Abstract
Objectives
Peripheral nerve injuries are a common occurrence, resulting in considerable patient suffering; it also represents a major economic burden on society. To improve treatment options following peripheral nerve injuries, scientists aim to find a way to promote Schwann cell (SC) myelination to help nerves to carry out their functions effectively. In this study, we investigated myelination ability of SCs, regulated by co‐culture with adipose‐derived stem cells (ASCs) or low‐intensity pulsed ultrasound (LIPUS), and synergistic effects of combined treatments.
Materials and methods
Schwann cells were co‐cultured with or without ASCs, and either left untreated or treated with LIPUS for 10 min/d for 1, 4 or 7 days. Effects of LIPUS and ASC co‐culture on pro‐myelination indicators of SCs were analysed by real‐time PCR (RT‐PCR), Western blotting and immunofluorescence staining (IF).
Results
Our results indicate that ASC‐SC co‐culture and LIPUS, together or individually, promoted mRNA levels of epidermal growth factor receptor 3 (EGFR3/ErbB3), neuregulin1 (NRG1), early growth response protein 2 (Egr2/Krox20) and myelin basic protein (MBP), with corresponding increases in protein levels of ErbB3, NRG1 and Krox20. Interestingly, combination of ASC‐SC co‐culture and LIPUS displayed the most remarkable effects.
Conclusion
We demonstrated that ASCs upregulated pro‐myelination indicators of SCs by indirect contact (through co‐culture) and that effects could be potentiated by LIPUS. We conclude that LIPUS, as a mechanical stress, may have potential in nerve regeneration with potential clinical relevance.
1. Introduction
Over 20 million Americans suffer from peripheral nerve injuries, resulting in a devastating decrease in patients’ quality of life and accounting for 150 billion dollars in health care costs annually.1, 2 Thus far, treatment options for peripheral nerve injuries are limited, but in vitro studies have shown that Schwann cells (SCs) may promote the regeneration of impaired nervous tissue.1 SCs are a subset of peripheral glial cells that promote peripheral nerve regeneration following injury by providing structural guidance and support via the release of neurotrophic factors.3, 4 SCs are considered one of the most promising tissue engineering methods for nerve regeneration and has been proposed for clinical application to remyelinate injured axons in the spinal cord.5, 6, 7, 8, 9
Several SCs proteins have been shown to be critical in facilitating their function. Neuregulin1 (NRG1) regulates all aspects of the SCs lineage, including proliferation, initial gliogenesis, ensheathment and myelination. High levels of type III NRG1 on the axonal membrane modulate the thickness of the myelin sheath and provide a signal for the induction of myelination.10, 11, 12 NRG1/ErbB (epidermal growth factor receptor/EGFR) signalling is a very important pathway for SCs response. After NRG1 binding, ErbB2/3 receptors phosphorylate specific tyrosine residues, and recruit specific adaptor proteins and enzymes. This results in the activation of a number of downstream signalling pathways (PI3K/Akt, Erk1/2, FAK, Rac/Cdc42 and so on). These pathways mediate specific SCs responses to the NRG1 signal, including survival, migration, proliferation and differentiation.13 Myelin basic protein (MBP) is a myelin‐associated protease existing in both CNS and PNS.14 MBP is critical in SCs synthesis of myelin, and therefore plays a crucial role in the early stages of myelination and maintenance of stable axon‐myelin interaction.15, 16 Egr2/Krox20 is a zinc finger protein of the evolutionarily conserved Egr family, and each member of this family (Egr1‐4) have very specific cell type‐dependent functions.17 Krox20 is an essential transcriptional activator that mediates SCs development and peripheral nerve myelination.18
Mechanical stimulation is effective in improving tissue regeneration in many cell types, including mesenchymal stem cells, osteoblasts, chondrocytes, epithelial cells and muscle cells.19, 20, 21, 22, 23, 24 Some studies on nerve regeneration showed that compressive strain and shear stress are harmful to injured nerve tissue, while tensile stress of appropriate strength and frequency is beneficial.25, 26, 27 Our team previously determined that both cyclic compressive and tensile loading downregulated expression of several important myelin‐related SCs genes, thus facilitating demyelination.28
Low‐intensity pulsed ultrasound (LIPUS) is a type of clinically effective mechanical stress that is approved to promote fracture healing by the Food and Drug Administration (FDA).29 Raso et al. demonstrated that sciatic functional index (SFI) and nerve fibre density were significantly higher in the group treated with ultrasound irradiation on sciatic nerve of rats under a controlled crush injury. They concluded that low‐intensity therapeutic ultrasound enhanced nerve regeneration.30 In addition, several in vitro studies have shown that LIPUS can enhance the proliferation of SCs, inhibit SCs apoptosis, and promote the secretion of neurotrophic factor.31, 32, 33, 34 According to the literature and our previous work, 1.0 MHz frequency, 20% duty ratio, and 20 mW/cm2 intensity are reasonable LIPUS parameters for cells that do not cause heat damage or proliferation.29, 35
In recent years, adipose‐derived stem cells (ASCs) have attracted public attention as a stem‐cell source due to its easy material availability and self‐renewal capacity.36, 37 Our previous work showed that co‐culturing of ASCs and SCs is an effective and practical way for ASCs neural transdifferentiation. ASCs secrete many different growth factors, including VEGF, HGF, bFGF and BDNF, which can stimulate blood vessel or nerve growth.38, 39, 40 Importantly, co‐culturing in Transwell plates without intercellular contacts proved to be effective via sharing growth media.41
Tissue engineering researchers are constantly looking for new ways to shorten healing time of a damaged nerve by promoting SCs function. Currently, there was no research to discuss SCs response to LIPUS under co‐culture conditions. Based on previous work, in the study we focused on evaluating the recovery capacity of nerve fibres after treatment with these two factors. We looked at the effectiveness of LIPUS and co‐culture individually, as well as their combinatorial effects for synergism in promoting neural repair.
