Abstract
Abstract. Oral mucositis is a common, dose‐limiting, acute toxicity of radiation therapy administered for the treatment of cancers of the head and neck. Accumulating data would suggest that the pathogenesis of mucositis is complex and involves the sequential interaction of all cell types of the oral mucosa, as well as a number of cytokines and elements of the oral environment. While a number of studies have reported on gene expression of particular cell types in response to radiation, the overall response of irradiated mucosa has only been evaluated in a limited way. The present study was undertaken to evaluate the expression of a target group of genes using RNA quantification assays and, more broadly, to assess patterns of mucosal gene expression using DNA microarray hybridization. Our results demonstrate the sequential upregulation of a series of genes that, when taken collectively, suggest an intricate functional interaction.
Keywords: Mucositis, gene expression, radiation, animal model
Introduction
Oral mucositis is a common, dose‐limiting, acute toxicity experienced by many patients receiving radiation or chemotherapy as treatment for cancer (Sonis 2000). In addition to pain and loss of function, mucositis‐induced ulcerations are a conduit for oral bacteria and often lead to bacteraemia or sepsis in granulocytopenic patients (Reuscher et al . 1998). Furthermore, the presence of mucositis is associated with increased use of a variety of healthcare resources that result in its being a driver of additional cost (Sonis et al . 2001).
It had been assumed that mucositis developed solely as the result of the nonspecific toxic effects of radiation or chemotherapy on the dividing cells of the oral basal epithelium. This paradigm no longer seems to be true as evidence has accumulated to suggest that the condition is more biologically complex than originally suspected (Sonis 1998). It appears that the pathogenesis of mucositis involves the sequential interaction of all of the cell and tissue types that comprise the oral mucosa, as well as tissue factors, cytokines and elements of the oral environment. Understanding the sequence and elements involved in the evolution of mucositis provides an opportunity to identify potential points for intervention. Consequently, mapping the sequence of gene expression or upregulation following mucosal challenge with radiation provides a mechanism to define those elements and to determine the order in which they are expressed.
It is clear that ionizing radiation effectively induces a sequenced array of genes that ultimately result in cell damage and/or death. Studies supporting this observation have been primarily performed in cell or tissue culture models and reveal rapid expression of early response genes c‐jun, c‐fos, and Erg‐1, activation of the NF‐κB family (Weichselbaum et al . 1994) and the 26s proteasome (Pajonk and McBride 2001), the hSNK gene (Shimizu‐Yoshida et al . 2001), and vascular adhesion molecules (Quarmby et al . 2000). In vivo studies have confirmed the upregulation of genes associated with endothelial cells (Mollàet al . 2001), and this finding may have pivotal consequences with respect to radiation‐induced intestinal injury (Paris et al . 2001). For the most part, radiation‐induced gene expression has been studied as an acute phenomenon, rarely going beyond 24 h. Nonetheless, the subsequent effects of early gene activation on later events have been postulated. It seems likely that radiation‐induced upregulation of early key genes causes a domino effect that results in the activation of other genes with consequent tissue damage.
The purpose of this investigation was to evaluate the expression of a target group of genes using RNA quantification assays and, more broadly, to assess patterns of mucosal gene expression using DNA microarray hybridization following acute radiation exposure. Reverse transcriptase‐polymerase chain reaction (RT‐PCR) was used to detect the expression of cyclin A, tumour necrosis factor‐alpha (TNF‐α), iNOS, p53, heat shock proteins 70 and 90 (Hsp70, Hsp90), and DNA polymerase δ. Gene selection for RT‐PCR analysis was based on the hypothesis that the target genes were all of potential significance in the development of radiation‐induced mucosal toxicity.
Materials and methods
Animals
Male golden Syrian hamsters (Harlan Sprague‐Dawley), aged 4‐6 weeks, and weighing between 70 g and 100 g were used. Animals were individually numbered, housed in small groups, and watered ad libitum. The Standing Committee on Animal Use of the Harvard Medical Area approved all protocols.
Radiation
Hamsters were anaesthetized with sodium pentobarbital and the left buccal cheek pouch mucosa was everted and secured. A protective lead shield was placed to expose the mucosa but cover the remainder of the animal. Animals were irradiated with a single dose of 35 Gy delivered to the targeted mucosa by a 250‐kV source at a focal distance of 50 cm and hardened with a 0.35‐mm Cu filtration system. This dose was selected because it consistently produces ulcerative mucositis in this model (Fig. 1).
