Abstract
Objectives
Protein kinases orchestrate activation of signalling cascades in response to extra‐ and intracellular stimuli for regulation of cell proliferation. They are directly involved in a variety of diseases, particularly cancers. Systems biology approaches have become increasingly important in understanding regulatory frameworks in cancer, and thus may facilitate future anti‐cancer discoveries. Moreover, it has been suggested and confirmed that high‐throughput virtual screening provides a novel, effective way to reveal small molecule protein kinase inhibitors. Accordingly, we aimed to identify kinase targets and novel kinase inhibitors.
Materials and methods
A series of bioinformatics methods, such as network construction, molecular docking and microarray analyses were performed.
Results
In this study, we computationally constructed the appropriate global human protein–protein interaction network with data from online databases, and then modified it into a kinase‐related apoptotic protein–protein interaction network. Subsequently, we identified several kinases as potential drug targets according to their differential expression observed by microarray analyses. Then, we predicted relevant microRNAs, which could target the above‐mentioned kinases. Ultimately, we virtually screened a number of small molecule natural products from Traditional Chinese Medicine (TCM)@Taiwan database and identified a number of compounds that are able to target polo‐like kinase 1, cyclin‐dependent kinase 1 and cyclin‐dependent kinase 2 in HeLa cervical carcinoma cells.
Conclusions
Taken together, all these findings might hopefully facilitate discovery of new kinase inhibitors that could be promising candidates for anti‐cancer drug development.
Introduction
Cancer is a major public health problem and one in four deaths in the United States is due to it. As recently estimated, numbers of people diagnosed with malignant disease will increase from 1.6 million in 2010 to 2.3 million in 2030 in the USA 1. Although cancer death rates in the USA reduced by 1.8% per year in men and 1.5% per year in women from 2005 to 2009, yet it is a leading cause of death in most parts of the world 2. Thus, fundamental breakthroughs in cancer research are urgently needed for identification of new target molecules and development of novel therapeutic techniques.
Protein kinases, enzymes that transfer phosphate groups from high‐energy molecules to others, control a series of cellular processes, including metabolism, transcription, differentiation, cell cycle progression and apoptosis 3. Accumulating studies have indicated that dysregulation and mutation of protein kinases are linked to various types of human diseases, most notably cancers 4. Understanding how to target protein kinases with small molecules has significantly benefited recent cancer drug discovery research studies, which have explored numerous approaches to target and inhibit protein kinase signalling 5. To date, approximately 30 distinct kinases have been recognized as drug targets for phase I clinical trials. More importantly, a growing number of kinase inhibitors have been approved by the Food and Drug Administration to be used in cancer therapeutics 6.
Apoptosis, referred as type I programmed cell death, is a normal process in which a cell undergoes a series of genetically programmed events that lead to cell death and disposal of its components 7. Induction of apoptosis is arguably the most potent defence against cancer, making it an ideal strategy for anti‐cancer therapy. Thus, better understanding to the molecular mechanisms of apoptosis encourages us to develop more rational approaches to treatment of cancer 8, 9.
Computer‐aided drug design is widely used in developing new pharmaceuticals, specially from traditional Chinese medicine. A number of research studies has proved that some traditional Chinese herbal compounds may exhibit remarkable anti‐tumour effects against various types of cancer cell lines 10. Virtual screening is a computational technique used in drug discovery to search libraries of small molecules, and has emerged as a reliable, cost‐effective and time‐saving technique for discovery of lead compounds 11. Hitherto, numerous potent kinase inhibitors have been identified using virtual screening, such as EGFR inhibitors, ALK inhibitors and PI3K inhibitors 12, 13, 14. As numbers of studies have benefited from discovery of various small molecule inhibitors by virtual screening, we thought that a high‐throughput discovery tools approach could be considered to be the most promising of methods in the quest for effective new small molecule kinase inhibitors.