2. Materials and methods
2.1. Cell culture
All animal experiments described in this report were approved by the Sichuan University Animal Care and Use Committee. In total, 50 Sprague‐Dawley rats were used. The ethics committee number of this experiment is WCCSIRB‐D‐2014‐081.
Rat ASCs (rASCs) are found in adipose tissue, which were obtained as subcutaneous fat from the groin of the rats (female, 2‐week old, Experimental Animal Center of Sichuan University, China). The fat tissue was carefully washed and minced, and then digested in collagenase type I (0.1% collagenase type I in α‐MEM medium without FBS) at 37°C water bath for an hour. The cells were then seeded on culture flasks (25 cm2) with total α‐MEM medium (α‐MEM medium supplemented with 10% FBS, 1% penicillin/streptomycin) and incubated at 37°C in a humidified atmosphere of 95% air and 5% CO2. After 5‐ to 7‐day culture, non‐adherent cells were removed and the adherent cells were cultured in a monolayer; the media was changed every 2–3 days, and the cells were subcultured when cells reached confluence (80%). We used the passage 3 cells and seeded them to culture plate (six‐well plate, 9.5 cm2/12‐well plate, 3.9 cm2) at an appropriate density in total supplemented α‐MEM medium.
Rat Schwann cell line RSC96 (ATCC, USA) was used as the source of SCs. SCs were seeded at an appropriate density in 6‐ or 12‐well plate with total low‐sugar DMEM medium (DMEM medium supplemented with 10% FBS, 1% penicillin/streptomycin).
The co‐culture condition was set up using polycarbonate membrane Transwell plates (Corning, NY, USA). The porous membrane used is optically transparent with a pore size 0.4 μm. In our experiments, the pore size is much smaller than the size of the ASCs body (approximately 80–160 μm), which greatly inhibits the migration of ASCs into the lower chamber. The ASCs (3 × 104/cm2) were cultured on the upper permeable membrane support and SCs (6 × 104/cm2) were cultured in lower plate chamber. Both cells were co‐cultured in total α‐MEM medium, so both cells were cultured in the same culture media without direct cell contact. The culture media was changed every 3 days.
There were four groups in our experimental conditions: group A, B, C and D. Group A and B were co‐cultured groups (A, co‐culture + LIPUS; B, co‐culture without LIPUS), group C was LIPUS treatment only (SCs culture + LIPUS) and group D was a control (SCs culture).
2.2. Ultrasound exposure
In this report, group A consisted of SCs co‐cultured with ASCs and exposed to LIPUS (co‐culture + LIPUS) (Fig. 1). Group B consisted of co‐cultured SCs without LIPUS (co‐culture without LIPUS). Group C consisted of SCs that underwent LIPUS without co‐culture (SCs culture + LIPUS), and group D (SCs culture) was control SCs cultured in α‐MEM media without any other treatment.
Figure 1.
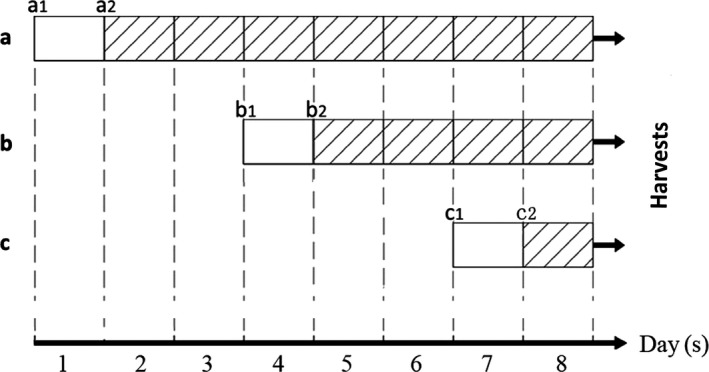
Experimental design to test RT‐PCR, Western blot and immunofluorescence. a, b and c represent 7‐day, 4‐day and 1‐day groups respectively, and each group contains group A (co‐culture + LIPUS), group B (co‐culture without LIPUS), group C (SCs culture + LIPUS) and group D as control (SCs culture). a1, b1 and c1 respectively display the cells of the third passage planted at culture plate with an appropriate density. a2, b2 and c2 indicate the SCs were cultured after 24 h of the cells planted at plate and group A and C were treated by LIPUS exposure (1.0 MHz frequency, 20% duty ratio, 20 mW/cm2 intensity) for 10 min every day. Finally, after 10 min LIPUS followed by incubation on the 8th day, cells were harvested by different methods
For group A and B, ASCs (P3) were plated on the upper membrane, and rat SCs were seeded on the plate chamber with α‐MEM media. Cells were then placed in a cell culture incubator (37°C, 5% CO2, 95% humidity). For group C and D, SCs alone were seeded on plates with α‐MEM media. After 24 hours, cells in the LIPUS exposure groups (groups A and C) were treated with ultrasound exposure; the first treatment was started either at the 1st, 4th and 7th day after cells adherence. After LIPUS exposure for 10 minutes per day for 1, 4 or 7 days individually, all treatments were finished on the eighth day. After the last ultrasound treatment, cells were incubated for 30 minutes and samples were collected (either mRNA or protein), or cells were fixed by 4% paraformaldehyde for immunofluorescence (IF).
The LIPUS device consists of an array of six transducers, each 30 mm in diameter. The LIPUS signal we used consisted of a 1.0 MHz and 20 mW/cm² at duty ratio of 20%. The plate was placed on the ultrasound transducer array conducted by a thin layer of coupling gel. All LIPUS treatments performed with the culture plates in the tissue culture incubator (37°C, 5% CO2, 95% humidity).