Figure 1.
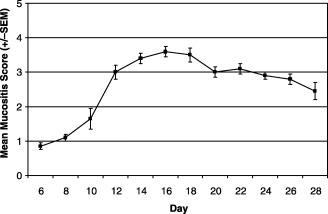
Typical mucositis curve following radiation of the hamster cheek pouch . Mucositis was induced with a single dose of radiation (35 Gy) administered on day 0. Irradiation specifically targeted the left buccal pouch mucosa using a lead shield to isolate it from the rest of the animal. Mucositis was scored on a scale ranging from 0 for normal, to 5 for severe ulceration using the following definitions: 0 = healthy pouch; 1 = erythema, but no mucosal erosion; 2 = severe erythema and vasodilation, superficial erosion may be present; 3 = ulcer formation at one or more sites with cumulative size of ulcers equal to or less than 25% of pouch surface area; 4 = ulceration with cumulative size equal to approximately 50% of pouch surface area; 5 = ulceration involving virtually all of pouch surface area.
RNA quantification assays
Cheek pouch samples were dissected at specified times and flash‐frozen immediately after culling. The tissue was homogenized in RNAsol solution (1 mL per cheek pouch) using an OMNI homogenizer with a 7‐mm probe at room temperature for about 1 min. Samples were extracted further by addition of 200 µL of chloroform, followed by vortexing for 15 min and set on ice for 5 min. Samples were centrifuged at 12 000 g for 15 min at 4 °C; the aqueous phase was collected (~1000 µL), an equal volume of phenol/chloroform/isoamyl alcohol added, inverted, and set on ice for 5 min. The samples were centrifuged at 12 000 g for 5 min at 4 °C. The aqueous phase was collected (~600 µL), an equal volume of chloroform added, the sample inverted, centrifuged at 12 000 g for 10 min at 4 °C, and the aqueous phase collected (~500 µL). The RNA was precipitated by addition of an equal volume of isopropanol (0 °C, 30‐45 min) and pelleted by centrifugation at 12 000 g for 15 min at 4 °C. The RNA pellet was washed with 75% ethanol and dried for 10 min. RNA was resuspended in 200 µL diethylpyrocarbonate (DEPC) in distilled H2O and quantified by absorbance at 250 nm. Approximately half of the sample (~300 µg) total RNA was adjusted to 0.5 m NaCl and applied to an oligo‐dT cellulose column, washed extensively and eluted in neutral buffered solution in the absence of salt. mRNA and total RNA samples were diluted to 500 ng/µL prior to reverse transcription and PCR amplification.
cDNA was prepared from a 1‐µg hamster mucosa biopsy by annealing random hexanucleotides (5 pm/mL) and poly‐A (100 µg/mL). Reverse transcription was performed using MMLV‐reverse transciptase (Stratagene, LaJolla, CA, USA) at 42 °C for 60 min. PCR amplification of cDNA was performed using oligonucleotide primer pairs specific for cyclin A, p53, DNA polymerase δ, hsp70, hsp90, TNF‐α, iNOS and calmodulin control, in separate reactions. PCR was performed using a Perkin‐Elmer 9600 thermocycler. Samples were visualized on a 1.5% agarose gel stained with ethidium bromide. Alternatively, samples were amplified in the presence of a gene‐specific dual fluorescence‐labelled probe (6‐carboxy fluorescein/6‐carboxy‐tetramethyl rhodamine; 5′ FAM; 3′ TAMRA) and products quantified on a cycle‐to‐cycle basis using a Perkin‐Elmer 7700 fluorogenic PCR instrument. This instrument allows for precise quantification of RNA concentration in a linear phase of the PCR reaction.