In this study, we used a series of bioinformatics methods to construct a kinase‐related apoptotic protein–protein interaction (PPI) network, and we identified several kinases as potential drug targets, including polo‐like kinase 1 (PLK1), cyclin‐dependent kinase 1 (CDK1) and cyclin‐dependent kinase 2 (CDK2) in HeLa cells. Subsequently, we further predicted several miRNAs that could potentially target and regulate expression of the above‐mentioned kinases. Ultimately, we carried out virtual screening from TCM@Taiwan database to find small molecules that potentially inhibit activities of kinases identified. In summary, these findings may provide new insights into identification of novel target kinases, as well as new inhibitors, which could arrest cell proliferation and induce apoptosis in neoplastic cells, as an effective and promising approach for cancer treatment.
Materials and methods
Data processing and network construction
To construct the global human PPI network, diverse sets of data were collected from five online databases, including human protein reference database 15, Biomolecular Object Network Databank 16, IntAct 17, MINT 18 and Biological General Repository for Interaction Datasets 19. Subsequently, a kinase‐related PPI network was established by extracting protein pairs having at least one protein interacting with a kinase. Similarly, a kinase‐related apoptotic PPI network was built on the criteria that there is at least one apoptotic protein (by Gene Ontology consortium), in a pair of proteins.
Microarray analyses of kinase‐related apoptotic genes
Proteins that interact with others often have similar gene expression patterns; thereby, genes that can co‐express should be more likely to interact with each other than genes that cannot co‐express. To test whether genes are co‐expressed or not, we carried out microarray data of HeLa cells treated with casiopeina Cas‐II‐gly to measure pair‐wise co‐expression level of intrinsic apoptosis triggered by oxidative stress (E‐GEOD‐41827) 20.
Targeted microRNA prediction
Recently, studies have revealed that a combination of methods might provide better understanding of complex regulatory mechanisms in which miRNAs are involved 21, 22. Thus, we carried out a prediction by sources of three algorithmically different methods, namely, TargetScan (stringent seed pairing, site number, site type, site context, option of ranking by likelihood of preferential conservation rather than site context) 23, DIANA LAB (hybridization energy threshold rules) 24 and MiRanda (moderately stringent seed pairing, site number, pairing to most of the miRNA) 25.
Molecular docking
Molecular structures of PLK1 (2YAC), polo‐like kinase 1 polo‐box domain (PLK1 PBD) (4E9C), CDK1 (1LC9) and cyclin‐dependent kinase 2 (CDK2) (4GCJ) were downloaded from Protein Database Bank (PDB) (http://www.rcsb.org/pdb/home/home.do). Then, we built a screening library from ZINC database TCM@Taiwan (http://zinc.docking.org/catalogs/tcmnp), the world's largest Traditional Chinese Medicine database (that contains 6595 small molecule natural products) to predict novel kinase inhibitors targeting PLK1, PLK1 PBD, CDK1 and CDK2.
USCF DOCK 6.4 program with AMBER force field parameters was used to dock pre‐generated conformations of natural products into PLK1, PLK1 PBD, CDK1 and CDK2 for a virtual screening of their inhibitors 26. We took advantage of flexible‐ligand docking to a rigid receptor with grid‐based scoring, in which ligands (small molecule compounds) were allowed to be structurally rearranged in response to these kinases. To make the results more accurate, data were scored twice. In docking processes, maximum number of orientations was set to 500 27. Subsequently, amber scoring function in DOCK 6.4 was used to re‐rank the top 100 small molecule compounds from previous grid‐based scoring. During amber score calculation, the PDB2PQR server was utilized to assign protonation state of PDB files with AMBER forcefield, and PROPKA was applied to maintain protonation state at PH = 7.0 28, 29, 30.
Results
Construction of a kinase‐related apoptotic PPI network
We computationally constructed a global human PPI network from databases, including 38988 protein pairs (9588 proteins) from human protein reference database, 8161 protein pairs (4129 proteins) from Biomolecular Object Network Databank, 44434 protein pairs (10408 proteins) from IntAct, 21455 protein pairs (7294 proteins) from MINT, and 82612 protein pairs (13059 proteins) from Biological General Repository for Interaction Datasets. Consequently, we identified 158664 unique protein pairs (15721 proteins). As the human PPI network is naturally complex for complicated connections amongst numerous signalling pathways, it is necessary to integrate high‐throughput data in a specific biological context such as cancer, to describe the network accurately and comprehensively 31. Based on the global human PPI network, we used 508 kinases to select protein pairs, which have at least one protein related to the kinases, to construct a kinase‐related PPI network. The established PPI network contained 22453 protein pairs (5407 proteins). Then, we identified 2062 apoptotic proteins from Gene Ontology database to extract from a kinase‐related PPI network, and further modified it into a kinase‐related apoptotic PPI network, which had 15001 protein pairs (4236 proteins) (Fig. 1) 32.
