Abstract
Genetic studies have identified mutations in key regulators of the Wnt/β-catenin pathway in a variety of cancers, most frequently in colon cancers. However, whether the pathway is activated in clinical cancer samples is not easily determined, and therefore it is useful to find markers that could be surrogates to show activation of the Wnt/β-catenin pathway. Gene expression profiles were analyzed in SW620, a colon cancer cell line in which β-catenin levels are stabilized as a consequence of truncated adenomatous polyposis coli and were compared with profiles of the same cells transfected with antisense oligodeoxynucleotides. Treatment of cells with β-catenin antisense oligodeoxynucleotides resulted in a decrease in the levels of axin2 and human naked cuticle (hnkd) mRNAs. Interestingly, the proteins encoded by both of these mRNAs are known inhibitors of the β-catenin pathway. In 30 human cell lines derived from different origins, axin2 and hnkd were expressed only in human colon cancer cell lines that are known to have activating mutations in the Wnt/β-catenin pathway. Further, levels of both axin2 and hnkd mRNA were also found to be elevated in about 65% of laser microdissected cells from human colon tumors compared with laser microdissected cells of normal morphology from the same patient samples. The increased expression of axin2 and hnkd correlated with truncations in adenomatous polyposis coli in the same patient samples. These results reveal that it is possible to detect activation of a carcinogenic pathway in human cancer samples with specific markers.
The Wnt/β-catenin pathway plays a pivotal role in animal development and tumorigenesis. Genetic studies have identified mutations in the key regulators of the pathway in human colon tumors that cause augmentation of Wnt/β-catenin signaling. For example, stabilizing mutations of β-catenin, a positive regulator, have been identified in a variety of cancers, whereas loss-of-function mutations of adenomatous polyposis coli (APC), axin1 and axin2, which are negative regulators, have also been identified in colorectal and hepatocellular cancers (1–5). Most prominently, mutations in APC have been detected in at least 80% of human colon tumors, indicating that activation of the Wnt/β-catenin pathway plays an important role in human colon tumorigenesis (6). Biochemical analysis has shown that activation of the Wnt/β-catenin pathway impairs the degradation process for β-catenin and leads to its accumulation in the cytoplasm. β-Catenin forms a heterodimeric complex with a member of the T cell factor (TCF)/lymphoid enhancer factor (LEF) family of transcription factors that translocates to the nucleus and subsequently regulates the expression of β-catenin target genes. When the pathway is activated, the expression levels and patterns of the Wnt/β-catenin target genes are altered. The findings that cyclin D1 and c-myc are β-catenin-inducible genes provide molecular evidence for the importance of this pathway in tumorigenesis (7, 8). However, because both these genes can also be regulated by the ras pathway (9, 10), we sought to identify additional Wnt/β-catenin pathway-regulated genes that would be more specific biomarkers for the activated pathway. Through analysis of gene expression profiles generated from the colon cancer cell line SW620 with reduced β-catenin protein levels resulting from antisense oligodeoxynucleotide (ODN) transfection, we found that axin2 and hnkd are β-catenin-induced target genes. Importantly, we extended this study to human colon tumor samples and found that the mRNA expression for axin2 and hnkd is up-regulated in colon tumors in which the Wnt/β-catenin pathway is activated. We propose that axin2 and hnkd may be used as biomarkers for the activated Wnt/β-catenin pathway in colon cancer diagnosis and drug development.
Methods
Antisense ODN Transfection.
SW620 cells were grown in DMEM supplemented with 10% FBS. Cells were transfected with antisense β-catenin ODN (AS ODN, 5′-ACTCAGCTTGGTTAGTGTGTCAGGC-3′) or reverse control ODN (RC ODN, 5′-CGGACTGTGTGATTGGTTCGACTCA-3′) to a final concentration of 100 nM by using a cationic peptoid reagent (11). Forty-eight hours posttransfection, cells were washed with PBS, and total RNA was prepared and reverse transcribed [RNeasy Mini kit, Qiagen (Chatsworth, CA); reverse transcription reaction kit, Perkin–Elmer]. Quantitative real-time analyses were performed with a LightCycler system from Roche Molecular Biochemicals. PCR primer sequences used are as follows: axin2F, 5′-TTATGCTTTGCACTACGTCCCTCCA-3′; axin2R, 5′-CGCAACATGGTCAACCCTCAGAC-3′; hnkdF, 5′-CTGGCTGCTACCACCATTGCGT-3′; hnkdR, 5′-CCAGGCCCAAATTGGGACGT-3′ (F, forward; R, reverse).