Gene expression profiling by DNA microarray hybridization
Mucosal biopsies were obtained from three animals at specified intervals following radiation. The tissue was flash frozen and stored at −70 °C. mRNA was separated from total RNA by oligo‐dT chromatography. Washed oligo‐dT resin (Amersham Biosciences Corp., Piscataway, NJ, USA) was incubated with total RNA in buffer containing 0.5 m NaCl. Resin was washed extensively in binding buffer, and mRNA eluted with buffer containing no NaCl. RNA was precipitated by the addition of 1 : 10 volume of 3 m sodium acetate and 2. 5 volumes of ethanol overnight at −20 °C. cDNAs were prepared and labelled with Cy3 or Cy5 fluorescent probes (under contract to Incyte Genomics, Inc. Palo Alto, CA, USA). Labelled cDNAs synthesized from treated and control RNA were hybridized to human UniGene V2.0 (9100 cDNAs) and mouse GEM 1 (8700 cDNAs) collections immobilized on a glass slide. The genes were represented only once in the panel, sequence verified and range in length from 500 bp to 3000 bp. The long gene template to which the cDNAs were hybridized markedly reduced the variability associated with hybridization conditions, but also reduced the ability to discriminate between closely related members of a gene family. A relatively large number of fluorescent‐labelled control cDNAs were mixed in specific ratios and spiked into the hybridization reaction to provide a robust performance control. Additionally, a series of ‘housekeeping’ genes of which expression is infrequently modulated were monitored. Data were transmitted via the internet and analysed using GEM Tools 2.4 (Incyte Genomics, Inc.) and Array Explorer 2 (Spotfire, Somerville, MA, USA) software running on an NT server.
Results
RNA quantification assays
RT‐PCR conditions were established for the measurement of cyclin A, DNA polymerase δ, p53, Hsp70, Hsp90, iNOS and TNF‐α gene expression. Detection and quantification of the individual RT‐PCR markers with respect to time was performed using the PE7700 fluorogenic PCR instrument for RNA obtained from two cheek pouch samples from animals 4, 8, 10, 12 and 15 days following radiation (Fig. 2). Specific amplified products for cyclin A, p53, DNA polymerase δ, hsp70 and hsp 90 were readily detectable using poly‐A+ RNA. Detection of the low levels for TNF‐α and iNOS were more variable, making accurate quantification of gene expression difficult. Expression of Hsp70 was maximal between days 8 and 10 (day 0 = day of radiation) and preceded peak mucositis by several days (peak mucositis day 14). By day 12, it had returned to baseline. The expression of Hsp90 gene peaked earlier at day 4, dipped on day 8, demonstrated a second ‘mini’ peak on day 10, and then reached baseline at day 12. Cyclin A expression peaked sharply on day 4 and then decreased steadily to baseline on day 12. In contrast, p53 expression remained at low levels until day 15, when it peaked sharply. Polymerase δ expression decreased from a low level of expression on day 0 to day 10 and, unlike p53, remained at low levels for the duration of the experiment.
Figure 2.
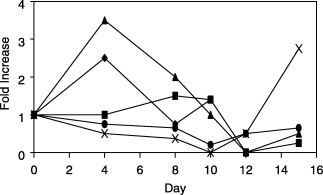
Fluorogenic RT‐PCR measurement of heat shock protein 70 (▪), heat shock protein 90 (◆), cyclin A (s), p53 (×), and polδ (•) gene expression with respect to time following radiation.
Microarray profiling
Mucosal samples were obtained 1, 5 and 10 days following radiation from three animals at each time point in the first study and at 1 h, 4 h, 8 h and 24 h in the second experiment. The analysis was limited to those genes demonstrating a differential expression ratio of 1.8 or greater.
Early marked expression of 10 genes was noted by 8 h after radiation: PCAF (PAF65), properdin, Hsp70, serum amyloid A1, small proline‐rich protein (SPRR), FK506‐binding protein, mitogen‐activated protein kinase (MAPK), serum‐inducible kinase (SNK), spermidine transferase and keratin 14 (Table 1). By 24 h, proteasome subunit α type 3, vimentin, G protein‐coupled receptor, and sarcolipin expression was seen. At 5 days, proteasome 26S, keratin 6b, keratin 17, zinc finger protein, adenosine deaminase, secretory leukoprotease inhibitor, and squamous cell antigen 1 (SCC AG‐1) were noted.
Table 1.