Figure 1.
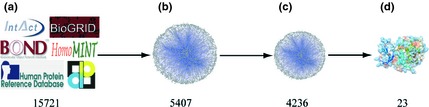
Construction of the kinase‐related apoptotic protein–protein interaction ( PPI ) network. (a) Global human PPI network (15721 proteins). (b) Kinase‐related PPI network (5407 proteins). (c) Kinase‐related apoptotic PPI network (4236 proteins). (d) Potential targets (23 proteins).
Identification of target kinases in HeLa cells
Of note, hub proteins can play more crucial roles in this kinase‐related apoptotic PPI network than those other non‐hub proteins, so we extracted target kinases based on high‐degree proteins. This method is similar to identification of novel cancer‐related genes 33. In total, there were 4236 proteins among those 15001 protein pairs in the kinase‐related apoptotic PPI network; we manually set the cut‐off degree as 100 and obtained 35 proteins, suggesting their pivotal roles in this sub‐network.
Subsequently, significance analysis of microarrays analysis was conducted with data of different expression profile from casiopeina Cas‐II‐gly‐treated HeLa cell apoptosis to indicate divergent expression genes between HeLa cells treated with casiopeina Cas‐II‐gly and untreated HeLa cells. We clarified that proteins identified as divergent expression functional hub proteins, depended on gene co‐expression profiles, thus playing their regulatory roles as potential targets in HeLa cells. According to the microarray data, we finally recognized 6 upregulated and 17 downregulated potential target kinases (shown in Fig. 2), such as PLK1, CDK1 and CDK2.
Figure 2.
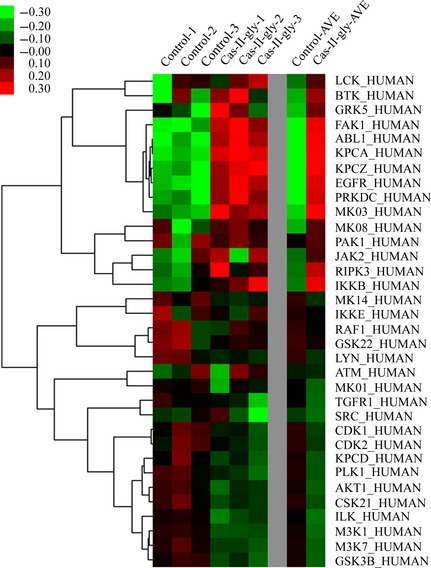
Microarray analyses of possible targets in HeLa cervical carcinoma cells.
Prediction of microRNAs targeting kinases
In this study, we predicted miRNAs that target PLK1, CDK1 and CDK2. Because of variation due to the different algorithms explored, combination of the results from several miRNA prediction approaches make the results more reliable 22. We combined these miRNAs into consensus results that some miRNAs were shown to target the above‐mentioned target kinases. For example CDK1, we obtained 1 miRNA through DIANA LAB, 24 miRNAs through MiRanda and 291 miRNAs through TargetScan. Then, we integrated these miRNAs into a combinatory result. Thus, only miR‐543 could negatively regulate CDK1. Similarly, PLK1 was predicted to be regulated by miR23a and miR23b, and CDK2 was predicted to be modulated by miR‐429, miR200c and miR200b (Fig. 3).
Figure 3.