2.3. RNA isolation and real‐time PCR (RT‐PCR)
We focused on transcriptional levels of ErbB3, NRG1, Krox20 and MBP in SCs by RT‐PCR. Total RNA was harvested on treatment day 8 as described previously. Initially, total SCs RNA was extracted using the Simply P Total Tissue⁄cell RNA Extraction Kit (Bioflux, Zhejiang, China) according to the manufacturer's protocol. Total RNA was reversed transcribed to cDNA by Prime Script RT reagent Kit with cDNA Eraser (Takara Bio, Tokyo, Japan) according to the manufacturer's protocol. PCR oligonucleotide primers were listed in Table 1. Gene expression was quantified with RT‐PCR, utilizing SYBR® Premix ExTaq™II (TliRNaseH Plus) (Takara).
Table 1.
Primer sequences of target genes and GAPDH for RT‐PCR assay
| Sequence (5′‐>3′) | |
|---|---|
| GAPDH | F:CCGTATCGGACGCCTGGTTA |
| R:CCGTGGGTAGAGTCATACTGGAAC | |
| ErbB3 | F:CCACATTGTACGGCTGCTAGG |
| R:TGGTCCCAATGTCTCACGGT | |
| NRG1 | F:CCTGTCAAACCCGTCAAGATACT |
| R:CTCCGCTTCCATAAATTCAATCC | |
| Krox20 | F:CCTACAATCCGCACCACCTG |
| R:GAACCTCCTGTCGCAACCCT | |
| MBP | F:GAAGTCGCAGAGGACCCAAGA |
| R:CTGCCTCCGTAGCCAAATCC |
Reactions were carried out on an ABI 7300 system (ABI, Foster City, CA, USA). Expression of GAPDH was used for standardization of RT‐PCR results, to compare mRNA levels of target genes in different amounts of samples. For each reaction, a melting curve was generated to rule out primer dimer formation and false priming followed by calculation of the 2−△△Ct to determine relative levels of mRNA. cDNA of each sample was examined using agarose electrophoresis.
2.4. Western blot
After treatment with LIPUS and/or co‐culture for 1, 4 and 7 days, SCs from the bottom layer were collected on day 8. The protein concentration was determined by BCA protein assay kit (Beyotime, China) according to the manufacturer's protocol. Equal amount of protein extracts were run on 10% SDS‐polyacrylamide gels at 80V for 20 minutes then 120V for 50 minutes, followed by transfer to a PVDF membrane at 15V for 30 minutes. The membrane was incubated with rabbit polyclonal antibody against ErbB3 (Santa Cruz Biotechnology, Santa Cruz, CA, USA), rabbit polyclonal antibody against NRG1 type III (Abcam, Cambridge, MA, USA), rabbit polyclonal rabbit polyclonal antibody against Krox20 (Abcam, Cambridge, MA, UK) or mouse antibody against GAPDH (Abcam, Cambridge, MA, USA) and incubated with anti‐rabbit IgG or anti‐mouse IgG Zhongshanjinqiao, Shanghai, China) secondary antibody. The protein bands were exposed using a scanner.
2.5. Immunofluorescence
To demonstrate distribution of ErbB3, NRG1 and Krox20 proteins, ASCs (1000/cm2) and SCs (2000/cm2) in a 12‐well plate and cultured with total α‐MEM medium for IF staining. After completing treatments followed by the 30‐min incubation, the experiment group cells and the control cells were then washed in PBS and fixed in cold 4% paraformaldehyde for 15 minutes at room temperature. Cells were then perforated by 0.5% Triton‐X and blocked in 0.5% bovine serum albumin for 15 minutes. Plates were subsequently incubated overnight at 4°C with rabbit polyclonal antibody against ErbB3 (Santa Cruz) (1:100), rabbit polyclonal antibody against NRG1 type III (Abcam, USA) (1: 200) and rabbit polyclonal antibody against Krox20 (Abcam, UK) (1:200). Sequentially, plates were incubated with secondary antibodies Alexa Fluor® 488 Donkey Anti‐Rabbit IgG (Invitrogen, Carlsbad, CA, USA) for 1 hour at room temperature and nuclei were stained with DAPI (Molecular Probes, Eugene, OR, USA) for 10 minutes.
2.6. Statistical analysis
We performed three or more independent sets of the experiments, and each experiment was run at least three times. Data are given as mean values ± SD. P values were calculated using two‐way ANOVA by graphpad prism 6.0. P<.05 was considered as statistically significant.
3. Results
3.1. Co‐culture and LIPUS together and individually increased mRNA expression of ErbB3, NRG1, Krox20 and MBP
After 1, 4 and 7 days of ultrasound exposure with or without co‐culture, mRNA expression of ErbB3, NRG1, Krox20 and MBP was detected by RT‐PCR (Fig. 2).
Figure 2.
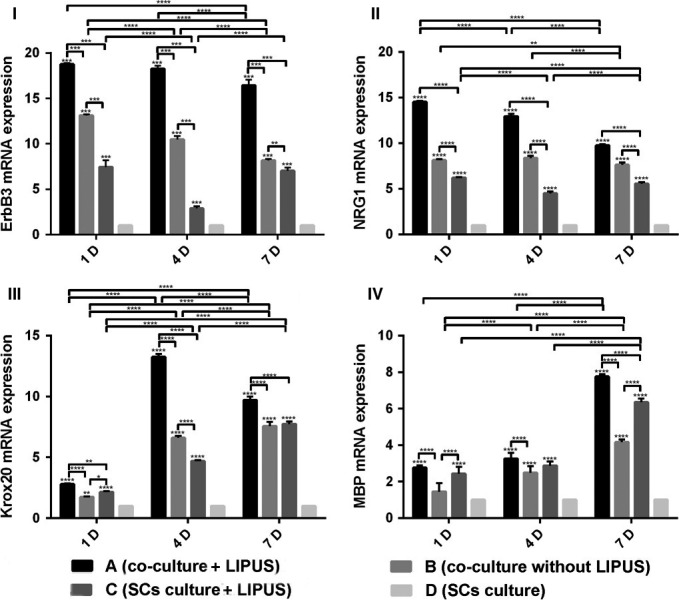
I, II, III and IV show RT‐PCR analysis of ErbB3, NRG1, Krox20 and MBP at different time points. A, B, C and D indicate groups, with group A (co‐culture + LIPUS), group B (co‐culture without LIPUS), group C (SCs culture + LIPUS) and group D as a control (SCs culture) respectively. Results demonstrated that co‐culture, ultrasound exposure or co‐culture with ultrasound exposure upregulated expression of all three genes of ErbB3, NRG1 and Krox20 compared to untreated control cultures. Method of statistical analysis was two‐way ANOVA in GraphPad Prism 6.0. *Represents significant difference from groups respectively (P<.05)
mRNA levels of ErbB3 in group A were significantly higher than all other groups at all time points, and group B and C were both significantly higher than the control at all time points. In addition, mRNA levels of ErbB3 in group B were significantly higher than group C at the time points of 4 and 7 days. With additional days of treatment, mRNA levels of ErbB3 in group A, B and C decreased.