Cell Fractionation and Western Blotting.
To prepare cytosolic fractions, cells were lysed in hypotonic buffer with a protein inhibitor mixture and passed through 27G1/2-gauge needles, as described (11). After centrifugation at 6,000 rpm for 10 min, supernatants were further separated in a Ultracentrifuge (Optima TLX, Beckman) at 100,000 rpm for 35 min at 4°C. The supernatant was subjected to Western blotting, and β-catenin was detected with a β-catenin mouse monoclonal antibody (Transduction Laboratories, Lexington, KY).
LEF-1 Luciferase Reporter Assay.
SW620 cells were grown in 12-well plates and transfected with LEF-1, LEF-1 reporter, and Renilla plasmids by using Lipofectamine Plus (GIBCO/BRL). Twenty-four hours posttransfection, relative luciferase activities were measured (Promega). 293 cells were grown in 12-well plates. Cells were transfected with expression constructs of LEF-1, LEF-1 reporter, Renilla, and β-galactosidase, hnkd, Wnt-1 plus β-galactosidase or Wnt-1 plus hnkd. Thirty-six hours posttransfection, relative luciferase activities were measured.
cDNA Microarray Analysis.
The cDNA microarray consisted of multiple chips containing 10,000 genes in total with duplicated spots/cDNAs on each chip. The probes were prepared and hybridized by using RNA that was extracted from SW620 cells, which were treated with β-catenin antisense and control ODN for 48 h (11). Data analysis was performed as described by E. Moler, M. Boyle, and F.M.R. (unpublished work).
Identification and Cloning of Human Naked.
A blast search was conducted with the nucleotide sequence of mouse Naked against the nonredundant GenBank human expressed sequence tag (EST) database and produced two hits: AI167910 and H55148. AI167910 was purchased from Research Genetics and sequenced. The EST clone showed extensive sequence homology to mouse Naked Cuticle (mnkd) but lacking the 5′ end of the gene. To obtain the full length sequence of hnkd, RNA was isolated from a human primary colon tumor tissue block (RNeasy Mini Kit, Qiagen) and reverse-transcribed [SMART rapid amplification of cDNA ends (RACE) cDNA Amplification Kit]. A 5′ RACE was performed by using 5′-CTTGCCGTTGTTGTCAAAGTC-3′ as the Gene Specific Primer according to the manufacturer's protocol with the exception that 1 M GC Melt (CLONTECH) was added to the reaction mixture. The PCR reaction was run at 94°C for 30 s, 58°C for 30 s, and 68°C for 2.0 min for 40 cycles followed by 72°C extension for 10 min. The resulting PCR product was cloned into the pCR4TOPO vector (Invitrogen) and sequenced. The PCR product, which contains the N-terminal fragment plus the 5′ untranslated region of human naked, was subcloned into the AI167910 construct to generate the full-length cDNA for hnkd. The coding region of human naked was found to be identical to the published sequence (12). The coding region of human Naked and mouse Naked are 85% identical at the nucleotide level and 87% identical at the protein level. The nucleotide sequence is 1,410 bp, which encodes a protein of 470 amino acids.
Promoter Analysis.
The putative promoters of axin2 and hnkd were determined by blast searches of the axin2 and hnkd cDNA sequence against the University of California, Santa Cruz genome assembly of the April 1, 2001 data freeze. axin2 was mapped to chromosome 17, whereas hnkd was mapped to chromosome 16. Further, the coding region of hnkd, the 5′ and 3′ untranslated regions are divided into 11 exons and can be mapped to an 86-kb region on chromosome 16.
Expression Profile of axin2 and hnkd in Human Cell Lines.
A collection of human cell lines, including Caco.2, SW480, SW620, Colo 320 DM, T84, HCT15, LS174T, LOVO, HT29, and HCT116 (colon cancer cell lines); 184B5 (primary breast cell line); MDA-MB-231, MDA-MB-435, Alab, MCF-7, and MDA-MB-468 (breast cancer cell lines); DU 145, LNCAP, WOCA, PC3, and GRDP2 (prostate cancer cell lines); SKOV3 and OVCAR3 (ovarian cancer cell lines); IMR90 (primary fibroblast cell line); 847 (SV40-transformed fibroblast cell line); HT1080 (fibrosarcoma cell line); A-431 (epidermoid carcinoma cell line); U373MG (glioblastoma cell line); NCI-H23 (non-small cell lung cancer cell line); and HMVEC (endothelial cell line), were grown in media as recommended by American Type Culture Collection. After extraction of total RNA, cDNA was synthesized with oligo dT primers and used in quantitative real-time PCR analysis to measure expression levels of axin2 and hnkd mRNA and both were normalized to the levels of actin. The following primers were used: actin-Forward, 5′-CGGGAAATCGTGCGTGACATTAAG-3′, and actin-Reverse, 5′-TGATCTCCTTCTGCATCCTGTCGG-3′.
