPDH mRNA expression at baseline and in PHx and sham operated control rats at 24 h post‐PHx. In both PHx and sham groups GAPDH expression were approximately 1.5‐fold greater than baseline expression (P < 0.001).
Figure 1.
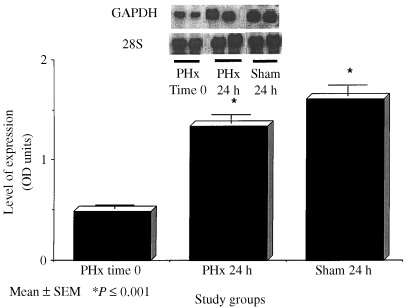
GAPDH mRNA expression as measured by Northern Blot analyses in the livers of rats at baseline, 24 h following 70% PHx and sham surgery. Data columns represent mean ± SEM. *P ≤ 0.05 versus baseline and sham (N = 4–6/group).
Subcellular fractionation
Changes in the abundance of GAPDH protein in specific subcellular compartments as detected by Western blot analyses at 24 h post‐PHx are shown in (2, 3). As shown in Fig. 2, greater than threefold increases in GAPDH protein were detected in the nuclear fractions of rats who had undergone a 70% PHx compared to sham and baseline levels (P < 0.05). While no differences in cytoplasmic concentration were detected (Fig. 2).
Figure 2.
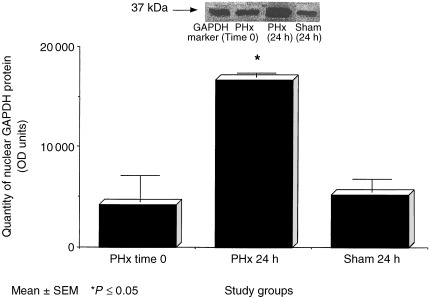
Nuclear content of GAPDH protein as measured by Western blot analyses in the livers of rats at baseline, 24 h following 70% PHx and sham surgery. Data columns represent mean ± SEM. *P ≤ 0.05 versus baseline and sham (N = 3–4/group).
Figure 3.
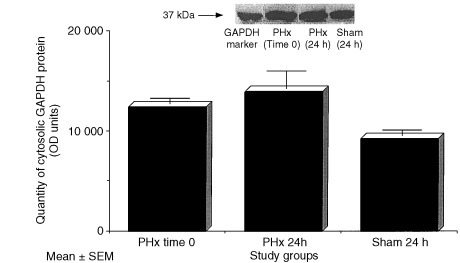
Cytosolic content of GAPDH protein as measured by Western blot analyses in the livers of rats at baseline, 24 h following 70% PHx and sham surgery. Data columns represent mean ± SEM (N = 3–4/group).
Refed/fasting study
Figure 4 provides the results of GAPDH mRNA expression following PHx or sham surgery in rats refed or fasted in the 8‐h post‐operative period. Refed sham operated rats had significantly higher GAPDH mRNA levels than fasted sham rats (P < 0.001), while animals who had undergone PHx had similar levels regardless of their refed or fasted state. Serum insulin concentrations were also determined (Fig. 5) and refed sham and PHx groups had significantly higher levels of insulin than their fasted counterparts (P < 0.01).
Figure 4.
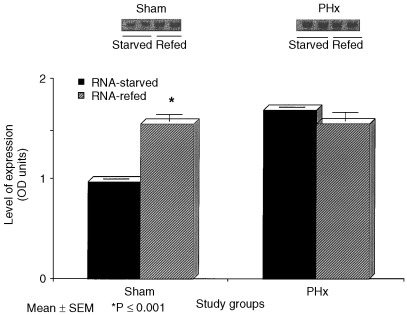
GAPDH mRNA expression as measured by Northern blot analyses in the livers of starved and refed rats 8 h following 70% and sham operations. Data columns represent mean ± SEM. *P ≤ 0.001 versus sham‐starved (N = 4/group).
Figure 5.
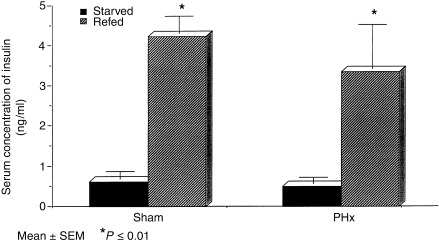
Serum insulin concentrations from starved and refed rats 8 h following 70% and sham operations. Data columns represent mean ± SEM. *P ≤ 0.01 versus sham‐starved and PHx‐starved (N = 3–4/group).