mRNA levels of NRG1 had the same trend as ErbB3, and mRNA levels of NRG1 in group A, B and C were significantly higher than in control groups respectively at all time points. Again, mRNA expression in group A was significantly higher than all other groups.
Transcription levels of the gene coding for Krox20 were all significantly higher in group A, B and C than that of control groups at the time points of 1, 4 and 7 days. Additionally, group A was higher than group B and C, but group B was significantly lower than group C at 1 day and higher at 4 days. Of note, global transcription levels of Krox20 were significantly higher at 4 days than 1 or 7 days.
With respect to MBP, we found that mRNA levels increased in groups A, B and C as time went on, with the peak mRNA levels at 7 days. The mRNA level in group A was higher than in group B and C at almost all time points, but there was no significant difference between 1 and 4 days for group A and C. Meanwhile, group B was lower than group C at 7 days.
3.2. Different treatment increased protein expression of ErbB3, NRG1 and Krox20 by Western blot
Western blot results showed that protein levels of ErbB3, NRG1 and Krox20 were consistent with the RT‐PCR results (Fig. 3). At all time points, protein expression of ErbB3, NRG1 and Krox20 was significantly higher in group A than all other groups. In comparing groups B and C, there was no clear trend regarding protein levels of Krox20, while group B had higher protein levels of ErbB3 and NRG1 than group C. Finally, the protein expressions of ErbB3 and NRG1 gradually declined with longer treatment time, while the protein expressions of Krox20 peaked at 4 days.
Figure 3.
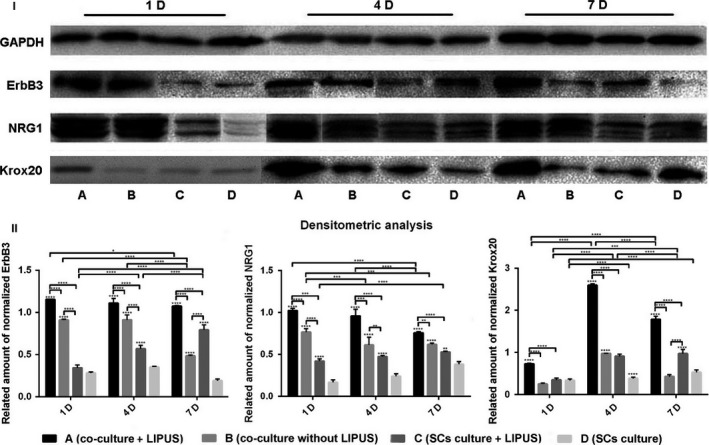
Protein expression of ErbB3, NRG1 and Krox20 were analysed by Western blot at different time points (I). A, B, C and D indicate groups: group A (co‐culture + LIPUS), group B (co‐culture without LIPUS), group C (SCs culture + LIPUS) and group D as a control (SCs culture). Co‐culture, ultrasound exposure or co‐culture with ultrasound exposure promoted expression of ErbB3, NRG1 and Krox20 compared to control at different time points, and combination of co‐culture with ultrasound exposure for SCs displayed the most remarkable effects(II). Method of statistical analysis chosen was two‐way ANOVA in graphpad prism 6.0. *Represents significant difference from groups respectively (P<.05)
3.3. Protein expression of ErbB3, NRG1 and Krox20 increased by IF after treatment
ErbB3, NRG1 and Krox20 are produced by SCs upon activation. IF was performed with anti‐ErbB3, anti‐NRG1 and anti‐Krox20 antibodies at different time points to validate protein expression levels as demonstrated by Western blot (Fig. 4).
Figure 4.
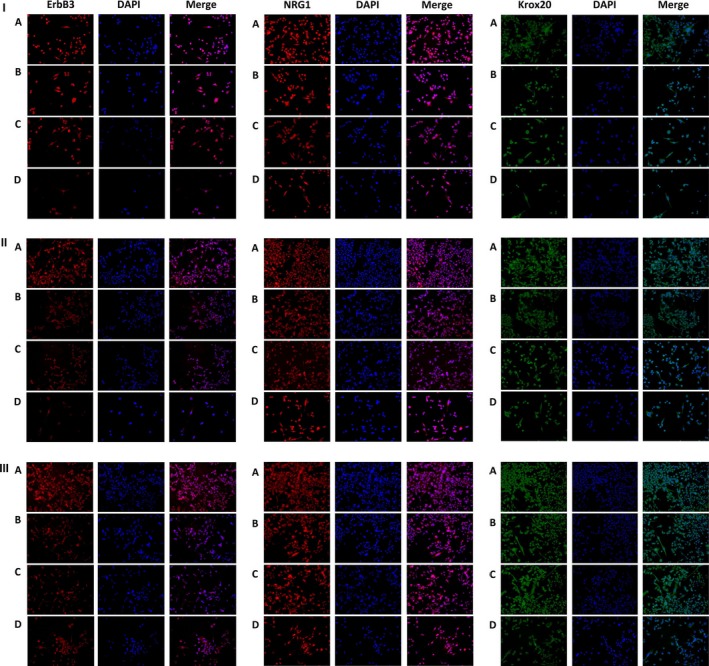
I, II and III show expression of ErbB3, NRG1 and Krox20 stained and analysed by immunofluorescence at 1, 4 and 7 d respectively. Co‐culture, ultrasound exposure or co‐culture with ultrasound exposure increased proteins expression of ErbB3, NRG1 and Krox20 compared to control at ll different time points, and the combination of co‐culture with ultrasound exposure for SCs displayed the most remarkable effect
Consistent with other experiments, protein expression levels of ErbB3, NRG1 and Krox20 by IF were higher in group A than any other group. At the same time, there was no determinate relationship between group B and C at different time points for each protein. Moreover, as treatment time increased, the Krox20 protein levels peaked at 4 days (like Western blot analysis), while the protein expression of ErbB3 and NRG1 gradually dropped (consistent with the Western blot analysis).