Amplification of RNA Isolated from Human Tissues.
For each specimen, about 1,000 colon cancer cells and 1,000 surrounding normal colon epithelial cells were isolated by laser-capture microdissection. Typically, 40 ng of mRNA was extracted from 1,000 captured cells. The mRNA was amplified according to Luo et al. (13) to yield 40 μg of RNA. Reverse-transcription reaction was performed by using RETROscrip kit (Ambion) with the random primers provided with the kit, followed by quantitative real-time PCR analysis normalizing axin2 and hnkd expression levels to β-glucuronidase (GUS-Forward, 5′-CCTTTTGCGAGAGAGATACT-3; GUS-Reverse, 5′-CCTTTAGTGTTCCCTGCTAG-3′).
Mutational Status of β-Catenin and APC in Patient Samples.
Two hundred nanograms of total RNA from the colon cancer patient samples was reverse-transcribed by using oligodT primers and amplified with a forward primer (β-cat-F, 5′-ATGGAACCAGAGAAAAGC-3′) and a reverse primer (β-cat-R, 5′-AAGGACTGAGAAAATCCCTG-3′). The PCR reaction was carried out for 30 cycles in the presence of [33P]dCTP; 94°C for 45 s, 60°C for 45 s, and 72°C for 45 s, and an extension at 72°C for 7 min. The PCR product was electrophoresed on a 12% MDE gel (FMC), dried, and autoradiographed. To detect truncations in APC, 200 ng of total RNA from colon tissue samples was reverse-transcribed with primer RT-APC (5′-GTATGGTTACAGATGAGGTTTTTCC-3′) and amplified (Advantage GC-rich kit, CLONTECH) with primers as described (14). The PCR products were in vitro transcribed and translated by using the T7-coupled reticulocyte lysate system (Promega). [35S]methione and [35S]cysteine were incorporated to detect translation products after SDS/PAGE and autoradiography.
Results
New β-Catenin Responsive Genes in SW620 Colon Cancer Cell Line.
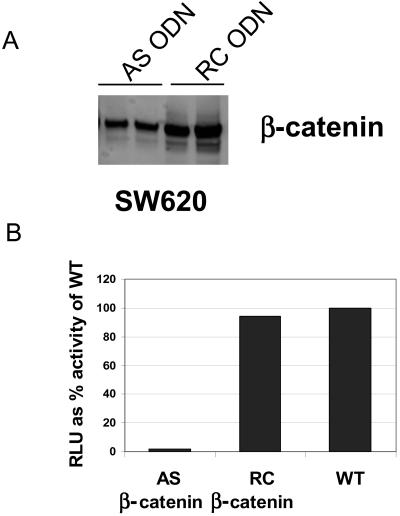
We used cDNA microarrays and quantitative PCR to identify β-catenin-regulated genes in SW620 cells, a colon cancer cell line in which β-catenin levels are stabilized as a consequence of truncated APC. When this cell line was transfected with β-catenin antisense ODN for 48 h, cytosolic β-catenin levels were reduced by 70% compared with cells transfected with control ODN (Fig. 1A). There was a concomitant reduction of 90% in the TCF/LEF-1 reporter activity (Fig. 1B). The mRNA isolated from cells transfected with antisense or RC ODN was reverse-transcribed, labeled with Cy3 and Cy5 fluorescent markers, and hybridized to an array with 25,344 cDNAs. The cDNAs spotted on the array originated either from normalized cDNA libraries or from purchased expressed sequence tags. Included in the 25,344 cDNAs were 11,547 cDNAs that matched National Center for Biotechnology Information (NCBI) GenBank entry with a blast score <1 × 10−50, within which there were 6,818 unique genes based on a blast score <1 × 10−80 and 3,343 cDNAs with no gene assignment. Also included in the 25,344 cDNAs were 6,910 novel sequences not present in the NCBI GenBank database. The total number of cDNAs does not match the number of genes because of the existence of multiple accession numbers for the same cDNA or duplicated sequences. We analyzed the microarray data for the expression of genes that correlated with the level of β-catenin (i.e., reduced by β-catenin antisense ODN) to specifically identify genes that are regulated by β-catenin. Only statistically significant ratios with a confidence level of 95% (P < 0.05) were included in the analysis. From the 77% of cDNAs that met the above criteria, ratios of antisense to RC ODN-treated cells with a value less than 0.5 or greater than 2.0 were considered significant evidence of gene regulation by β-catenin. Notably, axin2 was down-regulated 7-fold in cells that have reduced levels of β-catenin. Axin2 is the human homologue of mouse Conductin and rat Axil, both of which have been shown to be negative regulators of the Wnt/β-catenin pathway (15, 16). Axin2 also shares 45% homology to human Axin1, another negative regulator of the pathway (17). Furthermore, in the same microarray experiment c-myc, a recently validated β-catenin target gene (7), was reduced 4-fold. We were not able to see changes in cyclin D1 expression under these experimental conditions by either cDNA microarray or quantitative real-time PCR analysis. To further validate that expression of axin2 mRNA is regulated by β-catenin, we used real-time quantitative PCR analysis and showed that mRNA levels of axin2, when normalized to mRNA levels of glyceraldehyde-3-phosphate dehydrogenase (GAPDH), decreased by 90% in SW620 cells transfected with β-catenin AS ODN compared with cells transfected with control ODN (Fig. 2). Interestingly, no changes in the levels of axin1 mRNA were detected in the cells treated with β-catenin antisense ODN (data not shown).
