Abstract
Abstract. The Wilms’ tumour suppressor gene WT1 encodes multiple isoforms of a transcription factor essential for correct mammalian urogenital development. Maintenance of the correct isoform ratio is critical. In humans, perturbation of this ratio causes Frasier syndrome, which is characterized by developmental defects of the kidney and urogenital tract. Different WT1 isoforms are thought to regulate transcription and participate in mRNA processing, functions reflected by a complex sub‐nuclear distribution. However, the role of individual WT1 isoforms remains unclear and pathways leading to WT1 sub‐nuclear localization are completely unknown. Here we use cells expressing green fluorescent protein‐tagged WT1 to demonstrate that the two major WT1 isoforms occupy separate and dynamic intranuclear locations in which one isoform, WT1+KTS, preferentially associates with the nucleolus. The alternatively spliced zinc finger region is found to be critical for the initial sub‐nuclear separation of WT1 isoforms, but interactions between different isoforms influence the sub‐nuclear distribution of WT1. We illustrate how disruption of WT1 nuclear distribution might result in disease. This study contributes to the emerging picture of intranuclear protein trafficking.
INTRODUCTION
Correct control of gene expression by the Wilms’ tumour suppressor protein WT1 during mammalian development requires co‐ordinated expression of multiple WT1 isoforms (Hastie 1994; Little et al. 1999; Roberts 2005). Two alternative splices, exon 5 (51 bp) and a 9‐bp extension of exon 9, are present in the mammalian WT1 gene (Haber et al. 1991). The function of exon 5 is not well defined and alternative splicing of this region is absent in lower vertebrates (Miles et al. 2003). The variable splicing of exon 9, coding for the insertion of the amino acids lysine, threonine and serine (KTS) between the third and fourth zinc fingers, results in two major classes of WT1 isoform’ WT1+KTS and WT1−KTS. Disruption of the ratio of these isoforms occurs in Frasier syndrome caused by a heterozygous mutation in one of the exon 9 splice donor sites (Klamt et al. 1998). The consequent developmental defects of the kidney and urogenital tract indicate an absolute requirement for both WT1 isoforms. The creation of mice ablated for either WT1+KTS or WT1−KTS has provided direct evidence that these two WT1 splicing variants perform distinct functions in embryonic development (Hammes et al. 2001; Wagner et al. 2005). These findings validate in vitro studies that led to a hypothesis that WT1+KTS and WT1−KTS isoforms control separate aspects of gene expression (Bickmore et al. 1992; Caricasole et al. 1996; Englert 1998). The phenotypes of isoform‐specific knockout mice show that the WT1+KTS and WT1−KTS isoform functions are partially redundant, at least in the absence of the other isoform. The WT1−KTS isoforms are believed to act predominantly as transcription factors as they bind discrete DNA sequences with high affinity and co‐localize in cells with transcription factors (Larsson et al. 1995; Nakagama et al. 1995). In contrast, WT1+KTS isoforms display little affinity for consensus DNA sequences bound by WT1−KTS (Caricasole et al. 1996; Duarte et al. 1998; Laity et al. 2000) and there is evidence that the WT1+KTS isoform might play an additional role in mRNA processing. WT1+KTS isoforms were found to co‐localize predominately with nuclear domains rich in splicing factors and to associate preferentially with the essential splicing protein U2AF65 (Larsson et al. 1995; Davies et al. 1998). Davies et al. (1998) also showed that WT1 could be incorporated into active spliceosomes, suggesting that at least one WT1 isoform interacts with components of the splicing machinery. WT1+KTS was shown to associate with the U5 snRNP‐associated protein, p116 (Ladomery et al. 1999), and to stably bind transcripts in vivo (Ladomery et al. 2003). Niksic et al. (2004) showed that both classes of WT1 isoform shuttle between the nucleus and cytoplasm where WT1 protein is associated with ribonucleoprotein. Moreover, they reported that WT1 is associated with translating polysomes and they proposed an additional link between WT1 and the regulation of translation (Niksic et al. 2004).
The involvement of different WT1 isoforms in either transcription or mRNA processing makes WT1 a particularly attractive subject for analysing the nuclear distribution of proteins involved in gene expression. Dynamic models for spatial and temporal coordination of proteins involved in transcription and mRNA processing have emerged from observations of nuclear proteins (Misteli & Spector 1998; Gall et al. 1999; Lewis & Tollervey 2000; Gorski et al. 2006). Proteins are extremely mobile within the nucleoplasm (Misteli 2000; Phair & Misteli 2000; Dundr et al. 2004) and can move between nuclear domains where they might be stored or transiently assembled into complexes from which they are efficiently recruited to sites of active transcription and mRNA processing (Misteli et al. 1997). We have used fluorescent protein fusions to show that the two major WT1 isoforms move via different pathways around active nuclei and that the mechanism relies on the presence or absence of KTS insertion in the zinc finger region. We also uncover evidence of differential integration of the WT1+KTS isoform with nuclear sub‐domains involved in mRNA splicing.
MATERIALS AND METHODS
Plasmid construction
cDNA sequences encoding full length mouse WT1 isoforms including exon 5 were sub‐cloned as BamH1‐XbaI fragments from plasmid constructs described by Duarte et al. (1998) into pBlusescript KS+ (Stratagene, Cambridge, UK). Polymerase chain reaction (PCR) was used to include a 5′ BamH1 site and a‐3′ BglII site to fluorescent protein encoding sequences (both green fluorescent protein (GFP) and red fluorescent protein (RFP); Clontech, Basingstoke, UK) to enable subsequent cloning in‐frame at the 5′ end of the WT1 constructs. The fluorescent WT1 constructs were then cloned as BamH1‐XbaI fragments into the mammalian expression vector pcDNA3 (Invitrogen, Paisley, UK). A truncated zinc finger region was obtained from a plasmid encoding a stop mutant isoform of WT1 (F392 Stop) (Little & Wells 1997) and exchanged into pBluescript containing GFPWT1 fusions as an AvrII‐XbaI fragment before transferring to pCDNA3, where necessary DNA sequencing was used to verify the plasmid constructs. Details of PCR primers and conditions are available on request.
Cell culture and transient transfections
Cos7 cells, a fibroblastic cell line derived from monkey kidneys (ECACC, Wiltshire UK), M15 cells derived from the mouse mesonephros (Larsson et al. 1995), MCF 7 cells and Hela cells were all cultured at 37 °C with 5% CO2 in Dulbecco's modified Eagle's medium (DMEM, Sigma, St. Louis, MO, USA) supplemented with 10% foetal calf serum, penicillin (100 iµ/ml), streptomycin (0.1 mg/ml) and glutamine (2 mm) (Invitrogen). Most of the studies were performed in M15 cells, which express endogenous WT1, and in Cos7 cells, which do not. For transient transfections, the cells were plated at a density of 1.5 × 105 cells per 35‐mm dish and were transfected by addition of 2 µg plasmid DNA and 3 µl Fugene 6 (Roche, Basel, Switzerland). Actinomycin D (Sigma) was added at a concentration of 5 µg/ml for up to 3 h. Nuclear expression patterns were scored blindly for each isoform in at least 70 live cells from each of three or more separate transfections. For analysis of protein expression, the cells were washed in ice‐cold phosphate buffer saline (PBS) and treated with lysis buffer (10mm Tris‐HCl pH 8.0, 1 mm ethylenediaminetetraacetic acid pH 8.0, 150 mm NaCl and 0.65% NP40) containing a protease inhibitor cocktail (Roche) on ice for 20 min before harvesting and centrifugation at 5000 g for 15 min to remove cell debris. Total soluble protein in the cell lysates was measured using the Bio‐Rad protein assay reagent.