4. Discussion
Tissue engineering in neuroscience and regenerative medicine is the fabrication of controlled microenvironments for the study of axon guidance, with the goal of developing strategies to overcome nerve injury.35 Mechanical stress, including strain stress,42, 43, 44, 45 compressive stress,46 shear press15 and LIPUS, is getting more attention as a novel condition to manipulate nerve regeneration. Meanwhile, the use of co‐culture (shared culture media) as a method to induce stem cells to differentiation has become popular in recent years.41
In our experiment, we tested the individual and combinational function of mechanical stimulation (LIPUS) and co‐culture (ASCs with SCs) to determine if these treatment conditions’ effects on SCs myelination ability were synergistic. In this study, we gave group co‐culture and group SCs with or without LIPUS (1.0 MHz frequency, 20% duty ratio, 20 mW/cm2 intensity, 10 minutes every day) to obtain four different groups: group A (co‐culture with LIPUS), group B (co‐culture without LIPUS), group C (SCs culture with LIPUS) and group D (SCs culture). To assess the expression of SCs pro‐myelination indicators, we evaluated mRNA levels (by RT‐PCR) of ErbB3, NRG1, Krox20 and MBP, and protein levels (by Western blot and IF) of ErbB3, NRG1 and Krox20. These proteins are essential for SCs function and are considered reasonable markers for SCs activity.
NRG1 is a member of the neuregulin family and has at least 15 different isoforms from alternative splicing and multiple transcription start sites.47, 48 Structurally, all types NRG1 have a similar epidermal growth factor (EGF)‐like domain with distinct N‐terminal regions.49 Both membrane‐bound type I‐ and type III‐oriented isoforms are highly O‐glycosylated in close to the EGF‐like domain.50 Literature shows hypermyelination of peripheral nerves can be induced by neuronal overexpression of type III NRG1; however, overexpression of type I NRG1 does not affect peripheral myelination.12 In vivo models have confirmed the myelination effect of NRG1, as Wallerian degeneration increases the expression of NRG1 in SCs and mice overexpressing NRG1 type III in SCs only are capable of restoring normal myelination after peripheral axonal injury.51 Therefore, type III NRG1 plays a crucial role during SCs myelination and development, indicating this isoform is presented by the axon to the SCs. In vitro experiment, many aspects of SCs biology are affected by NRG1: (i) NRG1 increased glial; (ii) NRG1 is required for survival of cultured Schwann cell progenitors; (iii) NRG1 promotes migration and proliferation of Schwann cell precursors; and (iv) NRG1 is essential for myelination.10, 52
NRG1 as a key axonal signal controls many steps of SCs development through binding to ErbB2/3 receptors, including migration, proliferation and myelination.13 The two receptors of ErbB2 and ErbB3 are an unequal pair: ErbB3 directly binds NRG1 with high affinity.48 The NRG1/ErbB3 signalling pathway activates a variety of signalling pathways (PI3K/Akt, Erk1/2, FAK, Rac/Cdc42) with effects on many facets of SCs biology, including myelination, proliferation and migration.
Krox20, a zinc finger transcription factor, has been considered the master regulator of myelination which induces the expression of myelin‐related genes and coordinates the peripheral nerve myelination programme.53, 54 In fact, patients with mutations in Krox20 consistently have been shown to suffer from peripheral neuropathy,55 while loss of Krox20 function blocks peripheral myelination in mice.56 Thus, we assayed the levels of this protein to determine if global expression of myelin‐related proteins was increased.
Finally, we looked at MBP which is well known to play an important role in formation and maintenance of myelin sheaths and as a major structural component of compact myelin in the peripheral and central nervous systems.57, 58, 59 MBP is an abundant myelin protein and its absence in mutant rats results in a severe CNS hypomyelination.58, 59, 60 As the absence of other myelin proteins still allows the formation of myelination except MBP and MBP has been denoted as the “executive molecule of myelin”.60
Consequently, we chose these four signals (ErbB3, NRG1, Krox20 and MBP) to assess SCs myelination ability under conditions of co‐culture and LIPUS.
In this study, we found that individually or together, co‐culture (ASCs with SCs) and LIPUS treatment facilitated the mRNA expression of ErbB3, NRG1, Krox20 and MBP as well as increased the protein levels of ErbB3, NRG1 and Krox20 at different time points as evaluated by Western blot and IF. Thus, we concluded that LIPUS (as a kind of mechanical stress) can promote transcription and increased protein levels of myelination factors, and likely promote the myelination ability of SCs. Cong et al.61 measured the neurites length after LIPUS treatment to define the effect of LIPUS stimulation on neurons, and demonstrated that LIPUS increased elongation of neurites. Tsuang et al.32 designed a study to elucidate the effects of low‐intensity pulsed ultrasound on cultured SCs, and concluded that intervention with low‐intensity pulsed ultrasound could promote SCs proliferation, prevent cell death and keep adequate phenotype presentation for peripheral nerve recovery. Thus, our results are consistent with findings in the literature that LIPUS stimulation can potentiate SCs myelination ability.