Figure 1.
Reduction of cytosolic β-catenin levels and LEF-1 reporter activity in response to β-catenin antisense ODN transfection. (A) SW620 cell lysate was analyzed for cytosolic β-catenin levels by Western blotting 48 h posttransfection with β-catenin antisense and RC ODN in duplicated experiments. AS, antisense; RC, reverse control. (B) SW620 cells were transfected with β-catenin AS and RC ODN. After 24 h, the cells were transfected with reporter constructs as described in Methods. LEF-1 reporter activity was determined an additional 24 h later.
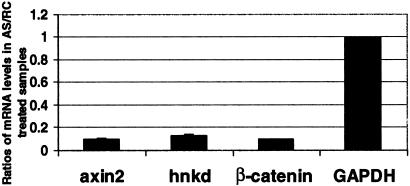
Figure 2.
axin2 and hnkd mRNA levels are reduced in response to β-catenin antisense ODN transfection. SW620 cells were transfected with β-catenin antisense ODN. Total RNA from the cells was reverse transcribed 48 h posttransfection. axin2, hnkd, and β-catenin mRNA expression levels were determined by quantitative real-time PCR and normalized to glyceraldehyde-3-phosphate dehydrogenase. The graph represents the average of three experiments.
Also included in this quantitative PCR analysis of the ODN-transfected cell line is the measurement of mRNA levels for hnkd, the human homologue of mnkd previously identified by us and others as a Dishevelled-binding protein that antagonizes Wnt signaling (12, 18). Similar to axin2, mRNA expression of hnkd, when normalized to mRNA levels of GAPDH, is reduced by 85% in cells transfected with β-catenin antisense ODN compared with control ODN transfected cells. These results, together with the results shown in Fig. 1, indicate that the mRNA levels of axin2 and hnkd are regulated by the amount of cytosolic β-catenin, suggesting that both are β-catenin target genes.
Human Nkd Antagonizes Wnt Signaling in Mammalian Cells.
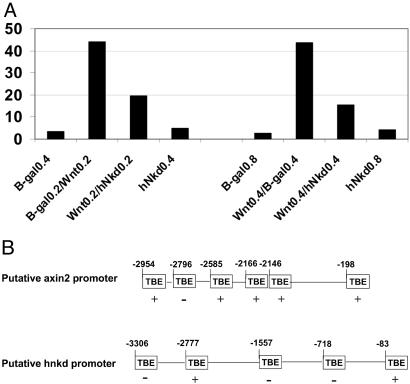
We cloned the human homologue of mNkd (see Methods). hNkd and mNkd share 87% identity in amino acid sequence and also have the identical EF-hand domain critical for the inhibitory function of mNkd in the Wnt/β-catenin pathway. To verify further that hNkd is functionally similar to Drosophila Nkd and mNkd, the role of hNkd in the Wnt/β-catenin pathway was tested in mammalian cell culture by using a Wnt-1 ligand responsive luciferase reporter. In multiple experiments, activation of the reporter was inhibited by 60% when Wnt-1 was coexpressed with hNkd in 293 cells (Fig. 3A). Thus, as previously described for its Drosophila and mouse homologues, human Nkd is also a negative regulator of the Wnt/β-catenin pathway (18, 19).