DISCUSSION
Previous studies have reported increased GAPDH mRNA expression in several types of cancer and transformed tumour cell lines (Tokunaga et al. 1987; Schek et al. 1988; Bhatia et al. 1994; Gong et al. 1996; Kim et al. 1998). In our own laboratory marked increases of GAPDH mRNA were observed in HCC compared to that seen in cirrhotic and normal liver tissue (Gong et al. 1996). Because GAPDH is also involved in transmembrane transport we tested the hypothesis that marked increases in GAPDH expression would be accompanied by increased levels of the enzymatic activity or protein in the sera of patients with HCC. However, the results of the present study indicate that neither GAPDH enzymatic activity nor protein could be detected in sera from patients with HCC or those with cirrhosis but no HCC or from healthy controls.
Intracellular GAPDH activity has been identified in the cytoplasmic and nuclear fraction of cells. Which fraction becomes dominant during cellular proliferation is unknown but such information could provide important insights into the role of GAPDH in regulated and unregulated or malignant growth states. In the cytoplasm, GAPDH binds to microtubule elements and plays an important role in endocytosis (Muronetz et al. 1994; Robbins et al. 1995). Cytosolic GAPDH has also been shown to bind to the AU rich regions in numerous RNA species and thereby influences the translation of these genes. Finally, there is the traditional role of GAPDH as a glycolytic enzyme, a process that is confined to the cytoplasm. Within the nucleus, GAPDH performs the following activities: (1) DNA repair, via uracil DNA glycosylase activity (Meyer‐Siegler et al. 1992a; Baxi & Vishwananatha 1995; McNulty & Tascanco 1995); (2) DNA replication, through its capacity to bind to Ap4A nucleotides (Baxi & Vishwananatha 1995); and (3) translational regulation, through its actions as a nuclear tRNA export protein (Nagy & Rigby 1995). The results of our study indicate that the nuclear fraction of GAPDH expression is preferentially up‐regulated in rapidly proliferating hepatocytes. This finding suggests that the principal role of increased GAPDH expression in proliferating hepatocytes is to regulate DNA synthesis and/or repair rather then to contribute to cellular metabolic activity. Similar results were described in human fibroblasts where enhanced fluorescence for GAPDH was detected in the nuclear and perinuclear regions of these cells during active proliferation (Cool & Sirover 1989). Further experiments must be performed to elucidate the precise role of this protein within the nucleus of rapidly proliferating cells.
In addition to differences in intracellular distribution, significant increases in GAPDH mRNA expression were observed during the post‐PHx period. To date, the majority of data documenting increases in GAPDH mRNA expression in rapidly proliferating cells were derived from studies involving malignant cells and/or tissues (Tokunaga et al. 1987; Schek et al. 1988; Bhatia et al. 1994; Gong et al. 1996; Kim et al. 1998). In non‐malignant cells, Goldsworthy et al. (1993) demonstrated increased GAPDH expression in the livers of rats following acute carbon tetrachloride exposure and recently, two other groups have noted changes in the expression of GAPDH in normal tissues during ontogenic (Calvo et al. 1997) and regenerative processes (Geerloff et al. 1999). Our results support these findings and collectively, suggest that GAPDH gene expression is regulated as a function of the proliferative state of cells (Meyer‐Siegler et al. 1992b; Mansur et al. 1993).
As evidenced by the results in sham‐operated rats, GAPDH expression is also regulated by non‐regenerative factors. Indeed, it is well established that GAPDH expression is influenced by the hormonal and nutritional state of the cell (Nasrin et al. 1990). This presumably relates to the multiple insulin responsive elements characterized in the GAPDH gene (Alexander‐bridges et al. 1992). Thus, it could be argued that the 20% sucrose solution provided to rats during the post‐operative period for nutritional support, may have been responsible for the increase in GAPDH expression during the course of the study. That similar increases were observed in fasted rats post‐PHx, however, argues against this possibility. The absence of an additive increase in GAPDH expression among refed rats subject to PHx may be explained by recent findings indicating a transient down regulation of the hepatic insulin receptor following hepatectomy in rats (Kogut et al. 1998).
Interestingly, despite increased GAPDH mRNA expression neither cytosolic nor nuclear GAPDH protein were elevated in the livers of sham operated animals at 24 h post‐surgery. This finding likely reflects post‐transcriptional modification of the GAPDH message.
In summary, the principal findings of this study indicates that the nuclear compartmentalization of GAPDH documented in malignant cell cultures also occurs in non‐malignant proliferating hepatocytes in vivo. The results also indicate that GAPDH mRNA expression is influenced by abdominal surgery and in the case of sham surgery, by the fasting versus refed state of the experimental animals. Finally, peripheral blood GAPDH levels hold little promise as a diagnostic tool for HCC in humans.