Co‐culture, which resulted in an increase of certain released growth factors in the media, also increased the mRNA expression of myelination factors (ErbB3, NRG1, Krox20 and MBP) along with the protein levels (NRG1, ErbB3 and Krox20). Adipose tissue is an accessible and affluent source of adult stem cells for tissue engineering. ASCs have the potential to differentiate into many different type cells in response to specific growth factors and environmental cues, including myocytes, neurons, adipocytes, chondrocytes and osteoblasts.62 It has been confirmed that ASCs can also boost tissue recovery through the delivery and localized secretion of cytokines.63, 64 Therefore, we concluded that shared culture media provides environmental cues via the secretion of growth factors or cytokines from ASCs, which primes SCs by upregulating pro‐myelination indicators. However, this study did not address which growth factors or cytokines from ASCs may be responsible for the enhancement of SCs pro‐myelination indicators expression, which is an important question that will be explored in the future. Furthermore, this study did not directly test the myelination ability of SCs, but upregulation of critical pro‐myelination factors has been considered a reasonable proxy.
We also demonstrated that the combination of co‐culture and LIPUS was significantly more effective than either individual treatment alone. We saw same trend of ErbB3, NRG1 and Krox20 by different treatments even when assayed by different methods, including RT‐PCR, Western blot and IF. Our results showed that mRNA and protein levels of myelination factors in group co‐culture with LIPUS were significantly higher than all the other groups at all time points, even though group co‐culture and group LIPUS alone augmented expression of these factors compared to the control. Thus, we propose that co‐culturing ASCs with SCs along with LIPUS treatment will maximize the myelination ability of SCs.
Moving forward, these results lay the foundation for future in vivo studies. We will investigate the effect of combination of LIPUS and co‐culture on myelination by transplanting ASCs and SCs together into rat wounds. After transplantation, we will periodically stimulate with LIPUS localized to the wound to help nerve regeneration. We hypothesize that ASCs will secret growth factors or cytokines to promote myelination of SCs and that effect could be potentiated by LIPUS treatment. In conclusion, our study demonstrated that ASCs co‐culture with SCs along with LIPUS stimulation synergistically act to promote the myelination ability of SCs.
Acknowledgements
This study was funded by grants from the National Natural Science Foundation of China (No. 81200810 and No. 81570987) and New Teacher Fund for Doctor Station of the Ministry of Education (No. 20120181120008).
Contributor Information
Xingmei Yang, Email: 65167950@qq.com.
Min Wang, Email: hxkqwangm@163.com.
References
- 1. Peng J, Wang Y, Zhang L, et al. Human umbilical cord Wharton's jelly‐derived mesenchymal stem cells differentiate into a Schwann‐cell phenotype and promote neurite outgrowth in vitro. Brain Res Bull. 2011;84:235–243. [DOI] [PubMed] [Google Scholar]
- 2. Grinsell D, Keating CP. Peripheral nerve reconstruction after injury: a review of clinical and experimental therapies. BioMed Res Int. 2014;2014:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wiberg M, Terenghi G. Will it be possible to produce peripheral nerves? Surg Technol Int. 2002;11:303–310. [PubMed] [Google Scholar]
- 4. Liu Y, Zhang Z, Qin Y, et al. A new method for Schwann‐like cell differentiation of adipose derived stem cells. Neurosci Lett. 2013;551:79–83. [DOI] [PubMed] [Google Scholar]
- 5. Rutkowski GE, Miller CA, Jeftinija S, Mallapragada SK. Synergistic effects of micropatterned biodegradable conduits and Schwann cells on sciatic nerve regeneration. J Neural Eng. 2004;1:151. [DOI] [PubMed] [Google Scholar]
- 6. Li Q, Jiang H, Liu K, et al. Nerve conduit filled with GDNF gene‐modified schwann cells enhances regeneration of the peripheral nerve. Microsurgery. 2006;26:116–121. [DOI] [PubMed] [Google Scholar]
- 7. Xu XM, Guénard V, Kleitman N, Bunge MB. et al. Axonal regeneration into Schwann cell‐seeded guidance channels grafted into transected adult rat spinal cord. J Comp Neurol. 1995;351:145–160. [DOI] [PubMed] [Google Scholar]
- 8. Tsai EC, Dalton PD, Shoichet MS, Tator CH. Synthetic hydrogel guidance channels facilitate regeneration of adult rat brainstem motor axons after complete spinal cord transection. J Neurotrauma. 2004;21:789–804. [DOI] [PubMed] [Google Scholar]
- 9. Ren YJ, Zhang S, Mi R, et al. Enhanced differentiation of human neural crest stem cells towards the Schwann cell lineage by aligned electrospun fiber matrix. Acta Biomater. 2013;9:7727–7736. [DOI] [PubMed] [Google Scholar]
- 10. Taveggia C, Zanazzi G, Petrylak A, et al. Neuregulin‐1 type III determines the ensheathment fate of axons. Neuron. 2005;47:681–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nave K‐A, Salzer JL. Axonal regulation of myelination by neuregulin 1. Curr Opin Neurobiol. 2006;16:492–500. [DOI] [PubMed] [Google Scholar]
- 12. Michailov GV, Sereda MW, Brinkmann BG, et al. Axonal neuregulin‐1 regulates myelin sheath thickness. Science. 2004;304:700–703. [DOI] [PubMed] [Google Scholar]