Time following radiation of initial gene expression (differential expression ratio > 1.8) as determined by microarray profiling
| Acute expression (8 h or less) |
| PCAF (PAF65) |
| Hsp 70 |
| FKBP (FK‐506) |
| Serum amyloid A1 |
| Properdin |
| Serum inducible kinase (SNK) |
| Small proline‐rich protein |
| Mitogen‐activated protein kinase (MAPK) |
| Spermidine acetyl transferase |
| Keratin 14 |
| Delayed expression (day 1) |
| Proteasome subunit α |
| Vimentin |
| G protein‐coupled receptor |
| Sarcolipin |
| Delayed expression (day 5) |
| Proteasome 26S subunit |
| Keratin 6b |
| Keratin 17 |
| Adenosine deaminase |
| Zinc finger protein |
| Secretory leukoprotease inhibitor |
| Squamous cell carcinoma Ag 1 (SCC AG‐1) |
Additionally, lesser expression of a large number of genes was also noted at various time points, but are not included in this analysis as they did not meet our inclusion criteria.
Discussion
The results of this study provide a preliminary picture of the sequence of mucosal gene expression following exposure to acute radiation and its relationship to normal tissue toxicity. Furthermore, analysis of the data supports the concept that mucosal injury is the consequence of damage to all mucosal tissue compartments, not just the epithelium. Finally, when mapped, the gene expression sequence may give clues as to the underlying mechanisms involved in stomatotoxicity.
Recent studies have shown that the pathogenesis of mucositis is multifactorial, involving the interaction of oral mucosal epithelial cells, endothelial cells, connective tissue, and the submucosal infiltrate (Sonis et al . 2000). The ability to elucidate the biological factors that initiate and promulgate the condition should provide opportunities for new interventions. In this study we used RT‐PCR to identify the extent of expression of particular genes thought to be of potential significance in mucositis, and also looked for unanticipated gene expression using microarray profiling. It is obvious, but important to note that in both instances, the RNA source was derived from a composite of mucosal tissue that included epithelium and submucosa. Consequently, the assignment of a particular gene to its most likely tissue or cell source was made secondarily based on earlier reports in the literature (Table 2). It is also noteworthy that the results are dependent on gene homologues between hamster and the test bed that was used. Evidence suggests that this assumption is likely to be correct.
Table 2.
Likely cell and or tissue source of expressed genes
| Gene | Cell or tissue origin |
|---|---|
| PCAF | Epithelium, endothelium, muscle |
| Hsp 70 | Ubiquitous |
| MAPK | Ubiquitous |
| FKBP1 (FK‐506) | Muscle and lymphocytes (T cells) |
| Serum amyloid A1 | Epithelium |
| Small proline‐rich protein | Epithelium |
| Properdin | Infiltrate |
| Proteasome subunit α | Ubiquitous |
| Vimentin | Epithelium and mesenchymal cells |
| G protein‐coupled receptor | Ubiquitous |
| Sarcolipin | Muscle |
| Proteasome 26S | Ubiquitous |
| Keratin 6b | Epithelium (suprabasal layer) |
| Keratin 17 | Epithelium |
| Keratin 14 | Epithelium (keratinocytes and basal cells) |
| Adenosine deaminase | Infiltrate |
| Zinc finger protein | Epithelium |
| Secretory leukoprotease inhibitor | Epithelium |
| Squamous cell carcinoma antigen 1 | Epithelium (spinous and granular layers) |
Our results show upregulation of a number of genes within 8 h of mucosal radiation. These included PCAF, Hsp 70, FK506‐binding protein, properdin, serum amyloid A1, small proline‐rich protein (SPRR), serum inducible kinase, MAPK, spermidine acetyl transferase and keratin 14. The rapidity with which genes are upregulated following ionizing radiation has been previously observed by a number of investigators, and, in some cases, reported to be within minutes of cell exposure to radiation. Because of the technical aspects of tissue removal and manipulation in this investigation, the earliest time point we studied was 1 hour after radiation. It is therefore possible that the peak expression of some genes had already passed by the time tissue was first sampled. Consequently, the upregulation of genes that were most proximate to radiation may have been missed so that their potential role in the subsequent sequence could not be assessed. Nonetheless, genes that were identified are consistent with earlier reports and their relationship appears to make sense with respect to the development of mucosal injury.