Figure 3.
hNkd antagonizes Wnt signaling in mammalian cells and the putative promoters of axin2 and hnkd contain TCF-binding elements. (A) hNkd inhibits Wnt-1 signaling in a luciferase reporter assay in 293 cells. The data represent the average of two experiments. RLU, relative luciferase unit. (B) Schematic representation of the genomic sequence upstream of the axin2 and hnkd coding region identified as described in Methods. The putative TCF sites are shown as TBE boxes and nucleotide residues relative to the starting codon are indicated. A perfect match (CTTTGA/TA/T) is labeled with +, and an inverted perfect match is labeled as −.
The Putative Promoter Sequences of axin2 and hnkd Contain TCF-Binding Elements.
The TCF/β-catenin complex regulates gene transcription through binding to TCF-binding elements (TBE) on DNA containing the core consensus sequence CTTTGA/TA/T. To determine whether β-catenin may regulate the transcription of axin2 and hnkd directly, we analyzed the genomic sequence upstream of the start codon. The putative promoter for axin2 is comprised of nucleotides −2,954 to −191 upstream of its coding region (see Methods). The putative promoter region for hnkd is defined as nucleotides −3306 to −76 upstream of the start of 5′-untranslated region we cloned (see Methods). We found six TBEs in the 3-kb putative axin2 promoter sequence and five TBEs in the putative hnkd promoter sequence that matched perfectly the core consensus sequence (Fig. 3B). This finding suggests that the β-catenin/TCF complex may regulate the promoters for axin2 and hnkd.
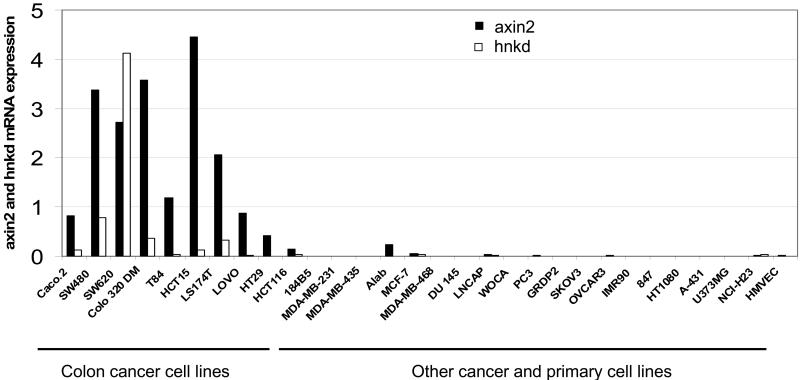
High Levels of axin2 and hnkd mRNA Expression Correlate with Mutations in APC in Colon Cancer Cell Lines.
To corroborate the findings that these two negative regulators are markers for the activated Wnt/β-catenin pathway, we measured axin2 and hnkd mRNA levels by quantitative real-time PCR analysis in a panel of 30 cancer and primary cell lines. These cell lines were derived from various tissue origins: breast, colon, prostate, kidney, ovary, lung, bone, brain, and endothelium. Interestingly, high expression levels of mRNA for axin2 and hnkd were observed exclusively in the colon cancer cell lines (Fig. 4). All of these colon cancer cell lines have been shown to harbor mutated APC or β-catenin (20). The cancer cells from origins other than colon, for example, breast cancer cell lines MCF-7, MDA-MD468, MDA-MB-231, and MDA-MB-435, do not have mutations in the mutation cluster region of APC (21) and do not show elevated expression of axin2 or hnkd. Together, these data provide evidence that the two newly identified β-catenin target genes, axin2 and hnkd, are markers for the activated Wnt/β-catenin pathway.
Figure 4.
axin2 and hnkd mRNA levels in human cell lines. Total RNA from a panel of 30 cell lines was reverse-transcribed, and axin2 and hnkd mRNA levels were determined by quantitative real-time PCR and normalized to actin. axin2 and hnkd mRNA levels are prominently elevated in human colon cancer cell lines.
axin2 and hnkd Are Up-Regulated in Human Colon Cancer Specimens.