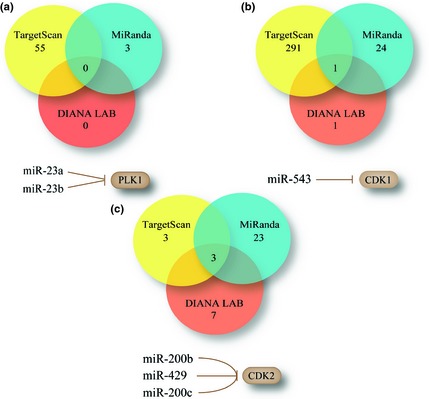
Prediction of microRNAs targeting polo‐like kinase 1 ( PLK 1), cyclin‐dependent kinase 1 ( CDK 1) and ( CDK 2) via combinational methods including Diana‐LAB, MiRanda and Targetscan.
Candidate small molecule compounds targeting kinases
Virtually, we screened the structure‐based candidate TCM compounds that could target PLK1, PLK1 PBD, CDK1 and CDK2. We retrieved the top 10 small molecule compounds that targeted the described kinases (shown in Table 1). Figure 4 presents four target small molecule compound complexes in the surface potential mode and stick‐ball model. For example, ZINC70455210, ZINC59586795, ZINC70455222, ZINC59586814, ZINC70455382, ZINC43278594, ZINC70451083, ZINC16051626, ZINC70455369, ZINC70454594 targeted PLK1. Detailed information of small molecule compounds targeting PLK1, PLK1 PBD, CDK1 and CDK2 in HeLa cells is shown in Table 1 and Table S1.
Table 1.
Small molecule compounds targeting PLK1, PLK1 PBD, CDK1 and CDK2 in HeLa cells
Figure 4.

Overall modelling of target kinase–small molecule compounds complexes. All complexes are presented in surface potential mode and in stick‐ball mode. In stick‐ball mode, small molecule compounds are shown as purple sticks.
Discussion
Network and systems biology strategies provide great insights for better understanding of the molecular mechanisms of human diseases. Such study may offer novel ideas for identification of potential drug targets for development of anti‐cancer therapy. To date, protein kinases have emerged as key regulators of all aspects of neoplasia, including cell proliferation, invasion, angiogenesis and metastasis. In the current study, we identified three kinases, PLK1, CDK1 and CDK2 that are important cell cycle regulators; their inhibition can lead to cell cycle arrest or apoptosis 34. It is well known that deregulation in the cell cycle is an essential component of tumour formation and progression.
To the best of our knowledge, we report for the first time that a kinase‐related apoptotic PPI network was built, which contained 4236 proteins from 15001 protein pairs. Then, we integrated degree and microarray analysis to identify 6 upregulated and 17 downregulated potential target kinases (including PLK1, CDK1 and CDK2) in HeLa cells. Subsequently, we virtually screened 6595 natural products from the TCM database and selected the top 10 small molecule compounds, which could target PLK1, PLK1 PBD, CDK1 and CDK2. Amber scores are also shown in Table 1; different ones are indicative of binding capabilities of these targets, in complex with diverse small molecule compounds. We inferred from this Table that PLK1 PBD had best affinity with ZINC70455210, amber score being −44.092584. Meanwhile, CDK1 in complex with ZINC70455381 had highest amber score −25.843222. Intriguingly, we found that ZINC70455210 could target PLK1 both at the ATP binding site and at the PLK1 PBD site. Similarly, ZINC59586795 could target PLK1 and CDK2; ZINC70454793 could target CDK1 and PLK1 PBD.
Polo‐like kinase 1, the best‐characterized member of the human PLK family, is a major regulator of the cell cycle, which controls entry into mitosis and regulates the spindle checkpoint. It is reported that PLK1 is essential for recovery from DNA damage‐induced G2/M arrest by activation of CDK1 35. A further study has demonstrated that PLK1 plays an essential role in mitosis, and PLK1 overexpression contributes to oncogenesis via promotion of chromosome instability and aneuploidy through checkpoint functions 36.
Polo‐like kinase 1 is overexpressed in a broad spectrum of cancer types and is often associated with poor prognosis 37. Interestingly, PLK1 offers two functionally crucial target sites within one molecule: an amino‐terminal catalytic kinase domain that is responsible for ATP‐binding and enzyme activation, and a unique carboxy‐terminal polo‐box domain (PBD) comprised of 2 polo‐boxes. A variety of studies has indicated that PLK1 PBD represents an attractive alternative target for development of PLK1 inhibitors 38. Additionally, numerous studies have demonstrated that small molecule inhibitors of PLK1 can inhibit tumour growth in mice, and these studies also indicated that PLK1 inhibition preferably kills cancer cells compared to normal cells 39. Thus, PLK1 presents an ideal target for development of specific small molecule inhibitors for cancer treatment while avoiding toxicity to normal tissues 40.