Western blotting
Cell lysates were boiled for 5 min in 2× sodium dodecyl sulphate (SDS) sample buffer (50 mm Tris‐HCl pH 6.8, 4% SDS, 10% glycerol, 0.006% bromophenol blue, 2%β‐mercaptoethanol) before being separated by electrophoresis on an 8% SDS‐polyacrylamide gel. They were then transferred to polyvinylidene difluoride (PVDF) membranes (Roche). The membranes were blocked in PBS containing 5% non‐fat dried milk for 1 h at room temperature before addition of the primary antibody and incubation, with agitation at 4 °C overnight. After multiple rinses with PBS, the membrane was incubated at 37 °C for 1 h in blocking buffer containing the appropriate alkaline phosphatase‐conjugated secondary antibody. Chemiluminescence detection was performed using disodium 3‐(4‐methoxyspiro){1,2‐dioxetane‐3,2′‐(5′‐chloro)tricyclo [3.3.13/7] decan}4‐yl phenyl phosphate (CSPD) substrate (Roche).
Immunofluorescence
For immunohistochemistry, the cells were grown in tissue culture wells attached to microscope slides (Invitrogen). To preserve fluorescence, the cells were fixed using paraformaldehyde (4% in PBS) by adding 1 ml of paraformaldehyde to 2 ml media in a 35‐mm dish and then mixing and incubating at room temperature for 5 min. The culture medium was then removed and the cells were fixed in paraformaldehyde for 10 min at room temperature before rinsing in PBS. For immunohistochemistry, slides were covered in blocking buffer (PBS containing 10% horse serum and 0.1% Tween) and were incubated at room temperature for 1 h. The cells were incubated overnight with the primary antibody diluted in blocking buffer at 4 °C before rinsing in PBS +0.1% Tween. Secondary antibodies were diluted in blocking buffer and were added to the slides before incubation at 37 °C for 1 h. Following further rinses with PBS, the cells were covered by VectaShield (Vector Laboratories, Burlingame, CA, USA) and were protected by cover slips. Nuclear localization of fluorescent WT1 isoforms in live cells was carried out in culture dishes using water immersion lenses (Nikon, Kingston, UK). All analyses were performed by standard fluorescence microscopy with an Eclipse 800 microscope (Nikon). Fluorescent and transmitted light images were documented using a C4880 cooled CCD camera (Hamamatsu, Photonics, Welwyn Garden City, UK) and processed using photoshop (Adobe, San Jose, CA, USA).
Antibodies
Antibodies used in this study recognized: WT1 (WT1 C19, Santa Cruz Biotechnologies Inc., Santa Cruz, CA, USA), GFP (mouse monoclonal, Clontech, Mountain View, CA, USA), Splicing factor Sc35 (Mouse monoclonal, Sigma), Myc epitope (mouse monoclonal, University of Bath), pKi67 (rabbit polyclonal, Dako Ltd., Ely, UK), B23 (B23 C19, Santa Cruz Biotechnologies Inc.) and PSP1 (rabbit polyclonal, Fox et al. 2002). The secondary antibodies were obtained from Vector Laboratories.
RESULTS
Expression of fluorescent WT1 fusion proteins
Vectors were constructed incorporating fluorescent proteins fused in‐frame to the 5′ end of cDNAs encoding mouse WT1+KTS or WT1−KTS isoforms (Fig. 1a). The integrity of the GFP‐tagged WT1+KTS and WT1−KTS proteins was analysed from whole cell extracts prepared from Cos7 cells transiently transfected with plasmids pGFPWT1+KTS or pGFPWT1−KTS (Fig. 1b). The fusion proteins were detected predominately as a single band of the predicted size (88 kDa) (lanes 5 and 6) in contrast to the 56‐kDa band of native WT1 isoforms transiently expressed in Cos7 cells (lanes 3 and 4) and endogenous WT1 expressed by M15 cells derived from mouse mesonephros (lane 1) (Larsson et al. 1995). As expected, the GFPWT1−KTS fusion retained sequence‐specific DNA binding activity in electrophoretic mobility shift assays (JR Dutton and A Ward, unpublished data).
Figure 1.
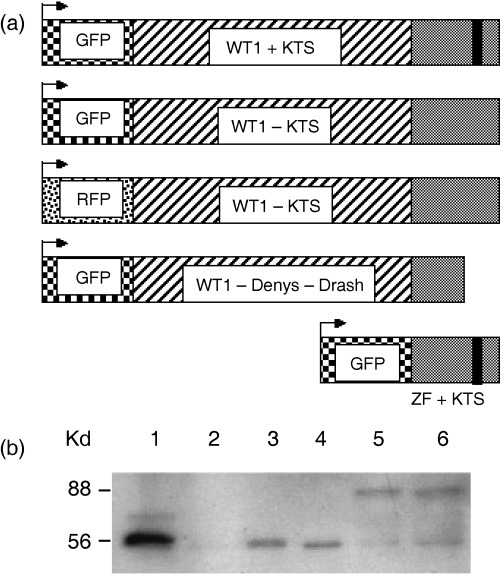
Construction of fusion protein expression plasmids and Western blot analysis of GFP‐tagged WT1 protein isolated from transfected cells. (a) Coding regions for fluorescent proteins GFP or RFP were cloned in‐frame at the 5′end of WT1 isoforms or the isolated zinc finger region in the expression vector pcDNA3. (b) Expression of WT1 and GFPWT1 isoforms in M15 and Cos7 cells. Transfected cells were harvested and equal protein amounts analysed on 8% polyacrylamide gels before transfer to PVDF membrane. Western blot analysis using WT1 antibody (C19) was used to detect the expression of WT1 and GFPWT1. Lane 1 = M15 cells expressing endogenous WT1 isoforms. Lane 2 = Cos7 cells expressing GFP only. Lanes 3 and 4 = Cos7 cells expressing native WT1 isoforms (WT1+KTS and WT1−KTS, respectively). Lanes 5 and 6 = Cos7 cells expressing GFPWT1 fusions (GFPWT1+KTS and GFPWT1−KTS, respectively; lower molecular weight bands in these lanes are presumed to be truncated GFPWT1 expression products).
In live cells, expression of GFPWT1+KTS and GFPWT1−KTS isoforms recapitulated the complex nuclear distribution patterns previously reported for non‐fusion WT1 proteins detected by antibody staining in fixed cells 48 hpt (hours post transfection) (Englert et al. 1995; Larsson et al. 1995). Figure 2a,b show the nuclear distribution of non‐fusion WT1 isoforms detected by antibodies in fixed cells 48 hpt whereas GFPWT1 fusions shown in Fig. 2c,d are in live cells observed with fluorescent microscopy. Notably, both wild‐type (Fig. 2a) and GFP‐fusion (Fig. 2c) forms of WT1+KTS were found to localize frequently to the nucleolus. On the other hand, this was rarely the case for wild‐type (Fig. 2b) and GFP‐fusion (Fig. 2d) forms of WT1−KTS.
Figure 2.