- 13. Newbern J, Birchmeier C. Nrg1/ErbB signaling networks in Schwann cell development and myelination. Semin Cell Dev Biol. 2010;21:922–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lutz D, Kataria H, Kleene R, et al. Myelin basic protein cleaves cell adhesion molecule L1 and improves regeneration after injury. Mol Neurobiol. 2016;53:3360–3376. [DOI] [PubMed] [Google Scholar]
- 15. Gupta R, Truong L, Bear D, Chafik D, Modafferi E, Hung CT. Shear stress alters the expression of myelin‐associated glycoprotein (MAG) and myelin basic protein (MBP) in Schwann cells. J Orthop Res. 2005;23:1232–1239. [DOI] [PubMed] [Google Scholar]
- 16. Rachana KS, Manu MS, Advirao GM. Insulin influenced expression of myelin proteins in diabetic peripheral neuropathy. Neurosci Lett. 2016;629:110–115. [DOI] [PubMed] [Google Scholar]
- 17. Wilkinson DG, Bhatt S, Chavrier P, Bravo R, Charnay P. Segment‐specific expression of a zinc‐finger gene in the developing nervous system of the mouse. Nature. 1989;337:461–464. [DOI] [PubMed] [Google Scholar]
- 18. Quintes S, Brinkmann BG, Ebert M, et al. Zeb2 is essential for Schwann cell differentiation, myelination and nerve repair. Nat Neurosci. 2016;19:1050–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Morgan EF, Salisbury Palomares KT, Gleason RE, et al. Correlations between local strains and tissue phenotypes in an experimental model of skeletal healing. J Biomech. 2010;43:2418–2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ogawa R. Mechanobiology of scarring. Wound Repair Regen. 2011;19:s2–s9. [DOI] [PubMed] [Google Scholar]
- 21. Sarraf C, Otto WR, Eastwood M. In vitro mesenchymal stem cell differentiation after mechanical stimulation. Cell Prolif. 2011;44:99–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Schwarz C, Wulsten D, Ellinghaus A, Lienau J, Willie BM, Duda GN. Mechanical load modulates the stimulatory effect of BMP2 in a rat nonunion model. Tissue Eng Part A. 2012;19:247–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Grottkau BE, Yang X, Zhang L, Ye L, Lin Y. Comparison of effects of mechanical stretching on osteogenic potential of ASCs and BMSCs. Bone Res. 2013;1:282–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yang X, Yang X, Cai X, Wang J, et al. Mechanical stretch inhibits adipogenesis and stimulates osteogenesis of adipose stem cells. Cell Prolif. 2012;45:158–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bueno FR, Shah SB. Implications of tensile loading for the tissue engineering of nerves. Tissue Eng Part B Rev. 2008;14:219–233. [DOI] [PubMed] [Google Scholar]
- 26. Ouyang H, Sun W, Fu Y, et al. Compression induces acute demyelination and potassium channel exposure in spinal cord. J Neurotrauma. 2010;27:1109–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Drake JD, Callaghan JP. Intervertebral neural foramina deformation due to two types of repetitive combined loading. Clin Biomech. 2009;24:1–6. [DOI] [PubMed] [Google Scholar]
- 28. Zhang L, Yang X, Yue Y, et al. Cyclic mechanical stress modulates neurotrophic and myelinating gene expression of Schwann cells. Cell Prolif. 2015;48:59–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yue Y, Yang X, Wei X, et al. Osteogenic differentiation of adipose‐derived stem cells prompted by low‐intensity pulsed ultrasound. Cell Prolif. 2013;46:320–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Raso VVM, et al. Can therapeutic ultrasound influence the regeneration of peripheral nerves? J Neurosci Methods. 2005;142:185–192. [DOI] [PubMed] [Google Scholar]
- 31. Chang CJ, Hsu SH, Lin FT, Chang H, Chang CS Low‐intensity‐ultrasound accelerated nerve regeneration using cell‐seeded poly (D, L‐lactic acid‐co‐glycolic acid) conduits: an in vivo and in vitro study. J Biomed Mater Res, Part B. 2005;75:99–107. [DOI] [PubMed] [Google Scholar]
- 32. Tsuang YH, Liao LW, Chao YH, et al. Effects of low intensity pulsed ultrasound on rat Schwann cells metabolism. Artif Organs. 2011;35:373–383. [DOI] [PubMed] [Google Scholar]
- 33. Park SC, Oh SH, Seo TB, Namgung U, Kim JM, Lee JH. Ultrasound‐stimulated peripheral nerve regeneration within asymmetrically porous PLGA/Pluronic F127 nerve guide conduit. J Biomed Mater Res, Part B. 2010;94:359–366. [DOI] [PubMed] [Google Scholar]
- 34. Zhang H, Lin X, Wan H, Li JH, Li JM. Effect of low‐intensity pulsed ultrasound on the expression of neurotrophin‐3 and brain‐derived neurotrophic factor in cultured Schwann cells. Microsurgery. 2009;29:479–485. [DOI] [PubMed] [Google Scholar]
- 35. Angle SR, Sena K, Sumner DR, Virdi AS. Osteogenic differentiation of rat bone marrow stromal cells by various intensities of low‐intensity pulsed ultrasound. Ultrasonics. 2011;51:281–288. [DOI] [PubMed] [Google Scholar]
- 36. Ba K, Yang X, Wu L, et al. Jagged‐1‐mediated activation of notch signalling induces adipogenesis of adipose‐derived stem cells. Cell Prolif. 2012;45:538–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tremp M, Salemi S, Gobet R, Sulser T, Eberli D. Adipose‐derived stem cells (ASCs) for tissue engineering. [DOI] [PubMed]