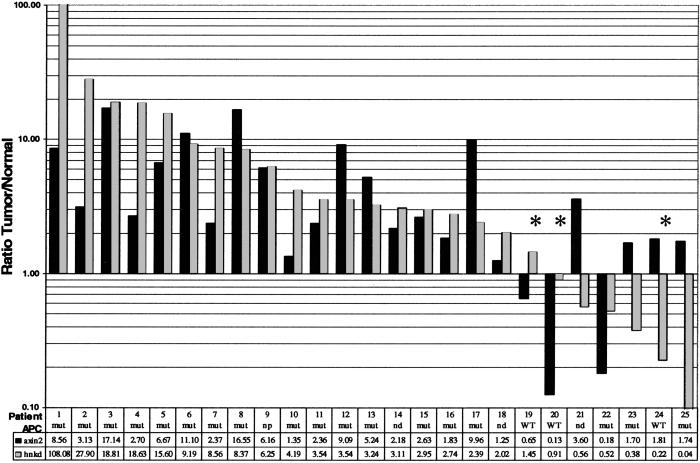
In colon cancer, truncations in APC result in elevated β-catenin protein levels in ≈80% of sporadic cancers (6). As described above, we have demonstrated in SW620 cells that reduction in levels of β-catenin lead to down-regulation of axin2 and hnkd mRNAs. This result, in combination with the high frequency of APC mutations in colon cancer patients, prompted us to investigate whether the levels of axin2 and hnkd were abnormal in patient colon tumors. Samples used for this study were obtained from patients that mostly presented with invasive and moderately differentiated tumors. In 50% of patients, the lymph nodes were positive for tumor cells, and 4 of 20 patients had liver metastases. In a gene profile analysis by using cDNA microarrays with probes from more than 30 colon cancer and patient-matched normal colon epithelium, we found that axin2 mRNA expression was elevated in a subgroup of patients greater than 2-fold (hnkd was not included on the array). Extending and confirming these studies, we showed by real-time quantitative PCR that the amount of axin2 and hnkd mRNA levels was increased in 63% and 72% of patients, respectively (Fig. 5). To correlate the mRNA level of axin2 and hnkd with mutations that activate the Wnt/β-catenin pathway, we determined the mutational status of both APC and β-catenin in the same patient samples by previously published methods. We used the protein truncation test for APC (14) and screened the N terminus of β-catenin by PCR-single-stranded conformational polymorphism (22). Of 25 patients analyzed for axin2 and hnkd expression, 19 had loss-of-function mutations in the APC gene, and three patients were wild type for APC in the mutation cluster region. Three patient samples with a loss-of-function mutation in the mutation cluster region of APC had an inexplicable decrease in axin2 and hnkd mRNA. No mutations in β-catenin were detected in the 24 of 25 patients (not determined for patient number 17). With few exceptions, we found that the tumor samples in which expression of axin2 and hnkd mRNA was increased over the normal control also harbored mutations in APC. The statistical significance of this correlation was tested with a two-tailed Student's t test by dividing the patients into two groups on the basis of the presence or absence of a mutation in the mutation cluster region of APC. We found a P value less than 0.001 for axin2 and less than 0.05 for hnkd (two sample unequal variance assumed). Taken together, the up-regulated expression of axin2 and hnkd correlates with the activated Wnt/β-catenin pathway in cell lines and human colon tumor specimens, indicating that axin2 and hnkd are markers for the activation of this pathway.
Figure 5.
axin2 and hnkd mRNA levels in human colon tumors. Total RNA from laser microdissected tissues was amplified and reversed transcribed as described in Methods. axin2 and hnkd mRNA levels were determined by quantitative real-time PCR and normalized to β-glucuronidase. The mutational status of APC in the patients was determined as described (Methods) and is indicated. Three patients that are wild type for APC are marked with *. The ratios of mRNA expression in tumor versus normal tissues for both axin2 and hnkd mRNA levels are shown in the table.
Discussion
Mutations in several components of the Wnt/β-catenin pathway have been found in a variety of cancers, most frequently in colon cancers. However, whether the pathway is activated in clinical cancer samples is not easily determined. Therefore, we initiated studies to compare Wnt pathway regulated genes in human colon tumor samples with those in colon cancer cell lines with the aim of identifying specific diagnostic/prognostic markers for the activation of the pathway. In cells transfected with β-catenin antisense ODN, the expression of axin2 and hnkd was found to be β-catenin regulated. To corroborate the data further, we identified multiple TBEs in the putative promoters of axin2 and hnkd, indicating that the two genes are likely to be direct targets of TCF/β-catenin regulated transcription. Furthermore, mRNA expression of axin2 and hnkd was increased in a panel of colon cancer cell lines with stabilized β-catenin mutations compared with cancer cell lines of noncolon cancer origin. Importantly, axin2 and hnkd mRNA levels were also significantly up-regulated in the colon tumor patient samples and with few exceptions, their overexpression correlated with APC mutations in the same colon tumor samples. These data suggest that the increased expression of the two genes are specific indicators of the activated Wnt/β-catenin pathway in colon cancer cell lines as well as in human colon cancer tissues.