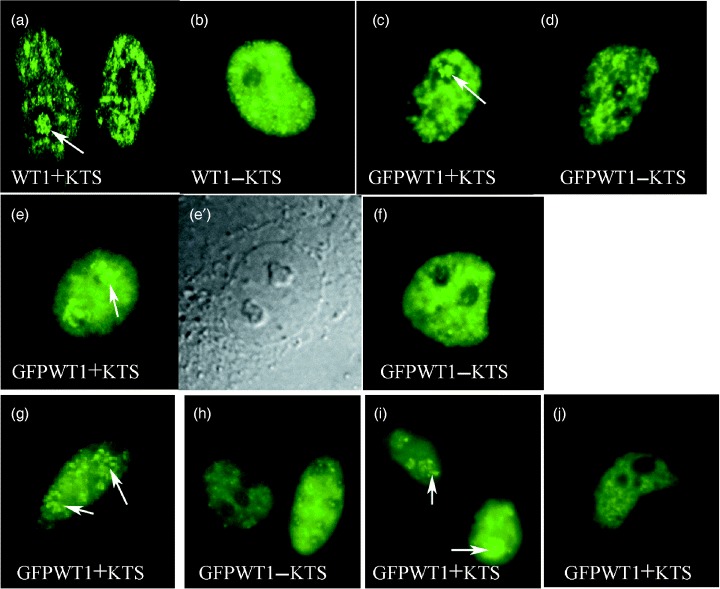
WT1 isoform distribution in mammalian cells (× 600). (a, b) Pattern of non‐fusion WT1 isoform expression in Cos7 cells 48 h after transfection identified using antiWT1 antibody staining. (a) WT1+KTS (b) WT1−KTS. (c–f) Pattern of fluorescent GFPWT1 isoform fusion proteins in live Cos7 cells. (e) Fluorescence showing diffuse/nucleolar distribution of GFPWT1+KTS in Cos7 cells 24 h after transfection. (e′) Phase contrast view of the nucleus in (e). (f) Diffuse/foci distribution of GFPWT1−KTS in Cos7 cells 24 h after transfection. (c, d) Steady state, discrete patterns of GFPWT1+KTS (c) and GFPWT1−KTS (d) Isoforms in Cos7 cells were similar at 48 hpt. (g) Diffuse/nucleolar (GFPWT1+KTS) and (h) diffuse/foci (GFPWT1−KTS) expression patterns were also seen in HeLa cells 7 h after transfection. (i,j) Diffuse/nucleolar expression of GFPWT1+KTS were seen in MCF7 cells 7 h after transfection (i) before dispersing to a mature discrete localization at 24 hpt (j). Arrows indicate localization of WT1+KTS to the nucleolus.
GFPWT1 fusion proteins are expressed in isoform‐specific patterns of nuclear distribution
We examined in further detail the sub‐nuclear distribution of GFP tagged WT1+KTS and –KTS isoforms following expression in Cos7 cells (Fig. 2, Table 1) There were clear differences in the nuclear distribution for each isoform at 24 and 48 h following transfection. All GFPWT1‐expressing nuclei displayed one of two basic distributions termed discrete or diffuse. Nuclei with a diffuse pattern possess a nucleoplasmic background expression of even intensity (Fig. 2e,f), whereas discrete expression describes nuclei with patterns of localized, irregular WT1 background staining (Fig. 2c,d). Nuclei with diffuse GFPWT1 expression were further subdivided if they displayed either multiple bright foci of WT1 expression superimposed on the diffuse background (Fig. 2f) or intense WT1 expression in nucleoli (Fig. 2e,e′). These nuclear patterns were referred to as diffuse/foci and diffuse/nucleolar.
Table 1.
Sub‐nuclear localization of GFPWT1 following transfection in Cos7 cells. Pattern of nuclear distribution after 24/48 h (percentage of total GFPWT1 expressing cells)
| WT1 isoform | Discrete | Diffuse | Diffuse/nucleolar | Diffuse/foci |
|---|---|---|---|---|
| +KTS (n = 419/255) | 22/28 | 21/44 | 57/28 | 0/0 |
| –KTS (n = 362/316) | 43/62 | 14/3 | 15/9 | 28/26 |
Analysing the expression of GFPWT1+KTS and GFPWT1−KTS isoforms in Cos7 cells showed that the nuclear distribution of WT1 is clearly determined by isoform type (Table 1). We were able to distinguish consistently, in blind experiments, between isoforms based solely on their nuclear distribution in live Cos7 cells. Strikingly, 57% of the cells expressing the GFPWT1+KTS isoform displayed diffuse/nucleolar distribution 24 h after transfection (Fig. 2e,e′) compared with 15% of GFPWT1−KTS expressing nuclei at this time point. Cell populations expressing the GFPWT1−KTS isoform were uniquely characterized by the diffuse/foci pattern of nuclear expression (Fig. 2f) with 28% of the nuclei showing this distribution 24 h after transfection. No cells expressing GFPWT1+KTS displayed this nuclear distribution.
The nuclear localization displayed by Cos7 cells expressing GFPWT1 isoforms was examined again 48 h after transfection (Fig. 2c,d). In nuclei expressing GFPWT1−KTS, the discrete distribution pattern increased from 43% at 24 h to 62% at 48 h. The nuclei displaying diffuse/nucleolar expression dropped to only 9% but the foci in a diffuse nucleoplasmic background were still seen in 26% of the nuclei counted. In GFPWT1+KTS isoform‐expressing cells, the discrete pattern of nuclear distribution was maintained (28% at 48 hpt, 22% at 24 hpt) but there was an increase in the simple diffuse pattern from 21% at 24 hpt to 44% at 48 hpt. However, the number of nuclei showing nucleolar expression in a diffuse background was reduced sharply from 57% to 28%. These results highlight the dynamic nature of the patterns of WT1 nuclear expression.
Nuclear localization of GFPWT1 isoforms in other mammalian cell lines
To illustrate that isoform determination of WT1 nuclear distribution was not restricted to Cos7 cells, we extended the analysis to other mammalian cell lines. This allowed, for the first time, live observation of single WT1 isoforms in the mouse kidney‐derived cell line M15 cells that also express endogenous WT1 protein. We transiently introduced the GFPWT1 isoform constructs into M15, MCF7 cells and also HeLa cells (Fig. 2g–j). Although the times during which the isoform specific patterns were visible differed between these cell types, it was evident that the GFPWT1 isoforms initially separated in nuclear distribution before dispersing to the complex, steady state pattern of nuclear sub‐localization described by previous antibody studies. The diffuse nucleolar expression of GFPWT1+KTS seen in Cos7 cells was observed in MCF7 and HeLa cells after 7 h (Fig. 2g,i) whereas at this time the diffuse foci expression of GFPWT1−KTS was seen in MCF7 cells (Fig. 2h). The subsequent dispersal of GFPWT1+KTS to a complex sub‐nuclear pattern is shown for MCF 7 cells after 24 h (Fig. 2j).
Co‐expression of both classes of fluorescent WT1 isoform perturbs the separate nuclear localization of WT1+KTS and WT1−KTS
An isoform fusion of WT1−KTS incorporating the red fluorescent protein (RFP) enabled us to distinguish live both GFPWT1+KTS and RFPWT1−KTS following co‐transfection of Cos7 cells. The nuclear localization of these fusion proteins was studied in cells that transiently co‐expressed both isoforms. The observed WT1 nuclear distributions were scored for being completely coincident where the green pattern of the GFPWT1+KTS overlapped exactly with that of the red RFPWT1−KTS (92%) (Fig. 3a,a′), or where the localization of the two patterns were different (8%) (Fig. 3b,b′). Strikingly, in the minority of nuclei where the two patterns were different, the discordant red and green fluorescent patterns reproduced the isoform‐specific distributions seen in cells transfected with single isoforms. The GFP+KTS isoform accumulated in a diffuse/nucleolar pattern and the RFP−KTS isoform in a pattern of nuclear foci (compare Fig. 2e,f with Fig. 3b,b′).
Figure 3.