References
- Alexander‐bridges M, Dugast I, Ercolani L, Kong XF, Giere L, Nasrin N (1992) Multiple insulin‐responsive elements regulate transcription of the GAPDH gene. Adv. Enzyme Regul 32, 149. [DOI] [PubMed] [Google Scholar]
- Assy N, Gong Y, Zhang M, Pettigrew M, Pashniak D, Minuk GY (1998) Use of proliferating cell nuclear antigen as a marker of liver regeneration after partial hepatectomy in rats. J. Lab. Clin. Med. 131, 251. [DOI] [PubMed] [Google Scholar]
- Baxi MD, Vishwananatha JK (1995) Uracil DNA glycosylase/ glyceraldehyde‐3‐phosphate dehydrogenase is an Ap4A binding protein. Biochemistry 34, 9700. [DOI] [PubMed] [Google Scholar]
- Bhatia P, Taylor R., Greenberg AH, Wright JA (1994) Comparison of glyceraldehyde‐3‐phosphate dehydrogenase and 28S‐ribosomal RNA gene expression as RNA loading controls for northern blot analysis of cell lines of varying malignant potential. Anal. Biochem. 216, 223. [DOI] [PubMed] [Google Scholar]
- Calvo EL, Boucher C, Coulombe Z, Morisset J (1997) Pancreatic GAPDH gene expression during ontogeny and acute pancreatits induced by caerulein. Biochem. Biophys. Res. Commun. 235, 636. [DOI] [PubMed] [Google Scholar]
- Cool BL, Sirover MA (1989) Immunocytochemical localization of the base excision repair enzyme uracil DNA glycosylase in quiescent and proliferating normal human cells. Cancer Res. 49, 3029. [PubMed] [Google Scholar]
- Dandliker WB, Fox JB (1955) Light Scattering of DL‐glyceraldehyde‐3‐phosphate dehydrogenase. J. Biol. Chem. 214, 275. [PubMed] [Google Scholar]
- Domena JD, Mosbaugh DW (1985) Purification of nuclear and mitochondrial uracil‐DNA glycosylase from rat liver. Identification of two distinct subcellular forms. Biochemistry 24, 7320. [DOI] [PubMed] [Google Scholar]
- Edwards DR, Parefett CL, Denhardt DT (1985) Transcriptional regulation of two serum‐induced RNA’s in mouse fibroblasts: equivalence of one species to B2 repetitive elements. Mol. Cell Biol. 11, 3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox JB, Dandliker WB (1956) A study of physical properties of glyceraldehyde‐3‐phosphate dehydrogenase. J. Biol. Chem 218, 53. [PubMed] [Google Scholar]
- Geerloff T, Geier A, Stieger B, Hagenbuch B, Meier PJ, Mateern S, Gartung C (1999) Differential expression of basolateral canalicular organic anion transporters during regeneration of rat liver. Gastroenterology 117, 1408. [DOI] [PubMed] [Google Scholar]
- Goldsworthy SM, Goldsworthy TL, Sprankle CS, Butter BE (1993) Variation in expression of genes used for normalization of Northern blots after induction of cell proliferation. Cell Prolif 26, 511. [DOI] [PubMed] [Google Scholar]
- Gong Y, Cui L, Minuk GY (1996) Comparison of glyceraldehyde‐3‐phosphate dehydrogenase and 28S‐ribosomal RNA gene expression in human hepatocellular carcinoma. Hepatology 23, 734. [DOI] [PubMed] [Google Scholar]
- Harris I, Perham RN (1963) Studies on glyceraldehyde‐3‐phosphate dehydrogenase. Biochem. J. 89, 60P. [Google Scholar]
- Higgins GM, Anderson RM (1931) Experimental pathology of liver: resection of liver of the white rat following partial surgical removal. Arch. Pathol. 12, 186. [Google Scholar]
- Ishitani R., Chaung D‐M (1996) Gylceraldehyde‐3‐phosphate dehydrogenase antisense oligodeoxynucleotides protect against cytosine arabinonucleoeoside‐induced apoptosis in culture cerebellar neurons in culture. Proc. Natl Acad. Sci. USA 93, 9937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishitani R., Kimura M, Sunaga K, Katsube T, Tankan M, Chuang DM (1996a) An antisense oligodeoxynucleotide to glyceraldehye‐3‐phosphate dehydrogenase blocks age induced apoptosis in mature cerebrocortical neurons on culture. J. Pharm. Exp Ther. 278, 447. [PubMed] [Google Scholar]