Significantly, Axin2 and hNkd are negative regulators of the Wnt/β-catenin pathway. On the basis of the 89% identity of Axin2 with its mouse and rat homologues, Conductin and Axil, respectively, Axin2 most likely participates in the rapid degradation of β-catenin in complex with APC and Gsk-3β, and the Axin-binding sites on APC are required for its function (15, 16, 23). In addition, mutations in the β-catenin-binding domains of Conductin abolished its ability to reduce cytosolic β-catenin levels (15). To date, mutations in axin2 have been identified only in colorectal cancers with defective mismatch repair (5).
hNkd, the human homologue of mouse Nkd (12), was shown here to inhibit the Wnt-1 activated LEF-1 luciferase reporter. Given its sequence similarity to mNkd, hNkd most likely elicits this inhibitory action upstream of β-catenin at the level of Dishevelled, as previously described (18, 24).
Clearly, in cancers in which the Wnt/β-catenin pathway is activated, the negative regulatory roles for axin2 and hnkd are either abolished or annulled by alternative overriding pathways. In the case of colon cancer, the stabilization of β-catenin is because of truncations in APC that eliminate axin-binding sites (17), and epistatically this defect is downstream of Dishevelled, previously shown to interact with Nkd (18, 24). Other studies have shown that the mRNAs for multidrug-resistant 1 gene (MDR1) and βTrCP, a ubiquitin-binding protein that assists in the degradation of β-catenin, are up-regulated in colon cancer patients (25, 26). However, no reports have described MDR1 as a component of the Wnt/β-catenin pathway, although it has been described as a β-catenin target gene.
In this study, we have identified axin2 and hnkd as two β-catenin regulated genes that are also negative regulatory components of the Wnt/β-catenin pathway. Levels of these genes may be used as surrogate markers to indicate the activated Wnt/β-catenin pathway in colon cancer tissues. The utility of these surrogates in other cancers awaits further study.
Acknowledgments
We thank Dr. R. Grosschedl (Max Planck Institute, Martinstried, Germany) for reagents; Drs. X. Meng, D. Den Otter, and L. Inge for technical assistance; and Drs. S. Harrison, J. Winter, G. Lamson, and E. Moler for helpful discussions. We are grateful to the Chiron cancer biology group for their input in this study.
Abbreviations
- ODN
oligodeoxynucleotides
- APC
adenomatous polyposis coli
- TCF
T cell factor
- LEF
lymphoid enhancer factor
- TBE
TCF-binding element
- RC ODN
reverse control ODN
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AY061883).
References
- 1.Polakis P. Curr Opin Genet Dev. 1999;9:15–21. doi: 10.1016/s0959-437x(99)80003-3. [DOI] [PubMed] [Google Scholar]
- 2.Groden J, Thliveris A, Samowitz W, Carlson M, Gelbert L, Albertsen H, Joslyn G, Stevens J, Spirio L, Robertson M, et al. Cell. 1991;66:589–600. doi: 10.1016/0092-8674(81)90021-0. [DOI] [PubMed] [Google Scholar]
- 3.Kinzler K W, Nilbert M C, Su L K, Vogelstein B, Bryan T M, Levy D B, Smith K J, Preisinger A C, Hedge P, McKechnie D, et al. Science. 1991;253:661–665. doi: 10.1126/science.1651562. [DOI] [PubMed] [Google Scholar]
- 4.Satoh S, Daigo Y, Furukawa Y, Kato T, Miwa N, Nishiwaki T, Kawasoe T, Ishiguro H, Fujita M, Tokino T, et al. Nat Genet. 2000;24:245–250. doi: 10.1038/73448. [DOI] [PubMed] [Google Scholar]
- 5.Liu W, Dong X, Mai M, Seelan R S, Taniguchi K, Krishnadath K K, Halling K C, Cunningham J M, Boardman L A, Qian C, et al. Nat Genet. 2000;26:146–147. doi: 10.1038/79859. [DOI] [PubMed] [Google Scholar]