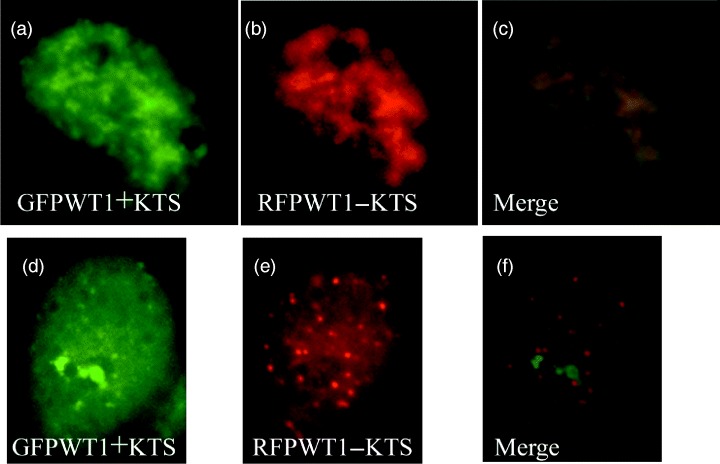
Isoform‐specific WT1 sub‐nuclear localization maintained in a subpopulation of cell co‐expressing multiple WT1 isoforms. Nuclei from Cos7 cells co‐transfected with GFPWT1+KTS (Green, a and d) and RFP tagged RFPWT1−KTS (Red, b and e). Most nuclei displayed co‐localization of the two isoforms (see merged image, c), but a small proportion of nuclei retained separate, isoform‐specific expression patterns (see merged image f).
WT1 isoforms localize independently in transcriptionally quiescent nuclei
Treatment of cells with actinomycin D renders nuclei transcriptionally quiescent and provides an opportunity to observe the positioning of nuclear proteins under less dynamic conditions. Cells transfected with GFPWT1+KTS or GFPWT1−KTS were incubated for 48 h and treated with actinomycin D before patterns of nuclear distribution were scored as described previously (Table 2). Transcriptional quiescence was confirmed using the previously reported re‐location of splicing factor SC35 (Zeng et al. 1997) (Fig. 4).
Table 2.
The effect of actinomycin D Treatment on sub‐nuclear localization of GFPWT1 following transfection in Cos7 cells. Pattern of nuclear distribution after actinomycin D Treatment (percentage of total GFPWT1 expressing cells)
| WT1 isoform | Discrete | Diffuse | Diffuse/nucleolar | Diffuse/foci |
|---|---|---|---|---|
| +KTS (n = 299) | 18 | 0 | 82 | 0 |
| –KTS (n = 327) | 33 | 0 | 4 | 63 |
Figure 4.
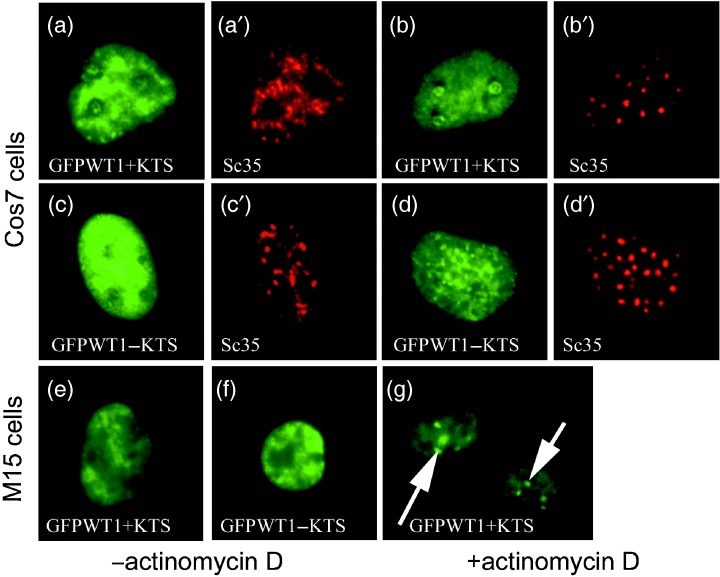
In transcriptionally quiescent cells, WT1 displays isoform‐specific nuclear distributions. Cos7 cells were transfected with GFPWT1 isoforms and incubated for 48 h before actinomycin D treatment was done to render them transcriptionally quiescent. The nuclear distribution of each GFPWT1 isoform and the splicing factor Sc35 were detected using GFP fluorescence (green) and texas‐red conjugated antibody to Sc35 (Red). GFPWT1+KTS and Sc35 before actinomycin D treatment (a and a′), GFPWT1‐KTS and Sc35 before actinomycin D treatment, (c and c′). GFPWT1+KTS and Sc35 after treatment (b and b′). GFPWT1−KTS and Sc35 after treatment (d and d′). 24 h after expression in M15 cells, the discrete nuclear distribution of GFPWT1+KTS (e) and GFPWT1‐KTS (f) were indistinguishable. However, following actinomycin D treatment, the +KTS isoform was again seen to re‐locate to the nucleolus (g, arrows).
In transcriptionally quiescent Cos7 nuclei, the two GFP‐tagged WT1 isoforms again displayed distinct, isoform‐specific sub‐nuclear distributions. The GFPWT1+KTS isoform was seen to re‐localize and accumulate within nucleolar structures with a diffuse dispersed nucleoplasmic background (Fig. 4b). Although this distribution appeared very similar to that seen following initial transfection of this isoform (Fig. 2a), in actinomycin D‐treated nuclei, the accumulations were present in compacted peri‐nucleolar structures. The pattern was also much more prevalent, being present in 82% of the GFPWT1+KTS expressing cells following treatment compared with the diffuse nucleolar localization seen in 28% of untreated nuclei after 48 h. There was no correlation in treated nuclei between the positions of re‐localized SC35 protein and the GFPWT1+KTS isoform (Fig. 4a,b). The GFPWT1−KTS isoform was also seen to adopt an isoform‐specific pattern of nuclear localization following actinomycin D treatment (Fig. 4d). In only a very small proportion of actinomycin D‐treated nuclei (4%), GFPWT1−KTS formed the diffuse/nucleolar pattern common in GFPWT1+KTS expressing cells, and 33% of GFPWT1−KTS expressing nuclei retained discrete patterns of WT1 expression. However, 63% of the nuclei exhibited a pattern of numerous small points of GFPWT1−KTS accumulation superimposed upon the nuclear background. This diffuse/foci pattern is similar to the foci seen in earlier experiments with this isoform (Fig. 2f). Once again the pattern of WT1−KTS specks did not co‐localize with the distribution of the re‐located splicing factor SC35.
When transfected into M15 cells that express endogenous WT1, the GFP‐tagged WT1 isoforms in this study also briefly displayed transient isoform‐specific nuclear localization seen in Cos7 cells (data not shown) before dispersing to a complex pattern of nuclear localization (Fig. 4e,f). However, when M15 cells expressing GFPWT1+KTS were treated with actinomycin D, the GFP‐tagged protein was again seen to accumulate in the nucleoli (Fig. 4g, arrows). This pattern was present in 82% of the nuclei counted (n = 300), having been almost entirely absent in untreated cells 24 h after transfection.
WT1+KTS co‐localizes with nucleolar associated proteins
To confirm the nucleolar accumulation of GFPWT1+KTS, we examined the co‐localization of this isoform with known nucleolar markers (Fig. 5). The nucleolar expression of GFPWT1+KTS was found to partially co‐localize with both pKi67 (Fig. 5a–c) and B23/nucleophosmin (Fig. 5d–f). pKi67 labels a fibrillarin‐deficient region of the dense fibrillar component of the nucleolus associated with rRNA transcript maturation. B23 is located mainly within the granular component where ribosome assembly occurs (Kill 1996).
Figure 5.