Our finding of early expression of MAPK is consistent with cell culture studies. The potential significance of this group of early response genes to effect the adaptation of tissue to radiation was suggested some time ago by Weichselbaum et al . (1994). They proposed that downstream regulation of effectors such as cytokines and growth factors was a possible consequence of early MAPK activation. The finding that TNF‐α and IL‐1β increase dramatically and correlate with mucosal injury would seem to support this hypothesis (Sonis et al . 2000). MAPK is an important regulator of the transcription factor AP‐1. AP‐1 may function in either pro‐apoptotic or protective roles within the cell (Whitmarsh and Davis 1996). Thus, it could provide an avenue leading to tissue injury. Furthermore, its role may be even more profound in that it may function with NF‐κB as a coactivator of a large number of genes (Shi et al . 1999) involved in mucosal injury such as the keratin genes (see below).
SPRR is typically associated with the cornified cell envelope of epithelial cells (Steinert et al . 1998) while Keratin 14 is normally expressed in mitotically active keratinocytes and in basal cells of normal epithelium (Munz et al . 2001). Hence the finding of these genes is not surprising. PCAF is normally expressed in all of the tissue compartments of the mucosa, including muscle. Increased PCAF complex expression is associated with p53. In fact, it has been reported that full transcriptional activity of p53 is dependent on coactivation with PCAF (Liu et al . 1999). The finding of late p53 activation in the present study may represent the manifestation of a time‐related phenomenon dependent, in part, on PCAF expression.
Hsp70 is ubiquitous in the tissues in which it is expressed. Its expression following radiation is well known, and has been hypothesized to have a cytoprotective function by altering the membrane signalling cascade leading to DNA damage (Lee et al . 2001). Clearly, with respect to radiation induced normal tissue injury, either its protective effect is overwhelmed, or Hsp70 functions differently. Hsp90 gene expression was marked when measured by RT‐PCR at day 4. Expression of Hsp90 has been associated with radiation‐induced apoptosis (Prasad et al . 1998) and may therefore be a significant early event leading to mucosal injury. Expression of cyclin A also peaked on day 4. The upregulation of cyclin A noted in our study contrasts with its down‐regulation reported by De Toledo et al . (1998). This difference may be the consequence of the timing of sampling (24‐48 h after radiation exposure), the cell type (diploid fibroblasts), or the controlled conditions studied (cultured cells).
Upregulation of G protein‐coupled receptor was noted by day 1 after radiation. The expression of this signal transduction mechanism with respect to radiation‐induced injury has yet to be defined. However, its activation might be viewed as a protective effort by the affected tissues as recent studies have suggested that platelet‐activating factor‐receptor (a G protein‐coupled receptor) can mediate an antiapoptotic effect (Southall et al . 2001). Of potential relevance is the finding that such activity occurs through an NF‐κB‐dependent process (Southall et al . 2001).
Serum amyloid A1 (SAA) is an acute phase protein typically associated with the liver (Suffredini et al . 1999). However, its presence in gastrointestinal mucosa has been reported (Ogawa et al . 2000). While SAA can be induced by a variety of stimuli including LPS and pro‐inflammatory cytokines, our finding of its activation in radiated mucosa appears to be unique. The role of SAA in mucosal injury remains to be defined, although it is chemoattractive for inflammatory cells (Xu et al . 1995).
Properdin is typically expressed by infiltrating inflammatory cells (Schwaeble and Reid 1999). As an identifiable inflammatory infiltrate is not typically seen until days after radiation in this model, it is more likely that properdin is either expressed by noninflammatory mucosal cells or by lymphocytes.
Keratins (K) are a group of cell‐type and differentiation‐specific intermediate filament proteins of the cytoskeleton that provide functional stability and structure for epithelial cells (Ma et al . 1997). Typically K5 and K14 are expressed in the basal layers of healthy stratified squamous epithelium (Ma et al . 1997). Thus, our finding of early K14 expression is consistent with its constituitive expression in normal mucosa. In contrast, K6b and K17 are not normally expressed in healthy epithelium (Navarro et al . 1995; McGowan and Coulombe 1998; Troy and Turksen 1999). Their upregulation 5 days following radiation is in agreement with reports of the increased expression of K6 and K17 following epithelial injury (Ma et al . 1997), and is similar to their expression in skin following exposure to ultraviolet light (Bayerl et al . 1995). Interestingly, as noted below, the upregulation of K6 and K14 appears to be under the transcriptional control of NF‐κB and AP‐1 (Ma et al . 1997).