- Ishitani R., Sunaga K, Hirano A, Saunders P, Katsube N, Chuang DM (1996b) Evidence that glyceraldehyde‐3‐phosphate dehydrogenase is involved in age‐induced apoptosis in mature cerebellar neurons in culture. J. Neurochem. 66, 928. [DOI] [PubMed] [Google Scholar]
- Kim JW, Kim SL, Han SM, Paik SY, Hur SY, Kim YW, Lee JM, Namkong SE (1998) Increased glyceraldehyde‐3‐phosphate dehydrogenase gene expression in human cervical cancers. Gynecol. Oncol. 71, 266. [DOI] [PubMed] [Google Scholar]
- Kogut KA, Nifong LW, Witt MJ, Krusch DA (1998) Hepatic insulin extraction after major hepatectomy. Surgery 4, 415. [PubMed] [Google Scholar]
- Mansur NR, Meyer‐Siegler K, Wurzer JC, Sirover MA (1993) Cell cycle regulation of the glyceraldehyde‐3‐phosphate dehydrogenase/ uracil DNA glycosylase gene in normal human cells. Nucl. Acid Res. 21, 993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNulty SE, Tascanco WA JR (1995) Transcripitional regulation of glyceraldehyde‐3‐phosphate dehydrogenase by 2,3,7,8‐tetrachlorodibenzenzo‐p‐dioxin. Biochem. Biophys. Res. Commun. 216, 165. [DOI] [PubMed] [Google Scholar]
- Meyer‐Siegler K, Mansur NR, Wurzer JC, Sirover MA (1992b) Proliferative dependent regulation of the glyceraldehyde‐3‐phosphate dehydrogenase/ uracil DNA glycosylase gene in human cells. Carcinogenesis 13, 2127. [DOI] [PubMed] [Google Scholar]
- Meyer‐Siegler K, Munro DJ, Seal G, Wurzer JC, Sirover MA (1992a) A human nuclear uracil DNA glycolase is the 37 kDa subunit of glyceraldehyde‐3‐phosphate dehydrogenase. Proc. Natl Acad. Sci. USA 88, 8460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muronetz VI, Wang ZX, Keith TJ, Knull HR, Srivastava DK (1994) Binding constants and stoichiometries of glyceraldehyde‐3‐phosphate dehydrogenase‐tubulin complexes. Arch. Biochem. Biophys. 313, 253. [DOI] [PubMed] [Google Scholar]
- Nagy E, Rigby WFC (1995) Glyceraldehyde‐3‐phosphate dehydrogenase selectively binds AU‐rich RNA in NAD+‐binding region (Rossman fold). J. Biol. Chem 270, 2755. [DOI] [PubMed] [Google Scholar]
- Nasrin N, Ercolani I, Denaro M, Kong XF, Kang I, Alexander M (1990) An insulin response element in the glyceraldehyde‐3‐phosphate dehydrogenase gene binds a nuclear protein induced by insulin in cultured cells and by nutritional manipulations in vivo . Proc. Natl Acad. Sci. USA 87, 5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins AR, Ward RD, Oliver C (1995) A mutation in glyceraldehyde‐3‐phosphate dehydrogenase alters endocytosis in CHO cells. J. Cell Biol. 130, 1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheek RM, Slater EC (1982) Glyceraldehyde‐3‐phosphare dehydrogenase from rabbit muscle. Methods Enzymol. 89, 305. [DOI] [PubMed] [Google Scholar]
- Schek N, Hall BL, Finn OJ (1988) Increased glyceraldehyde‐3‐phosphate dehydrogenase gene expression in human pancreatic adenocarcinoma. Cancer Res. 48, 6354. [PubMed] [Google Scholar]
- Singh R., Green MR (1993) Sequence‐specificity binding of transfer RNA by glyceraldehyde‐3‐phosphate dehydrogenase. Science 259, 365. [DOI] [PubMed] [Google Scholar]
- Tokunaga K, Nakamura Y, Sakata K, Fujimori K, Ohkubo M, Sawada K, Sakiyama S (1987) Enhanced expression of glyceraldehyde‐3‐phosphate dehydrogenase gene in human lung cancers. Cancer Res. 47, 5616. [PubMed] [Google Scholar]
- Velick S (1955) Glyceraldehyde‐3‐phosphate dehydrogenase from muscle. Methods Enzymol. 1, 401. [Google Scholar]