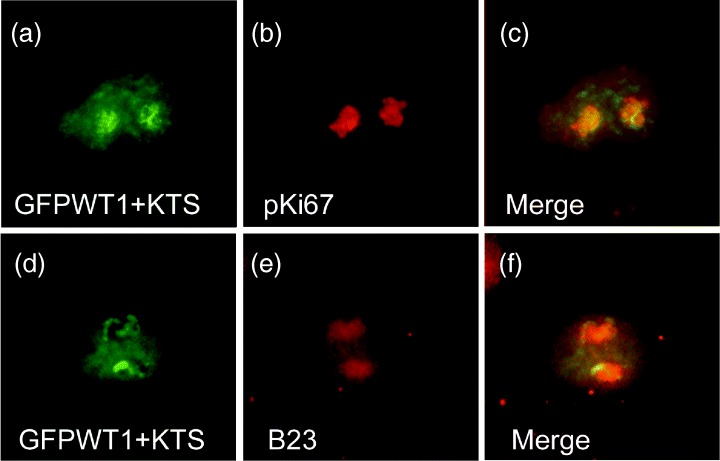
GFPWT1+KTS co‐localizes with known nucleolar proteins. The nucleolar associated expression of GFPWT1+KTS in Cos7 cells (a and d) was compared with the expression pattern of the nucleolar proteins pKi67 (b) and B23 (e) detected by texas‐red conjugated antibody staining. The extent of co‐localization is shown in merged images (c and f).
Alternative splicing of the zinc finger region controls the sub‐nuclear distribution of WT1 isoforms
To further investigate the mechanism that leads to separation of WT1 isoforms in the nucleus, we constructed fusion constructs encoding GFP fused directly to the isolated zinc finger region from the WT1+KTS and WT1−KTS isoforms. On expression in Cos7 cells, the GFPZF+KTS construct was found to accumulate exclusively in the nucleolus (Fig. 6a) but, in direct contrast to full‐length GFPWT1+KTS, was unable to disperse further to any other nuclear domains even after expression for over 48 h. Complete co‐localization with B23 (Fig. 6b–c) illustrates that the nucleolar distribution of the GFPZF+KTS construct was distinct from that of the full‐length GFPWT1+KTS (compare with Fig. 5d–f). A GFPZF‐KTS fusion was freely distributed in live Cos7 cells in a discrete pattern of nuclear expression (Fig. 6d) that reflects the mature expression pattern of full‐length GFPWT1‐KTS protein seen previously (Fig. 2d).
Figure 6.
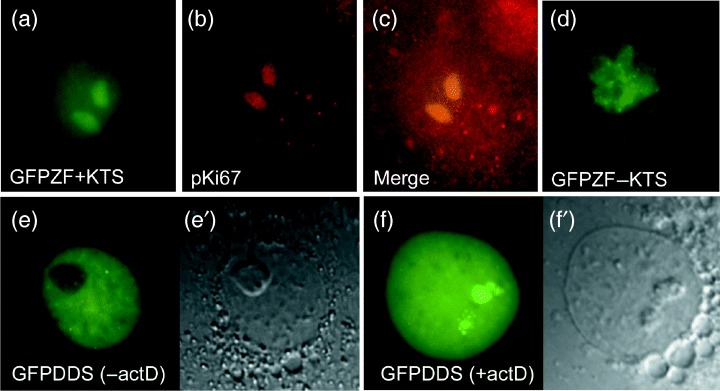
The zinc finger region determines sub‐nuclear distribution of WT1 isoforms. Nuclear distribution of fusions between GFP and only the WT1 zinc finger regions (GFPZF) in live Cos7 cells 48 hpt (a and d). Only nucleolar accumulation was seen in all cells expressing GFPZF+KTS (a). The nucleolar localization was confirmed using texas‐red conjugated antibody detection of B23 (b and merge c). Nuclei expressing GFPZF‐KTS all showed more complex, discrete patterns of expression (d). Abnormal nuclear distribution of GFPWT1 expressing a Denys‐Drash mutation truncated in zinc finger 3 (GFPDDS) at 24 hpt (e). (e′) shows a phase microscopy view of the nucleus in (e). Nuclear distribution of the GFPWT1 truncation mutant in Cos7 cells following actinomycin D treatment showing accumulation in the nucleolus (f, f′).
We extended this investigation of the sub‐nuclear localization properties of the zinc finger region to include the nuclear localization of a fusion of GFP to a full‐length WT1 construct containing a premature zinc finger truncation mutation found in some Denys‐Drash syndrome (DDS) patients. DDS has a phenotype distinct from Frasier syndrome and results from a wide spectrum of WT1 zinc finger mutations (reviewed by Little & Wells 1997). The mutation used here causes the protein to terminate within the third zinc finger so that the KTS insertion site and fourth zinc finger are missing. Fusions containing this mutation did not localize in a manner similar to either of the wild‐type isoforms (Fig. 6e). Interestingly, after 24 h of incubation, almost all the cells (93%n = 189) showed a diffuse nuclear distribution of the Denys‐Drash mutant protein. This percentage is closer to GFPWT1+KTS (78%) than GFPWT1−KTS (57%) isoforms at this time point. Twenty‐seven per cent of the nuclei displayed nucleolar accumulation within the diffuse background (compared with 59% for GFPWT1+KTS, 15% for GFPWT1−KTS) whereas 61% displayed a nuclear localization pattern characterized by regular dark patches within the diffuse nucleoplasmic staining, emphasizing exclusion of the mutant WT1 protein from nucleoli (Fig. 6e). This distribution is extremely uncommon in nuclei expressing either GFPWT1+KTS or GFPWT1−KTS. Intriguingly, however, following actinomycin D treatment, almost all the cells (93%n = 221) expressing the GFP‐Denys‐Drash fusion protein accumulated the truncated WT1 protein within nucleoli (Fig. 6f), a pattern characteristic of the WT1+KTS isoform.
GFPWT1+KTS trafficking is associated with nucleolar protein movement
A proteomic study of isolated nucleoli has identified paraspeckle protein 1 (PSP1) (Andersen et al. 2002). PSP1 labels a novel nuclear domain that contains RNA‐binding proteins and cycles between nuclear domains called paraspeckles and the nucleolus. In actinomycin D‐treated cells, PSP1 accumulates in peri‐nucleolar caps (Fox et al. 2002; Xie et al. 2006). These intriguing parallels with WT1+KTS suggested that we test each WT1 isoform for association with PSP1 in paraspeckles or the nucleolus to investigate if this protein might play a role in WT1 trafficking (Fig. 7). In Cos7 cells, PSP1 is found in both paraspeckles (Fig. 7a) and in compacted structures associated with nucleoli (Fig. 7d). Although there was no association with either GFPWT1+KTS or GFPWT1−KTS in PSP1 labelled paraspeckles (Fig. 7a–c), in cells where PSP1 was associated with nucleoli there was complete co‐localization with nucleolar associated GFPWT1+KTS (Fig. 7d–f).
Figure 7.