In the past decade, a number of structurally different small molecule compounds has been identified, to block PLK1 activity at the ATP binding site or that of PLK1 PBD 41. Among these, GCK461364, an ATP‐competitive inhibitor of PLK1, is under phase I clinical trials for patients with advanced solid malignancies 42. A further report has demonstrated that a natural product, thymoquinone and its synthetic derivative poloxin inhibit functions of PLK1 PDB in vitro, in HeLa cells 43.
Cyclin‐dependent kinase play essential roles in cell proliferation, which have stimulated considerable interest in development of inhibitors of these enzymes to suppress tumour growth. Uncontrolled cell proliferation, a hallmark of cancer, might result from dysregulation of cell cycle regulators 44. Previous studies have described that aberrant activation of CDK1 may contribute to tumourigenesis via phosphorylation and inhibition of FOXO1 transcription factor 45. A further study has reported that targeting the CDK1 pathway might be applied for treatment of FLT3ITD mutant acute myeloid leukaemias, suggesting that CDK1 inhibitors are currently under way to treat leukaemias, specially those resistant to FLT3 inhibitor therapies 46.
Previous reports have illustrated that CDK inhibitors are effective in regulating the cell cycle, and their potential value to treat cancer has been extensively studied 47. Abnormal expression of CDK1 has been observed in a variety of primary cancers, and CDK2 is dysregulated in various malignancies 48. Accordingly, they are recognized as important, emerging targets for anti‐cancer medication.
As a class of molecularly targeted therapies, protein kinase inhibitors have made a substantial beneficial impact on therapeutic care of cancer patients, providing improved quality of life for patients with advanced cancer and poor prognosis. Small molecule compounds of protein kinases typically prevent either auto‐phosphorylation of the kinase or subsequent phosphorylation of other protein substrates 5. In this work, screened PLK1, CDK1 and CDK2 small molecule kinase inhibitors potentially competed against the ATP binding pocket to potently block protein kinase activity and signal transduction. Currently, numerous compounds have been identified that strongly block PLK1 activity in an ATP‐competitive manner. Herein, we also virtually screened small molecule compounds targeting PLK1 PBD, a recently discovered protein domain, which mediated intracellular localization of PLK1. As PDB is unique to PLKs, its exploitation as an alternative docking site for new PLK inhibitors could overcome the hurdle posed by the conserved nature of the ATP‐binding site of Ser/Thr protein kinases, for development of mono‐specific inhibitors.
In summary, these above‐mentioned kinase inhibitors can be considered promising candidates for future cancer drug development. Although the top 10 potential small molecule compounds that target kinases were screened, these compounds still need further drug optimization with subsequent pre‐clinical and clinical trials before they can be accepted as targeting drugs.
MicroRNAs, a class of endogenously expressed, non‐coding RNAs, have been well known to regulate apoptotic pathways and cell proliferation in cancer. Because miRNAs play critical roles in tumourigenetic processes and disease‐specific expression, they are considered to be therapeutic targets and novel biomarkers. Our recent studies have illustrated that some relevant miRNAs could significantly regulate some sugar‐containing receptors, and thus inhibit downstream cancer‐related signalling pathways 21. Moreover, several miRNAs were identified to target three molecular switches between caspases and ATGs in MCF‐7 cells 49. In this study, we reported for the first time that several predicted miRNAs could target three kinases, PLK1, CDK1 and CDK2 in HeLa cells.