- 6.Polakis P. Curr Opin Genet Dev. 1995;5:66–71. doi: 10.1016/s0959-437x(95)90055-1. [DOI] [PubMed] [Google Scholar]
- 7.He T C, Sparks A B, Rago C, Hermeking H, Zawel L, da Costa L T, Morin P J, Vogelstein B, Kinzler K W. Science. 1998;281:1509–1512. doi: 10.1126/science.281.5382.1509. [DOI] [PubMed] [Google Scholar]
- 8.Tetsu O, McCormick F. Nature (London) 1999;398:422–426. doi: 10.1038/18884. [DOI] [PubMed] [Google Scholar]
- 9.Aktas H, Cai H, Cooper G M. Mol Cell Biol. 1997;17:3850–3857. doi: 10.1128/mcb.17.7.3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kerkhoff E, Houben R, Loffler S, Troppmair J, Lee J E, Rapp U R. Oncogene. 1998;16:211–216. doi: 10.1038/sj.onc.1201520. [DOI] [PubMed] [Google Scholar]
- 11.Haertel-Wiesmann M, Liang Y, Fantl W J, Williams L T. J Biol Chem. 2000;275:32046–32051. doi: 10.1074/jbc.M000074200. [DOI] [PubMed] [Google Scholar]
- 12.Wharton K A, Jr, Zimmermann G, Rousset R, Scott M P. Dev Biol. 2001;234:93–106. doi: 10.1006/dbio.2001.0238. [DOI] [PubMed] [Google Scholar]
- 13.Luo L, Salunga R C, Guo H, Bittner A, Joy K C, Galindo J E, Xiao H, Rogers K E, Wan J S, Jackson M R, Erlander M G. Nat Med. 1999;5:117–122. doi: 10.1038/4806. [DOI] [PubMed] [Google Scholar]
- 14.van der Luijt R, Khan P M, Vasen H, van Leeuwen C, Tops C, Roest P, den Dunnen J, Fodde R. Genomics. 1994;20:1–4. doi: 10.1006/geno.1994.1119. [DOI] [PubMed] [Google Scholar]
- 15.Behrens J, Jerchow B A, Wurtele M, Grimm J, Asbrand C, Wirtz R, Kuhl M, Wedlich D, Birchmeier W. Science. 1998;280:596–599. doi: 10.1126/science.280.5363.596. [DOI] [PubMed] [Google Scholar]
- 16.Yamamoto H, Kishida S, Uochi T, Ikeda S, Koyama S, Asashima M, Kikuchi A. Mol Cell Biol. 1998;18:2867–2875. doi: 10.1128/mcb.18.5.2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hart M J, de los Santos R, Albert I N, Rubinfeld B, Polakis P. Curr Biol. 1998;8:573–581. doi: 10.1016/s0960-9822(98)70226-x. [DOI] [PubMed] [Google Scholar]
- 18.Yan D, Wallingford J B, Sun T Q, Nelson A M, Sakanaka C, Reinhard C, Harland R M, Fantl W J, Williams L T. Proc Natl Acad Sci USA. 2001;98:3802–3807. doi: 10.1073/pnas.071041898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zeng W, Wharton K A, Jr, Mack J A, Wang K, Gadbaw M, Suyama K, Klein P S, Scott M P. Nature (London) 2000;403:789–795. doi: 10.1038/35001615. [DOI] [PubMed] [Google Scholar]
- 20.Rowan A J, Lamlum H, Ilyas M, Wheeler J, Straub J, Papadopoulou A, Bicknell D, Bodmer W F, Tomlinson I P. Proc Natl Acad Sci USA. 2000;97:3352–3357. doi: 10.1073/pnas.97.7.3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schlosshauer P W, Brown S A, Eisinger K, Yan Q, Guglielminetti E R, Parsons R, Ellenson L H, Kitajewski J. Carcinogenesis. 2000;21:1453–1456. [PubMed] [Google Scholar]
- 22.Bouzourene H, Gervaz P, Cerottini J P, Benhattar J, Chaubert P, Saraga E, Pampallona S, Bosman F T, Givel J C. Eur J Cancer. 2000;36:1008–1015. doi: 10.1016/s0959-8049(00)00036-8. [DOI] [PubMed] [Google Scholar]
- 23.Mai M, Qian C, Yokomizo A, Smith D I, Liu W. Genomics. 1999;55:341–344. doi: 10.1006/geno.1998.5650. [DOI] [PubMed] [Google Scholar]
- 24.Rousset R, Mack J A, Wharton K A, Jr, Axelrod J D, Cadigan K M, Fish M P, Nusse R, Scott M P. Genes Dev. 2001;15:658–671. doi: 10.1101/gad.869201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yamada T, Takaoka A S, Naishiro Y, Hayashi R, Maruyama K, Maesawa C, Ochiai A, Hirohashi S. Cancer Res. 2000;60:4761–4766. [PubMed] [Google Scholar]
- 26.Spiegelman V S, Slaga T J, Pagano M, Minamoto T, Ronai Z, Fuchs S Y. Mol Cell. 2001;5:877–882. doi: 10.1016/s1097-2765(00)80327-5. [DOI] [PubMed] [Google Scholar]