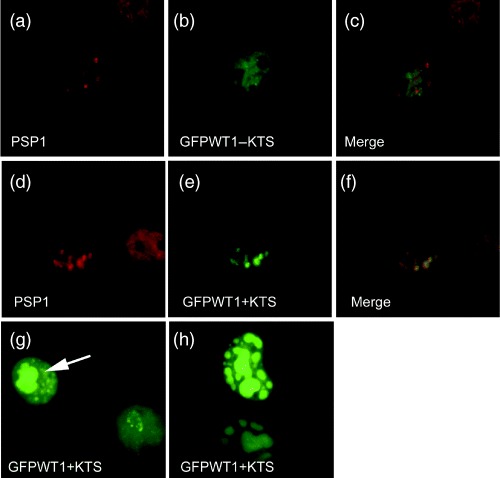
WT1+KTS nuclear distribution is linked to proteins associated with nucleolar protein trafficking. Using an antibody to PSP1, we determined distinct patterns of paraspeckle expression in Cos7 cells. PSP1 was expressed in speckles imposed on a diffuse nucleoplasmic background (a) or associated with nucleoli (d). The speckled pattern of PSP1 expression (a) did not co‐localize with diffuse/foci expression of GFPWT1‐KTS (b and merged c). Co‐localization observed between the nucleolar associated expression of PSP1 (d) and GFPWT1+KTS (e) is shown in the merged image (f). Ectopic p80coilin expression caused GFPWT1+KTS to accumulate in abnormal nucleoli (g, h). In Cos7 cells co‐expressing ectopic p80coilin and GFPWT1+KTS (g, arrow), the GFPWT1 accumulated in abnormally large nucleoli (arrow) compared to the typical diffuse/nuleolar pattern in an adjacent cell not co‐expressing ectopic p80coilin. (h) Two abnormal nucleoli 48 hpt. Abnormal nucleoli were large, bulbous and fragmented but still retain all GFPWT1+KTS expression
Cajal bodies are also implicated in trafficking proteins to the nucleolus (Sleeman & Lamond 1999a, 1999b). p80coilin Is a major protein component of Cajal bodies, and the ectopic expression of this protein in Cos7 cells caused GFPWT1+KTS to accumulate in abnormal nucleolar structures (Fig. 7g,h). Twenty‐four hours after co‐expression with ectopic p80coilin (confirmed by detection of the myc tag on p80coilin, not shown) in Cos7 cells, GFPWT1+KTS accumulated in larger abnormal nucleoli (arrow Fig. 7g) compared with the typical diffuse nucleolar pattern in cells not expressing ectopic p80coilin. Following a further 24 h, GFPWT1+KTS remained concentrated within abnormal enlarged, fragmented nucleoli (Fig. 7h).
The initial transient accumulation of GFPWT1+KTS in the nucleolus and subsequent re‐location there in quiescent nuclei is in direct contrast to the sub‐nuclear movement of the GFPWT1−KTS isoform. Correlation of GFPWT1+KTS with nucleolar PSP1 and the disruption of nucleolar WT1+KTS trafficking by ectopic p80coilin expression both suggest that the isoform‐specific, dynamic sub‐nuclear movement of WT1+KTS is intimately associated with specific pathways of nucleolar protein movement.
DISCUSSION
Here, we presented a detailed examination of WT1 nuclear distribution using fluorescent fusion proteins describing the separation of different isoforms into distinct sub‐nuclear domains. The observed isoform‐specific intranuclear trafficking pathways are consistent with different WT1 isoforms acting predominantly in either transcription (WT1‐KTS) or mRNA processing (WT1+KTS).
Interpretation of nuclear protein patterns such as WT1, visualized by antibody staining, or fluorescent proteins is complicated by the movement of associated proteins in a highly dynamic environment (Lewis & Tollervey 2000; Misteli 2000; Phair & Misteli 2000). The spatial and temporal coupling of transcription and mRNA splicing (Gall et al. 1999; Minvielle‐Sebastia & Keller 1999) suggest that if roles in transcription and splicing are carried out by different WT1 isoforms, they might often still display a coincident nuclear distribution similar to that described for RNA pol II and the splicing factor Sc35 (Zeng et al. 1997). This is consistent with reports of indistinguishable nuclear distributions for different WT1 isoforms, from which it is difficult to infer information regarding the function of the separate isoforms. In contrast, the use of GFP‐tagged WT1 isoforms in this study revealed novel information regarding isoform‐specific functions in cells expressing single labelled isoforms.
Expression patterns of GFPWT1 are interpreted here as illustrating transient stages in the co‐ordinated distribution of WT1 isoforms. Following transfection into cultured cells, GFPWT1−KTS initially showed a combination of nuclear distributions characterized by foci of expression, a pattern similar to that reported for WT1−KTS co‐localizing with transcription factors (Larsson et al. 1995). In contrast, nuclei expressing GFPWT1+KTS revealed a robust association of this isoform with the nucleolus. This initial separation of nuclear distribution patterns indicates that the different WT1 isoforms can enter divergent nuclear pathways consistent with integration into complexes with different functions. Importantly, similar isoform specific patterns were observed both by ourselves (See Fig. 2a) and others (Larsson et al. 1995) in cells expressing full‐length, wild‐type (non‐fusion) forms of WT1, including the localization of WT1+KTS to the nucleolus.
This sub‐nuclear, isoform‐specific separation was a general feature of GFPWT1 expression in the cell lines tested, although the isoform‐specific patterns were detected for only a limited period in some cell lines. This may be an inherent product of the dynamics of nuclear protein movement in different cell lines and types. In M15 cells that express endogenous WT1, isoform‐specific patterns of GFPWT1, although briefly observed on initial transfection, were indistinguishable after 24 h, matching the steady state patterns of WT1 expression detected in M15 nuclei by antibody staining (Larsson et al. 1995) and the patterns of GFPWT1 isoform expression in Cos7 cells 48 h after transfection. In M15 cells, the established nuclear trafficking of endogenous WT1 isoforms might result in more rapid resolution of the transient GFPWT1 isoform‐specific associations than in Cos7 cells that do not possess endogenous WT1 isoforms. This was supported by the obsevation that in contrast to single isoform expression, co‐expression of GFPWT1+KTS and RFPWT1−KTS resulted primarily in isoform co‐localization. In keeping with this, Hammes et al. (2001) reported changes in WT1 staining patterns in podocyte nuclei following single isoform ablation in mice, whereas in Xenopus oocytes, the co‐expression of WT1−KTS titrates WT1+KTS from Cajal body‐associated B snurposomes (Ladomery et al. 2003). Thus, formation of WT1 heterodimers and the ability of one isoform to alter the nuclear distribution of another may affect WT1 function.
The behaviour of GFPWT1 isoforms in actinomycin D‐treated cells provides further evidence that isoform‐specific distribution is an integral feature of WT1 nuclear expression. In cells rendered transcriptionally quiescent, transcription and splicing factors relocate from areas of activity to characteristic domains (Zeng et al. 1997). Under these conditions, GFPWT1 isoforms again adopt robust isoform‐specific patterns. The pattern of discrete foci formed by the GFPWT1–KTS isoform in Cos7 cells is identical to speckled domains reported for transcription factors in quiescent cells. In contrast, the pattern formed by GFPWT1+KTS in actinomycin D‐treated cells was not characteristic of some splicing factors (perhaps expected because of proposed WT1+KTS splice factor association) but was instead an accumulation restricted to a compacted component of the segregated nucleolus.
These results show that the mechanism of WT1 isoform segregation in the nucleus relies on the presence of KTS insertion between zinc fingers 3 and 4 to distinguish between isoforms. The DDS–GFP fusion lacking this region was not segregated in a pattern specific to either +KTS or –KTS isoforms, showing the requirement for an intact zinc finger region for correct sub‐nuclear distribution. In addition, fusions containing only the isolated +KTS zinc finger regions were unable to progress beyond the initial nucleolar association, showing that interactions with WT1 domains outside the zinc fingers are also important in the final distribution of WT1 in the nucleus. Therefore, although more commonly suggested as a mechanism for differentiating nucleic acid binding specificity between isoforms, the alternative splicing between fingers 3 and 4 may also provide the key to localizing individual WT1 isoforms in different nuclear compartments where they might perform distinct roles (for example, splicing or transcription).
The association shown between WT1+KTS and the nucleolus is intriguing because other components with established roles in mRNA splicing also associate with the nucleolus before integration into spliceosomes (Sleeman & Lamond 1999a). However, although WT1 can be incorporated into spliceosomes (Davies et al. 1998) and is present in nuclear ribonucleoprotein (RNP) particles (Ladomery et al. 1999; Morrison & Ladomery 2006), the identity of the isoforms present and the pathways of incorporation into these structures were previously unknown. The nucleolus is known to be a multifunctional organelle with roles in ribosome biogenesis, processing of some nuclear RNAs, sequestration of regulatory molecules and control of cell‐cycle progression and cellular ageing (Pederson 1998; Olson et al. 2000; Andersen et al. 2002; Leung & Lamond 2003). Other tumour suppressor proteins (including ARF, MDM2 and Rb) are known to act within the nucleolus to regulate cellular proliferation (Cavanaugh et al. 1995; Weber et al. 1999; Zhang & Xiong 1999). It is formally possible that the nucleolar association revealed in our study reflects a similar function of WT1+KTS.