Kinases offer a rich and diverse source of potential targets for blocking tumour growth and survival. Since approval of the first small‐molecule protein kinase inhibitor Gellvec@ (Imatinib), protein kinase inhibitors have made a substantially beneficial impact on therapeutic care of cancer patients. Promising in silico docking approaches may greatly expand the reservoir of molecules suitable for large‐scale screening. In recent years, beneficial features of many TCM small molecule compounds are becoming more recognized in devising new treatment options in cancer therapeutics. Because of their crucial roles and remarkable progress in screening small molecule compounds, an intriguing array of novel kinase inhibitors might soon make a significant contribution to the fight against cancer.
Conflicts of interest
The authors declare that they have no conflicts of interest.
Supporting information
Table S1. Detailed information of small molecule compounds targeting PLK1, PLK1 PBD, CDK1 and CDK2 in HeLa cells.
Fig. S1. Evaluation of microarray data from HeLa cells.
Acknowledgements
We are grateful to Dr. Jian Li (Ohio University), Dr. Bo Liu (Sichuan University), Miss Wen‐wen Li (UCL Institute of Ophthalmology) and Huai‐long Xu (Sichuan University) for providing constructive suggestions. This work was supported in part by the National Natural Science Foundation of China (nos 81173093, 30970643, J1103518, 81373311 and 31300674) and the Special Program for Youth Science and the Technology Innovative Research Group of Sichuan Province, China (no. 2011JTD0026).
Z. Shi, N. An and B. M. Lu contributed equally to this work.
References
- 1. Siegel R, DeSantis C, Virgo K, Stein K, Mariotto A, Smith T et al (2012) Cancer treatment and survivorship statistics, 2012. CA Cancer J. Clin. 62, 220–241. [DOI] [PubMed] [Google Scholar]
- 2. Siegel R, Naishadham D, Jemal A (2013) Cancer statistics, 2013. CA Cancer J. Clin. 63, 11–30. [DOI] [PubMed] [Google Scholar]
- 3. Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S (2002) The protein kinase complement of the human genome. Science 298, 1912–1934. [DOI] [PubMed] [Google Scholar]
- 4. Garber K (2006) The second wave in kinase cancer drugs. Nat. Biotechnol. 24, 127–130. [DOI] [PubMed] [Google Scholar]
- 5. Grant S (2009) Therapeutic protein kinase inhibitors. Cell. Mol. Life Sci. 66, 1163–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhang J, Yang PL, Gray NS (2009) Targeting cancer with small molecule kinase inhibitors. Nat. Rev. Cancer 9, 28–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ouyang L, Shi Z, Zhao S, Wang FT, Zhou TT, Liu B et al (2012) Programmed cell death pathways in cancer: a review of apoptosis, autophagy and programmed necrosis. Cell Prolif. 45, 487–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hanahan D, Weinberg RA (2011) Hallmarks of cancer: the next generation. Cell 144, 646–674. [DOI] [PubMed] [Google Scholar]
- 9. Shi Z, Li CY, Zhao S, Yu Y, An N, Liu YX et al (2013) A systems biology analysis of autophagy in cancer therapy. Cancer Lett. 337, 149–160. [DOI] [PubMed] [Google Scholar]
- 10. Yang SC, Chang SS, Chen CYC (2011) Identifying HER2 inhibitors from natural products database. PLoS One 6, e28793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Vyas V, Jain A, Jain A, Gupta A (2008) Virtual screening: a fast tool for drug design. Sci. Pharm. 76, 333–360. [Google Scholar]
- 12. Okamoto M, Kojima H, Saito N, Okabe T, Masuda Y, Furuya T et al (2011) Virtual screening and further development of novel ALK inhibitors. Bioorg. Med. Chem. 19, 3086–3095. [DOI] [PubMed] [Google Scholar]
- 13. Frédérick R, Mawson C, Kendall JD, Chaussade C, Rewcastle GW, Shepherd PR et al (2009) Phosphoinositide‐3‐kinase (PI3K) inhibitors: identification of new scaffolds using virtual screening. Bioorg. Med. Chem. Lett. 19, 5842–5847. [DOI] [PubMed] [Google Scholar]