We also noted upregulation of vimentin on day 5. Vimentin is a type III intermediate filament. While it is typically expressed in mesenchymal cells (Gilles et al . 1999), vimentin is transiently coexpressed with keratins in the basal and suprabasal layers of epithelial cells during wound healing (SunderRaj et al . 1992). Our finding of vimentin expression 5 days following radiation, concurrent with upregulation of K6 and K17, implies a pre‐emptive attempt by the mucosal epithelium to initiate healing prior to maximum injury. Based on the clinical course of mucositis in the model, it is clear that such an attempt fails to prevent ulceration adequately, although its effectiveness in limiting mucosal damage is unknown.
Expression of zinc finger protein at day 5 may suggest basal cell activity since some zinc finger proteins are cell specific. In particular, basonuclin is found in the basal layer of stratified squamous epithelium and is thought to be a cell‐type‐specific regulatory protein for rDNA transcription (Iuchi and Green 1999). Its strict association with the germinal elements of epithelium may suggest a role in renewal triggered by the radiation stress.
In previous studies, we have shown an upregulation of TNF‐α and IL‐1β as measured by TaqMan RT‐PCR at days 12 and 15 following radiation during the development of radiation‐induced oral mucositis (Sonis et al . 2000). Neither time point was included in the current analysis, but the absence of detection of either TNF‐α or IL‐1β expression earlier than day 12 is consistent with our original findings. Interestingly, although pro‐inflammatory cytokines have been implicated in radiation‐induced mucosal toxicity, their expression in proximity to maximal tissue damage may be more consistent with a manifestation or accelerator of injury rather than being truly causal. Consequently, we sought to determine a more proximal event in the initiation of radiation‐induced damage.
Upregulation of the proteasome subunit α type 3 was noted on day 1 and upregulation of proteasome 26S subunit was seen on day 5. The 26S subunit consists of seven α subunits, one of which was expressed on day 1. Proteasome 26S plays a critical role in the activation of the transcription factor NF‐κB by cleaving the inhibitor IκB (Marakov 2000). Upon cleaving, NF‐κB enters the nucleus where it is capable of upregulating a large number of genes, including those associated with biological defence processes, such as TNF‐α and IL‐1β, as well as genes which impact on apoptosis (Pahl 1999). It appears that NF‐κB is capable of activating both pro‐ and anti‐apoptotic genes (Bours et al . 2000). One would reason that, with respect to normal tissue toxicity, NF‐κB activation would be associated with a pro‐apoptotic outcome. While it is clear the ionizing radiation is capable of activating NF‐κB (Klahr 2001), its ultimate role in mucosal damage is unresolved. However, the potential role for NF‐κB in the development of mucositis is supported by the finding that upregulation of keratin 6b, keratin 17, and vimentin as seen in this study, are all mediated by NF‐κB.
Our results represent the first attempt to identify the sequence of gene activation in oral mucosa following exposure to an acute, stomatotoxic dose of ionizing radiation. It is clear that additional investigation will be necessary to better define this sequence and to identify its key components that result in tissue injury. Assignment of tissue‐ and cell‐specific gene activation to the components of the mucosa is also important if new opportunities for intervention are to be identified.