We now show that possession of the KTS insertion causes newly expressed GFPWT1+KTS protein to be initially imported to the nucleolus, where it could be combined with RNA and protein components of the splicing machinery. As with mature snRNPs that do not pass along the assembly pathway travelled by newly expressed Sm proteins (Sleeman & Lamond 1999a), recycling of mature splicing complexes containing incorporated WT1+KTS in actively transcribing cells may not require re‐association with the nucleolus. This might explain the absence of +KTS isoform‐specific patterns following prolonged WT1 expression.
Integration of splicing components via the nucleolus requires association with Cajal bodies (Sleeman et al. 1998; Sleeman & Lamond 1999a), with nuclear organelles called B snurposomes transporting some splicing proteins from Cajal bodies to the nucleolus (Gall et al. 1999). We have investigated whether similar interactions might be involved in the distribution of WT1+KTS to the nucleolus. Possession of KTS insertion results in preferential association of the WT1+KTS isoform with Cajal bodies (Larsson et al. 1995), and it is exclusively the WT1+KTS isoform that associates with Cajal bodies and B snurposomes in Xenopus oocytes (Ladomery et al. 2003). Disruption of the trafficking of p80coilin, the major component of Cajal bodies, results in intranucleolar accumulation of Cajal bodies and splicing components (Sleeman et al. 1998). Similarly, aberrant p80coilin expression in Cos7 cells causes trapping of GFPWT1+KTS in enlarged nucleolar structures consistent with correct Cajal body trafficking being required for sub‐nuclear distribution of WT1+KTS. GFPWT1+KTS also co‐localizes with intranucleolar Cajal bodies that are a feature of MCF 7 cells (data not shown). WT1+KTS exhibits strikingly similar behaviour to PSP1, a protein found in nucleolar protein trafficking paraspeckles. As for WT1+KTS, paraspeckles can normally be found adjacent to nuclear domains rich in splicing factors, interact dynamically with the nucleolus and re‐locate to peri‐nucleolar structures in transcriptionally quiescent cells (Fox et al. 2002; Xie et al. 2006). WT1 association with PSP1 was restricted to nucleolar regions where the two proteins fully co‐localized. Our data link WT1+KTS with two proteins known to associate with the nucleolus – p80 coilin and PSP1 – suggesting that common pathways will be involved in their nuclear distribution.
The theory that WT1 isoforms perform separate nuclear functions is consistent with studies of the Frasier syndrome and of mice in which individual WT1 isoforms were ablated (Hammes et al. 2001; Hastie 2001; Wagner et al. 2005). These studies also illustrate that neither isoform can fully compensate for the loss of the other. This may result from a combination of the WT1−KTS isoform being unable to contribute transcript processing usually determined by the WT1+KTS isoform and the WT1+KTS being unable to fully interact with WT1−KTS target genes. However, we propose that these deficiencies are perhaps not only a consequence of different nucleic acid binding affinities between WT1 isoforms but are also a result of differential sub‐nuclear targeting as well as the necessity for isoform interaction to fine tune this nuclear distribution and therefore WT1 function. The abnormal nuclear distribution of the Denys‐Drash mutant WT1 isoform seen in this study, together with other reports of WT1 nuclear re‐location following zinc finger mutation (Englert et al. 1995; Larsson et al. 1995), indicate that the zinc fingers play a role in the differential distribution of WT1 isoforms, which may be more complex than a simple consequence of loss of nucleic acid binding. Phenotypic imbalance resulting from such aberrant WT1 isoform targeting would compromise appropriate gene regulation and offers an alternative explanation for the action of some WT1 mutations in DDS (Little & Wells 1997; Davies et al. 1998).
ACKNOWLEDGEMENTS
The authors wish to thank the following: Nick Hastie for the gift of the M15 cell line; Angus Lamond, Judith Sleeman and Archa Fox for p80coilin constructs, anti‐PSP1 antibody and for helpful discussions and comments; and Robert Kelsh and Barbara Reaves for the critical review of this manuscript. This work was funded by the Wellcome Trust, grant no. 049475/Z/96/Z and a Medical Research Council studentship.
REFERENCES
- Andersen JS, Lyon CE, Fox AH, Leung AK, Lam YW, Steen H, Mann M, Lamond AI (2002) Directed proteomic analysis of the human nucleolus. Curr. Biol. 12, 1–11. [DOI] [PubMed] [Google Scholar]
- Bickmore WA, Oghene K, Little MH, Seawright A, Van Heyningen V, Hastie ND (1992) Modulation of DNA binding specificity by alternative splicing of the Wilms tumor wt1 gene transcript. Science 257, 235–237. [DOI] [PubMed] [Google Scholar]
- Caricasole A, Duarte A, Larsson SH, Hastie ND, Little M, Holmes G, Todorov I, Ward A (1996) RNA binding by the Wilms tumor suppressor zinc finger proteins. Proc. Natl. Acad. Sci. USA 93, 7562–7566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanaugh AH, Hempel WM, Taylor LJ, Rogalsky V, Todorov G, Rothblum LI (1995) Activity of RNA polymerase I transcription factor UBF blocked by Rb gene product. Nature 374, 177–180. [DOI] [PubMed] [Google Scholar]
- Davies RC, Calvio C, Bratt E, Larsson SH, Lamond AI, Hastie ND (1998) WT1 interacts with the splicing factor U2AF65 in an isoform‐dependent manner and can be incorporated into spliceosomes. Genes Dev. 12, 3217–3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte A, Caricasole A, Graham CF, Ward A (1998) Wilms’ tumour‐suppressor protein isoforms have opposite effects on IGF‐2 expression in primary embryonic cells, independently of p53 genotype. Br. J. Cancer 77, 253–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dundr M, Hebert MD, Karpova TS, Stanek D, Xu H, Shpargel KB, Meier UT, Neugebauer KM, Matera AG, Misteli T (2004) In vivo kinetics of Cajal body components. J. Cell Biol. 164, 831–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englert C (1998) WT1 – more than a transcription factor? Trends Biochem. Sci. 23, 389–393. [DOI] [PubMed] [Google Scholar]
- Englert C, Vidal M, Maheswaran S, Ge Y, Ezzell RM, Isselbacher KJ, Haber DA (1995) Truncated WT1 mutants alter the subnuclear localization of the wild‐type protein. Proc. Natl. Acad. Sci. USA 92, 11960–11964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox AH, Lam YW, Leung AK, Lyon CE, Andersen J, Mann M, Lamond AI (2002) Paraspeckles: a novel nuclear domain. Curr. Biol. 12, 13–25. [DOI] [PubMed] [Google Scholar]
- Gall JG, Bellini M, Wu Z, Murphy C (1999) Assembly of the nuclear transcription and processing machinery: Cajal bodies (coiled bodies) and transcriptosomes. Mol. Biol. Cell 10, 4385–4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorski SA, Dundr M, Misteli T (2006) The road much traveled: trafficking in the cell nucleus. Curr. Opin. Cell Biol. 18, 284–290. [DOI] [PubMed] [Google Scholar]