- 14. Li S, Sun X, Zhao H, Tang Y, Lan M (2012) Discovery of novel EGFR tyrosine kinase inhibitors by structure‐based virtual screening. Bioorg. Med. Chem. Lett. 22, 4004–4009. [DOI] [PubMed] [Google Scholar]
- 15. Mishra GR, Suresh M, Kumaran K, Kannabiran N, Suresh S, Bala P et al (2006) Human protein reference database – 2006 update. Nucleic Acids Res. 34, D411–D414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Alfarano C, Andrade C, Anthony K, Bahroos N, Bajec M, Bantoft K et al (2005) The biomolecular interaction network database and related tools 2005 update. Nucleic Acids Res. 33, D418–D424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kerrien S, Alam‐Faruque Y, Aranda B, Bancarz I, Bridge A, Derow C et al (2007) IntAct – open source resource for molecular interaction data. Nucleic Acids Res. 35, D561–D565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chatr‐Aryamontri A, Ceol A, Palazzi LM, Nardelli G, Schneider MV, Castagnoli L et al (2007) MINT: the Molecular INTeraction database. Nucleic Acids Res. 35, D572–D574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Winter AG, Wildenhain J, Tyers M (2011) BioGRID REST Service, BiogridPlugin2 and BioGRID WebGraph: new tools for access to interaction data at BioGRID. Bioinformatics 27, 1043–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Valencia‐Cruz AI, Uribe‐Figueroa LI, Galindo‐Murillo R, Baca‐López K, Gutiérrez AG, Vázquez‐Aguirre A et al (2013) Whole Genome Gene Expression Analysis Reveals Casiopeína‐Induced Apoptosis Pathways. PLoS One 8, e54664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shi Z, An N, Zhao S, Li X, Bao J, Yue B (2013) In silico analysis of molecular mechanisms of legume lectin‐induced apoptosis in cancer cells. Cell Prolif. 46, 86–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wuchty S, Arjona D, Li A, Kotliarov Y, Walling J, Ahn S et al (2011) Prediction of associations between microRNAs and gene expression in glioma biology. PLoS One 6, e14681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Friedman RC, Farh KKH, Burge CB, Bartel DP (2009) Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 19, 92–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Maragkakis M, Alexiou P, Papadopoulos GL, Reczko M, Dalamagas T, Giannopoulos G et al (2009) Accurate microRNA target prediction correlates with protein repression levels. BMC Bioinformatics 10, 295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Betel D, Wilson M, Gabow A, Marks DS, Sander C (2008) The microRNA. org resource: targets and expression. Nucleic Acids Res. 36, D149–D153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lang PT, Brozell SR, Mukherjee S, Pettersen EF, Meng EC, Thomas V et al (2009) DOCK 6: combining techniques to model RNA–small molecule complexes. RNA 15, 1219–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shi Z, Wang Z, Xu HL, Tian Y, Li X, Bao JK et al (2013) Modeling, docking and dynamics simulations of a non‐specific lipid transfer protein from Peganum harmala L. Comput. Biol. Chem. 47, 56–65. [DOI] [PubMed] [Google Scholar]
- 28. Li H, Robertson AD, Jensen JH (2005) Very fast empirical prediction and rationalization of protein pKa values. Proteins 61, 704–721. [DOI] [PubMed] [Google Scholar]
- 29. Dolinsky TJ, Nielsen JE, McCammon JA, Baker NA (2004) PDB2PQR: an automated pipeline for the setup of Poisson‐Boltzmann electrostatics calculations. Nucleic Acids Res. 32, W665–W667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Van Der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJ (2005) GROMACS: fast, flexible, and free. J. Comput. Chem. 26, 1701–1718. [DOI] [PubMed] [Google Scholar]