References
- Bayerl C, Taake S, Moll I, Jung EG (1995) Photodermatol. Photoimmunol. Photomed. 11, 149. [DOI] [PubMed] [Google Scholar]
- Bours V, Bentires‐Alj M, Hellin AC, Viatour P, Robe P, Dalhalle S, Benoit V, Merville MP (2000) Nuclear factor‐kappaB, cancer, and apoptosis. Biochem. Pharmacol. 60, 1085. [DOI] [PubMed] [Google Scholar]
- De Toledo SM, Azzam EI, Keng P, Laffrenier S, Little JB (1998) Regulation by ionizing radiation of CDC2, cyclin A, cyclin B, thymidine kinase, topoisomerase IIalpha, and RAD51 expression in normal human diploid fibroblasts is dependent on p53/p21Wafl . Cell Growth Differ. 9, 887. [PubMed] [Google Scholar]
- Gilles C, Polette M, Zahm J, Tournier J, Volders L, Foldart J, Birembaut P (1999) Vimentin contributes to human mammary epithelial cell migration. J. Cell Sci. 112, 4615. [DOI] [PubMed] [Google Scholar]
- Iuchi S, Green H (1999) Basonuclin, a zinc finger protein of keratinocytes and reproductive germ cells, binds to the rRNA gene promotor. Proc. Natl. Acad. Sci. 17, 9628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klahr S (2001) Urinary tract obstruction. Semin. Nephrol. 21, 133. [DOI] [PubMed] [Google Scholar]
- Lee S, Choi S, Lee K, Chung H, Kim T, Cho C, Lee Y (2001) Role of inducible heat shock protein 70 in radiation‐induced cell death. Cell Stress Chaperones 6, 273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Scolnick DM, Trievel RC, Zhang HB, Marmorstein R, Halazonetis TD, Berger SL (1999) P53 sites acetylated in vitro by PCAF and p300 are acetylated in vivo in response to DNA damage. Mol. Cell Biol. 19, 1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma S, Rao L, Freedbarg IM, Blumenberg MV (1997) Transcriptional control of K5, K6, K14, and K17 keratin genes by AP‐1 and NF‐κB family members. Gene Exp. 6, 361. [PMC free article] [PubMed] [Google Scholar]
- Marakov SS (2000) NF‐κB as a therapeutic target in chronic inflammation: recent advances. Mol. Med. Today 6, 441. [DOI] [PubMed] [Google Scholar]
- McGowan KM, Coulombe PA (1998) Onset of keratin 17 expression coincides with the definition of major epithelial lineages during skin development. J. Cell Biol. 143, 469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollà M, Gironella M, Salas A, Miquel R, Pérez‐Del‐Pulgar S, Conill C, Engel P, Biete A, Piqué JM, Panés J (2001) Role of P‐selectin in radiation‐induced intestinal damage. Int. J. Cancer 96, 99. [DOI] [PubMed] [Google Scholar]
- Munz B, Tretter YP, Hertel M, Engelhardt F, Alzheimer C, Werner S (2001) The roles of activins in repair processes of skin and the brain. Mol. Cell Endocrinol. 180, 169. [DOI] [PubMed] [Google Scholar]
- Navarro JM, Casatorres J, Jorcano JL (1995) Elements controlling the expression and induction of the skin hyperproliferation‐associated keratin K6. J. Biol. Chem. 270, 21362. [DOI] [PubMed] [Google Scholar]
- Ogawa H, Fukushima K, Sasaki I, Matsuno S (2000) Identification of genes involved in mucosal defense and inflammation associated with normal enteric bacteria. Am. J. Physiol. Gastrointest. Liver Physiol. 279, G492. [DOI] [PubMed] [Google Scholar]
- Pahl HL (1999) Activators and target genes for Rel/NF‐κB transcription factors. Oncogene 18, 6853. [DOI] [PubMed] [Google Scholar]
- Pajonk F, McBride WH (2001) Ionizing radiation affects 26s proteasome function associated molecular responses, even at low doses. Radiotherapy Oncol. 59, 203. [DOI] [PubMed] [Google Scholar]
- Paris F, Fuks Z, Kang A, Capodieci P, Juan G, Ehleiter D, Haimovitz‐Friedman A, Cordon‐Cardo C, Kolesnick R (2001) Epithelial apoptosis as the primary lesion initiating intestinal radiation damage in mice. Science 293, 293. [DOI] [PubMed] [Google Scholar]
- Prasad S, Soldatenkov VA, Srinivasarao G, Dritschilo A (1998) Identification of keratins 18,19 and heat‐shock protein 90 beta as candidate substrates of proteolysis during ionizing radiation‐induced apoptosis of estrogen‐receptor negative breast tumor cells. Int. J. Oncol. 13, 757. [DOI] [PubMed] [Google Scholar]