- Haber DA, Sohn RL, Buckler AJ, Pelletier J, Call KM, Housman DE (1991) Alternative splicing and genomic structure of the Wilms tumor gene WT1 . Proc. Natl. Acad. Sci. U S A 88, 9618–9622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammes A, Guo JK, Lutsch G, Leheste JR, Landrock D, Ziegler U, Gubler MC, Schedl A (2001) Two splice variants of the Wilms’ tumor 1 gene have distinct functions during sex determination and nephron formation. Cell 106, 319–329. [DOI] [PubMed] [Google Scholar]
- Hastie ND (1994) The genetics of Wilms’ tumor – a case of disrupted development. Annu. Rev. Genet. 28, 523–558. [DOI] [PubMed] [Google Scholar]
- Hastie ND (2001) Life, sex, and WT1 isoforms – three amino acids can make all the difference. Cell 106, 391–394. [DOI] [PubMed] [Google Scholar]
- Kill IR (1996) Localisation of the Ki‐67 antigen within the nucleolus. Evidence for a fibrillarin‐deficient region of the dense fibrillar component. J. Cell Sci. 109, 1253–1263. [DOI] [PubMed] [Google Scholar]
- Klamt B, Koziell A, Poulat F, Wieacker P, Scambler P, Berta P, Gessler M (1998) Frasier syndrome is caused by defective alternative splicing of WT1 leading to an altered ratio of WT1 +/‐KTS splice isoforms. Hum. Mol. Genet. 7, 709–714. [DOI] [PubMed] [Google Scholar]
- Ladomery MR, Slight J, McGhee S, Hastie ND (1999) Presence of WT1, the Wilm's tumor suppressor gene product, in nuclear poly (A) (+) ribonucleoprotein. J. Biol. Chem. 274, 36520–36526. [DOI] [PubMed] [Google Scholar]
- Ladomery M, Sommerville J, Woolner S, Slight J, Hastie N (2003) Expression in Xenopus oocytes shows that WT1 binds transcripts in vivo, with a central role for zinc finger one. J. Cell Sci. 116, 1539–1549. [DOI] [PubMed] [Google Scholar]
- Laity JH, Chung J, Dyson HJ, Wright PE (2000) Alternative splicing of Wilms’ tumor suppressor protein modulates DNA binding activity through isoform‐specific DNA‐induced conformational changes. Biochemistry 39, 5341–5348. [DOI] [PubMed] [Google Scholar]
- Larsson SH, Charlieu JP, Miyagawa K, Engelkamp D, Rassoulzadegan M, Ross A, Cuzin F, Van Heyningen V, Hastie ND (1995) Subnuclear localization of WT1 in splicing or transcription factor domains is regulated by alternative splicing. Cell 81, 391–401. [DOI] [PubMed] [Google Scholar]
- Leung AK, Lamond AI (2003) The dynamics of the nucleolus. Crit. Rev. Eukaryot. Gene Expr. 13, 39–54. [DOI] [PubMed] [Google Scholar]
- Lewis JD, Tollervey D (2000) Like attracts like: getting RNA processing together in the nucleus. Science 288, 1385–1389. [DOI] [PubMed] [Google Scholar]
- Little M, Holmes G, Walsh P (1999) WT1: what has the last decade told us? Bioessays 21, 191–202. [DOI] [PubMed] [Google Scholar]
- Little M, Wells C (1997) A clinical overview of WT1 gene mutations. Hum. Mutat. 9, 209–225. [DOI] [PubMed] [Google Scholar]
- Miles CG, Slight J, Spraggon L, O'Sullivan M, Patek C, Hastie ND (2003) Mice lacking the 68‐amino‐acid, mammal‐specific N‐terminal extension of WT1 develop normally and are fertile. Mol. Cell Biol. 23, 2608–2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minvielle‐Sebastia L, Keller W (1999) mRNA polyadenylation and its coupling to other RNA processing reactions and to transcription. Curr. Opin. Cell Biol. 11, 352–357. [DOI] [PubMed] [Google Scholar]
- Misteli T (2000) Cell biology of transcription and pre‐mRNA splicing: nuclear architecture meets nuclear function [In process citation]. J. Cell Sci. 113, 1841–1849. [DOI] [PubMed] [Google Scholar]
- Misteli T, Caceres JF, Spector DL (1997) The dynamics of a pre‐mRNA splicing factor in living cells. Nature 387, 523–527. [DOI] [PubMed] [Google Scholar]
- Misteli T, Spector DL (1998) The cellular organization of gene expression. Curr. Opin. Cell Biol. 10, 323–331. [DOI] [PubMed] [Google Scholar]
- Morrison AA, Ladomery MR (2006) Presence of WT1 in nuclear messenger RNP particles in the human acute myeloid leukemia cell lines HL60 and K562. Cancer Lett. 244, 136–141. [DOI] [PubMed] [Google Scholar]
- Nakagama H, Heinrich G, Pelletier J, Housman DE (1995) Sequence and structural requirements for high‐affinity DNA binding by the WT1 gene product. Mol. Cell Biol. 15, 1489–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niksic M, Slight J, Sanford JR, Caceres JF, Hastie ND (2004) The Wilms’ tumour protein (WT1) shuttles between nucleus and cytoplasm and is present in functional polysomes. Hum. Mol. Genet. 13, 463–471. [DOI] [PubMed] [Google Scholar]
- Olson MO, Dundr M, Szebeni A (2000) The nucleolus: an old factory with unexpected capabilities. Trends Cell Biol. 10, 189–196. [DOI] [PubMed] [Google Scholar]
- Pederson T (1998) The plurifunctional nucleolus. Nucl. Acids Res. 26, 3871–3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phair RD, Misteli T (2000) High mobility of proteins in the mammalian cell nucleus. Nature 404, 604–609. [DOI] [PubMed] [Google Scholar]
- Roberts SG (2005) Transcriptional regulation by WT1 in development. Curr. Opin. Genet. Dev. 15, 542–547. [DOI] [PubMed] [Google Scholar]
- Sleeman JE, Lamond AI (1999a) Newly assembled snRNPs associate with coiled bodies before speckles, suggesting a nuclear snRNP maturation pathway. Curr. Biol. 9, 1065–1074. [DOI] [PubMed] [Google Scholar]
- Sleeman JE, Lamond AI (1999b) Nuclear organization of pre‐mRNA splicing factors. Curr. Opin. Cell Biol. 11, 372–377. [DOI] [PubMed] [Google Scholar]
- Sleeman J, Lyon CE, Platani M, Kreivi JP, Lamond AI (1998) Dynamic interactions between splicing snRNPs, coiled bodies and nucleoli revealed using snRNP protein fusions to the green fluorescent protein. Exp. Cell Res. 243, 290–304. [DOI] [PubMed] [Google Scholar]
- Wagner N, Wagner KD, Hammes A, Kirschner KM, Vidal VP, Schedl A, Scholz. H (2005) A splice variant of the Wilms’ tumour suppressor Wt1 is required for normal development of the olfactory system. Development 132, 1327–1336. [DOI] [PubMed] [Google Scholar]
- Weber JD, Taylor LJ, Roussel MF, Sherr CJ, Bar‐Sagi D (1999) Nucleolar Arf sequesters Mdm2 and activates p53. Nat. Cell Biol. 1, 20–26. [DOI] [PubMed] [Google Scholar]
- Xie SQ, Martin S, Guillot PV, Bentley DL, Pombo A (2006) Splicing speckles are not reservoirs of RNA polymerase II, but contain an inactive form, phosphorylated on serine2 residues of the C‐terminal domain. Mol. Biol. Cell 17, 1723–1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng C, Kim E, Warren SL, Berget SM (1997) Dynamic relocation of transcription and splicing factors dependent upon transcriptional activity. EMBO J. 16, 1401–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Xiong Y (1999) Mutations in human ARF exon 2 disrupt its nucleolar localization and impair its ability to block nuclear export of MDM2 and p53. Mol. Cell 3, 579–591. [DOI] [PubMed] [Google Scholar]