- 31. Ng AC (2010) Integrative systems biology and networks in autophagy. Semin. Immunopathol. 32, 355–361. [DOI] [PubMed] [Google Scholar]
- 32. Fu LL, Zhao X, Xu HL, Wen X, Wang SY, Liu B et al (2012) Identification of microRNA‐regulated autophagic pathways in plant lectin‐induced cancer cell death. Cell Prolif. 45, 477–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Östlund G, Lindskog M, Sonnhammer EL (2010) Network‐based identification of novel cancer genes. Mol. Cell. Proteomics 9, 648–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lapenna S, Giordano A (2009) Cell cycle kinases as therapeutic targets for cancer. Nat. Rev. Drug Discov. 8, 547–566. [DOI] [PubMed] [Google Scholar]
- 35. van Vugt MA, Brás A, Medema RH (2004) Polo‐like kinase‐1 controls recovery from a G2 DNA damage‐induced arrest in mammalian cells. Mol. Cell 15, 799–811. [DOI] [PubMed] [Google Scholar]
- 36. Degenhardt Y, Lampkin T (2010) Targeting Polo‐like kinase in cancer therapy. Clin. Cancer Res. 16, 384–389. [DOI] [PubMed] [Google Scholar]
- 37. Takai N, Hamanaka R, Yoshimatsu J, Miyakawa I (2005) Polo‐like kinases (Plks) and cancer. Oncogene 24, 287–291. [DOI] [PubMed] [Google Scholar]
- 38. Strebhardt K, Ullrich A (2006) Targeting polo‐like kinase 1 for cancer therapy. Nat. Rev. Cancer 6, 321–330. [DOI] [PubMed] [Google Scholar]
- 39. Murugan RN, Park JE, Kim EH, Shin SY, Cheong C, Lee KS et al (2011) Plk1‐targeted small molecule inhibitors: molecular basis for their potency and specificity. Mol. Cells 32, 209–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rudolph D, Steegmaier M, Hoffmann M, Grauert M, Baum A, Quant J et al (2009) BI 6727, a Polo‐like kinase inhibitor with improved pharmacokinetic profile and broad antitumor activity. Clin. Cancer Res. 15, 3094–3102. [DOI] [PubMed] [Google Scholar]
- 41. McInnes C, Wyatt MD (2011) PLK1 as an oncology target: current status and future potential. Drug Discov. Today 16, 619–625. [DOI] [PubMed] [Google Scholar]
- 42. Olmos D, Barker D, Sharma R, Brunetto AT, Yap TA, Taegtmeyer AB et al (2011) Phase I study of GSK461364, a specific and competitive Polo‐like kinase 1 inhibitor, in patients with advanced solid malignancies. Clin. Cancer Res. 17, 3420–3430. [DOI] [PubMed] [Google Scholar]
- 43. Reindl W, Yuan J, Krämer A, Strebhardt K, Berg T (2008) Inhibition of polo‐like kinase 1 by blocking polo‐box domain‐dependent protein‐protein interactions. Chem. Biol. 15, 459–466. [DOI] [PubMed] [Google Scholar]
- 44. Malumbres M, Pevarello P, Barbacid M, Bischoff JR (2008) CDK inhibitors in cancer therapy: what is next? Trends Pharmacol. Sci. 29, 16–21. [DOI] [PubMed] [Google Scholar]
- 45. Liu P, Kao T, Huang H (2008) CDK1 promotes cell proliferation and survival via phosphorylation and inhibition of FOXO1 transcription factor. Oncogene 27, 4733–4744. [DOI] [PubMed] [Google Scholar]
- 46. Radomska HS, Alberich‐Jordà M, Will B, Gonzalez D, Delwel R, Tenen DG (2012) Targeting CDK1 promotes FLT3‐activated acute myeloid leukemia differentiation through C/EBPα. J. Clin. Invest. 122, 2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Malumbres M, Barbacid M (2009) Cell cycle, CDKs and cancer: a changing paradigm. Nat. Rev. Cancer 9, 153–166. [DOI] [PubMed] [Google Scholar]
- 48. Payton M, Chung G, Yakowec P, Wong A, Powers D, Xiong L et al (2006) Discovery and evaluation of dual CDK1 and CDK2 inhibitors. Cancer Res. 66, 4299–4308. [DOI] [PubMed] [Google Scholar]
- 49. Fu LL, Yang Y, Xu HL, Cheng Y, Wen X, Ouyang L et al (2013) Identification of novel caspase/autophagy‐related gene switch to cell fate decisions in breast cancers. Cell Prolif. 46, 67–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Detailed information of small molecule compounds targeting PLK1, PLK1 PBD, CDK1 and CDK2 in HeLa cells.
Fig. S1. Evaluation of microarray data from HeLa cells.