- Quarmby S, Hunter RD, Kumar S (2000) Irradiation induced expression of CD31, ICAM‐1 and VCAM‐1 in human microvascular endothelial cells. Anticancer Res. 20, 3375. [PubMed] [Google Scholar]
- Reuscher TJ, Sofedi A, Scravani S, Kaban L, Sonis ST (1998) The impact of mucositis on α‐hemolytic streptococcal infections in patients undergoing autologous bone marrow transplantation for hematologic malignancies. Cancer 82, 2275. [PubMed] [Google Scholar]
- Schwaeble WJ, Reid KBM (1999) Does properdin crosslink the cellular and the humoral immune response? Immunol. Today 20, 17. [DOI] [PubMed] [Google Scholar]
- Shi Q, Le X, Abbruzzese JL, Wang B, Mujaida N, Matsushima K, Huang S, Xiong Q, Xie K (1999) Cooperation between transcription factor AP‐1 and NF‐kappaB in the induction of interleukin‐8 in human pancreatic adenocarcinoma cells by hypoxia. J. Interferon Cytokine Res. 19, 1363. [DOI] [PubMed] [Google Scholar]
- Shimizu‐Yoshida Y, Sugiyama K, Rogounovitch T, Ohtsura A, Namba H, Saenko V, Yamashita S (2001) Radiation‐inducible hSNK gene is transcriptionally regulated by p53 binding homology element in human thyroid cells. Biochem. Biophys. Res. Commun. 30, 491. [DOI] [PubMed] [Google Scholar]
- Sonis ST (1998) Mucositis as a biologic process. A new hypothesis for the development of chemotherapy‐induced stomatotoxicity. Oral Oncol. 34, 39. [DOI] [PubMed] [Google Scholar]
- Sonis ST (2000) Oral complications of cancer therapy In: Holland JF et al Cancer Medicine, 5th edn, pp. 2371‐2379. Philadelphia: BC Decker. [Google Scholar]
- Sonis ST, Oster G, Fuchs H, Bellm L, Bradford WZ, Edelsberg J, Hayden V, Eilers J, Epstein JB, Leveque FG, Miller C, Peterson DE, Schubert MM, Spijkervet FK, Horowitz M (2001) Oral mucositis and the clinical and economic outcomes of hematopoietic stem‐cell transplantation. J. Clin. Oncol. 19, 2201. [DOI] [PubMed] [Google Scholar]
- Sonis ST, Peterson RL, Edwards LJ, Lucey CA, Wang L, Mason L, Login G, Ymamakawa M, Moses G, Bouchard P, Hayes LL, Bedrosian C, Dorner AJ (2000) Defining mechanisms of action of Interleukin‐11 on the progression of radiation‐induced oral mucositis in hamsters. Oral Oncol. 36, 373. [DOI] [PubMed] [Google Scholar]
- Southall MD, Isenberg JS, Nakshatri H, Yi Q, Pei Y, Spandau DF, Travers JB (2001) The platelet‐activating factor receptor protects epidermal cells from tumor necrosis factor (TNF) α and TNF‐related apoptosis‐inducing ligand‐induced apoptosis through an NF‐κB‐dependent process. J. Biol. Chem 49, 45548. [DOI] [PubMed] [Google Scholar]
- Steinert PM, Candi E, Kartasova T, Marekov L (1998) Small proline‐rich proteins are cross‐bridging proteins in the cornified cell envelopes of stratified squamous epithelia. J. Structural Bio. 122, 76. [DOI] [PubMed] [Google Scholar]
- Suffredini AF, Fantuzzi G, Badolato R., Oppenheim JJ, O'grady NP (1999) New insights into the biology of the acute phase response. J. Clin. Immuno. 19, 203. [DOI] [PubMed] [Google Scholar]
- SunderRaj N, Rizzo JD, Anderson SC, Gesiotto JP (1992) Expression of vimentin by rabbit corneal epithelial cells during wound repair. Cell Tissue Res. 267, 347. [DOI] [PubMed] [Google Scholar]
- Troy T, Turksen K (1999) In vitro characteristics of early epidermal progenitors isolated from keratin 14 (K14)‐deficient mice: Insights into the role of keratin 17 in mouse keratinocytes. J. Cell Physiol. 180, 409. [DOI] [PubMed] [Google Scholar]
- Weichselbaum RR, Hallahan D, Fuks Z, Kufe D (1994) Radiation induction of immediate early genes: effectors of the radiation‐stress response. Int. J. Radiat. Oncol. Biol. Phys. 30, 229. [DOI] [PubMed] [Google Scholar]
- Whitmarsh AJ, Davis RJ (1996) Transcription factor AP‐1 regulation by mitogen‐activated protein kinase signal transduction pathways. J. Mol. Med. 74, 589. [DOI] [PubMed] [Google Scholar]
- Xu L, Badolato R., Murphy WJ, Oppenheim JJ, Wang JM (1995) A novel biologic function for serum amyloid A. J. Immunol. 155, 1184. [PubMed] [Google Scholar]