- 38. Lopatina T, Kalinina N, Karagyaur M, et al. Adipose‐derived stem cells stimulate regeneration of peripheral nerves: BDNF secreted by these cells promotes nerve healing and axon growth de novo. PLoS ONE. 2011;6:e17899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rubina K, Kalinina N, Efimenko A, et al. Adipose stromal cells stimulate angiogenesis via promoting progenitor cell differentiation, secretion of angiogenic factors, and enhancing vessel maturation. Tissue Eng Part A. 2009;15:2039–2050. [DOI] [PubMed] [Google Scholar]
- 40. Traktuev D, Parfenova EV, Tkachuk VA, March KL. Adipose stromal cells–plastic type of cells with high therapeutic potential. Tsitologiia. 2005;48:83–94. [PubMed] [Google Scholar]
- 41. Liao D, Gong P, Li X, Tan Z, Yuan Q. Co‐culture with Schwann cells is an effective way for adipose‐derived stem cells neural transdifferentiation. Arch Med Sci. 2010;6:145–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Abe I, Ochiai N, Ichimura H, Tsujino A, Sun J, Hara Y. Internodes can nearly double in length with gradual elongation of the adult rat sciatic nerve. J Orthop Res. 2004;22:571–577. [DOI] [PubMed] [Google Scholar]
- 43. Pfister BJ, Iwata A, Meaney DF, Smith DH. Extreme stretch growth of integrated axons. J Neurosci. 2004;24:7978–7983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Iwata A, Browne KD, Pfister BJ, Gruner JA, Smith DH. Long‐term survival and outgrowth of mechanically engineered nervous tissue constructs implanted into spinal cord lesions. Tissue Eng. 2006;12:101–110. [DOI] [PubMed] [Google Scholar]
- 45. Hao H, Shreiber DI. Axon kinematics change during growth and development. J Biomech Eng. 2007;129:511–522. [DOI] [PubMed] [Google Scholar]
- 46. Gupta R, Rummler L, Steward O. The ABJS 2005 Marshall Urist Award: understanding the biology of compressive neuropathies. Clin Orthop Relat Res. 2005;436:251–260. [DOI] [PubMed] [Google Scholar]
- 47. Nave K‐A, Trapp BD. Axon‐glial signaling and the glial support of axon function. Annu Rev Neurosci. 2008;31:535–561. [DOI] [PubMed] [Google Scholar]
- 48. Birchmeier C, Nave KA. Neuregulin‐1, a key axonal signal that drives Schwann cell growth and differentiation. Glia. 2008;56:1491–1497. [DOI] [PubMed] [Google Scholar]
- 49. Rao SN, Pearse DD. Regulating axonal responses to injury: the intersection between signaling pathways involved in axon myelination and the inhibition of axon regeneration. Front Mol Neurosci. 2016;9:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Willem M. Proteolytic processing of neuregulin‐1. Brain Res Bull. 2016;10:361–365. [DOI] [PubMed] [Google Scholar]
- 51. Stassart RM, Fledrich R, Velanac V, et al. A role for Schwann cell‐derived neuregulin‐1 in remyelination. Nat Neurosci. 2013;16:48–54. [DOI] [PubMed] [Google Scholar]
- 52. Shah NM, Marchionni MA, Isaacs I, Stroobant P, Anderson DJ. Glial growth factor restricts mammalian neural crest stem cells to a glial fate. Cell. 1994;77:349–360. [DOI] [PubMed] [Google Scholar]
- 53. Nagarajan R, Svaren J, Le N, Araki T, Watson M, Milbrandt J. EGR2 mutations in inherited neuropathies dominant‐negatively inhibit myelin gene expression. Neuron. 2001;30:355–368. [DOI] [PubMed] [Google Scholar]
- 54. Ghislain J, Desmarquet‐Trin‐Dinh C, Jaegle M, Meijer D, Charnay P, Frain M. Characterisation of cis‐acting sequences reveals a biphasic, axon‐dependent regulation of Krox20 during Schwann cell development. Development. 2002;129:155–166. [DOI] [PubMed] [Google Scholar]
- 55. Warner LE, Mancias P, Butler IJ, et al. Mutations in the early growth response 2 (EGR2) gene are associated with hereditary myelinopathies. Nat Genet. 1998;18:382–384. [DOI] [PubMed] [Google Scholar]
- 56. Topilko P, Schneider‐Maunoury S, Levi G, et al. Krox‐20 controls myelination in the peripheral nervous system. Nature. 1994;371:796–799. [DOI] [PubMed] [Google Scholar]
- 57. Readhead C, Takasashi N, Shine HD, Saavedra R, Sidman R, Hood L. Role of myelin basic protein in the formation of central nervous system myelin. Ann N Y Acad Sci. 1990;605:280–285. [DOI] [PubMed] [Google Scholar]
- 58. Readhead C, Hood L. The dysmyelinating mouse mutations shiverer (shi) and myelin deficient (shi mld). Behav Genet. 1990;20:213–234. [DOI] [PubMed] [Google Scholar]
- 59. Boggs J. Myelin basic protein: a multifunctional protein. Cell Mol Life Sci. 2006;63:1945–1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kwiecien J, O'Connor LT, Goetz BD, Delaney KH, Fletch AL, Duncan ID. Morphological and morphometric studies of the dysmyelinating mutant, the Long Evans shaker rat. J Neurocytol. 1998;27:581–591. [DOI] [PubMed] [Google Scholar]
- 61. Cong R, Jia‐Mou L, Xin L. LIPUS enhance elongation of neurites in rat cortical neurons through inhibition of GSK‐3β. Biomed Environ Sci. 2010;23:244–249. [DOI] [PubMed] [Google Scholar]
- 62. Gimble J, Guilak F. Adipose‐derived adult stem cells: isolation, characterization, and differentiation potential. Cytotherapy. 2003;5:362–369. [DOI] [PubMed] [Google Scholar]
- 63. Miranville A, Heeschen C, Sengenès C, Curat CA, Busse R, Bouloumié A. Improvement of postnatal neovascularization by human adipose tissue‐derived stem cells. Circulation. 2004;110:349–355. [DOI] [PubMed] [Google Scholar]
- 64. Planat‐Benard V, Silvestre JS, Cousin B, et al. Plasticity of human adipose lineage cells toward endothelial cells physiological and therapeutic perspectives. Circulation. 2004;109:656–663. [DOI] [PubMed] [Google Scholar]


